Abstract
Auxin-resistant mutants have been useful for dissecting the mechanisms that underlie auxin-mediated biological processes. Here we report the isolation and molecular characterization of a novel auxin-resistant mutant in Arabidopsis (Arabidopsis thaliana). Like known mutated AUX/IAA transcription factors, the mutant described here displayed dominant resistance to exogenously supplied auxins (sirtinol, 2,4-dichlorophenoxyacetic acid, indole-3-acetic acid) and a host of pleiotropic phenotypes, including apical hook deformation, defects in lateral root development, reduced stature, and homozygous lethality. This mutant showed the same sensitivity to the ethylene precursor 1-aminocyclopropane carboxylic acid as wild-type plants, and retained the ability to induce IAA19 expression in response to exogenously supplied indole-3-acetic acid. To our surprise, these phenotypes were not caused by a mutation in an AUX/IAA gene, but rather a mutation in a tRNAala gene in which the anticodon was found changed from CGC to CAC. Such a change results in a tRNA that is charged with alanine but recognizes the second most highly used valine codon in Arabidopsis. Therefore, the observed phenotypes are likely the composite of stochastic mutations of many proteins, including downstream effectors.
The plant hormone auxin indole-3-acetic acid (IAA) has been implicated in almost every aspect of plant growth and development ranging from embryogenesis to senescence (for recent reviews on auxin biology and biochemistry, see Dharmasiri and Estelle, 2004; Willemsen and Scheres, 2004; Kepinski and Leyser, 2005a; Woodward and Bartel, 2005). Like any hormone, auxin exerts at least some of its effects by serving as a molecular rheostat for transcriptional regulation, both positive and negative, of a set of target genes. Auxin target genes are canonically defined by the presence of auxin response element (ARE) sequence motifs in their promoter regions, and many have been validated by recent genomic analyses (Ulmasov et al., 1995, 1997; Hagen and Guilfoyle, 2002; Goda et al., 2004; Liu et al., 2005). AREs recruit auxin response factor (ARF) homodimers that directly activate or repress transcription of the proximal gene (Ulmasov et al., 1995, 1997; Abel et al., 1996). Molecular interpretation of the auxin signal is a complex and incompletely defined process; however, the best models based on current data suggest that either (1) in a basal state, ARFs are sequestered as heterodimers by inhibitory subunits (AUX/IAA proteins) that are targeted for degradation in the presence of an auxin signal, liberating the ARFs to homodimerize, bind AREs, and in turn regulate gene expression (Hellman and Estelle, 2002), or (2) that ARFs are constitutively bound to AREs and recruit AUX/IAA proteins when auxin levels are low, repressing the normal function of the ARF (Tiwari et al., 2003). Regardless, degradation of AUX/IAA proteins in response to the auxin signal is a key step in signal propagation that has recently been shown to proceed by ternary complex formation between IAA, a given AUX/IAA protein, and the F-box protein TIR1 (transport inhibitor resistance 1; Dharmasiri et al., 2005; Kepinski and Leyser, 2005b).
Forward genetic screens for auxin-resistant mutants have identified a number of components in the IAA-signaling network including protein ubiquitin-labeling machinery, the dual-specificity phosphatase IBR5 (indole-3-butyric acid response 5), and an assortment of AUX/IAA proteins (for examples, see Leyser et al., 1993; Nagpal et al., 2000; Gray et al., 2001; Schwechheimer et al., 2001; Hellmann et al., 2003; Monroe-Augustus et al., 2003). Most of these screens are designed so as to select for mutants that are resistant to exogenously supplied auxin (usually a synthetic analog of IAA). That is, primary root growth persists when seeds are germinated and grown under cycling white light for several days on plates containing auxin, typically 100 nm of the synthetic auxin 2,4-dichlorophenoxyacetic acid (2,4-D). Any mutations that disrupt the uptake (or promote the export), transport, processing, or perception of the auxin are expected to confer auxin resistance. In addition, mutations in genes encoding for downstream effectors that participate in long-range network feedback mechanisms could also potentially confer auxin resistance, but to date none have been reported with the possible exception of the dual-specificity phosphatase IBR5, whose function has yet to be fully elucidated (Monroe-Augustus et al., 2003).
Here we report the phenotypic characterization and cloning of a novel auxin-resistant mutant in Arabidopsis (Arabidopsis thaliana). Like the phenotypes caused by the reported mutations in AUX/IAA genes, the phenotypes associated with the mutant described here segregate in dominant fashion and include resistance to several auxins (IAA, 2,4-D, and sirtinol), apical hook deformation, reduced stature, and defects in lateral root development. Similar to axr5/iaa1 plants (Yang et al., 2004), the rosette leaves of this mutant were more rounded with shortened petioles, but it is distinct in that it appears to retain sensitivity to ethylene and the mutation is homozygous lethal. Surprisingly, positional cloning on the basis of 2,4-D resistance revealed that the mutation linked to the observed phenotypes was not found in an AUX/IAA gene, but in the anticodon of a tRNAala. The mutation transforms this tRNAala into a tRNA that remains charged with Ala but now recognizes a Val codon (GUG), which by codon bias analysis is found to be the second most highly utilized Val codon in Arabidopsis. The presumed molecular effect of this mutation is that some population of Vals in the proteome encoded by GUG will be replaced by Ala in a stochastic way, and that the bulk composite confers auxin resistance and other auxin-related phenotypes. Yanagida and colleagues have recently recovered similar tRNAala anticodon mutations from Schizosaccharomyces pombe while screening for suppressors of mutations in the Cdc16 subunit of the anaphase-promoting complex (Kimata and Yanagida, 2004). These mutants, while suppressing the anaphase-promoting complex phenotype (arrest at the metaphase to anaphase transition), conferred dominantly segregating phenotypes at anaphase spindle elongation that were lethal at restrictive temperatures because of chromosome missegregation at cell division (Kimata and Yanagida, 2004).
RESULTS
Identification of a Dominant Auxin-Resistant Mutant That Is Homozygous Lethal
We screened the M2 population of ethyl methanesulfonate-mutagenized Columbia (Col) seeds for plants that were resistant to 20 μm sirtinol to identify auxin-resistant mutants (Zhao et al., 2003; Cheng et al., 2004; Dai et al., 2005). When the mutant described here was backcrossed to wild-type Col or outcrossed to Landsberg erecta, the resulting F1 populations of plants were resistant to both sirtinol and 2,4-D, indicating that the auxin resistance segregates in dominant fashion. However, we later found that this mutant was originally isolated in the presence of background mutations as successive backcrosses reduced the number of homozygous progeny recovered from self-fertilizing heterozygous plants. Ultimately, the homozygous mutant was determined to be embryonic lethal.
Phenotypic Analyses
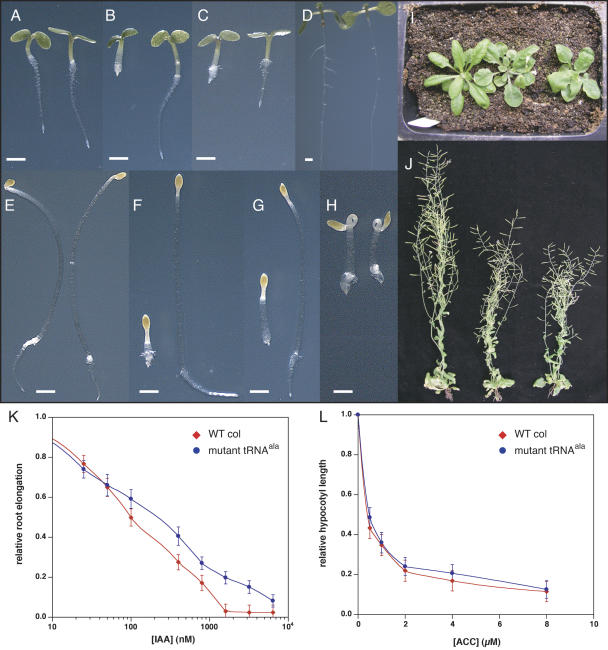
All analyses/comparisons were performed with heterozygous mutant plants, which were confirmed by genotyping (below). Three-day-old light-grown mutant and wild-type seedlings are compared in Figure 1, A to C. At this stage, the difference between wild-type and heterozygous mutant plants is subtle when grown on unsupplemented Murashige and Skoog (MS) medium and consists only of a more flattened architecture of the cotyledons in the mutant, so that they are perpendicular to the hypocotyl (Fig. 1A). Though subtle, genotyping progeny of self-fertilized heterozygous plants grown on unsupplemented MS and scored on this basis confirmed that this trait was linked to the mutation (72/72). Resistance to sirtinol, 2,4-D, and IAA, as judged by primary root elongation in the presence of typically inhibiting concentrations of these compounds, is also readily discerned in the mutant at this stage (Fig. 1, B, C, and K). Finally, after 7 d on unsupplemented MS, mutant seedlings were found to have a defect in lateral root development (Fig. 1D).
Figure 1.
Phenotypic analysis of an auxin-resistant tRNAala anticodon mutant from Arabidopsis. Seedling phenotypes under the given conditions are shown in sections A to H, wild type on the left, heterozygous mutant on the right. A to C, Three-day-old light-grown seedlings on 0.5× MS (A), 0.5× MS supplemented with 10 μm of sirtinol (B), or 100 nm of the synthetic auxin 2,4-D (C). D, Seven-day-old light-grown seedlings on 0.5× MS. E to H, Three-day-old dark-grown seedlings on 0.5× MS (E), 0.5× MS supplemented with 2 μm sirtinol (F), 250 nm 2,4-D, or 2 μm of the ethylene precursor ACC (G). Later stage phenotypes are shown in I and J, wild type on the left, heterozygous mutant in the center, and heterozygous recapitulation line on the right. I, Fully formed rosette leaf stage. J, Mature plants. Hormone response curves for IAA and ACC are provided in sections K and L, respectively.
In the dark, the mutant lacked the normal apical hook structure and was found to be resistant to sirtinol and 2,4-D in both the root and hypocotyl tissues (Fig. 1, E–G). Somewhat surprisingly, the mutant appears to retain full sensitivity to the ethylene precursor 1-aminocyclopropane-1-carboxlyic acid (ACC; Fig. 1, H and L).
As the plants mature, more phenotypes are evident in the mutant. The rosette leaves are more rounded and the petioles are shorter in mutant and recapitulation line (20 lines generated, below) plants (Fig. 1I), and all lines are reduced in stature relative to wild type (Fig. 1J).
Molecular Cloning and Recapitulation
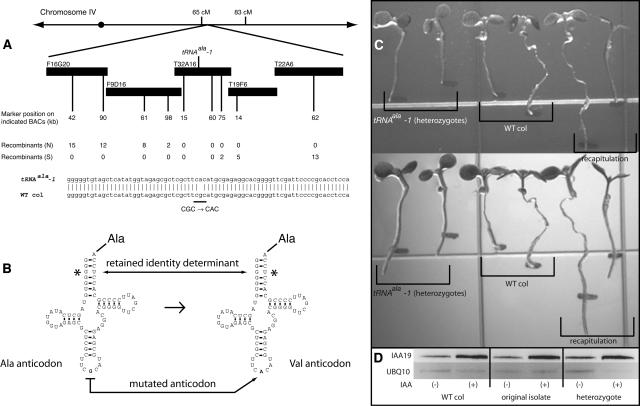
Linkage analysis placed the mutated gene in the middle of the bottom arm of chromosome IV, and fine mapping narrowed the interval to an 80-kb region between markers F9D16-98 and T32A16-75 (Fig. 2A). DNA sequencing of all open reading frames in this region identified a single G-to-A transition in the anticodon (CGC to CAC) of a tRNAala encoded by the gene At4g23915. This change results in a tRNA that is charged with Ala but expected to recognize a codon for Val (GUG). It should be noted that unlike other tRNAs, which have multiple determinants located in the arms and core of the structure, sometimes including the anticodon, the indicated (*) G3-U70 wobble conserved among tRNAalas (Fig. 2B) is the only major identity determinant for tRNAala identity (Hou and Schimmel, 1988; McClain and Foss, 1988). Therefore, we only expect to be able to obtain similar results with mutations in other tRNAalas. However, as a caveat, previous studies have focused mostly on Escherichia coli, Saccharomyces cerevisiae, and Thermus thermophilus, which differ significantly from one another at the level of tRNA identity determinacy, and, therefore, it is conceivable that other, non-Ala-charging, tRNA mutants could be recovered from genetic screens in Arabidopsis (for review, see Giegé et al., 1998).
Figure 2.
Cloning of the mutant tRNAala and recapitulation of auxin resistance. A, Mapping data. BAC, Bacterial artificial chromosome; cM, centimorgan, the unit of genetic distance describing the recombination frequency between two loci as 1%/cM. The observed G-to-A transition in the tRNA gene is highlighted. B, Unlike other tRNAs, which have multiple determinants located in the arms and core of the structure, the indicated (*) G3-U70 wobble conserved among tRNAalas is the only major identity determinant (Hou and Schimmel, 1988; McClain and Foss, 1988). Therefore, we expect the documented phenotypes to occur only with this or other mutated tRNAalas. C, Recapitulation. The top section is a picture of seedlings (right two, tRNAala mutant heterozygotes; center two, wild type; left two, recapitulation lines [text]) immediately after transfer to 0.5× MS vertical plates supplemented with 100 nm 2,4-D; the bottom section shows the same seedlings after 24 h continued growth. D, IAA19 induction. RT-PCR shows that IAA19 expression is induced by IAA (10 μm) in the mutant tRNAala lines. UBQ10 expression is not regulated by IAA and is included as a control.
To confirm that this mutation was responsible for the observed phenotypes, we transformed wild-type Col plants with a construct harboring the mutated tRNAala under the transcriptional control of its natural promoter. This was sufficient to both confer auxin resistance and recapitulate the adult phenotypes (Figs. 1, I and J, and 2C).
Effect of the Mutated tRNAala on Auxin-Induced Gene Expression
One of the hallmarks of the auxin-mediated transcriptional response is the induction of AUX/IAA genes. IAA19 expression is robustly induced during the seedling stage (Nakamura et al., 2003), so we asked whether this was compromised in the mutated tRNAala lines reported here. Using reverse transcription (RT)-PCR, seedlings of both the original isolate (which was homozygous for the mutation) and of the backcrossed heterozygous line showed IAA19 induction in response to IAA comparable to that observed in wild-type Col seedlings (Fig. 2D). This is minimally consistent with a downstream effect giving rise to the observed phenotypes, as opposed to a defect at or before transcription.
Arabidopsis tRNA Bioinformatics and Codon Bias Analysis
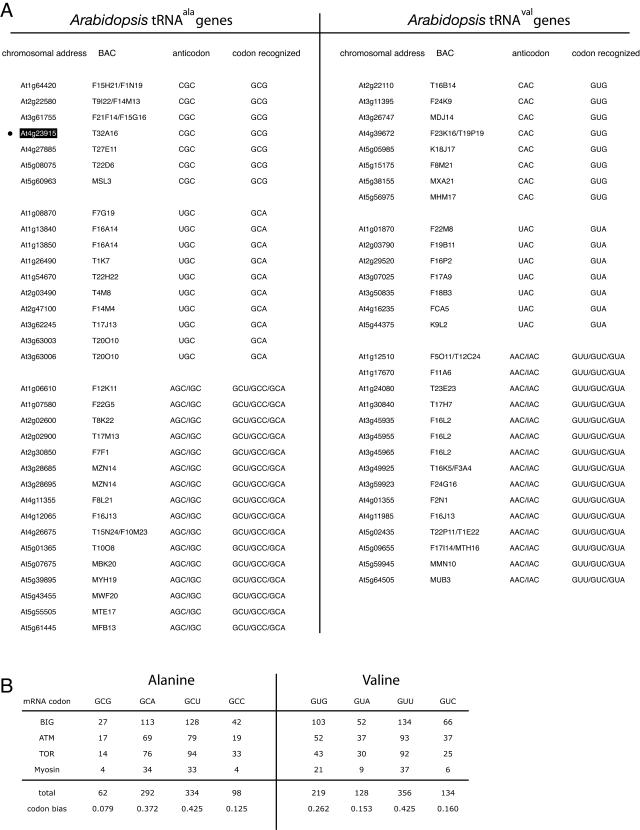
tRNAscan-SE analysis of the Arabidopsis genome previously indicated the presence of 630 standard tRNAs, no selenocysteine (UCA) or suppressor (CUA, UUA) tRNAs, one tRNA of unknown isotype, and eight predicted pseudogenes (http://lowelab.ucsc.edu/GtRNAdb/Athal/; Lowe and Eddy, 1997). Of those, 33 recognize a codon for Ala (using CGC [7], UGC [10], and AGC/IGC [16] anticodons) and 30 recognize a codon for Val (using CAC [8], UAC [7], and AAC/IAC [15] anticodons). Their genomic locations and anticodon and codon recognition distribution patterns are shown in Figure 3A. Using a set of genes that encode for large proteins with high Ala and Val content (BIG [gb|AF507018], ATM [ref|NM_114689], TOR [ref|NM_103891], and myosin [ref|NM112886]; 786 total Ala codons, 837 total Val codons), we estimated the codon bias in Arabidopsis. There appears to be a very strong bias for certain Ala codons (GCU>GCA≫GCC>GCG), and a weaker but still easily measured bias in Val codon usage (GUU>GUG>GUA≈GUC; Fig. 3B). In the mutant described here, a tRNAala that recognizes the codon GCG is transformed into a tRNA that is charged with Ala but that recognizes the Val codon GUG; therefore, a tRNA that is presumably used only very sparingly (14 copies are encoded to recognize approximately 8% of Ala codons) is converted to a tRNA that could be used more frequently (16 copies of the CAC anticodon tRNA for Val are encoded to recognize approximately 26% of Val codons), and serves as a mutator. Interestingly, only one copy of the mutated tRNAala in the presence of 16 copies of GUG recognizing wild-type tRNAval is sufficient to give rise to the phenotypes documented here, and two copies are lethal. As a caveat, it is not trivial to determine the expression pattern of the various tRNAs (because many of the different genes have identical nucleotide sequences, and their inherent hairpin structures are not amenable to RT-PCR) and to know whether they all contribute equally to the pool.
Figure 3.
Informatic analysis of tRNAala and tRNAval and codon usage bias in Arabidopsis. A, The tRNAala and tRNAval genes in Arabidopsis. Note that there are three types of tRNAs encoded to recognize four types of codons in each instance. The gene cloned in this study is highlighted and indicated (•). B, Codon usage bias analysis. A total of 786 Ala codons and 837 Val codons were analyzed.
DISCUSSION
Here we have shown that a mutation in the anticodon of a tRNAala can in many ways mimic the phenotypes observed in several reported AUX/IAA gene mutants. In both cases, mutant phenotypes are dominant and include resistance to exogenously supplied auxin, reduced stature, and defects in lateral root development. The phenotypes associated with the mutant tRNA reported here are perhaps most similar to those recently reported with axr5/iaal, which also include more rounded rosette leaves and shortened petioles (Yang et al., 2004).
The precise reasons for the similar phenotypes is not clear at present; however, in both cases (the mutant tRNA versus AUX/IAA mutants) the observed phenotypes are likely to be composites of downstream effects. With the mutant tRNA, we expect that a steady-state level of many mutated proteins contributes to the phenotypes, though a more direct effect or a major effect on one or more specific components cannot be ruled out. For example, 15 different AUX/IAA proteins contain at least one Val residue encoded by GTG within their respective domain II segments, the genetic region where all recovered AUX/IAA mutations have occurred. Therefore, Val to Ala substitutions in AUX/IAA proteins could contribute to auxin resistance, but at this time it is not clear whether that is a major determinant because (1) none of these Val residues are invariant among AUX/IAA proteins in Arabidopsis, and (2) mapped mutations have occurred only in DNA sequences encoding either a Gly residue or one of two Pro in that region. With AUX/IAA mutants, many genes are expected to be misregulated with respect to the auxin signal, and it is not known exactly how the observed pleiotropic phenotypes arise. However, given the apparent sensitivity to ACC and the potent IAA-mediated induction of IAA19, it seems likely that the mutated tRNA exerts its greatest influence over proteins encoded by downstream effector genes of the auxin pathway rather than previously identified components such as those that comprise protein degradation machinery (all of which exhibit resistance to ACC) and thus may be a useful tool for studying these auxin regulated pathways.
One such pathway is auxin-regulated cell division. It has recently been reported that transgenic Arabidopsis lines carrying a Ran-binding protein 1c RNAi knockdown construct were defective in lateral root development but auxin hypersensitive, suggesting that Ran-binding protein 1c functions to suppress certain auxin effects yet regulates mitosis in root tips (Kim et al., 2001). Given that many mutations to emerge from genetic screens, including the mutated tRNA described here, are defective in lateral root development but auxin resistant, the interface of auxin signaling and mitosis is expected to be complex. However, recent experiments in S. pombe have shown that tRNAala anticodon mutations can confer specific dominant defects in late mitosis due to abnormal spindle dynamics in that organism (Kimata and Yanagida, 2004). Therefore, the mutant described here might be able to be used for studying the auxin-signaling/mitosis interface, and it provides a handle to leverage information regarding translation quality control networks in Arabidopsis, a virtually unexplored avenue for research.
MATERIALS AND METHODS
Mutagenesis and the Sirtinol Resistance Screen
Ethylmethane sulfonate-mutagenized Arabidopsis (Arabidopsis thaliana) Col M2 seeds were purchased from Lehle Seeds. Genetic screens for sirtinol-resistant mutants were performed as previously described (Zhao et al., 2003; Cheng et al., 2004; Dai et al., 2005). In brief, M2 seeds were germinated on 0.5× MS/agar plates supplemented with 20 μm sirtinol and allowed to grow under cycling white light (16 h light/8 h darkness) for 6 d, after which time seedlings were scored for resistance to sirtinol on the basis of primary root elongation, then transplanted directly to soil to set seeds.
Positional Cloning and Recapitulation
The mutant tRNAala described here was cloned by a map-based strategy (Lukowitz et al., 2000) on the basis of its resistance to the synthetic auxin 2,4-D. Polymorphic sequence markers were designed with reference to the polymorphisms between the Col and Landsberg ecotypes as defined by Monsanto (accessed by http://www.arabidopsis.org). The observed phenotypes were recapitulated by cloning a genomic fragment containing the mutated tRNA and its promoter elements into the binary vector pZP211, and then transforming that construct to wild-type Arabidopsis (Col) plants. The primers used to amplify the approximately 1.0-kb fragment were 5′-GGGGTACCACAAAGCTTTTACGGGTTGTTCCAGTTT-3′ and 5′-TTTCTGCAGTTTTACGTCAAATTCTTTAATTCACTT-3′. Seeds from transformed plants were sown on 0.5× MS containing 30 μg/mL kanamycin, stratified for 2 d at 4°C, then allowed to germinate and grow under cycling white light for 6 d. Those plants that were resistant to kanamycin were transferred to vertical MS plates supplemented with 100 nm 2,4-D and allowed to grow for an additional 24 h. Root elongation was observed in all plants containing the kanamycin-resistant transgene.
Hormone Response Assays
For auxin, 5-d-old seedlings were transferred from 0.5× MS plates to vertical 0.5× MS plates supplemented with varying concentrations of IAA. The primary root tips were marked, and then seedlings were allowed to grow under cycling white light for an additional 2 d, after which continued root elongation was quantified using the public domain NIH Image program (developed at the United States National Institutes of Health and available on the Internet at http://rsb.info.nih.gov/nih-image). All data points are derived from a minimum of 10 plants.
For ethylene, seeds were sown on 0.5× MS plates containing varying concentrations of the ethylene precursor ACC. These seeds were stratified for 2 d at 4°C, germinated under white light for exactly 2 h, and then the plates were wrapped in foil for exactly 72 h, after which time hypocotyl length was quantified with NIH Image. All data points are derived from a minimum of 15 plants.
IAA19 Induction
Five-day-old seedlings grown on 0.5× MS plates were immersed in 0.5× MS liquid medium supplemented with IAA to a final concentration of 10 μm for 2 h. Seedlings were then harvested and total RNA was prepared using the Rneasy Plant mini kit (Qiagen) following the instructions provided by the manufacturer. Following first-strand cDNA synthesis, transcripts from IAA19 and UBQ10 were analyzed by PCR using the following primer combinations: IAA19, 5′-TGGTGACAACTGCGAATACG-3′ and 5′-TCACTCGTCTACTCCTCTAG-3′; and UBQ10, 5′-GTCCTCAGGCTCCGTGGTG-3′ and 5′-TGCCATCCTCCAACTGCTTTC-3′.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number At4g23915.
Acknowledgments
We thank Ben Greener for help with genotyping analyses.
This work was supported by the National Institutes of Health (grant no. 1RO1GM68631 to Y.Z.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yunde Zhao (yzhao@biomail.ucsd.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.068700.
References
- Abel S, Ballas N, Wong L-M, Theologis A (1996) DNA elements responsive to auxin. Bioessays 18: 647–654 [DOI] [PubMed] [Google Scholar]
- Cheng Y, Dai X, Zhao Y (2004) AtCAND1, a HEAT-repeat protein that participates in auxin signaling in Arabidopsis. Plant Physiol 135: 1020–1026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai X, Hayashi K, Nozaki H, Cheng Y, Zhao Y (2005) Genetic and chemical analyses of the action mechanisms of sirtinol in Arabidopsis. Proc Natl Acad Sci USA 102: 3129–3134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dharmasiri N, Dharmasiri S, Estelle M (2005) The F-box protein TIR1 is an auxin receptor. Nature 435: 441–445 [DOI] [PubMed] [Google Scholar]
- Dharmasiri N, Estelle M (2004) Auxin signaling and regulated protein degradation. Trends Plant Sci 9: 302–308 [DOI] [PubMed] [Google Scholar]
- Giegé R, Sissler M, Florentz C (1998) Universal rules and idiosyncratic features in tRNA identity. Nucleic Acids Res 26: 5017–5035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goda H, Sawa S, Asami T, Fujioka S, Shimada Y, Yoshida S (2004) Comprehensive comparison of auxin-regulated and brassinosteroid-regulated genes in Arabidopsis. Plant Physiol 134: 1555–1573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray WM, Kepinski S, Rouse D, Leyser O, Estelle M (2001) Auxin regulates SCF(TIR1)-dependent degradation of AUX/IAA proteins. Nature 414: 271–276 [DOI] [PubMed] [Google Scholar]
- Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49: 373–385 [PubMed] [Google Scholar]
- Hellman H, Estelle M (2002) Plant development: regulation by protein degradation. Science 297: 793–797 [DOI] [PubMed] [Google Scholar]
- Hellmann H, Hobbie L, Chapman A, Dharmasiri S, Dharmasiri N, del Pozo C, Reinhardt D, Estelle M (2003) Arabidopsis AXR6 encodes CUL1 implicating SCF E3 ligases in auxin regulation of embryogenesis. EMBO J 22: 3314–3325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou YM, Schimmel P (1988) A simple structural feature is a major determinant of the identity of a transfer RNA. Nature 333: 140–145 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005. a) Plant development: auxin in loops. Curr Biol 15: R208–R210 [DOI] [PubMed] [Google Scholar]
- Kepinski S, Leyser O (2005. b) The Arabidopsis F-box protein TIR1 is an auxin receptor. Nature 435: 446–451 [DOI] [PubMed] [Google Scholar]
- Kim SH, Arnold D, Lloyd A, Roux SJ (2001) Antisense expression of an Arabidopsis Ran binding protein renders transgenic roots hypersensitive to auxin and alters auxin-induced root growth and development by arresting mitotic progress. Plant Cell 13: 2619–2630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimata Y, Yanagida M (2004) Suppression of a mitotic mutant by tRNA-Ala anticodon mutations that produce a dominant defect in late mitosis. J Cell Sci 117: 2283–2293 [DOI] [PubMed] [Google Scholar]
- Leyser HM, Lincoln CA, Timpte C, Lammer D, Turner J, Estelle M (1993) Arabidopsis auxin-resistance gene AXR1 encodes a protein related to ubiquitin-activating enzyme E1. Nature 364: 161–164 [DOI] [PubMed] [Google Scholar]
- Liu F, Vantoai T, Moy LP, Bock G, Linford LD, Quackenbush J (2005) Global transcription profiling reveals comprehensive insights into hypoxic response in Arabidopsis. Plant Physiol 137: 1115–1129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe TM, Eddy SR (1997) tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Nucleic Acids Res 25: 955–964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukowitz W, Gillmor CS, Scheible WR (2000) Positional cloning in Arabidopsis: why it feels good to have a genome initiative working for you. Plant Physiol 123: 795–805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClain WH, Foss K (1988) Changing the identity of a tRNA by introducing a G-U wobble pair near the 3′ acceptor end. Science 240: 793–796 [DOI] [PubMed] [Google Scholar]
- Monroe-Augustus M, Zolman BK, Bartel B (2003) IBR5, a dual-specificity phosphatase-like protein modulating auxin and abscisic acid responsiveness in Arabidopsis. Plant Cell 15: 2979–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123: 563–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura A, Higuchi K, Goda H, Fujiwara MT, Sawa S, Koshiba T, Shimada Y, Yoshida S (2003) Brassinolide induces IAA5, IAA19, and DR5, a synthetic auxin response element in Arabidopsis, implying a cross talk point of brassinosteroid and auxin signaling. Plant Physiol 133: 1843–1853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwechheimer C, Serino G, Callis J, Crosby WL, Lyapina S, Deshaies RJ, Gray WM, Estelle M, Deng XW (2001) Interactions of the COP9 signalosome with the E3 ubiquitin ligase SCFTIRI in mediating auxin response. Science 292: 1379–1382 [DOI] [PubMed] [Google Scholar]
- Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15: 533–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Liu ZB, Hagen G, Guilfoyle TJ (1995) Composite structure of auxin response elements. Plant Cell 7: 1611–1623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulmasov T, Murfett J, Hagen G, Guilfoyle TJ (1997) Aux/IAA proteins repress expression of reporter genes containing natural and highly active synthetic auxin response elements. Plant Cell 9: 1963–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willemsen V, Scheres B (2004) Mechanisms of pattern formation in plant embryogenesis. Annu Rev Genet 38: 587–614 [DOI] [PubMed] [Google Scholar]
- Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot (Lond) 95: 707–735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Lee S, So JH, Dharmasiri S, Dharmasiri N, Ge L, Jensen C, Hangarter R, Hobbie L, Estelle M (2004) The IAA1 protein is encoded by AXR5 and is a substrate of SCF(TIR1). Plant J 40: 772–782 [DOI] [PubMed] [Google Scholar]
- Zhao Y, Dai X, Blackwell HE, Schreiber SL, Chory J (2003) SIR1, an upstream component in auxin signaling identified by chemical genetics. Science 301: 1107–1110 [DOI] [PubMed] [Google Scholar]