Abstract
In the fully sequenced Arabidopsis (Arabidopsis thaliana) genome, many gene models are annotated as “hypothetical protein,” whose gene structures are predicted solely by computer algorithms with no support from either expressed sequence matches from Arabidopsis, or nucleic acid or protein homologs from other species. In order to confirm their existence and predicted gene structures, a high-throughput method of rapid amplification of cDNA ends (RACE) was used to obtain their cDNA sequences from 11 cDNA populations. Primers from all of the 797 hypothetical genes on chromosome 2 were designed, and, through 5′ and 3′ RACE, clones from 506 genes were sequenced and cDNA sequences from 399 target genes were recovered. The cDNA sequences were obtained by assembling their 5′ and 3′ RACE polymerase chain reaction products. These sequences revealed that (1) the structures of 151 hypothetical genes were different from their predictions; (2) 116 hypothetical genes had alternatively spliced transcripts and 187 genes displayed polyadenylation sites; and (3) there were transcripts arising from both strands, from the strand opposite to that of the prediction and possible dicistronic transcripts. Promoters from five randomly chosen hypothetical genes (At2g02540, At2g31270, At2g33640, At2g35550, and At2g36340) were cloned into report constructs, and their expressions are tissue or development stage specific. Our results indicate at least 50% of hypothetical genes on chromosome 2 are expressed in the cDNA populations with about 38% of the gene structures differing from their predictions. Thus, by using this targeted approach, high-throughput RACE, we revealed numerous transcripts including many uncharacterized variants from these hypothetical genes.
The first plant genome project, the whole-genome sequencing of Arabidopsis (Arabidopsis thaliana), was completed at the end of 2000 (Arabidopsis Genome Initiative, 2000). However, the goal of the project is not only the collection of the complete sequence of the Arabidopsis genome but also understanding the function of each gene. This is the major goal of the National Science Foundation 2010 projects, and this challenge has been taken up by many Arabidopsis researchers (Chory et al., 2000). Identifying each gene in the fully sequenced Arabidopsis genome and uncovering their function will provide crucial information for biologists to understand plant physiology, genetics, development, and evolution. One approach to achieve this goal, which has been taken by several groups (Ceres, Stanford Genome Center, Salk Institute, Plant Gene Expression Center, RIKEN, and Institut National de la Recherche Agronomique/Genoplante), is to obtain full-length cDNA sequence for every gene in the genome by large-scale sequencing.
The group of genes/gene products with the most obscure functions, from the annotation of the Arabidopsis genome produced by The Institute for Genomic Research (TIGR), is called “hypothetical proteins.” These genes and their products are predicted solely by computer algorithms such as Genscan (Burge and Karlin, 1997), Genemark.hmm (Lukashin and Borodovsky, 1998), and various splice-site prediction programs (Uberbacher and Mural, 1991; Hebsgaard et al., 1996; Brendel and Kleffe, 1998). At the time of genome completion, 5,690 of the annotated genes were designated as “hypothetical.” However, with increased database content and improved annotation methods, this number decreased over time so that the final TIGR annotation release (Version 5, February 2004) contained only 2,248 hypothetical proteins among the 26,207 annotated open reading frames (ORFs).
The lack of expressed sequence tag (EST)/cDNA matches from Arabidopsis or protein homologs from other species indicates that there is no experimental evidence for the expression, structure, or function of these genes and also suggests that if they are expressed, their expression is likely localized, transient, at low levels, or under specific conditions. Thus, large-scale, undirected cDNA cloning and sequencing projects may have less chance of recovering them. In contrast, we applied a more specific and sensitive PCR-based targeted approach, high-throughput RACE (Frohman et al., 1988), to investigate the expression of all hypothetical genes on chromosome 2 under a variety of growth conditions to obtain cDNA sequences of expressed genes. Although several reports based on amplicon or oligonucleotide array hybridization and massively parallel signature sequencing (MPSS) showed that many genes annotated as “hypothetical proteins” were expressed (Kim et al., 2003; Yamada et al., 2003; Meyers et al., 2004; Redman et al., 2004), their cDNA sequences could not be recovered by these methods. In contrast, the cDNA sequences of expressed hypothetical genes can be obtained by our approach and are of great value for validating the predicted gene structures and laying a foundation for the analysis of their functions in vivo. In a previous pilot study, 138 out of 169 hypothetical genes tested were expressed in our cDNA populations with full-length cDNA sequences obtained from 16 hypothetical genes analyzed (Xiao et al., 2002). In this study, we developed a high-throughput RACE method and examined all the hypothetical genes on Arabidopsis chromosome 2 (797 when the work was initiated). We expanded our cDNA populations from six to 11 to increase the likelihood of detecting expression of these hypothetical genes. As a result, we recovered numerous transcripts from annotated hypothetical genes including many examples of alternate splicing and multiple polyadenylation sites, transcripts arising from both strands, transcripts arising only from the strand opposite to that of the prediction, and possible dicistronic transcripts. These results provide a statistically more significant data set for comparing genes structures predicted ab initio by computer programs with those supported by experimental evidence. To begin an investigation of the function of some of these hypothetical genes, we generated green fluorescent protein (GFP) and β-glucuronidase (GUS) gene reporter constructs of five randomly chosen genes (At2g02540, At2g31270, At2g33640, At2g35550, and At2g36340). The expression patterns of these five promoter-reporter constructs display highly localized tissue and/or developmental stage specificity, providing an explanation for their low abundance in all EST collections sequenced to date. To our knowledge, this is the first large-scale study focusing on annotated hypothetical genes by high-throughput RACE.
RESULTS
Generation, Sequencing, and Analysis of RACE Products
In this study, we tested all of the 797 genes from chromosome 2 that were annotated as hypothetical (Supplemental Table I). To increase the RACE success rate, the cDNA populations were expanded from the six tissues/treatments in our previous study (Xiao et al., 2002) to a total of 11 populations including tissue exposed to cold; heat; 2,4-dichlorophenoxyacetic acid (2,4-D); hydrogen peroxide (H2O2); UV; indole-3-acetic acid (IAA); salt and pathogens (Xanthomonas campestris pv campestris and Pseudomonas syringae); and tissue from callus, roots, and young seedlings, as well as sampling at different time points for some treatments (see “Materials and Methods”). To assess the transcript diversity of this set of samples, each RNA was labeled and hybridized to the whole-genome Affymetrix chip designed by TIGR (Redman et al., 2004). Collectively, 17,501 genes were assigned a “present” call in one or more tissues and 4,880 genes were not detected in any RNA samples by Microarray Analysis Suite 5.0 (J.C. Redman and C.D. Town, unpublished data).
RACE reactions were performed on pooled cDNA populations. Nested RACE reactions were performed after first-round RACE to increase specificity, and their products were cloned. For each targeted gene, 12 colonies from each end (5′ or 3′ RACE) were screened by PCR using gene-specific primers, and positive clones were sequenced. Among the 797 hypothetical genes on chromosome 2, insert-positive clones were obtained from 506 genes (Supplemental Table I). These were sequenced from both ends and produced a total of 11,237 good reads. The sequences clustered and assembled using the Program to Assemble Spliced Alignments (PASA; Haas et al., 2003), which first aligns each sequence read to the finished Arabidopsis genome and then assembles each cluster of overlapping reads into a minimal number of transcript isoforms consistent with the collection of sequences. The entire flow scheme and outcome of the RACE process is shown in Figure 1. Only 399 targets yielded clones whose sequences matched the initial target gene and can thus unambiguously be called “expressed” in our new cDNA populations (Fig. 1; Supplemental Table I). For the remaining 107 RACE-positive target genes, either no sequence was generated or the sequences did not match the correct target. Among the 4,880 genes not detected on the Affymetrix chip, 327 genes were tested in our RACE experiments and 160 were found to be expressed. As expected, our PCR-based method is more sensitive than Affymetrix hybridization.
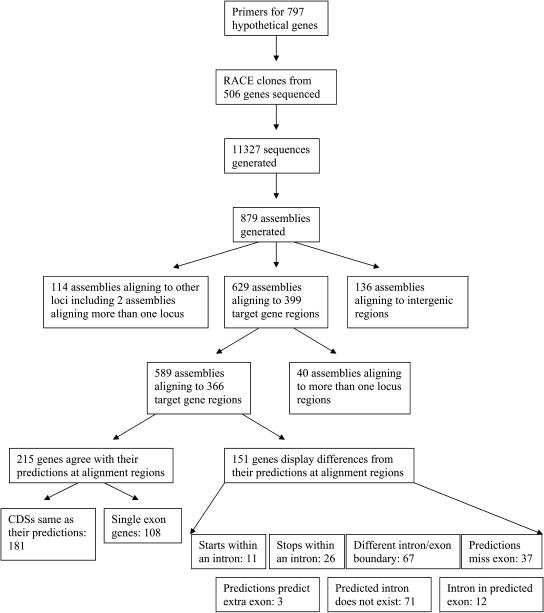
Figure 1.
Flow scheme and outcome of the RACE process. The figure shows the numbers of genes for which primers were designed, the success rate for cloning and sequencing, and the outcome in terms of numbers of assemblies produced and their relationship to the original gene predictions.
Analysis of Gene Structures
Assemblies Aligning to Individual Target Loci
As shown in Figure 1, there are 589 PASA assemblies that align to 366 single-target genes with 40 assemblies matching regions that span more than one locus (see below). Among 366 single-target matches, 215 (58.7%) genes have at least one transcript whose structure is identical to its prediction over the alignment region. Of these, the structures of 181 genes with ORFs are the same as predictions, adding only 5′ and 3′ untranslated regions (UTRs) to the original prediction (Fig. 2A). For the other 34 genes the RACE assemblies do not cover the entire predicted ORF, suggesting that they may not represent the complete transcript. Of the 215 matched genes, 108 consist of single exons.
Figure 2.
Examples of comparison of experimentally derived cDNA sequences to the predictions. A to H, Different types of relationships between the predicted gene structure (identified by AGI identifier: At2g#####) and that inferred from RACE sequence (Assembly). The continuous upper line indicates the uninterrupted genomic sequence, the second line the spliced alignment of the predicted hypothetical gene model to the genomic sequence, and subsequent lines the spliced alignments of RACE-derived sequence. Vertical dotted lines highlight differences in splice site locations. All the predicted gene structures are annotation Version 1. The GenBank accession numbers are At2g02320 assembly, AY168990; At2g33400 assembly, AY144106; At2g10850 assembly, AY501348; At2g01240 assembly, AY500325; At2g03020 assembly, AY219083; At2g06800 assembly, AY464642; At2g01960 assembly, AY168991; and At2g04850 assembly, DQ069802.
The expressed sequences from the other 151 genes (41.3%) reveal gene structures that are different from their predictions, with 11 genes having predicted start codons within regions that are actually introns (Fig. 2B) and 26 genes with predicted stop codons in regions that are actually introns (Fig. 2C). Sixty-seven genes display intron/exon boundary or boundaries that are different from their predictions (Fig. 2D). Thirty-seven gene predictions fail to identify existing exon/exons (Fig. 2E), and three genes are predicted with additional exons (Fig. 2F). The predicted introns from 71 genes are not supported by the cDNA sequences obtained (Fig. 2G), and 12 genes display introns within their predicted exons (Fig. 2H). For a particular gene whose structure is different from its prediction, there are usually discrepancies in more than one of the above categories. Specific examples of each type of discrepancy are shown in Figure 2.
Alternate Splicing and Polyadenylation
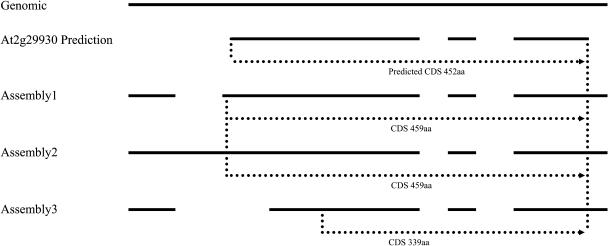
In our analysis, if any one of a gene's splice isoforms is consistent with the prediction, the gene is flagged only as “alternatively spliced transcript,” not “different from prediction.” Among our target genes, alternatively spliced transcripts were found from 116 target regions. The alternative transcripts can be classified as showing an alternative donor site (31 cases), an alternative acceptor site (37 cases), unspliced intron (71 cases), and exon skipping (six cases). For a particular gene, the alternatively spliced transcripts may fall into two or more categories. For example, there are three different assemblies from gene At2g29930 that represent alternatively spliced transcripts (Fig. 3). The differences among them are in the structure of the first intron. One assembly (assembly 1) has an intron in the 5′ UTR and an in-frame ATG 21 bp upstream of the predicted initiation codon (ATG site). Therefore, the ORF encoded by assembly 1 is seven amino acids larger than predicted. However, assembly 2 has an unspliced first intron that does not cause a change in the encoded protein, while assembly 3 displays a different intron acceptor site creating a larger intron that results in a shorter protein. The longest ORF in each of the three transcripts is different from the predicted ORF, but all are in the same reading frame. Overall, in our study, about 29% (116/399) of the hypothetical genes have alternatively spliced transcripts.
Figure 3.
Example of an alternatively spliced transcript. Notation for spliced alignments as for Figure 2. Vertical dotted lines indicate the starts and stops of CDS. Horizontal arrowed, dotted lines indicate the CDS direction of transcription and the sizes of the CDS. The predicted gene structures are annotation Version 1. The GenBank accession numbers are assembly1, AY231412; assembly2, AY231413; and assembly3, AY231414.
In addition, polyadenylation sites were identified in 187 genes with multiple polyadenylation sites from 83 genes. The distance between different polyadenylation sites was from 1 to 543 bp. Only one poly(A) was found from 104 genes; two poly(A)s were found from 41 genes; three poly(A)s were found from 27 genes; four poly(A)s were found from 11 genes; and only four genes were found with five poly(A) sites. All the poly(A) sites, even the two from At2g16140 that are 543 bp apart, occur after the stop codon so that none of these alternative polyadenylation events affects the protein sequences. All the genes identified with poly(A) site(s), the poly(A) location, and the distance of each poly(A) from the stop codon are shown in Supplemental Table II.
Transcripts from the Opposite Strand
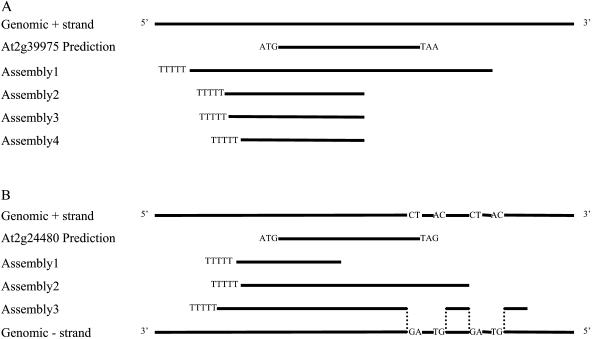
From 20 targeted regions, transcripts were obtained from the strands opposite from their predictions. For example, in the region of hypothetical gene At2g39975, a poly(A) tail is present in each of four assemblies formed. However, the alignment of assemblies with genomic and predicted At2g39975 coding sequence (CDS) reveals poly(T) tails aligning to the 5′ end of the At2g39975 prediction, which clearly suggests that these transcripts arise from the opposite strand (Fig. 4A). Another example is in the region of At2g24480. Not only do all assemblies display poly(T) sequences that align to the 5′ end of the predicted gene (At2g24480), but one assembly (no. 3) also contains two introns, whose splice-site orientations (GT-AG) demonstrate that this transcript is from the opposite strand (Fig. 4B). The other cases are similar to these two and were judged by the same criteria.
Figure 4.
Examples of transcripts from strands opposite to the original prediction. Solid lines are genomic sequences in genomic strands or exon sequences in prediction and assemblies. Dotted lines show the alignment of intron/exon boundaries. Splice site, GT-AG or CT-AC, is shown in genomic strands. Poly(T) tails are shown as TTTTT in assemblies. Start and stop codons are shown in predicted CDS. The predicted gene structures are annotation Version 1. None of the opposite-strand transcripts shown here has a good ORF. The GenBank accession numbers are transcript on the opposite strand of At2g39975 (assembly1), DQ069850; transcript on the opposite strand of At2g24480 (assembly2), DQ069848; and transcript on the opposite strand of At2g24480 (assembly3), DQ069849.
Transcripts from Both Strands
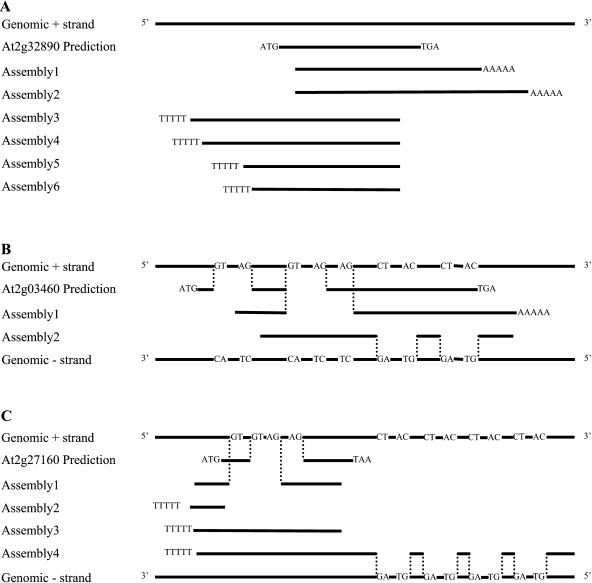
At three targeted regions (At2g32890, At2g03460, and At2g27160), transcripts from both strands were found. At the At2g32890 locus (Fig. 5A), six assemblies align to the predicted gene and genomic sequences. Among them, two contain poly(A) tails at the 3′ end of the At2g32890 prediction, suggesting that their direction of transcription is the same as predicted for the At2g32890 gene. However, the other four assemblies display poly(T) tails that align to the 5′ end of the prediction, indicating that these transcripts are derived from the other strand (Fig. 5A). Two different assemblies were obtained from the predicted At2g03460 gene region (Fig. 5B). For assembly 1, the orientation of intron splice sites (GT-AG) and the poly(A) tail demonstrate that the direction of transcription is the same as predicted for At2g03460, although the structure is different because the acceptor site of intron 2 is 37 bp downstream from its prediction and the acceptor site of intron 1 is also different. By contrast, the orientation of the splice sites shown by the alignment of assembly 2 to the genome indicates that this transcript is derived from the other strand (Fig. 5B). A similar situation occurs in the region of hypothetical gene At2g27160 (Fig. 5C). One assembly (no. 1) displays an intron whose boundaries are different from those in the prediction but whose transcription is in the same direction as its prediction (Fig. 5C). All the other assemblies (nos. 2, 3, and 4) have poly(T) tracts aligning to the predicted 5′ end of At2g27160, and one of them (no. 4) predicts four introns, indicating that these three assemblies are all transcribed from the opposite strand (Fig. 5C). Thus, although the transcripts obtained from these three regions may not be full-length cDNAs, the poly(A) tails and the orientations of the intron splice sites clearly demonstrate that the transcripts recovered from these regions are from both strands.
Figure 5.
Three examples of transcripts recovered from both strands. Notation as for Figure 4. Poly(A) or poly(T) tails are show as AAAAA or TTTTT in assemblies. The predicted gene structures are annotation Version 1. The GenBank accession numbers are At2g32890 assembly2, DQ069836; At2g32890 assembly4 (opposite-strand transcript), DQ069843; At2g03460 assembly1, AY501354; At2g03460 assembly2 (opposite-strand transcript), DQ069851; At2g27160 assembly1, DQ069824; and At2g27160 assembly4 (opposite-strand transcript), DQ069842.
Assemblies Aligning to More Than One Locus
Forty assemblies at 28 regions align to more than one locus, encompassing a total of 57 loci, 36 of which are from our target list. At seven of these regions, the single-aligned assembly simply overlaps the neighboring gene on the other strand, an observation that is not uncommon in either Arabidopsis or other organisms. The other 33 assemblies that align to two or more genes at 21 regions may indicate either the existence of dicistronic transcripts or a necessity for merging of existing gene models.
Dicistronic Transcripts
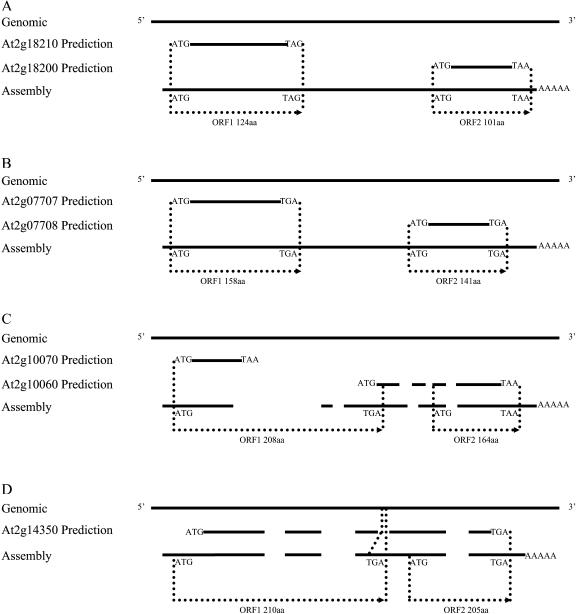
At four target gene regions, dicistronic transcripts were recovered. The assembly from At2g18200 actually covers the nearby At2g18210 region (Fig. 6A). Two ORFs found in this transcript are identical to the ORFs predicted for At2g18210 and At2g18200, and the distance between them is 819 bp (Fig. 6A). Similarly, the transcript obtained from At2g07708 is extended to the At2g07707 region and the two ORFs within this transcript, separated by 582 bp, are identical to At2g07708 and At2g07707, respectively (Fig. 6B). The orientation of both transcripts was confirmed by their poly(A) tails. Another dicistronic transcript was obtained from assembling sequences from the At2g10060 RACE products and contains two ORFs of 208 and 164 amino acids separated by 279 bp. Both gene structures and the encoded ORFs differ from those of the predictions (Fig. 6C). The transcript from the At2g14350 locus covers only the predicted region without any extension to other loci. However, the At2g14350 prediction has four introns, while the assembly contains only three introns, skipping the smallest predicted intron (9 bp) that contains an in-frame stop codon. Consequently, two ORFs are encoded by this single transcript (Fig. 6D). The first ORF is 210 amino acids and starts 9 bp (three amino acids) upstream of the predicted At2g14350 start site because there is a 1-bp difference between our recovered cDNA sequence and the genomic sequence. This difference was supported by two different clones and confirmed by manually checking their chromatograms. The second ORF contains 205 amino acids and starts at 45 bp downstream from the stop site of the first ORF and ends at the stop site predicted for At2g14350 (Fig. 6D). Both ORFs are in the same reading frame as the original prediction.
Figure 6.
Four genomic regions that give rise to dicistronic transcripts. Notation as for Figure 4. The start and stop codons and lengths of each ORF are shown. The stop codon of ORF1 in the region of At2g14350 (D) lies within the third intron of the originally predicted hypothetical gene. The predicted gene structures are annotation Version 1. The GenBank accession numbers are At2g18200 assembly, DQ069798; At2g07708 assembly, DQ069799; At2g10070 assembly, DQ069800; and At2g14350 assembly, AY227649.
Gene Merging
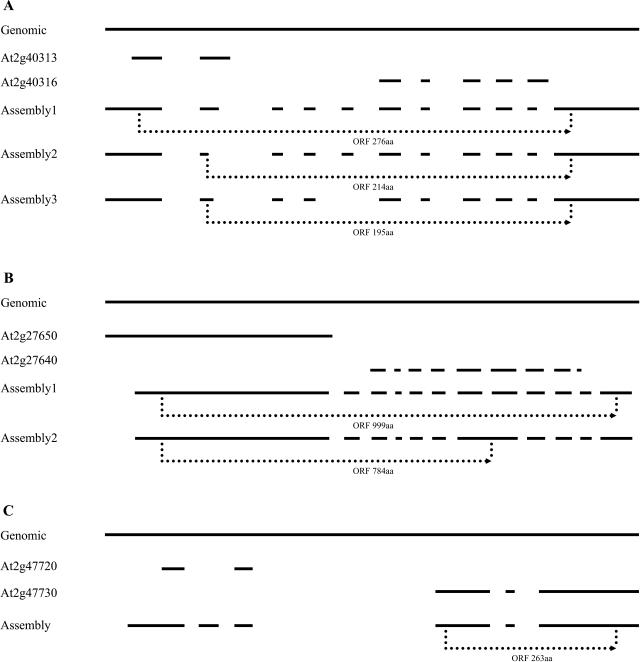
At three target regions, assemblies aligning to more than one gene suggest that the existing gene models should be merged because each of the transcripts obtained contain only one uninterrupted ORF (Fig. 7). The assemblies from At2g40316 cover the nearby gene At2g40313 and alternatively spliced transcripts were detected in this region (Fig. 7A). The three different transcript forms all have continuous ORFs with 276, 214, or 195 amino acids, respectively, strongly suggesting that these genes should be merged. Indeed, At2g40313 was merged into At2g40316 in later versions of the Arabidopsis genome annotation based on our data. A similar situation is found in the region of At2g27640 and At2g27650 (Fig. 7B). Each of the two transcripts obtained covers both regions, and they have long ORFs of 999 and 784 amino acids, respectively. At2g27640 was also merged with At2g27650 in later versions of annotation. In the region of At2g47720, the transcript from this hypothetical gene actually completely covers the nearby gene At2g47730, annotated as glutathione S-transferase (Fig. 7C). However, the longest ORF from this transcript is the same as At2g47730. The portion of the transcript covering the predicted At2g47720 region contains an unpredicted exon, which results in a small potential ORF (64 amino acids) at this part of the transcript that is different from the predicted At2g47720 ORF (72 amino acids; Fig. 7C). Thus, this portion of the transcript might be just the UTR region. The remaining 14 assemblies span more than one gene but extend only partially into nearby genes (data not shown), thus not permitting clear conclusions.
Figure 7.
Examples of gene models that require merging. Notation as for Figure 4. In each case, a single transcript encompasses both predicted ORFs and can be translated as a single protein. The GenBank accession numbers are At2g40316 assembly1, DQ069793; At2g40316 assembly2, AY231418; At2g40316 assembly3, DQ069794; At2g27650 assembly1, DQ069795; At2g27650 assembly2, DQ069796; and At2g47720 assembly, DQ069797.
Promoter-Reporter Fusion Expression Patterns from Five Hypothetical Genes
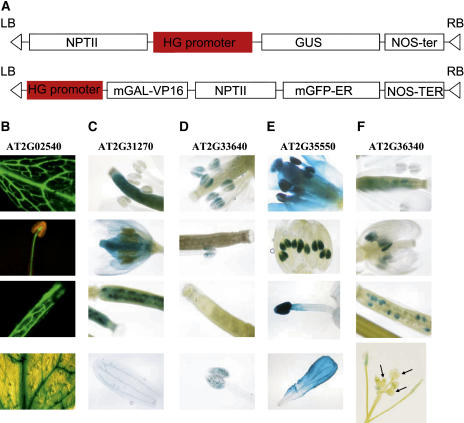
Five genes (At2g02540, At2g31270, At2g33640, At2g35550, and At2g36340) were chosen at random for a pilot study of expression patterns, in which their presumptive promoter regions, approximately 1.2-kb upstream sequence from ATG site, were cloned into GUS or GFP reporter constructs (pYXT1 and pYXT2; Fig. 8A) and transformed into Arabidopsis. Three independent transgenic lines were obtained from At2g02540 and At2g36340 and two independent lines from each of At2g31270, At2g33640, and At2g35550. Their expression patterns were examined (Fig. 8, B–F). The At2g02540 promoter-reporter construct is expressed in vascular tissues of leaf and different parts of the flower, and in siliques, but is not expressed in roots and old stems (Fig. 8B). Reporter constructs from At2g31270, At2g33640, At2g35550, and At2g36340 are all expressed in flowers, but in different parts and at different developmental stages (Fig. 8, C–F). The At2g31270 promoter-reporter fusion construct is expressed in carpels and petals (Fig. 8C). The At2g33640 construct is expressed exclusively in pollen (Fig. 8D). The At2g35550 construct is expressed in carpels, stamens (especially in the anther), and petals (Fig. 8E). The At2g36340 construct is expressed in young anthers and some young seeds in siliques (Fig. 8F), but not in mature pollen and in some but not all young flowers (Fig. 8F, arrows). Thus, the promoter-reporter fusion constructs from all five hypothetical genes tested exhibit tissue-specific and/or developmental stage-specific expression patterns.
Figure 8.
Expression patterns of tested hypothetical genes A, Constructs developed for testing the expression of hypothetical genes. RB and LB, T-DNA right and left borders; NPTII, kanamycin resistance gene; HG promoter, hypothetical gene promoter; NOS-ter, nos terminator; mGAL-VP16, GAL4-VP16 gene with modified codon usage; mGFP-ER, modified GFP with increased fluorescent properties and targeted to the endoplasmic reticulum. B, At2g02540 is expressed in vascular tissue. C, At2g31270 is expressed in carpels and petals. D, At2g33640 is expressed in pollen. E, At2g35550 is expressed in carpels, stamens, young anthers, and petals. F, At2g36340 is expressed in young anthers and some young seeds. Arrows indicate absent expression in three flowers.
DISCUSSION
In our study, we examined the structure and expression of all 797 genes on Arabidopsis chromosome 2 that were originally annotated as hypothetical (Version 1 data). To do this, we developed a high-throughput method with increased specificity that included automated primer design, the use of nested primers, more restricted screening for insert-positive colonies, and a set of scripts for semiautomated sequence assembly and analysis. As a result, a great deal of useful information was obtained from this study.
Expression Analysis
In an attempt to recover more expressed hypothetical genes, we expanded the cDNA populations and simplified the RACE process by pooling them together while at the same time increasing the input of each component cDNA so as not to compromise the concentration of each transcript in this complex pool. Without the specific expression-testing step (Xiao et al., 2002), expression is determined by the generation of RACE product(s) that match the intended target. The overall expression frequency observed (50%; 399/797) is lower than in our previous report (82% [137/169]; Xiao et al., 2002). This is most likely due to the smaller number of clones screened as well as to false priming during RACE of the complex pool as compared to the tissue-by-tissue assay. The numbers of hypothetical genes that are reported to be expressed by hybridization methods range from 37% for the whole-genome tiling array (Yamada et al., 2003) to 81% for the chromosome 2 amplicon array (Kim et al., 2003), with the Affymetrix ATH1 array reporting an intermediate value (60%; Redman et al., 2004). Despite some limitations of our high-throughput RACE protocol, this PCR-based method is still more sensitive than hybridization-based assays for the detection of gene expression. Among the hypothetical genes represented on, but not detected by, the ATH1 Affymetrix chip, 327 genes were included in our RACE experiments and 160 were found to be expressed. With the whole-genome tiling array (Yamada et al., 2003), only 190 of the 399 genes found expressed in our study were called present. Comparing our results to transcription data generated by MPSS from callus, inflorescence, leaves, root, and silique (Meyers et al., 2004), 170 out of the 399 genes for which transcripts were detected in this study have MPSS expression support.
Gene Structure Analysis
Insert-positive clones from RACE reactions were obtained for 506 out of the 799 genes attempted. However, the sequences generated matched only 399 target gene regions. For the remaining 107 gene targets, either the sequencing reactions failed or the sequences generated did not match the right target genes. Clearly some mispriming and amplification of nontarget regions coupled with false-positive clone identification occurred during our experiments. Among the 366 individual hypothetical genes matched by our sequences with full-length or partial cDNAs, about 38% of the predicted gene structures have some degree of inaccuracy. This is similar to an earlier study in which incorporation of 5,000 full-length cDNA sequences into the Arabidopsis genome annotation revealed that 33% of the gene models needed modifications (Haas et al., 2002). In our study, the number of incorrect gene models may be underestimated, since approximately half of the cDNA sequences are likely incomplete due either to failed RACE reactions or short sequence reads.
Alternate Splicing
Previous reports based on full-length cDNA and EST alignments have suggested that 10% or fewer Arabidopsis genes are alternately spliced (Haas et al., 2002, 2005; Zhu et al., 2003). However, in this study, 116 out of 399 expressed genes (approximately 29.0%) display alternatively spliced transcripts, a number consistent with but statistically more reliable than that reported previously (four out of 16; Xiao et al., 2002) and not much less than the value of 38% reported for the human genome (Brett et al., 2000; Kan et al., 2001; Modrek et al., 2001). The hypothetical genes that we examined lacked EST or cDNA support most likely because of low expression levels, localized or transient expression, or expression only under certain biological conditions. However, when a specific targeted approach was used in our experiments, a high level of sequence coverage was generated at many of the target regions. Although not all sequences matched their target regions, there were 8,757 sequences aligning to 399 target regions, corresponding to an average coverage of 22 reads per region, which facilitated the identification of multiple splice isoforms at particular regions. Use of a diverse pool of cDNA populations could also contribute to the higher percentage of alternative-splicing isoforms observed in our study, since it is well accepted that different tissues or biological conditions may produce different transcript variants. Lazar and Goodman (2000) showed that one alternative-splicing isoform of SR1 in Arabidopsis could play a role in cellular adaptation to a high-temperature environment. In pumpkin (Cucurbita maxima), light can regulate the alternative splicing of the hydroxypyruvate reductase gene (Mano et al., 1999, 2000). Recently, genome-wide analysis showed that environmental stress conditions significantly affected alternative-splicing profiles in Arabidopsis (Iida et al., 2004). Although many reports about alternative splicing of individual genes demonstrated that alternatively spliced transcripts have important biological functions in different plants (Zhou et al., 1998; Mano et al., 1999; Hartung and Puchta, 2000; Lazar and Goodman, 2000; Asakura et al., 2002; Jasinski et al., 2002; Quesada et al., 2003; Savaldi-Goldstein et al., 2003), it is unknown at this point which alternative-splicing transcripts in our study have real biological function and which are just misspliced products, especially in the 71 cases of intron retention that has been reported as a major phenomenon in alternative splicing in Arabidopsis (Ner-Gaon et al., 2004). Therefore, we believe that the high percentage of alternatively spliced transcripts found in our study are not specific to hypothetical genes but are observed because of the facets of our experimental approach discussed above.
Multiple Poly(A) Sites
Among the 187 genes for which polyadenylated transcripts were recovered, 83 genes (about 44%) display more than one poly(A) site. Multiple polyadenylation sites have been reported previously (Dean et al., 1986; Graber et al., 1999; Lupold et al., 1999; Hartung and Puchta, 2000; Magnotta and Gogarten, 2002). Alternative splicing of FCA gene transcripts leads to alternative polyadenylation, and interactions between the products of these splice isoforms control Arabidopsis flowering time (Quesada et al., 2003; Simpson et al., 2003). However, none of the alternate polyadenylation events observed in this study affect the coded-protein sequence. Searching of the sequences 300 bp upstream of each poly(A) site revealed that only about 27% (89/331) of the sequences contained the consensus AATAAA poly(A) signal, indicating that this signal (AAUAAA) is not consistently utilized in plants (Graber et al., 1999). Further investigation will be required to determine whether any of these alternate polyadenylation events has functional significance.
Unusual Transcripts
In addition to expected transcripts from the targeted hypothetical genes, some unusual transcripts were found, including transcripts from the strand opposite to the targeted prediction, transcripts from both strands at a particular locus, and transcripts covering more than one locus.
Transcripts from the Opposite Strand
From 20 regions, the transcripts recovered are not from the strands on which the predicted genes are located but from the opposite strand, as confirmed by the presence of poly(A) tails or the orientation of splice sites (Fig. 4; see Supplemental Fig. 1 for a detailed explanation). In every case, the ORFs in these opposite-strand transcripts are shorter than 100 amino acids. In contrast, the predicted ORFs on the other strand are always longer, explaining why they would be favored by the gene prediction programs. These opposite-strand transcripts may be noncoding transcripts (MacIntosh et al., 2001; Jones, 2002), or they may simply encode small proteins (Wen et al., 2004; for review, see Xia, 2004). Furthermore, the existence of opposite-strand transcripts does not preclude the possibility that there is also a transcript from the predicted strand that simply was not detected in our experiment, an idea supported by the fact that four out of these 20 gene regions do show both sense and antisense transcription in the MPSS data (Meyers et al., 2004). Among the remaining 16 genes, 14 do not show any expression by MPSS and the other two show only sense expression. At the whole-genome level, Yamada et al. (2003) detected 1,846 annotated genes with only antisense expression, including eight of the 20 regions found in our study.
Transcriptions from Both Strands
At three target regions (At2g32890, At2g03460, and At2g27160), transcripts were obtained from both strands (Fig. 5). As with the transcripts from opposite strands only, the transcripts from the strands opposite from predicted genes have short or nonexistent ORFs. The overlap regions of the pair of transcripts are 95 bp from the At2g32890 region, 695 bp from the At2g03460, and 505 bp from the At2g27160 region. Although antisense transcripts have been found in plants previously, their occurrences and functions are still unclear (Schmitz and Theres, 1992; Dolfini et al., 1993; Cock et al., 1997; Quesada et al., 1999; for review, see Terryn and Rouze, 2000). One report demonstrated that activation of a retrotransposon could change the expression of adjacent genes and result in transcripts from both strands in wheat (Triticum aestivum), in which the transcription from the long terminal repeat of the retrotransposon could produce transcription from the antisense strand of the adjacent gene (Kashkush et al., 2003). However, there are no retrotransposons or transposons on either side of our three targeted genes. The genome-wide transcription study in Arabidopsis also showed 3,027 annotated genes with sense and antisense expression (Yamada et al., 2003). In that study, sense expression of At2g32890 was detected in three out of four RNA populations, and its antisense expression was detected in all four populations. However, neither sense nor antisense expression of At2g03460 or At2g27160 was detected in their study. Similarly, MPSS analysis also detected transcription from both strands of At2g32890, but only sense expression of At2g03460 and At2g27160 was detected.
Dicistronic Transcripts
The transcripts found from four targeted hypothetical gene regions (At2g18200, At2g07708, At2g10060, and At2g14350) show clear dicistronic characteristics (Fig. 6) as defined by (1) two distinct and nonoverlapping coding regions contained within a single processed transcript; and (2) each CDS is more than 100 amino acids. There are likely more dicistronic transcripts in the Arabidopsis genome because, even in our study, there are 33 assemblies matching two loci that extend partially to nearby loci. Additionally, there are at least 20 examples of transcripts corresponding to two adjacent genes found in TIGR's latest annotation process (Haas et al., 2005; ftp://ftp.tigr.org/pub/data/a_thaliana/ath1/DATA_RELEASE_SUPPLEMENT/polyCistronicTranscripts.txt.gz). The mechanism by which the downstream ORF could be translated is still unclear. In the four cases described here, the presence of ATG codons (out of frame) in the intercistronic regions argues against the ribosome scanning mechanism, which requires the absence of any ATG codons in the intercistronic region (Levine et al., 1991). Another mechanism of dicistronic transcript translation is that the presence of an internal ribosome entry site (IRES; Peabody and Berg, 1986) could initiate the translation of the second CDS, which may be more applicable to our cases. IRESs have been identified from different viruses, and they vary in structural, sequence, length, and functional requirements (Belsham and Sonenberg, 2000; Pestova et al., 2001; Dorokhov et al., 2002). To date, except for some reports about the expression of dicistronic constructs made by virus sequences (Zijlstra and Hohn, 1992; Urwin et al., 2000; Toth et al., 2001; Jaag et al., 2003), there is only one report about functional dicistronic transcripts in plants (Garcia-Rios et al., 1997), in which tomPRO1 locus encodes γ-glutamyl kinase and γ-glutamyl phosphate reductase separated by only 5 bp (Garcia-Rios et al., 1997). Coordinate regulation of this pair of genes as a single operon is consistent with their functions in the same biological pathway and suggests that the pairs of hypothetical genes on the dicistronic transcripts reported here may also have related functions. Therefore, functional study of the ORFs in each dicistronic transcript is needed. The intercistronic regions from all four cases in our study are from 45 to 819 bp and do not have obvious consensus sequences among them except for a 10-bp consensus sequence ATCACATGGT obtained from Multiple Em for Motif Elicitation (Bailey and Elkan, 1994). However, except for the shortest 45-bp intercistronic sequence, the other three sequences contain numerous palindrome sequences that might form the hairpin structures that are typical for IRES. More experiments such as protein expression analysis are needed to confirm this possibility. In addition to two proteins being coded by dicistronic transcripts, there is a possibility that a cis-acting selenocysteine insertion sequence (SECIS) located at the 3′ UTR region translates stop codon (UGA) into selenocysteine and results in a fusion protein (Berry et al., 1991; Rother et al., 2001; Howard et al., 2005; for review, see Low and Berry, 1996). An algorithm is now available for identification of SECIS elements (Zhang and Gladyshev, 2005). However, SECISs were not found by searching the sequences of three dicistronic transcripts with TGA stop codon at first ORF (http://genome.unl.edu/SECISearch.html).
Expression Patterns of Hypothetical Genes Tested
Expression patterns from five randomly chosen hypothetical genes (At2g02540, At2g31270, At2g33640, At2g35550, and At2g36340) were examined. The reporter construct driven by the promoter of At2g02540 is expressed in young vascular tissue of above-ground parts of Arabidopsis (Fig. 8B). Several genes that are expressed in vascular tissues belong to the homeodomain-Leu zipper (HD-ZIP) class of transcription factors (Mattsson et al., 2003; for review, see Ye et al., 2002) or have promoters containing a domain recognized by Leu zipper (HD-ZIP) factors (Ayre et al., 2003). However, At2g02540 apparently does not have an HD-ZIP domain but has a domain homology to a group of plant transcription factors named as ZF-HD for zinc-finger homeodomain proteins that might be involved in the mesophyll-specific expression of the C4 phosphoenolpyruvate carboxylase gene in C4 species of the genus Flaveria (Windhovel et al., 2001).
The other four promoter-reporter constructs tested from At2g31270, At2g33640, At2g35550, and At2g36340 are all expressed in different parts of the floral organ at different development stages (Fig. 8). According to the Arabidopsis ABC model of flower development (Coen and Meyerowitz, 1991), At2g31270, which gave strong expression in carpel (whorl 4), should belong to group C. The protein encoded by At2g31270 is similar to the CDT1 protein that promotes DNA replication in yeast (Saccharomyces cerevisiae; Nishitani et al., 2000) and was recently identified as an Arabidopsis CDT1 protein affecting leaf cell division and cell proliferation (Castellano Mdel et al., 2004). Detection of At2g31270 expression in young flowers is consistent with this function. However, Castellano Mdel et al. (2004) also detected its expression in leaf and root. The differences in the length of promoter used and the fusion reporter protein may account for the variation. The promoter-reporter construct of At2g33640 is expressed in pollen, a part of the stamen (whorl 3), and should belong to the group C or B. At2g33640 has a DHHC zinc-finger domain that was first isolated from Drosophila (Mesilaty-Gross et al., 1999), which might be involved in protein-protein or protein-DNA interactions (Putilina et al., 1999). The promoter-reporter construct of At2g35550 is expressed in petals (whorl 2) and stamens (whorl 3) but not in carpels (whorl 4), which should be in gene group B. The protein sequence encoded by At2g35550 has a domain that matches a protein from soybean (Glycine max), GBP, which binds to GAGA element dinucleotide repeat DNA (Sangwan and O'Brian, 2002) and is likely involved in DNA binding. The Gbp gene is expressed in soybean leaves and is induced in symbiotic root nodules (Sangwan and O'Brian, 2002). Interestingly, the promoter-reporter construct of At2g36340 shows expression in young anthers and some young seeds in the siliques, but not in the pollen or some young flowers even in the same inflorescence (Fig. 8F, arrows). At2g36340 was recently reported as a member of a new gene family in Arabidopsis that encodes a nuclear protein with DNA-binding activity that is regulated by KNAT1 (Curaba et al., 2003). However, the expression pattern of At2g36340 detected here is different from the expression pattern of GeBP gene, the typical member of this gene family. GeBP is expressed in the apical meristem and young leaf primordial, but not in flowers (Curaba et al., 2003). This might be due to the fact that At2g36340 was identified as a GeBP family member only by amino acid sequence similarity (Curaba et al., 2003). Although none of the five genes tested are still annotated as “hypothetical protein” in the current TIGR annotation, their biological functions in Arabidopsis are still unclear. To understand these genes, including in which cell types, at which developmental stages they are expressed, and which genes they regulate, more detailed experiments, such as sections, in situ hybridization, knockout, and overexpression are definitely needed. Overall, the promoter-reporter constructs from all five hypothetical genes tested have very localized and/or transient expression patterns providing a likely explanation for their absence from EST or cDNA collections.
MATERIALS AND METHODS
Selection of hypothetical genes, experimental concept, methods of construction of cDNA populations, and RACE reactions are the same as our previous study (Xiao et al., 2002) except that nested PCR was used in the RACE reactions according to the instructions of the Marathon cDNA amplification kit (BD Biosciences Clontech).
Development of a High-Throughput Method
Based on our previous results, a high-throughput pipeline was developed to clone and analyze cDNA sequences of hypothetical genes. The first advance was the development of a Perl script for automatic primer design that combines Primer3 (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3_www.cgi) and BLASTn programs. The primer design script functions as follows: (1) accepts multifasta format input sequence; (2) uses the Primer3 program to design outside primer pair; (3) blasts designed primers against TIGR ATH1 cDNA database to check their uniqueness; and (4) if the outside primers are accepted, Primer3 is used to design a nested primer pair omitting the uniqueness check. This script outputs outer and nested primers for each gene and product sizes based on predicted hypothetical gene structures. The criteria for primer design are that they should be 18- to 35-nt long (optimum 25 nt) having 20% to 80% GC content with melting temperature ≥ 70°C, which enables touchdown PCR. The primers were designed to give a 200- to 500-bp overlap between the 5′ and 3′ RACE products so that the 5′ RACE sequences and 3′ RACE sequences could be assembled together. The second protocol adjustment is the use of 96-well format for all PCR reactions, cloning reactions, transformations into Escherichia coli, and colony screening. For each target gene, 12 clones from 5′ RACE and 3′ RACE cloning reaction were screened by PCR for insert, and all insert-positive clones were sequenced from both ends. A sequence analysis script automatically (1) retrieves sequences for each gene from the sequence database and assembles them using TIGR Assembler (Sutton et al., 1995); (2) retrieves predicted CDS sequence and genomic sequence for each gene from the annotation database; and (3) aligns genomic, predicted CDS, and assembled experimental cDNA sequences together using the dds/gap2 alignment program (Huang et al., 1997) for manual inspection/validation. The entire set of sequences was subsequently mapped to the Arabidopsis genome by PASA for final analysis. This high-throughput method has greatly increased the speed of cloning and analysis of cDNA of hypothetical genes. The primer design script and sequence analysis scripts are available upon request.
Plant Material
Arabidopsis (Arabidopsis thaliana) ecotype Columbia-0 seeds/plants were subjected to a variety of treatments as described below. Callus, roots, and young plants and the treatments of cold and heat were obtained as described in the previous study (Xiao et al., 2002). Pathogen-treated plants in this study were a mixture of Xanthomonas-treated and Pseudomonas-treated plants. Before infection, the plants were grown at 25°C and 8-h photoperiod for 21 d. The leaves were inoculated with fresh cultures, and aerial plant parts were harvested 24-h later for Xanthomonas infection and 12-h later for Pseudomonas infection. For UV treatment, soil-grown plants as above were exposed to 400,000 μj/cm2 or 800,000 μj/cm2 UV light. Aerial plant parts were harvested at 2, 6, 16, and 30 h following treatment. The treatments with salt, 2,4-D, IAA, and H2O2 all were completed with liquid-cultured plants. Sterile seeds were put at 4°C for 48 h and then inoculated into 200 mL of sterile 0.5× Murashige and Skoog salts (Murashige and Skoog 1962), 10 g/L Suc, 0.5× Vitamins-Glycine mix, pH adjusted to 5.8 with KOH. Plants were grown under a 24-h photoperiod with shaking at 100 rpm. Treatments were carried out by adding the chemical challenge to the liquid at its designated final concentration at approximately 14-d postgermination. All tissue (whole plant) was harvested at indicated intervals posttreatment. Treatment concentrations and collection time points are as follows: 250 mm NaCl harvested at 1, 3, 6, and 24 h; 50 μm 2,4-D harvested at 1, 3, 6, and 24 h; 1.0 μm IAA harvested at 1 and 3 h; and 5 mm and 25 mm H2O2 harvested at 1, 3, 8, and 24 h. For all treatments with multiple time points, equal masses of tissue were combined for total RNA isolation with the exception of IAA treatments, for which RNA was isolated for each individual time point and then equal masses of RNA mixed for mRNA isolation.
Vectors Construction, Transformation, and Plant Growth
To create the Gateway-compatible GUS reporter construct pYXT1 from binary vector pBI121, pBI121 was first digested with HindIII and the ends blunted using Klenow. Then, SmaI was used to release the 35S promoter fragment from pBI121. The large fragment lacking the 35S promoter (pBI121 backbone) was then recovered from an agarose gel and dephosphorylated with calf intestinal alkaline phosphatase. Finally, the Gateway reading frame A fragment (Invitrogen) was ligated into the pBI121 backbone and transformed into DB3.1 competent cells (Invitrogen). The plasmid with the correct orientation of the reading frame A insertion is pYXT1.
The Gateway-compatible GFP reporter construct, pYXT2, was made from the binary vector pET-15GAL4-VP16UASmGFP5ER (Bougourd et al., 2000) as follows. First, pET-15GAL4-VP16UASmGFP5ER was digested with SacII. The small fragment (about 3.5 kb) was recovered and cloned into the SacII site in pGEM5Zf (+; Promega) to form an intermediate plasmid. The large fragment was recovered and reserved for religation in the future. The intermediate plasmid was digested with BamHI, dephosphorylated by calf intestinal alkaline phosphatase, then ligated to the Gateway reading frame B (RfB) fragment (Invitrogen) and transformed into DB3.1 competent cells (Invitrogen). Colonies with the correct orientation of RfB insertion were digested with SacII again and the 5.3-kb fragment containing RfB was recovered. This 5.3-kb fragment was ligated into the large fragment obtained from the SacII digestion of pET-15GAL4-VP16UASmGFP5ER. The plasmid with the correct orientation of the SacII insertion containing RfB is pYXT2. All cloning junctions in the pYXT1 and pYXT2 constructs were confirmed by sequencing.
Amplification of the promoter fragments with attB sites, as well as BP and LR reactions, were completed according to the protocols in the Gateway Cloning Technology booklet (Invitrogen). Arabidopsis (Columbia ecotype) genomic DNA was used to amplify promoter fragments of five hypothetical genes. The primers used include gene-specific sequence as well as the attB site sequence for BP cloning and are as follows: At2g02540, upstream primer, AAAAAGCAGGCTTTTGTTTTGGGTGAATATGAAAATCTT, downstream primer, AGAAAGCTGGGTGCCCACCTCCACTATTACCATAACTA; At2g31270, upstream primer, AAAAAGCAGGCTGATCTAGATCAGATTCTTGGTATCA, downstream primer, AGAAAGCTGGGTAATCCATCACCAATCGTTTCTTCGA; At2g33640, upstream primer, AAAAAGCAGGCTTTCCCAGACATACCATAAGAAGCAA, downstream primer, AGAAAGCTGGGTGAAATGTGTGAGCTGGAAGTTGCCA; At2g35550, upstream primer, AAAAAGCAGGCTACTATAGCAACCTGTTCAAGAGACG, downstream primer, AGAAAGCTGGGTAGTCCTTGTTTGCGTTTGTAGCAGA; and At2g36340, upstream primer, AAAAAGCAGGCTATGCTGTACTCTCGATGGTATTCCT, downstream primer, AGAAAGCTGGGTTGAGTGTGTCATCCGAGTTGGTGTC.
The amplified promoter fragments with attB sites were introduced into the pDONR207 entry vector (Invitrogen) using a BP cloning reaction and then transferred into pYXT1 or pYXT2 destination vectors with an LR cloning reaction. Resulting constructs were transformed into Arabidopsis using Agrobacterium tumefaciens (strain GV3101) and transformants were selected on kanamycin, according to standard methods (Clough and Bent, 1998). Three independent transformations were done for each construct.
Sequence data from this article can be found in the GenBank/EMBL data libraries, and accession numbers are shown in Supplemental Table III.
Supplementary Material
Acknowledgments
We thank all members of the plant group at TIGR for their help, especially Bin Feng for his computational support and Robin Buell, Francoise Thibaud-Nissen, and Beverly Underwood for critical reading of the manuscript.
This work was supported by the National Science Foundation (grant no. DBI–9813586).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Yong-Li Xiao (yxiao@tigr.org).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.063479.
References
- Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815 [DOI] [PubMed] [Google Scholar]
- Asakura N, Nakamura C, Ishii T, Kasai Y, Yoshida S (2002) A transcriptionally active maize MuDR-like transposable element in rice and its relatives. Mol Genet Genomics 268: 321–330 [DOI] [PubMed] [Google Scholar]
- Ayre BG, Blair JE, Turgeon R (2003) Functional and phylogenetic analyses of a conserved regulatory program in the phloem of minor veins. Plant Physiol 133: 1229–1239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey TL, Elkan C (1994) Fitting a mixture model by expectation maximization to discover motifs in biopolymer. In R Altman, D Brutlag, P Karp, R Lathrop, D Searls, eds, Proceedings of the Second International Conference on Intelligent Systems for Molecular Biology (ISMB-94). AAAI Press, Stanford, CA, pp 28–36A [PubMed]
- Belsham GJ, Sonenberg N (2000) Picornavirus RNA translation: roles for cellular proteins. Trends Microbiol 8: 330–335 [DOI] [PubMed] [Google Scholar]
- Berry MJ, Banu L, Chen YY, Mandel SJ, Kieffer JD, Harney JW, Larsen PR (1991) Recognition of UGA as a selenocysteine codon in type I deiodinase requires sequences in the 3′ untranslated region. Nature 353: 273–276 [DOI] [PubMed] [Google Scholar]
- Bougourd S, Marrison J, Haseloff J (2000) Technical advance: an aniline blue staining procedure for confocal microscopy and 3D imaging of normal and perturbed cellular phenotypes in mature Arabidopsis embryos. Plant J 24: 543–550 [DOI] [PubMed] [Google Scholar]
- Brendel V, Kleffe J (1998) Prediction of locally optimal splice sites in plant pre-mRNA with applications to gene identification in Arabidopsis thaliana genomic DNA. Nucleic Acids Res 26: 4748–4757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brett D, Hanke J, Lehmann G, Haase S, Delbruck S, Krueger S, Reich J, Bork P (2000) EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett 474: 83–86 [DOI] [PubMed] [Google Scholar]
- Burge C, Karlin S (1997) Prediction of complete gene structures in human genomic DNA. J Mol Biol 268: 78–94 [DOI] [PubMed] [Google Scholar]
- Castellano Mdel M, Boniotti MB, Caro E, Schnittger A, Gutierrez C (2004) DNA replication licensing affects cell proliferation or endoreplication in a cell type-specific manner. Plant Cell 16: 2380–2393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chory J, Ecker JR, Briggs S, Caboche M, Coruzzi GM, Cook D, Dangl J, Grant S, Guerinot ML, Henikoff S, et al (2000) National Science Foundation-sponsored workshop report: “The 2010 Project” functional genomics and the virtual plant: blueprint for understanding how plants are built and how to improve them. Plant Physiol 23: 423–426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough SJ, Bent AF (1998) Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16: 735–743 [DOI] [PubMed] [Google Scholar]
- Cock JM, Swarup R, Dumas C (1997) Natural antisense transcripts of the S locus receptor kinase gene and related sequences in Brassica oleracea. Mol Gen Genet 255: 514–524 [DOI] [PubMed] [Google Scholar]
- Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353: 31–37 [DOI] [PubMed] [Google Scholar]
- Curaba J, Herzog M, Vachon G (2003) GeBP, the first member of a new gene family in Arabidopsis, encodes a nuclear protein with DNA-binding activity and is regulated by KNAT1. Plant J 33: 305–317 [DOI] [PubMed] [Google Scholar]
- Dean C, Tamaki S, Dunsmuir P, Favreau M, Katayama C, Dooner H, Redbrook J (1986) mRNA transcripts of several plant genes are polyadenylated at multiple sites in vivo. Nucleic Acids Res 5: 2229–2240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dolfini S, Consonni G, Mereghetti M, Tonelli C (1993) Antiparallel expression of the sense and antisense transcripts of maize alpha-tubulin genes. Mol Gen Genet 241: 161–169 [DOI] [PubMed] [Google Scholar]
- Dorokhov YL, Skulachev MV, Ivanov PA, Zvereva SD, Tjulkina LG, Merits A, Gleba YY, Hohn T, Atabekov JG (2002) Polypurine (A)-rich sequences promote cross-kingdom conservation of internal ribosome entry. Proc Natl Acad Sci USA 99: 5301–5306 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohman MA, Dush MK, Martin GR (1988) Rapid production of full-length cDNAs from rare transcripts: amplification using a single gene-specific oligonucleotide primer. Proc Natl Acad Sci USA 85: 8998–9002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Rios M, Fujita T, LaRosa PC, Locy RD, Clithero JM, Bressan RA, Csonka LN (1997) Cloning of a polycistronic cDNA from tomato encoding gamma-glutamyl kinase and gamma-glutamyl phosphate reductase. Proc Natl Acad Sci USA 94: 8249–8254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graber JH, Cantor CR, Mohr SC, Smith TF (1999) In silico detection of control signals: mRNA 3′-end-processing sequences in diverse species. Proc Natl Acad Sci USA 96: 14055–14060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK Jr, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al (2003) Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res 31: 5654–5666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas BJ, Volfovsky N, Town CD, Troukhan M, Alexandrov N, Feldmann KA, Flavell RB, White O, Salzberg SL (2002) Full-length messenger RNA sequences greatly improve genome annotation. Genome Biol 3: RESEARCH0029 [DOI] [PMC free article] [PubMed]
- Haas BJ, Wortman JR, Ronning CM, Hannick LI, Smith RK Jr, Maiti R, Chan AP, Yu C, Farzad M, Wu D, et al (2005) Complete reannotation of the Arabidopsis genome: methods, tools, protocols and the final release. BMC Biol 3: 7–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartung F, Puchta H (2000) Molecular characterisation of two paralogous SPO11 homologues in Arabidopsis thaliana. Nucleic Acids Res 28: 1548–1554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebsgaard SM, Korning PG, Tolstrup N, Engelbrecht J, Rouze P, Brunak S (1996) Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res 24: 3439–3452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howard MT, Aggarwal G, Anderson CB, Khatri S, Flanigan KM, Atkins JF (2005) Recoding elements located adjacent to a subset of eukaryal selenocysteine-specifying UGA codons. EMBO J 24: 1596–1607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Adams MD, Zhou H, Kerlavage AR (1997) A tool for analyzing and annotating genomic sequences. Genomics 46: 37–45 [DOI] [PubMed] [Google Scholar]
- Iida K, Seki M, Sakurai T, Satou M, Akiyama K, Toyoda T, Konagaya A, Shinozaki K (2004) Genome-wide analysis of alternative pre-mRNA splicing in Arabidopsis thaliana based on full-length cDNA sequences. Nucleic Acids Res 32: 5096–5103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaag HM, Kawchuk L, Rohde W, Fischer R, Emans N, Prufer D (2003) An unusual internal ribosomal entry site of inverted symmetry directs expression of a potato leafroll polerovirus replication-associated protein. Proc Natl Acad Sci USA 100: 8939–8944 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jasinski S, Perennes C, Bergounioux C, Glab N (2002) Comparative molecular and functional analyses of the tobacco cyclin-dependent kinase inhibitor NtKIS1a and its spliced variant NtKIS1b. Plant Physiol 130: 1871–1882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones L (2002) Revealing micro-RNAs in plants. Trends Plant Sci 7: 473–475 [DOI] [PubMed] [Google Scholar]
- Kan Z, Rouchka EC, Gish WR, States DJ (2001) Gene structure prediction and alternative splicing analysis using genomically aligned ESTs. Genome Res 11: 889–900 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kashkush K, Feldman M, Levy AA (2003) Transcriptional activation of retrotransposons alters the expression of adjacent genes in wheat. Nat Genet 33: 102–106 [DOI] [PubMed] [Google Scholar]
- Kim H, Snesrud EC, Haas B, Cheung F, Town CD, Quackenbush J (2003) Gene expression analyses of Arabidopsis chromosome 2 using a genomic DNA amplicon microarray. Genome Res 13: 327–340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazar G, Goodman HM (2000) The Arabidopsis splicing factor SR1 is regulated by alternative splicing. Plant Mol Biol 42: 571–581 [DOI] [PubMed] [Google Scholar]
- Levine F, Yee JK, Friedmann T (1991) Efficient gene expression in mammalian cells from a dicistronic transcriptional unit in an improved retroviral vector. Gene 108: 167–174 [DOI] [PubMed] [Google Scholar]
- Low SC, Berry MJ (1996) Knowing when not to stop: selenocysteine incorporation in eukaryotes. Trends Biochem Sci 21: 203–208 [PubMed] [Google Scholar]
- Lukashin AV, Borodovsky M (1998) GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res 26: 1107–1115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupold DS, Caoile AG, Stern DB (1999) Polyadenylation occurs at multiple sites in maize mitochondrial cox2 mRNA and is independent of editing status. Plant Cell 11: 1565–1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacIntosh GC, Wilkerson C, Green PJ (2001) Identification and analysis of Arabidopsis expressed sequence tags characteristic of non-coding RNAs. Plant Physiol 127: 765–776 [PMC free article] [PubMed] [Google Scholar]
- Magnotta SM, Gogarten J (2002) Multi site polyadenylation and transcriptional response to stress of a vacuolar type H+-ATPase subunit A gene in Arabidopsis thaliana. BMC Plant Biol 2: 3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mano S, Hayashi M, Nishimura M (1999) Light regulates alternative splicing of hydroxypyruvate reductase in pumpkin. Plant J 17: 309–320 [DOI] [PubMed] [Google Scholar]
- Mano S, Hayashi M, Nishimura M (2000) A leaf-peroxisomal protein, hydroxypyruvate reductase, is produced by light-regulated alternative splicing. Cell Biochem Biophys 32: 147–154 [DOI] [PubMed] [Google Scholar]
- Mattsson J, Ckurshumova W, Berleth T (2003) Auxin signaling in Arabidopsis leaf vascular development. Plant Physiol 131: 1327–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mesilaty-Gross S, Reich A, Motro B, Wides R (1999) The Drosophila STAM gene homolog is in a tight gene cluster, and its expression correlates to that of the adjacent gene ial. Gene 231: 173–186 [DOI] [PubMed] [Google Scholar]
- Meyers BC, Vu TH, Tej SS, Ghazal H, Matvienko M, Agrawal V, Ning J, Haudenschild CD (2004) Analysis of the transcriptional complexity of Arabidopsis thaliana by massively parallel signature sequencing. Nat Biotechnol 22: 1006–1011 [DOI] [PubMed] [Google Scholar]
- Modrek B, Resch A, Grasso C, Lee C (2001) Genome-wide detection of alternative splicing in expressed sequences of human genes. Nucleic Acids Res 29: 2850–2859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol Plant 15: 473–497 [Google Scholar]
- Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R (2004) Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J 39: 877–885 [DOI] [PubMed] [Google Scholar]
- Nishitani H, Lygerou Z, Nishimoto T, Nurse PX (2000) The Cdt1 protein is required to license DNA for replication in fission yeast. Nature 404: 625–628 [DOI] [PubMed] [Google Scholar]
- Peabody DS, Berg P (1986) Termination-reinitiation occurs in the translation of mammalian cell mRNAs. Mol Cell Biol 6: 2695–2703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova TV, Kolupaeva VG, Lomakin IB, Pilipenko EV, Shatsky IN, Agol VI, Hellen CU (2001) Molecular mechanisms of translation initiation in eukaryotes. Proc Natl Acad Sci USA 98: 7029–7036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Putilina T, Wong P, Gentleman S (1999) The DHHC domain: a new highly conserved cysteine-rich motif. Mol Cell Biochem 195: 219–226 [DOI] [PubMed] [Google Scholar]
- Quesada V, Macknight R, Dean C, Simpson GG (2003) Autoregulation of FCA pre-mRNA processing controls Arabidopsis flowering time. EMBO J 22: 3142–3152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quesada V, Ponce MR, Micol JL (1999) OTC and AUL1, two convergent and overlapping genes in the nuclear genome of Arabidopsis thaliana. FEBS Lett 461: 101–106 [DOI] [PubMed] [Google Scholar]
- Redman JC, Haas BJ, Tanimoto G, Town CD (2004) Development and evaluation of an Arabidopsis whole genome Affymetrix probe array. Plant J 38: 545–561 [DOI] [PubMed] [Google Scholar]
- Rother M, Resch A, Gardner WL, Whitman WB, Bock A (2001) Heterologous expression of archaeal selenoprotein genes directed by the SECIS element located in the 3′ non-translated region. Mol Microbiol 40: 900–908 [DOI] [PubMed] [Google Scholar]
- Sangwan I, O'Brian MR (2002) Identification of a soybean protein that interacts with GAGA element dinucleotide repeat DNA. Plant Physiol 129: 1788–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savaldi-Goldstein S, Aviv D, Davydov O, Fluhr R (2003) Alternative splicing modulation by a LAMMER kinase impinges on developmental and transcriptome expression. Plant Cell 15: 926–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz G, Theres K (1992) Structural and functional analysis of the Bz2 locus of Zea mays: characterization of overlapping transcripts. Mol Gen Genet 233: 269–277 [DOI] [PubMed] [Google Scholar]
- Simpson GG, Dijkwel PP, Quesada V, Henderson I, Dean C (2003) FY is an RNA 3′ end-processing factor that interacts with FCA to control the Arabidopsis floral transition. Cell 113: 777–787 [DOI] [PubMed] [Google Scholar]
- Sutton G, White O, Adams MD, Kerlavage AR (1995) TIGR Assembler: a new tool for assembling large shotgun sequencing projects. Genome Sci Technol 1: 9–19 [Google Scholar]
- Terryn N, Rouze P (2000) The sense of naturally transcribed antisense RNAs in plants. Trends Plant Sci 5: 394–396 [DOI] [PubMed] [Google Scholar]
- Toth RL, Chapman S, Carr F, Santa Cruz S (2001) A novel strategy for the expression of foreign genes from plant virus vectors. FEBS Lett 489: 215–219 [DOI] [PubMed] [Google Scholar]
- Uberbacher EC, Mural RJ (1991) Locating protein-coding regions in human DNA sequences by a multiple sensor-neural network approach. Proc Natl Acad Sci USA 88: 11261–11265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urwin P, Yi L, Martin H, Atkinson H, Gilmartin PM (2000) Functional characterization of the EMCV IRES in plants. Plant J 24: 583–589 [DOI] [PubMed] [Google Scholar]
- Wen J, Lease KA, Walker JC (2004) DVL, a novel class of small polypeptides: overexpression alters Arabidopsis development. Plant J 37: 668–677 [DOI] [PubMed] [Google Scholar]
- Windhovel A, Hein I, Dabrowa R, Stockhaus J (2001) Characterization of a novel class of plant homeodomain proteins that bind to the C4 phosphoenolpyruvate carboxylase gene of Flaveria trinervia. Plant Mol Biol 45: 201–214 [DOI] [PubMed] [Google Scholar]
- Xia Y (2004) Peptides as signals. In A Fleming, ed, Intercellular Communication in Plants. Blackwell Publishing, Oxford, pp 27–48
- Xiao YL, Malik M, Whitelaw CA, Town CD (2002) Cloning and sequencing of cDNAs for hypothetical genes from chromosome 2 of Arabidopsis. Plant Physiol 130: 2118–2128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K, Lim J, Dale JM, Chen H, Shinn P, Palm CJ, Southwick AM, Wu HC, Kim C, Nguyen M, et al (2003) Empirical analysis of transcriptional activity in the Arabidopsis genome. Science 302: 842–846 [DOI] [PubMed] [Google Scholar]
- Ye Z-H, Freshour G, Hahn MG, Burk DH, Zhong R (2002) Vascular development in Arabidopsis. Int Rev Cytol 220: 225–256 [DOI] [PubMed] [Google Scholar]
- Zhang Y, Gladyshev VN (2005) An algorithm for identification of bacterial selenocysteine insertion sequence elements and selenoprotein genes. Bioinformatics 21: 2580–2589 [DOI] [PubMed] [Google Scholar]
- Zhou DX, Kim YJ, Li YF, Carol P, Mache R (1998) COP1b, an isoform of COP1 generated by alternative splicing, has a negative effect on COP1 function in regulating light-dependent seedling development in Arabidopsis. Mol Gen Genet 257: 387–391 [DOI] [PubMed] [Google Scholar]
- Zhu W, Schlueter SD, Brendel V (2003) Refined annotation of the Arabidopsis genome by complete expressed sequence tag mapping. Plant Physiol 132: 469–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zijlstra C, Hohn T (1992) Cauliflower mosaic virus gene VI controls translation from dicistronic expression units in transgenic Arabidopsis plants. Plant Cell 4: 1471–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.