Abstract
We have isolated and characterized the Nectarin IV (NEC4) protein that accumulates in the nectar of ornamental tobacco plants (Nicotiana langsdorffii × Nicotiana sanderae var LxS8). This 60-kD protein has a blocked N terminus. Three tryptic peptides of the protein were isolated and sequenced using tandem mass spectroscopy. These unique peptides were found to be similar to the xyloglucan-specific fungal endoglucanase inhibitor protein (XEGIP) precursor in tomato (Lycopersicon esculentum) and its homolog in potato (Solanum tuberosum). A pair of oligonucleotide primers was designed based on the potato and tomato sequences that were used to clone a 1,018-bp internal piece of nec4 cDNA from a stage 6 nectary cDNA library. The remaining portions of the cDNA were subsequently captured by 5′ and 3′ rapid amplification of cDNA ends. Complete sequencing of the nec4 cDNA demonstrated that it belonged to a large family of homologous proteins from a wide variety of angiosperms. Related proteins include foliage proteins and seed storage proteins. Based upon conserved identity with the wheat (Triticum aestivum) xylanase inhibitor TAXI-1, we were able to develop a protein model that showed that NEC4 contains additional amino acid loops that are not found in TAXI-1 and that glycosylation sites are surface exposed. Both these loops and sites of glycosylation are on the opposite face of the NEC4 molecule from the site that interacts with fungal hemicellulases, as indicated by homology to TAXI-I. NEC4 also contains a region homologous to the TAXI-1 knottin domain; however, a deletion in this domain restructures the disulfide bridges of this domain, resulting in a pseudoknottin domain. Inhibition assays were performed to determine whether purified NEC4 was able to inhibit fungal endoglucanases and xylanases. These studies showed that NEC4 was a very effective inhibitor of a family GH12 xyloglucan-specific endoglucanase with a Ki of 0.35 nm. However, no inhibitory activity was observed against other family GH10 or GH11 xylanases. The patterns of expression of the NEC4 protein indicate that, while expressed in nectar at anthesis, it is most strongly expressed in the nectary gland after fertilization, indicating that inhibition of fungal cell wall-degrading enzymes may be more important after fertilization than before.
Many higher plants have evolved unique strategies for successful fertilization and propagation that rely on animal-mediated pollen transfer. A metabolically rich nectar reward is one of the most important factors in this strategy. The attraction of these pollinators results in a more efficient transfer of pollen from flower to flower, increasing genetic out-crossing and fecundity. The nectar produced by most plants is a rich combination of substances, primarily Suc, Glc, and Fru. However, other carbohydrates occur in some nectars (Baker and Baker, 1981). All 20 normal amino acids have been identified in various nectars (Baker and Baker, 1973). Other substances include organic acids (Baker and Baker, 1975), terpenes (Ecroyd et al., 1995), alkaloids (Deinzer et al., 1977), flavonoids (Rodriguez-Arce and Diaz, 1992), glycosides (Roshchina and Roshchina, 1993), vitamins (Griebel and Hess, 1940), phenolics (Ferreres et al., 1996; Cabras et al., 1999), oils (Vogel, 1969), and five proteins, termed nectarins (Carter et al., 1999; Thornburg et al., 2003).
We have previously characterized four of the five nectar proteins (Carter et al., 1999; Carter and Thornburg, 2000, 2003, 2004a, 2004c); however, Nectarin IV (NEC4) had remained elusive until this study. Our earlier work has led us to the conclusion that the nectar proteins are present in nectar to protect the gynoecium and developing embryos therein from microbial attack caused by nonsterile visiting pollinators (Thornburg et al., 2003). This observation is based on the identification of a novel nectar-contained biochemical pathway, termed the Carter-Thornburg nectar redox cycle (Carter and Thornburg, 2004b). This pathway generates high levels of hydrogen peroxide, and its primary function is to maintain nectar in an axenic state.
In this study, we have isolated and characterized the cDNA encoding NEC4 and have demonstrated that the NEC4 protein functions as a xyloglucan-specific fungal endoglucanase-inhibitor protein (XEGIP). This confirms that this nectar protein is also a defense-related protein. Because NEC4 was the last of the nectar proteins to be isolated and characterized, these studies have permitted us to complete the characterization of the entire tobacco nectar proteome and have allowed us to demonstrate that the primary function of each of the nectar proteins is in plant defense or stress responses.
RESULTS
Identification of NEC4
Because automated Edman degradation of NEC4 isolated from SDS-PAGE gels does not identify any N-terminal sequence, we had previously concluded that the NEC4 N terminus is apparently blocked (Carter and Thornburg, 2004a). Therefore, to obtain sequence information to permit the cloning of the NEC4 cDNA, we used mass spectrometry (MS) sequencing of NEC4 tryptic peptides. To generate these sequences from NEC4, crude nectar was run on an SDS-PAGE and the protein bands corresponding to NEC4 were excised and subjected to tryptic peptide tandem mass spectroscopy (MS/MS) sequencing. Peptide sequences were derived by comparisons of the masses of the b- and y-series ions. This analysis identified one peptide with a mass of 1,712.85 amu (ISLPSQFSAEFSFPR; Table I) that matched the identity of a tomato (Lycopersicon esculentum) protein (AAN87262). This tomato protein is a XEGIP (Qin et al., 2003). Subsequently, two additional peptides (VAAVAPFKVCFDSR and LGFTSSILFR) were identified. These peptides matched a potato (Solanum tuberosum) protein (AAP84703) that is the potato homolog of the tomato XEGIP.
Table I.
Peptides sequenced from tryptic digest of NEC4
| Number | Peptide 1 | Peptide 2 | Peptide 3 |
|---|---|---|---|
| Tobacco NEC4 | ISLPSQFSAEFSFPR | LGFTSSILFR | VASVAPFKV–CFDSR |
| AY155579a | ISLPSQFSAEFSFPR | LGFTSSILFR | VAAVA-FKVCKCFDSR |
| Location in AY155579 | 187–201 | 415–424 | 325–338 |
GenBank accession number for the tomato XEGIP.
Cloning of the NEC4 cDNA
To isolate the NEC4 cDNA, we utilized a strategy that was based upon the common identities of the tomato and potato XEGIP homologs. ClustalW pileup analysis of the gene sequences encoding the proteins AAN87262 and AAP84703 (accession nos. AY155579 and AY321357) permitted us to identify conserved nucleotide sequences and to design a pair of conserved oligonucleotide primers for PCR amplification. These oligonucleotides were used to generate a PCR product from an ornamental tobacco (Nicotiana langsdorffii × Nicotiana sanderae var LxS8) nectary cDNA library. The PCR product was cloned into pGEM-T. This partial cDNA clone (pRT537) was sequenced to generate additional oligonucleotides used together with oligonucleotides designed from library vector sequences to amplify the 5′ and 3′ ends of the cDNA from a stage 6 nectary cDNA library. After the first round and a nested round of PCR, the 3′ RACE identified two independent nearly full-length clones that contained the 3′ ends of the NEC4 clone (pRT540 and 541). These 3′ ends were >99% identical. They differed by three single nucleotide changes (C845, G994, and T1094 of pRT540 were T845, A994, and C1094 in pRT541) and a single 32-bp deletion in the 3′ untranslated region of pRT540 relative to pRT541 at positions 1,298 to 1,330. The single nucleotide changes were all located in the coding region and each of them results in amino acid changes: Asp-289, Val-339, and Val-372 in pRT540 were Val-289, Thr-339, and Ala-372 in pRT541. The polyA addition sites also differed between these two clones; pRT540 was 48 nucleotides longer than pRT541. These cDNA differences may represent recently diverged, closely related genes that are similarly expressed; however, because the plants used in this study were derived from an interspecific cross between two closely related tobacco species, the two sequences may represent the NEC4 sequences from these two species. The 5′ RACE identified a number of short, identical clones, one of which was selected as pRT544.
Analysis of the cDNA
Comparisons of the isolated NEC4 cDNA sequences with the 5′ and 3′ RACE sequences permitted us to generate a full-length NEC4 cDNA. This sequence was deposited in GenBank (accession no. AY898762). This 1,618-nucleotide composite cDNA contains no significant hairpins or repeat elements. The 5′ and 3′ untranslated regions are 36 and 237 nucleotides, respectively. The cDNA encodes a 437-amino acid protein with high identity to the tomato and potato XEGIPs. The NEC4 protein shares 88.8% identity with the potato clone (AY321357) and 88.3% identity with the tomato clone (AY155579). The potato and tomato clones share 96.6% identity. To further confirm that the NEC4 cDNA encoded the NEC4 protein, NEC4 was isolated from floral nectar and subjected to tryptic peptide mass fingerprinting. This analysis (Table II) confirmed the presence of additional peptides that were encoded by the NEC4 cDNA. Together with the sequences of the three peptides that were sequenced from the NEC4 protein, this confirms that 28.7% of the NEC4 coding region corresponds to amino acid sequences in the mature NEC4 protein.
Table II.
MALDI-TOF identification of additional NEC4 peptides
| Peptide
|
Mass
|
Position
|
Sequence
|
|
|---|---|---|---|---|
| Observed | Predicted | |||
| 1 | 502.213 | 502.298 | 263–266 | INQK |
| 2 | 676.399 | 676.003 | 332–337 | TIVSTR |
| 3 | 1,673.838 | 1,673.861 | 33–46 | DASTLQYLTQIHQR |
| 4 | 1,712.839 | 1,712.864 | 179–193 | ISLPSQFSAEFSFPR |
| 5 | 1,737.866 | 1,737.896 | 207–222 | GVVLFGDGPYSFLPNR |
| 6 | 2,734.434 | 2,734.349 | 287–310 | ISTVNPYTILETSIYNAVTNFFVK |
NEC4 N Terminus
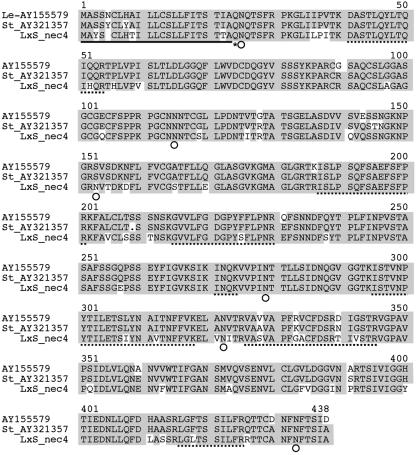
Examination of the N-terminal sequence of the NEC4 protein (McGeoch, 1985; Nakai and Kanehisa, 1991) revealed that the NEC4 N terminus likely contains a 22-amino acid signal sequence (Fig. 1). This is consistent with the extracellular localization of the NEC4 protein. This sequence is also similar to the signal sequences of each of the other tobacco nectarins. After cleavage of the N-terminal signal sequence at Ala-22, the resulting N-terminal amino acid is Gln (Gln-23), which is consistent with the presence of a pyroglutamyl-blocked N terminus that is refractory to Edman degradation. Together with the identification of almost 30% of the mature NEC4 coding region from the NEC4 protein (Fig. 1; Tables I and II), the finding that the NEC4 clone should also have a blocked N terminus convinced us that the NEC4 cDNA did indeed encode the NEC4 protein from the nectar of ornamental tobacco.
Figure 1.
Alignment of the amino acid sequences of NEC4 with the tomato and potato XEGIPs AY155579 and AY321357, respectively. Shaded sequences show identity between the clones. The N-terminal signal sequence is indicated by a single underline. The N-terminal Gln is indicated by an asterisk. Sites of N-glycosylation are indicated by white circles. Tryptic peptides identified either by sequencing or by MALDI MS fingerprinting are indicated by dashed underlines.
NEC4 Model
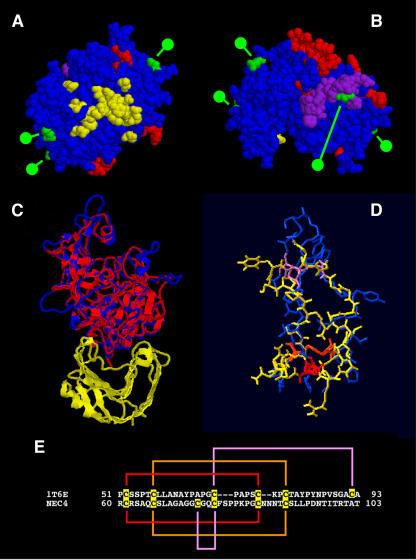
Using the amino acid sequence of the NEC4 protein, we searched the Protein Data Bank (PDB) to identify similar proteins. One protein, the wheat (Triticum aestivum) xylanase inhibitor (PDB no. 1T6E), showed 29.1% identity and 53.4% similarity to the protein sequence predicted from the NEC4 cDNA. Using the coordinates of the wheat xylanase inhibitor TAXI-1, a model was built by threading the NEC4 amino acid sequence through those coordinates. The model was optimized using the backbone atoms only, and, after two rounds of structural alignment, the optimization procedure produced a usable model. This model is based on 399 amino acids of the NEC4 sequence from Lys-31 to Cys-428, and contained 1,292 protein backbone atoms that showed a root mean square (RMS) deviation of 0.57 Å from the wheat xylanase inhibitor. The models are shown in Figure 2. The most disordered regions in the tobacco model relative to the wheat inhibitors corresponded to six loops caused by insertions of between three to 10 amino acids in the tobacco sequence (shown as red residues in Fig. 2, A and B). These loops are not present in the wheat xylanase inhibitor. If we excluded these loops from the NEC4 model, the RMS deviation of the remaining 1,228 backbone atoms dropped to 0.40 Å, indicating that the remainder of the molecular conformation is well conserved between the two proteins. Subsequently, a second model was built using the wheat xylanase inhibitor complexed with the Aspergillus niger xylanase I (1T6G). In this second model, the optimized NEC4 protein model was overlaid onto the structure of the wheat xylanase inhibitor (Fig. 2C). This model also contained 399 amino acids of the optimized NEC4 protein sequence and the 1,300 NEC4 backbone atoms that showed a RMS deviation of 0.61 Å with the wheat inhibitor.
Figure 2.
NEC4 protein models based on the structure of the TAXI-1. A and B, Model based on threading of NEC4 sequence through the coordinates of 1T6E. Residues shown in red represent the loops found in the tobacco NEC4 but not in TAXI-1. Residues shown in green indicate the locations of putative N-glycosylation sites. Shown in yellow are NEC4 residues that are homologous to TAXI-I residues that are within 4 Å of its ligand (an A. niger xylanase) as identified from the model shown in C. Residues shown in purple indicate the location of the pseudoknottin domain. A, Bottom of the NEC4 model looking up from the hemicellulase ligand (assuming that NEC4 binds its XEG ligand with a geometry that is similar to the binding of xylanase by TAXI-I). B, Top of the same NEC4 model. C, Second model based on the replacement of the TAXI-1 molecule of 1T6G with the refined NEC4 model; the ribbon diagram of the A. niger xylanase is in yellow and the ribbon diagram of TAXI-1 is in red. This is overlaid with the NEC4 Cα backbone (in blue). D, Knottin domain of TAXI-1 in blue overlaid with the pseudoknottin domain of NEC4 (in yellow). The three disulfide linkages are color coded (red, orange, and purple). E, Amino acid sequences of the knottin domain of TAXI-1 and the corresponding pseudoknottin domain of NEC4. Cys residues are highlighted and disulfide linkages are color coded to match those shown in D.
The yellow region of Figure 2A shows the amino acids that are homologous to the TAXI-1 xylanase-binding site. The loops (red) and sites of glycosylation (green) are also shown on the model (Fig. 2, A and B). Interestingly, all of the loops and carbohydrates are on the opposite face of the NEC4 molecule from the region that is homologous to the TAXI-1 xylanase-binding site. Thus, we expect that these loops and glycosylation sites would not interfere with interactions with the glycosylhydrolase ligand, assuming its interaction with NEC4 is similar to the TAXI-1 xylanase interaction. Alternatively, the possibility exists that these loops could be alternative binding sites for other glycosylhydrolases.
The TAXI-1 molecule contains a knottin-like domain (Sansen et al., 2004). While this domain is generally conserved in the NEC4 molecule, the amino acid sequence of TAXI-1 and NEC4 differ in this region, resulting in an alteration of the disulfide bridge pattern and the formation of a pseudoknottin domain in NEC4. As shown in Figure 2E, the Ala-65 and Cys-92 of TAXI-1 are changed to Cys-66 and Ala-102 in NEC4. Based upon the results of our model analysis, there is an apparent change of disulfide bridges from the ABCABC pattern found in classical knottin domains to an ABCCAB pattern found in NEC4. Still, the overall features of the knottin domain are conserved in the NEC4 molecule. Figure 2D shows this domain from NEC4 overlaid on the TAXI-1 knottin domain. There is generally good alignment in the structures. The NEC4 pseudoknottin domain is shown in purple in Figure 2B. Like the novel loops and putative glycosylation sites, the conserved knottin domain is surface exposed and is located on the opposite face of the NEC4 molecule from the region that is homologous to the TAXI-1 xylanase-binding site. Whether the NEC4 pseudoknottin domain maintains knottin domain functions is unknown.
Further, like TAXI-1, the NEC4 molecule also has overall structural homology with aspartic proteases (Sansen et al., 2004). These proteases contain a pair of highly conserved Asp-Thr-Gly motifs at the active site. In the NEC4 model, these residues are Asp-37-Leu-38-Gly-39 and Ser-269-Thr-270-Val-271, suggesting that the proteolytic activity has not been conserved. This is consistent with our earlier findings that the protein profile of nectar is quite stable and does not show proteolytic degradation with time.
NEC4 Glycosylation
The mature protein sequence contains six potential sites for N-linked glycosylation. These are Asn-24, Asn-114, Asn-152, Asn-278, Asn-321, and Asn-432. The difference between the observed molecular mass of the NEC4 protein (60 kD) and that predicted from the amino acid sequence (44.6 kD) suggests that at least some of these potential sites must be glycosylated. Four of these N-glycosylation sites are present in the NEC4 model, and each of them is surface exposed as would be expected if they are glycosylated (Fig. 2, A and B, green). Consistent with this is the finding that the NEC4 protein binds to a concanavalin A affinity column (C. Carter and R.W. Thornburg, unpublished data). These results are similar to those obtained for XEGIP isolated from suspension-cultured tomato cells (Qin et al., 2003).
NEC4 Phylogenetic Relationships
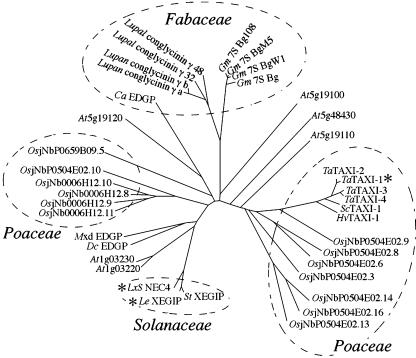
BLAST analysis of the amino acid sequence predicted from the NEC4 cDNA identified a large family of NEC4 homologs each having an E-score of < e−20. As was previously observed, three main classes of genes were identified in this analysis: XEGIP-related proteins, TAXI-related proteins, and seed storage proteins (York et al., 2004). We performed a phylogenetic analysis of this gene family using the online ClustalW tool (http://clustalw.genome.jp). From the multiple sequence alignment, we developed the phylogenetic tree shown in Figure 3, using the neighbor-joining method (Saitou and Nei, 1987). This analysis demonstrates that the NEC4 homologs fall into several clades that generally distinguish plant families. The NEC4 homologs are from the Solanaceae, the Fabaceae, and the Poaceae cluster. However, various Arabidopsis (Arabidopsis thaliana) homologs are dispersed throughout this gene family tree.
Figure 3.
Unrooted neighbor-joining tree for the NEC4-related genes. Highly homologous (E scores < 1e−20) protein sequences were aligned using ClustalW and an unrooted nonjoining tree was developed. The NEC4, tomato XEGIP, and wheat TAXI-1 gene products (marked by asterisks) have been shown to inhibit fungal hemicellulases. Three plant families, Solanaceae, Poaceae, and Fabaceae, are indicated by dashed circles. The protein sequences were derived from the following gene sequences: LxS NEC4 (ornamental tobacco), accession number AY898762; StXEGIP (potato), AY321357; LeXEGIP (tomato), AY155579; DcEDGP (carrot), D14550; Mxd EDGP (apple), AY347867; At1g03220 (Arabidopsis), AY085016; At1g03230 (Arabidopsis), AF332411; At5g19100 (Arabidopsis), NM_121915; At5g19110 (Arabidopsis), NM_121916; At5g19120 (Arabidopsis), AY055793; At5g48430 (Arabidopsis), AB020745; OsjNb0006J12.8 (rice), AC120991.3; OsjNb0006J12.9 (rice), AC120991.3; OsjNb0006J12.10 (rice), AC120991.3; OsjNb0006J12.11 (rice), AC120991.3; OsjNb0504E02.3 (rice), NM_190067; OsjNb0504E02.6 (rice), NM_190069; OsjNb0504E02.8 (rice), NM_190071; OsjNb0504E02.9 (rice), NM_190072; OsjNb0504E02.10 (rice), NM_190073; OsjNb0504E02.13 (rice), NM_190075; OsjNb0504E02.14 (rice), NM_190076; OsjNb0504E02.16 (rice), NM_190078; OsjNbP0659B09.5 (rice), AP005886.3; CaEDGP (chickpea), AJ299060; Lupal conglutin γ clone 32 (white lupin), AJ297490; Lupal conglutin γ clone 48 (white lupin), AJ297568; Lupan conglutin γa (narrow leaved blue lupin), X65601; Lupan conglutin γb (narrow leaved blue lupin), X65601; Gm7SBg (soybean), AB084260; Gm7SBg108 (soybean), X16469; Gm7SBPM5 (soybean), U59425.1; Gm7SBgW1 (soybean), D16107; HvXI-1 (barley), AJ581529; ScXI-1 (rye), AJ581532; TaXI-1 (wheat), AB114626; TaXI-2 (wheat), AJ438880; TaXI-3 (wheat), AB178471; and TaXI-4 (wheat), AB114628.
Based upon this analysis, we can derive several features about the biology of this gene family. First, the 13 rice (Oryza sativa) homologs identify two islands of tandemly repeated genes within the rice genome. One is present on the bacterial artificial chromosome clone 006J12 (four sequences). The other is present on the bacterial artificial chromosome clone 504E02 (eight sequences). Similarly, five of the six Arabidopsis NEC4 homologs are clustered in a pair of tandem arrays on chromosomes 1 and 5.
Second, the gene sequences from the Fabaceae were previously identified as seed storage proteins in lupins (Lupinus sp.), chickpea (Cicer arietinum), and soybeans (Glycine max; York et al., 2004). Thus, the expression of this diverse gene family is clearly not limited to nectar. Further, the grass family, Poaceae, is wind pollinated, rather than insect pollinated, and it does not produce nectar. Therefore, these NEC4 homologs must be expressed in different tissues, suggesting that the expression of members of this gene family in nectar may be the exception. It is interesting to note that both the soybean basic 7S glubulins and the lupine conglutins are cleaved at a Ser^Ser site, resulting in two peptides of 28 and 16 kD (Kagawa and Hirano, 1989; Kolivas and Gayler, 1993; Watanabe and Hirano, 1994). This Ser^Ser site is conserved among all of the protein sequences from the Fabaceae, but is missing from all of the other genes. Based upon the structural model presented above, this Ser^Ser site is predicted to be located on a surface loop that appears to be readily accessible for interactions with plant proteases.
Finally, NEC4 shows significant homology with the wheat xylanase inhibitors TAXI-1 through TAXI-4 (E-scores < e−37). The identification of hemicellulase inhibitor activity in gene products in widely separated clades of this phylogenetic tree (NEC4 and TAXI-1) suggests that the ability to inhibit fungal glycan hydrolases may be a common feature of all members of this gene family and that inhibition of fungal hemicellulases may be a much more important feature of plant defense than has been previously recognized. Further, the identification of NEC4-related sequences from six different plant families (Apiaceae, Brassicaceae, Fabaceae, Poaceae, Rosaceae, and Solanaceae) confirms that these genes are widely dispersed throughout the angiosperms.
Purification of NEC4
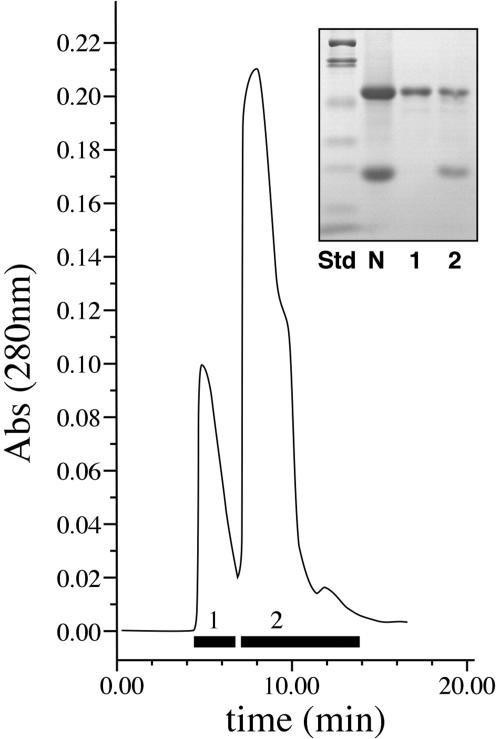
Because NEC4 has such high identity with a known xyloglucan-specific endoglucanase inhibitor, it was important to determine whether the NEC4 protein also can act as a glucanse inhibitor. Therefore, we purified the NEC4 to homogeneity so that we could test it for XEGIP and other inhibitory activities. The simplicity of the nectar proteome (only five proteins accumulate in nectar; Carter et al., 1999) and the known sequences of all of the nectar proteins permitted us to design a simple, one-step HPLC purification method. As outlined in “Materials and Methods,” concentrated nectar was treated to exchange the buffer and to remove free sugars. This treated nectar was applied to a cation exchange HPLC column and eluted with 10 mm HEPES, pH 8.3. Under these conditions, each of the other Nectarin proteins is positively charged and binds to the column, while NEC4 is negatively charged and elutes in peak 1 with the flow-through column volume (Fig. 4). Peak 2 of this profile contains NEC5 at 65 kD (NEC5 and NEC4 have nearly identical SDS-PAGE profiles; Carter and Thornburg, 2004a), difficult to see minor peaks of NEC3 and NEC2 at 40 and 35 kD, respectively, and NEC1 at 29 kD.
Figure 4.
HPLC purification of NEC4. Fractions that were pooled are shown by the bars at the bottom of the partial column profile. The inset shows an SDS-PAGE profile: lane 1, protein standards (myosin, 200 kD; β-galactosidase, 116 kD; phosphorylase b, 97 kD; bovine serum albumin, 66 kD; ovalbumen, 45 kD; carbonic anhydrase, 31 kD; soybean trypsin inhibitor, 21.5 kD; and lysozyme, 14.4 kD); lane 2, raw nectar; lane 3, proteins from pool 1; lane 4, proteins from pool 2.
NEC4 Is a XEGIP
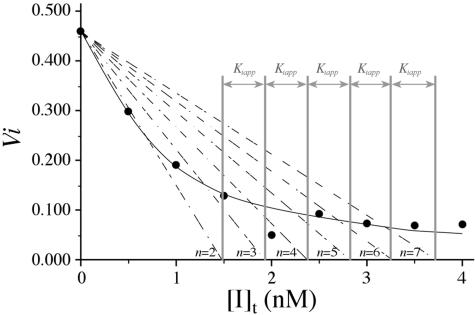
To test for inhibition, aliquots of purified NEC4 were incubated with a xyloglucan-specific endoglucanase (XEG) from Aspergillus aculeatus (Pauly et al., 1999). In preliminary studies, the NEC4 protein showed strong inhibition of the XEG. The inhibition constant (Ki) for the association of XEG with the tightly bound NEC4 was determined using a modification of the graphic technique of Dixon (1972; Segel, 1975). This method requires low protein concentrations to ensure that significant portions of both the enzyme and inhibitor are in the free (uncomplexed) form in the incubation solution. Therefore, XEG activity was measured at an enzyme concentration of approximately 1 nm and plotted versus inhibitor concentration (0–10 nm). The data were fit (Fig. 5) to a theoretical function corresponding to the ideal curve described by Dixon (1972; Segel, 1975) for competitive inhibition (see “Materials and Methods”). The calculated inhibition constant (Ki = 0.35 nm) for the interaction of XEG and NEC4 was not significantly different from that determined by Qin et al. (2003) for the interaction of XEG with XEGIP from tomato cell cultures. Thus, NEC4 has potent XEG inhibition activity, confirming that this protein is an XEGIP. Because of its homology to TAXI-I, which inhibits family GH11 endoxylanases, purified NEC4 protein was also assayed for its ability to inhibit these enzymes. In spite of this homology, the purified NEC4 protein did not inhibit any of the GH10 and GH11 xylanases tested.
Figure 5.
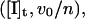 where n is an integer. The distance (0.45 nm) between successive intercepts of these lines with the [I]t axis is an estimation of Ki,app. The competitive inhibition constant Ki is calculated as follows:
where n is an integer. The distance (0.45 nm) between successive intercepts of these lines with the [I]t axis is an estimation of Ki,app. The competitive inhibition constant Ki is calculated as follows: 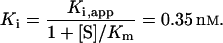 |
Expression of NEC4
To evaluate the temporal and spatial expression patterns of the NEC4 gene, we examined the levels of NEC4 mRNA by reverse transcription (RT)-PCR. mRNA was isolated from a number of floral and nonfloral plant organs, reverse transcribed, and used as a template for PCR. As a control, we used a pair of glyceraldehyde-3-P dehydrogenase (GAPDH) oligonucleotides. The GAPDH mRNA showed an equal level of expression in each of the organs examined (Fig. 6, all lanes). In contrast, the NEC4 oligonucleotides showed expression only in the nectary tissues. No other floral organ, nor roots, stems, or leaves, showed expression of the NEC4 mRNA. Temporally, NEC4 showed no expression at the earliest stages of nectary development (Fig. 6, lanes 1 and 2). By stage 9 (presecretory, ripening nectaries), a low level of expression was observed (Fig. 6, lane 3). This level of expression increased with further development until a moderate level was observed by stage 12 (mature nectaries at anthesis; Fig. 6, lane 4). Surprisingly, the level of NEC4 showed an increased expression after pollination. Figure 6, lane 5, shows an extremely high level of expression 48 h after fertilization.
Figure 6.
RT-PCR expression of NEC4. Top, RT-PCR performed using oligos nec4bF and nec4bR specific for NEC4. Bottom, RT-PCR performed using oligos GAP2F and GAP2R specific for GAPDH. Lane 1, Nectary stage 2; lane 2, nectary stage 6; lane 3, nectary stage 9; lane 4, nectary stage 12 (anthesis); lane 5, nectary 48 h postfertilization; lane 6, ovary; lane 7, stem; lane 8, floral tube; lane 9, root; lane 10, anther/filament; lane 11, stigma/style; lane 12, sepal; lane 13, leaf; lane 14, petal; and lane 15, petiole.
DISCUSSION
We have isolated and characterized a 64-kD protein, NEC4, from the nectar of ornamental tobacco plants. This protein is unique among the known nectar proteins. It has a blocked N terminus. To generate protein sequence in preparation for cloning, we used MS sequencing of NEC4 tryptic peptides. Three peptides were identified that closely matched a tomato protein (AAN87262) or its potato homolog (AAP84703). Alignment of the cDNAs encoding these two proteins permitted the identification of conserved regions that were used to generate oligonucleotide primers. PCR was performed on an ornamental tobacco nectary cDNA library to isolate a partial cDNA clone. 5′ and 3′ RACE were used to generate a full-length clone, which was deposited in GenBank (accession no. AY898762).
NEC4 Structure
The N-terminal sequence of the NEC4 protein likely contains a 22-amino acid signal sequence that, when cleaved, results in an N-terminal Gln residue (Gln-23). It is of interest to note that N-terminal Gln residues are frequently converted into pyroglutamate by specific Gln cyclase enzymes (Oberg et al., 1998; Schilling et al., 2003). The inability to generate N-terminal sequence from the mature protein by Edman degradation methods implies the presence of a pyroglutamate N terminus of NEC4. Each of the seven most closely related NEC4 homologs also retains a mature N-terminal Gln residue, implying that a blocked N terminus may be a common feature of each of these close homologs.
Based upon similarities with the wheat xylanase inhibitor (TAXI-1), we were able to model the structure of the NEC4 protein. The models produced in this study permitted us to identify a number of structural features that may be related to NEC4 function. We found that the NEC4 protein contains six small loops caused by the insertion of between three to 10 amino acids that are not found in the TAXI-1 molecule. We also identified the sites of glycosylation on the model. Each of these loops and glycosylation sites are all located well away from the region that is homologous with the TAXI-1 xylanase-binding site and would not be expected to interfere with glycohydrolase binding. This model also permitted us to identify a modified knottin domain that was also located well away from the glycohydrolase-binding site.
Knottin domains have been proposed to be sites mediating protein-protein interactions (Smith et al., 1998). This suggests a rather appealing antimicrobial mechanism. Nicotiana nectar contains only five proteins, two of which, NEC1 and NEC5, produce high levels of hydrogen peroxide (Carter and Thornburg, 2000, 2004a). If the knottin domain mediates interactions with other nectar proteins, then the NEC4 protein could bind to a fungal glycosylhydrolase, serving as a bridging molecule to bring one of the hydrogen peroxide-generating systems into close proximity with the fungus, thereby mediating a targeted antimicrobial response. However, determining whether such a mechanism is functioning in nectar will require further experimentation.
NEC4 Phylogenetic Relationships
The identification of closely related NEC4 homologs demonstrated that NEC4 belongs to a moderately sized multigene family. Rice contains 13 NEC4 homologs and Arabidopsis contains six homologs. Based upon the genome sequences of rice and Arabidopsis, we observed that the NEC4 family members are located on islands of tandemly repeated genes, as is found with other plant defense genes (Wei et al., 2002; Ferrari et al., 2003).
As was previously observed, three main classes of genes were identified in this analysis: XEGIP-related proteins, TAXI-related proteins, and seed storage proteins (York et al., 2004). The wide diversity of the related gene sequences indicates that most of the NEC4 homologs have no relationship with nectar physiology. Indeed, the expression of these NEC4 homologs varies significantly. The first family member discovered, carrot EDGP, is foliage expressed and is wound inducible (Satoh et al., 1992). The Arabidopsis NEC4 homolog (At5g19120) is expressed in foliage and is induced by salicylic acid and methyl jasmonate, but is unaffected by ethylene (Schenk et al., 2000). That homolog is also not induced by infection with the fungus Alternaria brassicicola. The more closely related NEC4 homolog (At1g03220) is also foliage expressed and appears to be light induced and repressed by iron deficiency (expression viewer analysis of the public data from the Stanford Microarray Database; http://www.arabidopsis.org). A Brassica campestris NEC4 homolog is expressed in guard cell protoplasts (Kwak et al., 1997). Others are seed expressed (Fabaceae) or are expressed in wind-pollinated species (Poaceae) that do not produce nectar. This implies that the nectar-specific expression of the tobacco NEC4 may have been co-opted from an alternate expression pattern during the evolution of ornamental tobacco. Several Arabidopsis NEC4 homologs have recently been demonstrated to be apoplastic (Borderies et al., 2003; Boudart et al., 2005), so the alternative expression of such a gene under the control of a strong nectary-specific promoter could have brought about the NEC4 phenotype.
NEC4 Function
NEC4 was purified from ornamental tobacco nectar using a one-step HPLC method. The NEC4 protein was subsequently shown to tightly bind a GH12 family XEG. The glycosylhydrolase family GH12 (Coutinho and Henrissat, 1999) includes many fungal proteins capable of degrading the cellulosic and hemicellulosic components of plant cell walls. XEG is a family GH12 enzyme that specifically degrades xyloglucans, the predominant hemicelluloses in the primary cell walls of dicotyledonous angiosperms. Xyloglucans bind to cellulose microfibrils via strong, noncovalent polysaccharide interactions, forming a cross-linked xyloglucan-cellulose network in the primary cell walls of most plants. A similar network composed of cellulose and xylan is present in the cell walls of grasses (Carpita and Gibeaut, 1993). These networks are thought to provide mechanical strength to the primary cell wall and their disruption significantly weakens the cell wall. Thus, fungal proteins that hydrolyze hemicellulosic polysaccharides may represent a group of pathogenesis factors that have evolved to facilitate fungal invasion.
Endoglucanases represent a large family of fungal proteins that degrade plant cell walls (Collins et al., 2005). Specifically, they degrade the xyloglucans that are the predominant hemicelluloses of plant cell walls. The hemicelluloses generally coat the cellulosic microfibrils via strong, noncovalent interactions and bridge interactions with other cell wall polysaccharides (Carpita and Gibeaut, 1993). Disruption of these interactions significantly weakens the mechanical properties of the cell wall and thus fungal endoglucanases represent a group of pathogenesis factors evolved to permit fungal invasion. We hypothesize that the NEC4 XEGIP functions to inhibit endoglucanases expressed by pathogenic fungi attempting to infect and colonize plant tissues analogously to the inhibition of pathogenic polygalacturonases by plant PGIPs (Powell et al., 2000).
The finding that NEC4 could inhibit GH12 endoglucanases, but was unable to inhibit GH11 xylanases, is intriguing. Family GH11 xylanases have overall folds that are similar to GH12 endoglucanases. In contrast, family GH10 xylanases have a different folding pattern than either the GH11 or GH12 hydrolases and are inhibited by a different wheat protein called XIP-I (Payan et al., 2004). Thus, in spite of its homology to TAXI-I, failure to inhibit either the GH10 or the GH11 xylanases indicates that NEC4 is distinct from both TAXI-I and XIP-I in its ligand-binding specificity.
NEC4 Expression
Because NEC4 is a potent inhibitor of fungal endoglucanases, the pattern of NEC4 expression suggests a unique feature about the biochemistry of this protein. The protein is increasingly expressed as the nectary develops. However, it is most highly expressed after fertilization. Thus, it appears that inhibition of fungal hemicellulases may be more important after fertilization than it is at anthesis. Examination of other plant organs indicates that the NEC4 protein is not expressed in any other plant organ tested. Thus, the NEC4 promoter may be very valuable for future studies to express novel genes during the late stages of nectary development.
The Complete Tobacco Nectar Proteome
The nectar of ornamental tobacco contains a limited array of five proteins. NEC4 was the last of the tobacco nectar proteins to be identified and characterized. Thus, the characterization of NEC4 as an inhibitor of XEG completes the analysis of the Nicotiana nectar proteome. The analysis of these novel proteins has permitted us to identify several unique biochemical functions of nectar (Table III). First, we have identified that the nectar of Nicotiana plants contains very high levels of hydrogen peroxide (Carter and Thornburg, 2000). This hydrogen peroxide is produced by two nectar proteins, NEC1, a superoxide dismutase (Carter and Thornburg, 2000), and NEC5, a Glc oxidase (Carter and Thornburg, 2004a). One potential problem that results from the accumulation of high levels of hydrogen peroxide in the presence of metal ions (and nectar does contain metal ions; Heinrich, 1989) is the production of hydroxyl free radicals (Rowley and Halliwell, 1983; Halliwell and Gutteridge, 1999). Fortunately, nectar also contains an antioxidant, ascorbate that can detoxify these free radicals. NEC3 and possibly NEC5 participate in maintaining high levels of ascorbate in nectar (Carter and Thornburg, 2004a, 2004c). This pathway has been termed the Carter-Thornburg nectar redox cycle (Carter and Thornburg, 2004b).
Table III.
The complete tobacco nectar proteome
| Protein | Demonstrated Biochemical Activity | Function in Nectar | Reference |
|---|---|---|---|
| NEC1 | Superoxide dismutase | Generates hydrogen peroxide | Carter and Thornburg (2000) |
| NEC2 | Breakdown product of NEC3 | Probably nonfunctional | |
| NEC3 | Bifunctional carbonic anhydrase and monodehydroascorbate reductase | Provides a pH-balanced meal to pollinators and recycles nectar ascorbate to protect from hydroxyl free radicals | Carter and Thornburg (2004c) |
| NEC4 | Xyloglucan-specific endoglucanase inhibitor protein (XEGIP) | Prevents fungal invasion of the gynoecium; also protects postfertilization | This article |
| NEC5 | Glucose oxidase and possible dehydroascorbate reductase | Generates hydrogen peroxide; may recycle nectar ascorbate to protect from hydroxyl free radicals | Carter and Thornburg (2004a); C. Carter and R.W. Thornburg, unpublished data |
We have previously proposed that a heretofore unrecognized function of the nectary is the protection of the gynoecium from pathogen invasion (Thornburg et al., 2003). The finding of a complex defense system in nectar suggests a novel function that has been a previously unrecognized function of nectar, that the secreted nectar not only functions to attract insect pollinators, but it also functions to protect the gynoecium from microbial invasion of the metabolite-rich nectar (Thornburg et al., 2003). This finding is similar to the finding that other defense-related proteins also accumulate in other floral organs and are also thought to protect flowers and presumably the gynoecium from infection (Lotan et al., 1989; Lay et al., 2003). Our identification of NEC4 as a potent inhibitor of fungal endoglucanases provides strong support for this hypothesis. Further, the expression pattern of NEC4 mRNA implies that the nectary continues this protective function even after pollination has already occurred.
MATERIALS AND METHODS
Plants
The line of ornamental tobacco plants used in this study was derived from an interspecific cross between Nicotiana langsdorffii and Nicotiana sanderae (Kornaga et al., 1997). Both of these species are diploid and belong to the Alatae section of tobacco. These plants are both male and female fertile, but are largely self-incompatible. We propagate them vegetatively to ensure a stable nectar/nectary phenotype. These plants were previously used to study a genetic instability as well as the tobacco nectar proteins (Carter et al., 1999; Carter and Thornburg, 2000). The conditions for growth and the methods for collecting and processing nectar were described previously (Carter et al., 1999). Flower stages were determined according to Koltunow et al. (1990).
Peptide Sequence Identification
Bands from an SDS-PAGE gel containing NEC4 were excised and processed for MS analysis as described previously (Wang et al., 2000). Resultant peptides were then subjected to MS/MS analyses.
MALDI-TOF
Tryptic peptides (1 μL) were mixed with 1 μL α-cyano-4-hydroxycinnamic acid (10 mg/mL in ethanol/acetonitrile; 1:1 [v/v]) and analyzed on an Applied Biosystems Q-STAR MS/MS mass spectrometer.
Database Searches
For peptide sequence identification, the Internet-based MASCOT MS/MS ions search tool (http://www.matrixscience.com/cgi/search_form.pl?FORMVER=2&SEARCH=MIS) was used to manually search against the National Center for Biotechnology Information (NCBI) nonredundant database as described in Carter et al. (2004). Search parameters were set as follows: Taxonomy, Viridiplantae; enzyme, trypsin; variable modifications, acetyl (N terminus); oxidation (M); phospho (ST); phospho (Y); pyro-glu (N-terminal E); pyro-glu (N-terminal Q); mass values, monoisotopic; protein mass, unrestricted peptide; mass tolerance, ± 150 ppm; fragment mass tolerance, ± 0.2 D max; missed cleavages, 1; instrument type, MALDI-QUAD-TOF.
rDNA Methods
Cloning methods were conducted either according to the manufacturer's directions or by accepted methods (Sambrook et al., 1989). mRNA was isolated from nectary tissues using standard methods (Ausubel et al., 1987) and reverse transcribed into first-strand cDNA using Moloney murine leukemia virus reverse transcriptase (Promega). PCR was performed according to the methods outlined in Erlich (1989). When designing oligonucleotides for RT-PCR, cDNA sequences were compared with genomic sequences from both similar and different species to construct primers that spanned putative intron-splice sites so as to eliminate erroneous signals from genomic DNA. RT-PCR primers were always pretested with genomic DNA to ensure that no PCR bands were synthesized.
Nectary cDNA Library
For the preparation of the nectary cDNA library, total RNA was isolated (Chomczynski and Sacchi, 1987) from the nectaries of ornamental tobacco flowers at stage 6 of floral development. PolyA mRNA was prepared using the polyATract mRNA isolation kit (Promega). The purified PolyA mRNA was then used to create a cDNA library using the SMART cDNA library construction kit according to the manufacturer's instructions (CLONTECH).
cDNA Library Screening
After protein identification based on MALDI-TOF and MASCOT MS/MS ion search, the XEGIP homologs from tomato (Lycopersicon esculentum; AY155579) and potato (Solanum tuberosum; AY321357) were aligned using ClustalW. Primers were designed against two highly homologous regions, one located near the 5′ end (Nec4F, 5′-GCCATTCTTTTGTGTTCTCT-3′) and the other (Nec4R, 5′-GTCCAACTCTTGTACTTCCA-3′) about 1 kb downstream from the forward primer in the tomato and potato sequences. Using this pair of primers, plasmid DNA isolated from a nectary stage 6 cDNA library was PCR amplified. The PCR-amplified product was eluted from agarose gel and ligated into pGEM-T and maintained in Escherichia coli DH-5α.
5′ and 3′ RACE
The remaining 5′ end of the cDNA was captured by amplification from plasmid DNA isolated from a stage 6 cDNA library with Nec4R1 (5′-CTGAAGCTGTACTCACTGGA-3′) and T7-1 primers (5′-CGCGTAATACGACTCACTATAG-3′). The amplified product was diluted 100-fold and used as the template for a second, nested PCR amplification using Nec4R2 (5′-TGGTGGATTTGTGTGAGATA-3′) and T7-D primers (5′-GAGTCAGTGAGCGAGGAA-3′). Resulting PCR products were cloned and maintained in DH-5α.
For amplification of the remaining 3′ end of the cDNA, plasmid DNA isolated from the stage 6 nectary cDNA library was amplified using Nec4F2 (5′-GGGTTGATTGTGACCAAGGT-3′) and M13-R (5′-CAGGAAACAGCTATGACC-3′) oligonucleotides. The amplified product was diluted 100-fold and used as template for nested amplification using Nec4F3 (5′-GGGAGAACCCTCTTCTGAAT-3′) and oligo(dT)25V (5′-TTTTTTTTTTTTTTTTTTTTTTTTTV-3′). PCR products were processed for cloning as for 5′ RACE.
Expression Patterns of NEC4
Plant mRNA was isolated from various floral and nonfloral organs using the SV total RNA isolation kit (no. Z3500) according to the manufacturer's directions (Promega). This RNA was used for first-strand cDNA synthesis using the StrataScript first-strand cDNA synthesis kit (no. 200420; Stratagene). The cDNA was aliquoted and stored at −70°C until use. RT-PCR was performed using the first-strand cDNA and oligonucleotides specific for the coding region of NEC4, nec4bF (5′-GGCTGGTCTTGGTAGGACAAGAATAT-3′) and nec4bR (5′-GATTGTCAATGGACAACAAAGTTGTG-3′). The LxS8 GAPDH mRNA (expressed sequence tag clone 101G06) isolated from an expressed sequence tag study of nectary-expressed genes was used as an internal control. GAPDH oligonucleotides GAP2F (5′-TTGAAGGGTGGTGCCAAGAAA-3′) and GAP2R (5′-CATTTATGACCTTTGCCAAGGGAG-3′) were specific for the GAPDH coding regions. PCR products were run on a 1.5% agarose gel and visualized using ethidium bromide.
Isolation of NEC4
Ten milliliters of nectar were collected over a 1-h time frame and were maintained on ice during this process. Following collection, the nectar was passed through a 0.2-μm filter and then subjected to concentration through Centricon YM-10 centrifugal columns at 4,000g until the retentate volume was reduced to about 1/20. The concentrated nectar was then diluted 10-fold with 10 mm HEPES buffer, pH 7.3, and recentrifuged to concentrate the volume to less than 100 μL. The retentate was again diluted 20-fold and used as an HPLC sample. Aliquots (750 μL) of this sample were injected into HPLC in buffer A (10 mm HEPES, pH 7.3) and analyzed through a cation exchange column CM300-silver (250 mm × 4.6 mm i.d.). The elution profile included the first 20 min with buffer A. This was followed by a linear gradient from 100% buffer A to 100% buffer B (10 mm HEPES, pH 7.3, 400 mm NaCl) followed by another 30 min with 100% buffer B. The flow rate was kept constant at 0.5 mL/min. The elution profile was monitored at 280 nm. Fractions pertaining to the various peaks were separately pooled and concentrated by Centricon YM-10 centrifugal columns. Protein concentrations were estimated by Bradford (1976). SDS-PAGE was performed according to the method of Laemmli (1970). Standards were broad-range Mr standards (Bio-Rad).
NEC4 Modeling
Protein models were developed using SWISS MODEL (Peitsch, 1995, Schwede et al., 2003). PDB coordinates for the crystal structures of the Triticum aestivum xylanase inhibitor I (TAXI-1) either alone (1T6E) or in complex with the Aspergillus niger xylanase I (1T6G) were used to develop the NEC4 models (Sansen et al., 2004). The authors would like to thank Dr. A. Rabijns for sharing the TAXI-1 coordinates prior to their publication. Refinement and analysis of these models was performed with DeepView Swiss-PdbViewer (Guex and Peitsch, 1997). Figures were generated using RasMol (Sayle and Milner-White, 1995).
Enzyme Inhibition Assay
The purified NEC4 protein was diluted with buffer C (50 mm sodium acetate, pH 4.7) to yield a 50.0 nm solution, as determined using an extinction coefficient (ɛ) of 82,123 m−1cm−1 (Qin et al., 2003). XEG, generously provided by Novozymes A/S, was purified as previously described (Pauly et al., 1999). Aliquots (0.4–8.0 μL) of the nectar protein solution were each combined with XEG (1 μL, 34.7 nm) in buffer C to bring an initial volume of 200 μL. After a 10-min incubation at room temperature, 200 μL of a mixture containing 100 μL of 4 mg mL−1 solution of tamarind xyloglucan in buffer C, and 1 μg of bovine serum albumin in buffer C, were added to bring the final incubation volume to 400 μL, such that the final xyloglucan concentration was 1 mg mL−1, the final XEG concentration was approximately 1 nm, and the final NEC4 concentration was in the range 0 to 1.0 nm. Two controls were set up, one in the absence of both proteins (B) and one in the absence of only the NEC4 protein (A). Control A was used to determine v0 (the reaction velocity in the absence of inhibitor), and control B was used as a blank during the PAHBAH assay (Lever, 1972). Following incubation at 37°C for 24 h, the concentration of reducing residues generated by XEG activity was determined by the PAHBAH assay using cellobiose as a standard.
The resulting data were fit to a theoretical curve in which each value of the dependent variable vi (the reaction velocity in the presence of inhibitor) is a function of the independent variable [I]t (the total inhibitor concentration), and the parameters Ki,app (the apparent inhibition constant) and [E]t (the total enzyme concentration), as illustrated in figure III-29 of Segel (1975). Then, for each inhibitor concentration [I]t, one can define the parameter n, such that 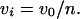 A diagonal line passing through the points (0, v0) and ([I]t, vi) will intercept the [I]t axis at a value
A diagonal line passing through the points (0, v0) and ([I]t, vi) will intercept the [I]t axis at a value 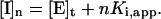 For graphical convenience, the original formulation of this approach limited n to integer values, allowing Ki,app to be estimated as the distance between successive intercepts. Examination of figure III-29 of Segel (1975) indicates that the x intercept [I]n of the line corresponding to each value of n is described by the equation:
For graphical convenience, the original formulation of this approach limited n to integer values, allowing Ki,app to be estimated as the distance between successive intercepts. Examination of figure III-29 of Segel (1975) indicates that the x intercept [I]n of the line corresponding to each value of n is described by the equation:
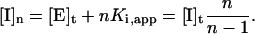 |
Upon rearrangement, it is clear that this equation is quadratic in n:
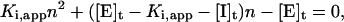 |
and n can be recovered by the quadratic formula:
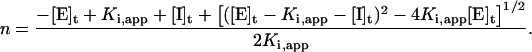 |
A theoretical data curve was constructed by calculating theoretical (not necessarily integer) values for n based on the independent variable [I]t and the adjustable parameters Ki,app and [E]t. That is, n was determined for each experimental [I]t, allowing the corresponding theoretical value of vi,theor to be calculated. The calculated values of vi,theor were compared to the corresponding experimental value of vi, and trial values of Ki,app and [E]t were adjusted until the RMS value of the residuals was minimized.
Several xylanases were tested for inhibition by adding xylanase to a solution containing substrate (xylan) and a stoichiometric excess of the NEC4 protein and colorimetrically measuring the enzyme-catalyzed increase in reducing residues. Enzymes that were assayed included Trichoderma viride xylanase M1 (Megazyme International Ireland), which is a family GH11 xylanase, three family GH11 xylanases from Magnaporthe grisea that were heterologously expressed in Pichia pastoris, and one family GH10 xylanase purified from M. grisea strain CP987 (Wu et al., 1997).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AY898762.
Acknowledgments
We thank Dr. Natasha Raikhel and Songqin Pan for technical assistance and the generous use of instrumentation housed in the W.M. Keck Proteomics Laboratory, Center for Plant Cell Biology, University of California, Riverside, and Novozymes A/S, who generously provided the xyloglucan-specific endoglucanase used in this research.
This work was supported by the National Science Foundation (grant no. IBN–0235645); the Carver Trust, the Hatch Act, and State of Iowa Funds; a visiting fellowship from the Higher Education Commission of Pakistan (to S.M.S.N.); and in part by funds from the U.S. Department of Energy (DOE; grant no. DE–FG02–96ER20220) and from the DOE-funded Center for Plant and Microbial Complex Carbohydrates (grant no. DE–FG02–93ER20097).
This paper is dedicated with grateful appreciation to Professor C.A. “Bud” Ryan, Institute of Biological Chemistry, Washington State University, on the occasion of his 74th birthday, September 29, 2005.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Robert W. Thornburg (thorn@iastate.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.065227.
References
- Ausubel FM, Brent R, Kingston RE, Moore DD, Smith JA, Seidman JG, Struhl K (1987) Current Protocols in Molecular Biology. John Wiley & Sons, New York
- Baker HG, Baker I (1973) Some anthecological aspects of the evolution of nectar-producing flowers, particularly amino acid production in the nectar. In VH Heywood, ed, Taxonomy and Ecology. Proceedings of an International Symposium Held at the Department of Botany, University of Reading, Vol 5. Academic Press, London, pp 243–264
- Baker HG, Baker I (1975) Studies of nectar-constitution and pollinator-plant coevolution. In LE Gilbert, PH Raven, eds, Coevolution of Animals and Plants. Symposium V, First International Congress of Systematic and Evolutionary Biology, Boulder, Colorado, August 1973. University of Texas Press, Austin, TX, pp 100–140
- Baker HG, Baker I (1981) Chemical constituents of nectar in relation to pollination mechanisms and phylogeny. In MH Nitecki, ed, Biochemical Aspects of Evolutionary Biology. University of Chicago Press, Chicago, pp 131–171
- Borderies G, Jamet E, Lafitte C, Rossignol M, Jauneau A, Boudart G, Monsarrat B, Esquerre-Tugaye MT, Boudet A, Pont-Lezica R (2003) Proteomics of loosely bound cell wall proteins of Arabidopsis thaliana cell suspension cultures: a critical analysis. Electrophoresis 24: 3421–3432 [DOI] [PubMed] [Google Scholar]
- Boudart G, Jamet E, Rossignol M, Lafitte C, Borderies G, Jauneau A, Esquerre-Tugaye MT, Pont-Lezica R (2005) Cell wall proteins in apoplastic fluids of Arabidopsis thaliana rosettes: identification by mass spectrometry and bioinformatics. Proteomics 5: 212–221 [DOI] [PubMed] [Google Scholar]
- Bradford MM (1976) A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principles of protein-dye binding. Anal Biochem 72: 248–254 [DOI] [PubMed] [Google Scholar]
- Cabras P, Angioni A, Tuberoso C, Floris I, Reniero F, Guillou C, Ghelli S (1999) Homogentisic acid: a phenolic acid as a marker of strawberry-tree (Arbutus unedo) honey. J Agric Food Chem 47: 4064–4067 [DOI] [PubMed] [Google Scholar]
- Carpita NC, Gibeaut DM (1993) Structural models of primary cell walls in flowering plants: consistency of molecular structure with the physical properties of the walls during growth. Plant J 3: 1–30 [DOI] [PubMed] [Google Scholar]
- Carter C, Graham R, Thornburg RW (1999) Nectarin I is a novel, soluble germin-like protein expressed in the nectar of Nicotiana sp. Plant Mol Biol 41: 207–216 [DOI] [PubMed] [Google Scholar]
- Carter C, Pan S, Zouhar J, Avila EL, Girke T, Raikhel NV (2004) The vegetative vacuole proteome of Arabidopsis thaliana reveals predicted and unexpected proteins. Plant Cell 16: 3285–3303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Thornburg RW (2000) Tobacco Nectarin I: purification and characterization as a germin-like, manganese superoxide dismutase implicated in the defense of floral reproductive tissues. J Biol Chem 275: 36726–36733 [DOI] [PubMed] [Google Scholar]
- Carter C, Thornburg RW (2003) The nectary-specific pattern of gene expression is regulated by multiple promoter elements in the tobacco Nectarin I promoter. Plant Mol Biol 51: 451–457 [DOI] [PubMed] [Google Scholar]
- Carter C, Thornburg R (2004. a) Tobacco Nectarin V is a flavin-containing berberine bridge enzyme-like protein with glucose oxidase activity. Plant Physiol 134: 460–469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter C, Thornburg RW (2004. b) Is the nectar redox cycle a floral defense against microbial attack? Trends Plant Sci 9: 320–324 [DOI] [PubMed] [Google Scholar]
- Carter C, Thornburg RW (2004. c) Tobacco Nectarin III is a bifunctional enzyme with monodehydroascorbate reductase and carbonic anhydrase activities. Plant Mol Biol 54: 415–425 [DOI] [PubMed] [Google Scholar]
- Chomczynski P, Sacchi N (1987) Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem 162: 156–159 [DOI] [PubMed] [Google Scholar]
- Collins T, Gerday C, Feller G (2005) Xylanases, xylanase families and extremophilic xylanases. FEMS Microbiol Rev 29: 3–23 [DOI] [PubMed] [Google Scholar]
- Coutinho PM, Henrissat B (1999) Carbohydrate-active enzymes: an integrated database approach. In HJ Gilbert, G Davies, B Henrissat, B Svensson, eds, Recent Advances in Carbohydrate Bioengineering. The Royal Society of Chemistry, Cambridge, pp 3–12
- Deinzer ML, Thompson PA, Burgett DM, Isaacson DL (1977) Pyrrolizidine alkaloids: their occurrence in honey from tansy ragwort (Senecio jacobaea L.). Science 195: 497–499 [DOI] [PubMed] [Google Scholar]
- Dixon M (1972) The graphical determination of Km and Ki. Biochem J 129: 197–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ecroyd CE, Franich RA, Kroese HW, Steward D (1995) Volatile constituents of Dactylanthus taylorii flower nectar in relation to flower pollination and browsing by animals. Phytochemistry 40: 1387–1389 [Google Scholar]
- Erlich HA, editor (1989) PCR Technology: Principles and Applications for DNA Amplification. Stockton Press, New York
- Ferrari S, Vairo D, Ausubel FM, Cervone F, De Lorenzo G (2003) Tandemly duplicated Arabidopsis genes that encode polygalacturonase-inhibiting proteins are regulated coordinately by different signal transduction pathways in response to fungal infection. Plant Cell 15: 93–106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreres F, Andrade P, Gil MI, Tomas Barberan FA (1996) Floral nectar phenolics as biochemical markers for the botanical origin of heather honey. Z Lebensm-Unters-Forsch 202: 40–44 [Google Scholar]
- Griebel C, Hess G (1940) The vitamin C content of flower nectar of certain Labiatae. Z Lebensm-Unters-Forsch 79: 168–171 [Google Scholar]
- Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18: 2714–2723 [DOI] [PubMed] [Google Scholar]
- Halliwell B, Gutteridge J (1999) Free Radicals in Biology and Medicine. Oxford University Press, New York
- Heinrich G (1989) Analysis of cations in nectars by means of a laser microprobe mass analyser (LAMMA). Beitr Biol Pflanz 64: 293–308 [Google Scholar]
- Kagawa H, Hirano H (1989) Sequence of a cDNA encoding soybean basic 7S globulin. Nucleic Acids Res 17: 8868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolivas S, Gayler KR (1993) Structure of the cDNA coding for conglutin gamma, a sulphur-rich protein from Lupinus angustifolius. Plant Mol Biol 21: 397–401 [DOI] [PubMed] [Google Scholar]
- Koltunow AM, Truettner J, Cox KH, Walroth M, Goldberg RB (1990) Different temporal and spatial gene expression patterns occur during anther development. Plant Cell 2: 1201–1224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornaga T, Zyzak DV, Kintinar A, Baynes J, Thornburg R (1997) Genetic and biochemical characterization of a “lost” unstable flower color phenotype in interspecific crosses of Nicotiana sp. WWW J Biol 2: 8 [Google Scholar]
- Kwak JM, Kim SA, Hong SW, Nam HG (1997) Evaluation of 515 expressed sequence tags obtained from guard cells of Brassica campestris. Planta 202: 9–17 [DOI] [PubMed] [Google Scholar]
- Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227: 680–685 [DOI] [PubMed] [Google Scholar]
- Lay F, Brugliera F, Anderson M (2003) Isolation and properties of floral defensins from ornamental tobacco and petunia. Plant Physiol 131: 1283–1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lever M (1972) New reaction for colorimetric determination of carbohydrates. Anal Biochem 47: 273–279 [DOI] [PubMed] [Google Scholar]
- Lotan T, Ori N, Fluhr R (1989) Pathogenesis-related proteins are developmentally regulated in tobacco flowers. Plant Cell 1: 881–887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGeoch DJ (1985) On the predictive recognition of signal peptide sequences. Virus Res 3: 271–286 [DOI] [PubMed] [Google Scholar]
- Nakai K, Kanehisa M (1991) Expert system for predicting protein localization sites in Gram-negative bacteria. Proteins Struct Funct Genet 11: 95–110 [DOI] [PubMed] [Google Scholar]
- Oberg KA, Ruysschaert JM, Azarkan M, Smolders N, Zerhouni S, Wintjens R, Amrani A, Looze Y (1998) Papaya glutamine cyclase, a plant enzyme highly resistant to proteolysis, adopts an all-beta conformation. Eur J Biochem 258: 214–222 [DOI] [PubMed] [Google Scholar]
- Pauly M, Andersen LN, Kaupinen S, Kofod LV, York WS, Albersheim P, Darvill AG (1999) A xyloglucan-specific endo-β-1,4-glucanase from Aspergillus aculeatus: expression cloning in yeast, purification, and characterization of the recombinant enzyme. Glycobiology 9: 93–100 [DOI] [PubMed] [Google Scholar]
- Payan F, Leone P, Porciero S, Furniss C, Tahir T, Williamson G, Durand A, Manzanares P, Gilbert HJ, Juge N, et al (2004) The dual nature of the wheat xylanase protein inhibitor XIP-I: structural basis for the inhibition of family 10 and family 11 xylanases. J Biol Chem 279: 36029–36037 [DOI] [PubMed] [Google Scholar]
- Peitsch MC (1995) Protein modeling by e-mail. Biotechnology 13: 658–660 [Google Scholar]
- Powell AL, van Kan J, ten Have A, Visser J, Greve LC, Bennett AB, Labavitch JM (2000) Transgenic expression of pear PGIP in tomato limits fungal colonization. Mol Plant Microbe Interact 13: 942–950 [DOI] [PubMed] [Google Scholar]
- Qin Q, Bergmann CW, Rose JK, Saladie M, Kolli VS, Albersheim P, Darvill AG, York WS (2003) Characterization of a tomato protein that inhibits a xyloglucan-specific endoglucanase. Plant J 34: 327–338 [DOI] [PubMed] [Google Scholar]
- Rodriguez-Arce AL, Diaz N (1992) The stability of beta-carotene in mango nectar. J Agric Univ PR 76: 101–102 [Google Scholar]
- Roshchina VV, Roshchina VD (1993) The Excretory Function of Higher Plants. Springer-Verlag, Berlin
- Rowley D, Halliwell B (1983) Formation of hydroxyl radicals from hydrogen peroxide and iron salts by superoxide- and ascorbate-dependent mechanisms: relevance to the pathology of rheumatoid disease. Clin Sci (Lond) 64: 649–653 [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4: 406–425 [DOI] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular Cloning: A Laboratory Manual, Ed 2. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Sansen S, De Ranter CJ, Gebruers K, Brijs K, Courtin CM, Delcour JA, Rabijns A (2004) Structural basis for inhibition of Aspergillus niger xylanase by Triticum aestivum xylanase inhibitor-I. J Biol Chem 279: 36022–36028 [DOI] [PubMed] [Google Scholar]
- Satoh S, Sturm A, Fujii T, Chrispeels JJ (1992) cDNA cloning of an extracellular dermal glycoprotein of carrot and its expression in response to wounding. Planta 188: 432–438 [DOI] [PubMed] [Google Scholar]
- Sayle RA, Milner-White EJ (1995) RASMOL: biomolecular graphics for all. Trends Biochem Sci 20: 374–376 [DOI] [PubMed] [Google Scholar]
- Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci USA 97: 11655–11660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schilling S, Manhart S, Hoffmann T, Ludwig HH, Wasternack C, Demuth HU (2003) Substrate specificity of glutaminyl cyclases from plants and animals. Biol Chem 384: 1583–1592 [DOI] [PubMed] [Google Scholar]
- Schwede T, Kopp J, Guex N, Peitsch MC (2003) SWISS-MODEL: an automated protein homology-modeling server. Nucleic Acids Res 31: 3381–3385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segel IH (1975) Enzyme Kinetics: Behavior and Analysis of Rapid Equilibrium and Steady State Enzyme Systems. John Wiley & Sons, New York
- Smith GP, Patel SU, Windass JD, Thornton JM, Winter G, Griffiths AD (1998) Small binding proteins selected from a combinatorial repertoire of knottins displayed on phage. J Mol Biol 277: 317–332 [DOI] [PubMed] [Google Scholar]
- Thornburg RW, Carter C, Powell A, Rizhsky L, Mittler R, Horner HT (2003) A major function of the tobacco floral nectary is defense against microbial attack. Plant Syst Evol 238: 211–218 [Google Scholar]
- Vogel S (1969) Flowers offering fatty oil instead of nectar (abstract no. 229). In Abstracts of the Papers Presented at the XI International Botanical Congress, August 24-September 2, 1969 and the International Wood Chemistry Symposium, September 2-4, 1969. International Botanical Congress, Seattle
- Wang Y, Sun J, Chitnis P (2000) Proteomic study of the peripheral proteins from thylakoid membranes of the cyanobacterium Synechocystis sp. PCC 6803. Electrophoresis 21: 1746–1754 [DOI] [PubMed] [Google Scholar]
- Watanabe Y, Hirano H (1994) Nucleotide sequence of the basic 7S globulin gene from soybean. Plant Physiol 105: 1019–1020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Wing RA, Wise RP (2002) Genome dynamics and evolution of the Mla (powdery mildew) resistance locus in barley. Plant Cell 14: 1903–1917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S-C, Ham K-S, Darvill AG, Albersheim P (1997) Deletion of two endo-β-1,4-xylanase genes reveals additional isozymes secreted by the rice blast fungus. Mol Plant-Microbe Interact 10: 700–708 [Google Scholar]
- York WS, Qin Q, Rose JK (2004) Proteinaceous inhibitors of endo-beta-glucanases. Biochim Biophys Acta 1696: 223–233 [DOI] [PubMed] [Google Scholar]