Abstract
Calvin cycle enzymes glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and phosphoribulokinase (PRK) form together with the regulatory peptide CP12 a supramolecular complex in Arabidopsis (Arabidopsis thaliana) that could be reconstituted in vitro using purified recombinant proteins. Both enzyme activities were strongly influenced by complex formation, providing an effective means for regulation of the Calvin cycle in vivo. PRK and CP12, but not GapA (A4 isoform of GAPDH), are redox-sensitive proteins. PRK was reversibly inhibited by oxidation. CP12 has no enzymatic activity, but it changed conformation depending on redox conditions. GapA, a bispecific NAD(P)-dependent dehydrogenase, specifically formed a binary complex with oxidized CP12 when bound to NAD. PRK did not interact with either GapA or CP12 singly, but oxidized PRK could form with GapA/CP12 a stable ternary complex of about 640 kD (GapA/CP12/PRK). Exchanging NADP for NAD, reducing CP12, or reducing PRK were all conditions that prevented formation of the complex. Although GapA activity was little affected by CP12 alone, the NADPH-dependent activity of GapA embedded in the GapA/CP12/PRK complex was 80% inhibited in respect to the free enzyme. The NADH activity was unaffected. Upon binding to GapA/CP12, the activity of oxidized PRK dropped from 25% down to 2% the activity of the free reduced enzyme. The supramolecular complex was dissociated by reduced thioredoxins, NADP, 1,3-bisphosphoglycerate (BPGA), or ATP. The activity of GapA was only partially recovered after complex dissociation by thioredoxins, NADP, or ATP, and full GapA activation required BPGA. NADP, ATP, or BPGA partially activated PRK, but full recovery of PRK activity required thioredoxins. The reversible formation of the GapA/CP12/PRK supramolecular complex provides novel possibilities to finely regulate GapA (“non-regulatory” GAPDH isozyme) and PRK (thioredoxin sensitive) in a coordinated manner.
Life of photosynthetic organisms depends on a finely controlled balance between light reactions of photosynthesis and photosynthesis-dependent metabolism. Signals involved in the maintenance of such balance include thioredoxins and a number of biochemical factors, including pyridine nucleotides, several metabolites, as well as pH and magnesium ions (Wolosiuk et al., 1993). In the Calvin cycle, five out of 11 enzymes are prominently sensitive to these signals (glyceraldehyde-3-phosphate dehydrogenase [GAPDH], Fru bisphosphate and sedoheptulose bisphosphate phosphatases, phosphoribulokinase [PRK], and Rubisco via Rubisco activase), and their activities are strongly regulated in vivo by light/dark conditions (Ruelland and Miginiac-Maslow, 1999; Schürmann and Jacquot, 2000; Dai et al., 2004; Buchanan and Balmer, 2005). Moreover, redox regulation in chloroplasts is not restricted to carbon reduction, and the whole plastid metabolism seems to be controlled by the redox status of thioredoxins, as suggested by redox proteomic analysis (for review, see Buchanan and Balmer, 2005).
Besides the fine and complex tuning of individual enzyme activities, protein-protein interactions also contribute to the overall regulation of photosynthetic metabolism. Supramolecular complexes of Calvin cycle enzymes with different compositions and stoichiometries, possibly interacting with thylakoids, have been widely documented (Müller, 1972; Wara-Aswapati et al., 1980; Clasper et al., 1991; Rault et al., 1993; Süss et al., 1993; Graciet et al., 2004; Dani and Sainis, 2005). The presence of GAPDH and PRK in the aggregates was a common trait of all these reports, which were eventually supported by discovery of the small peptide CP12 (Pohlmeyer et al., 1996; Wedel and Soll, 1998).
CP12 is a small protein of 75 amino acids including four Cys separated by eight residues such that two short loops are believed to be generated by the formation of two internal disulfides. A supramolecular complex including GAPDH, CP12, and PRK has been described in different photosynthetic organisms (Wedel et al., 1997; Wedel and Soll, 1998; Scheibe et al., 2002; Graciet et al., 2003a; Tamoi et al., 2005). CP12 has no enzymatic activity and seems to act as a scaffold protein within these complexes.
In higher plants photosynthetic GAPDH is formed by two types of subunits (A and B), giving rise to two isozymes A4 and AnBn (Cerff, 1979; Pupillo and Faggiani, 1979; Brinkmann et al., 1989). B-subunits are very similar to A-subunits but contain an additional C-terminal extension (CTE) homologous to the C-terminal end of CP12 (Pohlmeyer et al., 1996). This 30-amino acid tail contains a conserved couple of redox-sensitive Cys and is responsible for the regulatory properties of AnBn isoforms, including the capacity to form stable A8B8 oligomers in the presence of NAD(H) (Baalmann et al., 1996; Li and Anderson, 1997; Sparla et al., 2002). Consistently, the A4 isozyme has no CTE and is constitutively activated (Scagliarini et al., 1998). In spinach (Spinacia oleracea) chloroplasts, hexadecameric GAPDH coexists with GAPDH/CP12/PRK complexes, and both regulatory systems seem to contribute to dark/light regulation of Calvin cycle in vivo (Scheibe et al., 2002).
Several genes potentially implicated in the GAPDH/CP12/PRK complex are present in the genome of Arabidopsis (Arabidopsis thaliana). GAPDH is coded by two duplicated genes (GapA-1 and A-2) and one GapB gene, PRK is coded by a single PRK gene, and CP12 by two closely related genes (CP12-1 and CP12-2) and a third divergent one (CP12-3; Marri et al., 2005; Fig. 1). GapA-1, GapB, PRK, and CP12-2 were found to be coordinately expressed under different conditions in Arabidopsis leaves (Marri et al., 2005). In this work, we used recombinant A4-GAPDH (GapA-1), CP12 (CP12-2), and PRK of Arabidopsis to reconstitute the GAPDH/CP12/PRK complex under given conditions. Enzyme activities in the complex were found to be strongly inhibited, demonstrating that a constitutively activated enzyme such as A4-GAPDH could be regulated by the reversible formation of a supramolecular complex with PRK and CP12. It is proposed that GAPDH/CP12/PRK supramolecular complexes occur in chloroplasts in the dark to ensure strong down-regulation of the Calvin cycle. Conditions leading to complex destabilization and enzyme reactivation were also identified and appear to be representative of the general resurgence of photosynthetic metabolism at the onset of light.
Figure 1.
Multiple alignment of Arabidopsis CP12s (CP12-1, At2g47400; CP12-2, At3g62410; CP12-3, At1g76650; according to Marri et al. [2005]). The alignment was performed by ClustalW. The transit peptide as predicted by ChloroP is shown on a gray field. Conserved Cys are on a black field. The sequence of recombinant CP12-2 retained at the N terminus four residues from the His tag. Therefore, the N terminus of recombinant CP12-2 started with GSHM (not shown), followed by AAPEGG….
RESULTS
As previously shown in other plant species (Wedel et al., 1997; Scheibe et al., 2002), GAPDH/CP12/PRK complexes could be detected in Arabidopsis by gel filtration of NAD-treated chloroplast proteins. GAPDH and PRK activities coeluted at high molecular mass (600–750 kD) together with a 20-kD protein recognized by antibodies raised against spinach CP12 (data not shown). These fractions are likely to contain a mixture of complexes of similar size, including A4-GAPDH/CP12/PRK, A2B2-GAPDH/CP12/PRK, and A8B8-GAPDH oligomers (Scheibe et al., 2002). In order to study the GAPDH/CP12/PRK complex in a simplified system, we tried to reconstitute the supramolecular complex from isolated recombinant components.
Expression, Purification, and Redox Properties of Arabidopsis Recombinant GAPDH, CP12, and PRK
Each single component of the GAPDH/CP12/PRK complex was heterologously expressed in Escherichia coli. For the sake of simplicity, GAPDH was expressed as the simplest “non-regulatory” isozyme constituted by A-subunits only (GapA). Recombinant GapA of Arabidopsis was purified until it displayed a single band of 36 kD in SDS-PAGE (Fig. 2A). Purified GapA was redox insensitive when treated with thioredoxins but was found to be inactivated by oxidized dithiothreitol (DTT) and other oxidants, including H2O2. This effect could not be reverted by reductants and seemed to depend on irreversible oxidation of catalytically essential Cys-149 (M. Zaffagnini and S. Lemaire, personal communication).
Figure 2.
SDS-PAGE of GapA, PRK, and CP12 of Arabidopsis expressed in E. coli and purified to homogeneity. Gels were stained with Coomassie Brilliant Blue R-250. A, GapA and PRK were treated with sample buffer including reductant (5 mm DTT) and loaded on a 12.5% polyacrylamide gel. B, Reduced (rd) and oxidized (ox) CP12 samples were obtained by incubation for 2 h at 25°C with equimolar concentrations of prereduced or preoxidized thioredoxin, respectively (see "Materials and Methods"). Samples were then boiled for 3 min in sample buffer with no reductants, and the proteins were separated on 15% polyacrylamide gels. Thioredoxin migrated below the 14.4-kD marker. The apparent molecular mass of CP12 (arrowheads) shifted from 20 to 16 kD depending on redox conditions.
Although spinach PRK was reported to be reluctant to expression as an active enzyme in E. coli (Brandes et al., 1996), about 2 mg of active Arabidopsis PRK per liter of liquid culture were routinely obtained when bacterial cells were grown at 30°C. Purified PRK migrated as a single 38-kD band in SDS-PAGE (Fig. 2A). Redox sensitivity of Arabidopsis PRK showed a midpoint redox potential of −330 mV at pH 7.9, within the range of Em measured for PRK of other plant species (Hirasawa et al., 1999; Hutchinson and Ort, 2000). Residual activity of the enzyme equilibrated with oxidized DTT and thioredoxin accounted for 25% of the activity of the fully reduced enzyme (see Fig. 4).
Figure 4.
Activities of GapA and PRK as free enzymes, as enzymes embedded in complexes, and after complex treatment with several potential effectors. A, Activity expressed as percentage of the full activity of free tetrameric GapA (NADPH dependent). GapA/CP12 and GapA/CP12/PRK complexes were obtained under the same conditions as described in legends to Figures 3C and 3D, respectively. B, PRK activity was assayed and expressed as a percentage of fully reduced PRK. PRK oxidation was obtained by 3 h incubation at 25°C with 25 mm oxidized DTT. The ternary complex with GapA and CP12 was obtained as in Figure 3D. C, The GapA/CP12/PRK complex was reconstituted and chromatographed as in Figure 3D, re-equilibrated with 100 mm Tricine-NaOH, pH 7.9, in the absence of NAD, and incubated under different conditions (0.2 mm NAD; 0.5 mm ribulose-5-P; 2 mm ATP; 0.2 mm NADP; 43 μm BPGA; 5 mm reduced DTT plus 1 μg/mL thioredoxin). BPGA (43 μm) was produced at equilibrium by the reaction of phosphoglycerate kinase with 3 mm 3-phosphoglycerate and 2 mm ATP (initial concentrations). After 1 h incubation at 4°C with different effectors, the NADPH activity of GapA was assayed and expressed as percentage of the activity of GapA before complex reconstitution. An aliquot of the incubated sample was also loaded on a gel filtration column to check the aggregation state of the proteins. The stars indicate those conditions that did not lead to complex dissociation (NAD and ribulose-5-P; see also Fig. 5). D, Same conditions as in C except that PRK activity was assayed and expressed as percentage of the activity of the fully reduced enzyme. Ru5P, Ribulose-5-P.
Recombinant CP12-2 was expressed as a fusion protein with a His tag at the N terminus of the transit peptide cleavage site as predicted by ChloroP (Emanuelsson et al., 1999). The protein was purified by nickel affinity chromatography and the tag removed by proteolysis. Despite the calculated molecular mass of 8.5 kD, purified CP12 migrated in SDS-PAGE as a peptide of 20 kD after incubation with reduced thioredoxins and 16 kD after incubation with oxidized thioredoxins, indicating that disulfide formation led to a major conformational change that appeared to be retained even under denaturating conditions (Fig. 2B).
In Vitro Reconstitution of Binary and Ternary Complexes
The capability of isolated GAPDH, CP12, and PRK to bind to each other protein was tested, and supramolecular complexes were detected by gel filtration chromatography. Different possible conformations of single protein components were compared: Holo-GapA was tested as either NADP or NAD complex, and CP12 and PRK were tested as either reduced or oxidized proteins. On the whole, 12 possible combinations of binary complexes and eight combinations of ternary complexes could be envisaged, but only a few were found to be productive in terms of formation of supramolecular complexes.
Free GapA is a tetramer of identical subunits with an apparent mass of 120 kD in the presence of either NAD or NADP (Fig. 3A). PRK is a dimer with an estimated mass of 110 kD under oxidizing conditions (Fig. 3A), and apparently smaller (97 kD) when reduced (data not shown). Although the peaks of GapA and PRK overlapped, both were fully separated from the 29-kD peak of oxidized CP12 and the 35-kD peak of reduced CP12 (Fig. 3A). Since CP12 is an intrinsically unstructured protein (Graciet et al., 2003a) while the gel filtration column was calibrated with globular proteins, these determinations of CP12 molecular mass may be highly overestimated, making the polymerization state of native CP12 uncertain.
Figure 3.
In vitro reconstitution of binary (GapA/CP12) and ternary (GapA/CP12/PRK) complexes. A, Gel filtration (Superdex 200) of purified GapA, PRK (oxidized), and CP12 (either oxidized or reduced). Oxidized and reduced proteins were obtained by 3 h incubation at 25°C in the presence of 25 mm reduced or oxidized DTT. The four samples were individually loaded on the column and run under identical conditions. The absorption patterns at 280 nm were normalized and superimposed. Column equilibration buffer was 50 mm Tris-HCl, pH 7.5, 150 mm KCl, 1 mm EDTA; volume of loaded samples was 0.2 mL; and flow rate was 0.5 mL min−1. Column calibration is reported at the bottom of the figure. Estimated molecular masses of samples were 120 kD (GapA), 110 kD (PRK, oxidized), 35 kD (CP12, reduced), and 29 kD (CP12, oxidized). B, Same type of chromatography as in A, except that the sample loaded on the column was a mixture of equimolar GapA and CP12 (oxidized) on subunit basis, incubated for 2 h at 4°C in the presence of 0.5 mm NADP before loading. Column equilibration buffer was as in A with the addition of 0.2 mm NADP. Insert, Western blots showing that anti-GAPDH polyclonal antibodies recognize GapA only in the peak corresponding to 120 kD, and anti-CP12 antibodies recognize CP12 only in the low Mr peak (29 kD). Stars indicate the column fractions (0.35 mL) which were concentrated and loaded on the gel for western blotting. C, Same experiments as in B except that NAD substituted NADP in both incubation and column buffers. The GapA peak shifted to the left to an apparent molecular mass of 150 kD. The elution pattern of B is superimposed for easy comparison. Insert, Western blots showing that anti-CP12 antibodies recognized a 16-kD peptide coeluting with GapA tetramers (36 kD in SDS-PAGE). D, Same experiment as in C except for the addition, in the incubation buffer, of equimolar PRK (oxidized) on subunit basis. Insert, Immunoblots showing that the peak at 640 kD contained GapA, CP12, and PRK.
Incubation of GapA with PRK at equimolar ratio (subunit basis) failed to result in formation of a binary complex, regardless of the type of pyridine nucleotide bound to GapA or the redox state of PRK (data not shown). No complex was also detected when GapA was incubated with NADP and oxidized CP12 (1:1 subunit ratio). Both proteins eluted as isolated moieties and specific antibodies could not detect any CP12 interacting with GapA (Fig. 3B). On the contrary, in the presence of NAD instead of NADP, the peak of GapA displayed an apparent increase in size of 30 kD, while the peak of free CP12 dropped to low levels (Fig. 3C). Immunoblots confirmed that under these conditions most of CP12 coeluted with GapA, although inhibition of GapA activity was negligible (Fig. 4A). Replacing oxidized CP12 with reduced CP12 had the effect of abolishing the interaction between the two proteins, even in the presence of NAD (data not shown).
PRK was itself unable to bind CP12 under any redox conditions (data not shown). On the other hand, oxidized PRK quantitatively formed a supramolecular complex of about 640 kD when incubated with GapA-NAD and oxidized CP12 (Fig. 3D). Immunoblots demonstrated that all three partner proteins coeluted in the high Mr peak. Formation of the complex was prevented by reduction of PRK (data not shown). Interestingly, formation of the GapA/CP12/PRK complex led to dramatic inhibition of the activity of both enzymes. Within the complex, PRK was 12-fold less active than the free oxidized counterpart and 50-fold less active than the reduced enzyme (Fig. 4B). In a similar mood, the NADPH-dependent activity of GapA embedded in the complex was 5-fold lower than for free enzyme (Fig. 4A), whereas the NADH-activity remained unchanged.
In Vitro Dissociation of Binary and Ternary Complexes
The supramolecular GapA/CP12/PRK complex isolated in the presence of NAD proved to be quite stable. Buffer exchange to remove excess of NAD did not affect complex stability, and reloading of the complex on the column equilibrated without NAD led to negligible dissociation.
Ligands of either GapA or PRK were tested as possible effectors of complex dissociation. The PRK substrate ribulose-5-P had no significant effect on complex stability or enzyme activities (Figs. 4 and 5). Incubations with NADP or ATP dissociated the complex, and a major peak of about 120 kD, including both GapA and PRK free proteins, was observed. The peak of free CP12 was hardly detectable under these conditions, partially due to the low molar extinction coefficient of this small protein (Fig. 5). While dissociating the complex, ATP and NADP stimulated the activity of both GapA (NADPH dependent, 2-fold) and PRK (3- to 4-fold). However, the activity of both GapA and PRK released from the complex was much lower than the activity displayed by the enzymes before complex formation (Fig. 4). Full recovery of GapA activity could be achieved by further incubation with the substrate 1,3-bisphosphoglycerate (BPGA; produced by ATP, 3-phosphoglycerate, and phosphoglycerate kinase). The BPGA-producing mixture also dissociated the complex directly (Fig. 5), thereby activating GapA (6-fold) at maximal levels. Complex dissociation by BPGA also resulted in PRK activation (7-fold) yet without reaching the activity of the oxidized free enzyme (Fig. 4, B and D). Reduced DTT and thioredoxins quantitatively dissociated the complex and fully activated PRK. GapA was activated only 2-fold by reducing conditions, similar to the effect of NADP or ATP. Independent of the effector used to destabilize the GapA/CP12/PRK complex, full activity of GapA was always recovered by further incubation with BPGA, while full PRK activity required DTT (Fig. 4). In no case did dissociation of the ternary complex lead to binary complexes of whatsoever composition, as indicated by the elution volumes of released proteins. Complex dissociation invariably gave rise to GapA, CP12, and PRK free proteins (Fig. 5).
Figure 5.
Effect of GapA and PRK ligands on GapA/CP12/PRK complex stability. The GapA/CP12/PRK complex was reconstituted and chromatographed as in Figure 3D, re-equilibrated with 100 mm Tricine-NaOH, pH 7.9, in the absence of NAD, and incubated under different conditions (A, 0.2 mm NAD; B, 0.5 mm ribulose-5-P; C, 2 mm ATP; D, 0.2 mm NADP; E, 43 μm BPGA; F, 5 mm reduced DTT plus 1 μg/mL thioredoxin) before loading on the gel filtration column (Superdex 200). The column equilibration buffer included different effectors as reported in the figure. The elution volumes of GapA/CP12/PRK (640 kD), GapA and PRK (110–120 kD), and CP12 (29–35 kD) are indicated. Under conditions of complex disruption (ATP, NADP, BPGA, or reductants), the CP12 peak was hardly detectable, partly due to the lower molar extinction coefficient in comparison with either GapA or PRK (see “Materials and Methods”).
DISCUSSION
Enzymatic supramolecular complexes in photosynthetic organisms have long been investigated, but their physiological meaning is still a matter of debate (Gontero et al., 2002). Like in other cell compartments characterized by strong metabolic activity (Goodsell, 1991), the protein concentration of the stroma is very high and chloroplastic enzymes are necessarily subjected to continuous interactions in vivo. However, we do not know whether protein-protein interactions commonly result in the formation of supramolecular complexes with specific organization and function. The existence of a complex network of interactions within the proteome has recently been documented in many organisms (Gavin and Superti-Furga, 2003) and analogous networks of protein interactions may be suspected to exist in plants as well. Among photosynthetic enzymes, GAPDH and PRK catalyze two nonconsecutive reactions of the Calvin cycle and have been proposed to physically interact in vivo together with a scaffold protein known as CP12 (Wedel et al., 1997; Graciet et al., 2004). The reversible formation of this complex has been proposed to contribute to the regulation of the Calvin cycle in vivo (Wedel and Soll, 1998; Scheibe et al., 2002; Graciet et al., 2004; Tamoi et al., 2005).
In oxygenic photosynthetic organism thioredoxins, pyridine nucleotides and metabolites play an important role in regulating the Calvin cycle in dark/light transitions (Wolosiuk et al., 1993; Buchanan and Balmer, 2005). PRK is strongly regulated by thioredoxins in higher plants and green algae (Porter et al., 1988; Graciet et al., 2004), but apparently is less redox sensitive in cyanobacteria and diatoms (Kobayashi et al., 2003; Michels et al., 2005). GAPDH is finely regulated by thioredoxins, NAD(P)(H), and BPGA in higher plants (Pupillo and Giuliani Piccari, 1975; Wolosiuk and Buchanan, 1978; Trost et al., 1993; Baalmann et al., 1995; Sparla et al., 2002), but it does not appear to be regulated in lower photosynthetic organisms. In fact, in green unicellular algae and in cyanobacteria, the GAPDH is composed of a single type of subunit related to subunit A of higher plants (Figge et al., 1999). This subunit lacks the pair of redox-sensitive Cys of subunits B and is therefore insensitive to regulatory effectors including thioredoxins (Sparla et al., 2002).
CP12 is a redox-sensitive protein widely distributed in oxygenic photosynthetic organisms that is able to interact with both GAPDH and PRK (Pohlmeyer et al., 1996; Wedel et al., 1997). In those organisms that do not contain an autonomously regulated GAPDH, CP12 may provide a means to regulate this activity in concert with PRK (Wedel and Soll, 1998; Graciet et al., 2004; Tamoi et al., 2005). In Chlamydomonas reinhardtii, for instance, native GAPDH could be purified as a stable complex with CP12 (Graciet et al., 2003b). PRK steadily interacted with the GAPDH/CP12 binary complex, and activities of both enzymes were inhibited compared to free (activated) counterparts (Graciet et al., 2003a, 2003b).
The relevance of the GAPDH/CP12 interaction in lower photosynthetic organisms is strengthened by the existence of an autonomously regulated GAPDH in higher plants. This GAPDH isoform contains B-subunits resulting from the fusion of GapA with the C-terminal end of CP12 (Pohlmeyer et al., 1996). As a result, B-subunits are regulated by thioredoxins, pyridine nucleotides, and BPGA, while A-subunits are not (Baalmann et al., 1996; Li and Anderson, 1997; Sparla et al., 2002). The C-terminal extension of GapB is suspected to interact with the coenzyme binding site of the protein, thereby leading to a specific down-regulation of the NADPH-dependent enzyme activity (Sparla et al., 2005). The high sequence similarity between CTE and CP12 (Graciet et al., 2004) suggests that both peptides interact with GAPDH in the same way.
Despite the evolution of the autonomous regulation of GAPDH (CTE independent) from an ancient CP12-dependent system, CP12 genes are present in multiple copies in higher plant genomes, and CP12-dependent regulation of GAPDH and PRK seems to be conserved up to higher photosynthetic organisms. In this work, we show that in Arabidopsis the homomeric A4 isozyme of photosynthetic GAPDH (GapA) can form a complex with CP12 (GapA/CP12) and this binary complex can further polymerize by interacting with PRK to give rise to the ternary complex GapA/CP12/PRK (Fig. 6). The molecular mass of the reconstituted GapA/CP12/PRK complex was about 640 kD, similar to the 550- to 600-kD complexes previously detected in spinach chloroplasts (Clasper et al., 1991; Wedel et al., 1997; Scheibe et al., 2002). A complex composition of two GapA tetramers (2 × 120 kD), two PRK dimers (2 × 110 kD), and two CP12 (29 or 2 × 29 kD, depending on whether native CP12 is a monomer or a dimer; Fig. 3), as tentatively proposed by others (Wedel and Soll, 1998; Graciet et al., 2004), might apply to the Arabidopsis complex as well, but different technical approaches are needed to precisely define this stoichiometry.
Figure 6.
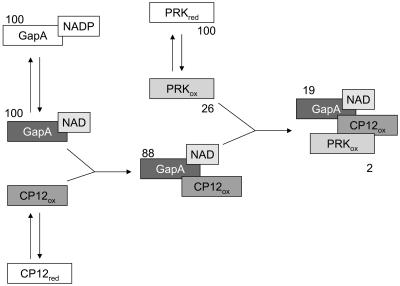
Schematic representation of the sequential formation of GapA/CP12 and GapA/CP12/PRK complexes. The percentage activity of GapA (NADPH dependent) and PRK in respect to free, fully activated enzymes is reported by numbers close to each enzyme form. The scheme indicates which enzyme forms participate in complex formation but does not represent the stoichiometry of complexes.
The formation of complexes involving CP12 in Arabidopsis was promoted by specific conditions. Only the interaction between GapA bound to NAD and CP12 in the oxidized state led to a stable binary complex (Fig. 6). The effect of CP12 on GapA activity was negligible, in contrast with the inhibition observed in the GapA/CP12 complex of Chlamydomonas (Graciet et al., 2003b). In Arabidopsis, strong inhibition of GapA activity was only observed in the presence of oxidized PRK, when a stable GapA/CP12/PRK complex could be reconstituted (Fig. 6). Albeit intrinsically insensitive to oxidized thioredoxins and pyridine nucleotides (Baalmann et al., 1996; Scagliarini et al., 1998; Sparla et al., 2002), GapA could thus be effectively regulated by a CP12/PRK-dependent mechanism, provided NAD and oxidizing conditions were applied. Moreover, although oxidation of free PRK by oxidized thioredoxins resulted in 80% loss of activity, complexation with GapA/CP12 caused a further, almost complete PRK inhibition (Fig. 6).
Both oxidized/reduced thioredoxins and NAD/NADP ratios increase in chloroplasts in the dark (Muto et al., 1980; Heineke et al., 1991; Buchanan and Balmer, 2005), suggesting that the formation of the GapA/CP12/PRK complex in vivo may contribute to the kinetic down-regulation of the Calvin cycle in the dark. Similar oscillations of pyridine nucleotides were also measured under dark/light conditions in Synechococcus PCC7942. In this cyanobacterium, a GapA/CP12/PRK complex was detected in vitro under conditions reproducing the cellular NAD/NADPH ratio measured in the dark. In this organism at least, pyridine nucleotides seem to play a major role in regulating the Calvin cycle via CP12 (Tamoi et al., 2005).
The contrasting effects of NAD and NADP on GapA/CP12/PRK complex formation and consequent enzyme inhibition are consistent with the structural similarity between CP12 and the C-terminal extension of GAPDH B-subunits. In A2B2-GAPDH, binding of NAD leads to enzyme aggregation to A8B8 oligomers and specific inhibition of the NADPH-dependent activity. The process is slow, reverted by NADP, and strictly dependent on the C-terminal extension of B-subunits (Pupillo and Giuliani Piccari, 1975; Baalmann et al., 1996; Sparla et al., 2002). Clearly, the process of GapA/CP12/PRK complex formation shares several features with this system, further supporting the evolutionary derivation of regulated B-subunits of photosynthetic GAPDH from a fusion between ancient A-subunits and the C-terminal portion of CP12 (Pohlmeyer et al., 1996).
Dissociation of the GapA/CP12/PRK complex of Arabidopsis occurred under several conditions with variable effects on enzyme activities. Reduced thioredoxins led to complex disruption and total recovery of full PRK activity. Although GapA was found as a free tetramer after complex dissociation by reductants, the activity was only partially recovered. NADP and ATP behaved similarly to reductants in dissociating the complex and partially activating GapA. Full activation of GapA required BPGA incubation. This result was puzzling since GapA, which lacks the CTE, was known to be insensitive to common activators of AB-GAPDH, including BPGA (Cerff, 1979; Baalmann et al., 1996; Scagliarini et al., 1998; Sparla et al., 2002). Therefore, we propose that dissociation of the GapA/CP12/PRK complex by reductants, NADP, or ATP released tetrameric GapA in a state reminiscent of the inhibited conformation the protein had within the complex, being rescued by its substrate BPGA alone. PRK behaved somehow similarly, as the activity of the enzyme released by the complex by NADP, ATP, or BPGA was still much lower than the activity of free oxidized enzyme. A similar “imprinting” effect was reported for both GAPDH and PRK of Chlamydomonas (for review, see Graciet et al., 2004).
Although in higher plants both AB-GAPDH and PRK can be directly regulated by thioredoxins and metabolites in the absence of CP12, the formation of a GapA/CP12/PRK supramolecular complex provides new potentialities for the regulation of the Calvin cycle in dark/light conditions. First, in the absence of CP12, GapA would be constitutively activated, and this might not be compatible with the need to silence the Calvin cycle in the dark. Second, CP12-mediated regulation of GAPDH and PRK provides a novel way to coordinately regulate both enzyme activities. Within the complex, GapA becomes sensitive to molecules (primarily BPGA, but also thioredoxins, ATP, and NADP) that do not affect the activity of free, isolated enzyme at all; much in the same way, complexed PRK becomes sensitive to GapA substrates (NADP, BPGA) while the free enzyme is only sensitive to thioredoxins. The CP12-dependent, coordinated regulation of GAPDH and PRK may be a major requirement for an effective modulation of the Calvin cycle in light/dark conditions. As a first confirmation of this hypothesis, a Synechococcus mutant in which the CP12 gene was disrupted showed limited growth in light/dark cycle but normal growth under continuous light (Tamoi et al., 2005).
MATERIALS AND METHODS
Plant Material
Arabidopsis (Arabidopsis thaliana) plants were grown on sterile soil (humus:perlite 3:1) for 1 month at 22°C, under 15-h-dark/9-h-light cycle in a growth chamber. Leaves were collected after 15 h of dark. One gram of green tissue was homogenized on ice with 30 mL of Xpl buffer (330 mm sorbitol, 50 mm HEPES, pH 7.5, 2 mm EDTA, 1 mm MgCl2), filtered through a Miracloth net, and centrifuged at 1,200g for 10 min. The pellet was resuspended in 1.5 mL of Xpl buffer, and chloroplasts were separated on a Percoll gradient, as described by Weigel and Glazebrook (2002). Arabidopsis chloroplast proteins were precipitated with 80% ammonium sulfate and resuspended in Xpl buffer without sorbitol but with the addition of 0.2 mm NAD. The sample was incubated for 2 h on ice before loading on the gel filtration column equilibrated with 50 mm Tris-HCl, pH 7.5, 150 mm KCl, 1 mm EDTA, and 0.2 mm NAD.
Expression Vectors
Arabidopsis cDNAs coding for the A-subunit of GAPDH (GapA-1, At3g26650), CP12 (CP12-2, At3g62410), and PRK (At1g32060) were transferred into a pET28a(+) expression vector (Novagen) using the following PCR primers: GapA forward (NcoI site), 5′-TGTGACCATGGCCAAGC-3′, reverse (BamHI site), 5′-CAAGGATCCCTCACTTC-3′; CP12 forward (NdeI site), 5′-CGCATATGGCAGCACCGG-3′, reverse (BamHI site), 5′-AGGATCCTGATCGCTTCAG-3′; and PRK forward (NcoI site), 5′-AGAAACCATGGTGATCGGAC-3′, reverse (BamHI site), 5′-TTGGATCCGTTTGTTTTAGGC-3′.
Specific endonuclease sites (underlined) were introduced at the 5′ and 3′ ends of the cDNA sequences. PCR-amplified fragments were digested with endonucleases, purified, and ligated into a predigested pET28a(+) vector. In the CP12 construct only, the cDNA sequence for CP12 was in frame with a His tag and a cleavable thrombin site. The coding sequence for mature CP12-2 was established after alignment of the three Arabidopsis CP12s (CP12-1, At2g47400; CP12-2, At3g62410; CP12-3, At1g76560; Marri et al., 2005) and prediction of transit peptides by ChloroP (Emanuelsson et al., 1999; Fig. 1).
Recombinant plasmids, amplified into Escherichia coli HB101 cells, were sequenced before transformation of E. coli BL21(DE3) cells.
Expression and Purification of Recombinant Proteins
Heterologous expression and purification of recombinant GapA was performed as described by Sparla et al. (2005). Expression and purification of CP12-2 was performed as by Sparla et al. (1999). After cleavage of the His tag by thrombin, four amino acids of the thrombin cleavage site (GSHM) remained attached to the N terminus of CP12 (AAPEG…; Fig. 1). Resulting pure proteins were desalted in 100 mm Tricine-NaOH, pH 7.9, and stored at −20°C.
An overnight culture of E. coli BL21(DE3) cells, harboring the pET28-PRK expression plasmid, was transferred to fresh LB medium supplied with kanamycin (50 μg/mL) and grown for 6 to 8 h at 30°C. When the optical density at 600 nm reached 0.7, expression was induced by addition of 0.4 mm isopropylthio-β-galactoside. Fifteen hours after induction, the cells were collected by centrifugation. The resulting pellet was washed with 25 mm potassium phosphate, pH 7.5, 1 mm EDTA, 10 mm β-mercaptoethanol (buffer A), and spun down again before storage at −70°C. The same GapA-purification protocol comprising two anion-exchange chromatographic steps (Sparla et al., 2005) was adopted to purify recombinant PRK. Pure PRK was desalted in 100 mm Tricine-NaOH, pH 7.9, and stored at −20°C.
Purified proteins were quantified by absorbance at 280 nm. Molar extinction coefficients at 280 nm were derived from the sequence of each monomer: ɛGapA = 36,250 m−1, ɛCP12 = 8,370 m−1, and ɛPRK = 29,360 m−1.
Enzyme Activity Assays and Redox Titrations
GAPDH activity was assayed as described by Sparla et al. (2002). PRK activity was assayed as described by Porter et al. (1986). Redox titrations of purified GapA and PRK were performed according to Hutchinson and Ort (1995) after desalting the purified proteins in 100 mm Tricine-NaOH, pH 7.9. In this buffer, enzymes were incubated for 3 h at 25°C with equimolar concentrations of recombinant thioredoxin from E. coli (Sigma) and 20 mm DTT in different dithiol/disulfide ratios, as described by Sparla et al. (2002). Following incubation, GAPDH and PRK activities were assayed and results were fit by nonlinear regression (CoStat, CoHort Software) to the Nernst equation with an n value of 2 (Hirasawa et al., 2000) and analyzed as described (Sparla et al., 2002).
Electrophoresis and Immunoblotting
Purified samples of GapA, CP12, and PRK were examined by vertical SDS-PAGE on 12.5% acrylamide gels.
Reduced and oxidized CP12 were obtained by incubating the samples for 2 h at 25°C with equimolar concentrations of prereduced or preoxidized thioredoxin, respectively. Prereduced and preoxidized thioredoxins were prepared by incubation for 2 h at 25°C with 20 mm reduced or oxidized DTT followed by washing out the DTT by ultrafiltration (Centricon YM3). Samples were then boiled for 3 min in sample buffer with no reductants and the proteins were separated on 15% acrylamide gels. Gels were stained with Coomassie Brilliant Blue R-250.
Fractions obtained from gel filtration columns were concentrated by ultrafiltration (Centricon YM3), run on denaturing 12.5% acrylamide gels, and electroblotted (Sammy-dry cell; Schleicher-Schuell) on nitrocellulose membranes. The membranes were stained with Red Ponceau before incubation with rabbit antiserum raised against spinach (Spinacia oleracea) CP12, spinach GapA, and Arabidopsis PRK, kindly provided by Renate Scheibe (University of Osnabrueck), and peroxidase-conjugated secondary antibodies. Primary and secondary antibodies were diluted 1:2,000 and 1:10,000, respectively. Blots were developed by chemiluminescence according to standard procedures.
Supramolecular Complex Reconstitution and Gel Filtration
Samples containing different combinations of purified recombinant GAPDH, PRK, and CP12 under different conditions were analyzed by gel filtration to detect the in vitro reconstitution of binary and ternary complexes. Oxidized and reduced forms of PRK and CP12 were obtained by incubation for 3 h at 25°C with 25 mm oxidized DTT or 25 mm reduced DTT, respectively, followed by buffer exchange to 100 mm Tricine-NaOH, pH 7.9, through ultrafiltration (Centricon YM10 and YM3 for PRK and CP12, respectively). GapA was also equilibrated in buffer 100 mm Tricine-NaOH, pH 7.9. In order to in vitro reconstitute the supramolecular complexes, purified proteins were incubated for 2 h at 4°C at equimolar ratios (subunit basis) under different conditions as described in the text.
Gel filtration analysis was performed on a Superdex 200 HR10/30 column connected to an ÅKTA Purifier system (General Electric Healthcare). The column was equilibrated with 50 mm Tris-HCl, pH 7.5, 150 mm KCl, 1 mm EDTA, and 0.2 mm NAD(P) or 2 mm DTT as specified in the figure legends. The volume of loaded samples was 0.2 mL, and fractions of 0.35 mL were collected. The column was calibrated as done by Sparla et al. (2002).
Complex Dissociation and Recovery of Enzyme Activity
The GapA/CP12/PRK supramolecular complex eluted from the Superdex 200 column in the presence of 0.2 mm NAD was collected and equilibrated with 100 mm Tricine-NaOH, pH 7.9, in the absence of NAD. Potential dissociating agents as described in “Results” were incubated with the complex for 1 h at 25°C. A steady-state concentration 43 μm BPGA was obtained in a mixture of 3 mm 3-phosphoglycerate, 2 mm ATP, and 5 units mL−1 of rabbit muscle phosphoglycerate kinase (Sigma). Following incubation, samples were reloaded on the Superdex 200 column equilibrated with 100 mm Tricine-NaOH, pH 7.9, in the absence of effectors. GAPDH and PRK activities were measured immediately before the addition of effectors and immediately before loading the samples on the gel filtration column.
Acknowledgments
We thank Renate Scheibe for useful discussions and for the kind gift of antisera.
This work was supported by the Ministero dell'Istruzione, dell'Università e della Ricerca (PRIN 2003).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Francesca Sparla (sparla@alma.unibo.it).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.068445.
References
- Baalmann E, Backhausen JE, Rak C, Vetter S, Scheibe R (1995) Reductive modification and non-reductive activation of spinach chloroplast NADP-glyceraldehyde-3-phosphate dehydrogenase. Arch Biochem Biophys 324: 201–208 [DOI] [PubMed] [Google Scholar]
- Baalmann E, Scheibe R, Cerff R, Martin W (1996) Functional studies of chloroplast glyceraldehyde-3-phosphate dehydrogenase subunits A and B expressed in Escherichia coli: formation of highly active A4 and B4 homotetramers and evidence that the aggregation of the B4 complex is mediated by the B-subunit carboxy terminus. Plant Mol Biol 32: 505–513 [DOI] [PubMed] [Google Scholar]
- Brandes HK, Larimer FW, Hartman FC (1996) The molecular pathway for the regulation of phosphoribulokinase by thioredoxin f. J Biol Chem 271: 3333–3335 [DOI] [PubMed] [Google Scholar]
- Brinkmann H, Cerff R, Salomon M, Soll J (1989) Cloning and sequence analysis of cDNAs encoding the cytosolic precursors of subunits GapA and GapB of chloroplast glyceraldehyde-3-phosphate dehydrogenase from pea and spinach. Plant Mol Biol 13: 81–94 [DOI] [PubMed] [Google Scholar]
- Buchanan BB, Balmer Y (2005) Redox regulation: a broadening horizon. Annu Rev Plant Biol 56: 187–220 [DOI] [PubMed] [Google Scholar]
- Cerff R (1979) Quaternary structure of higher plant glyceraldehyde-3-phosphate dehydrogenases. Eur J Biochem 94: 243–247 [DOI] [PubMed] [Google Scholar]
- Clasper S, Easterby JS, Powls R (1991) Properties of two high-molecular mass forms of glyceraldehyde-3-phosphate dehydrogenase from spinach leaf, one of which also possesses latent phosphoribulokinase activity. Eur J Biochem 202: 1239–1246 [DOI] [PubMed] [Google Scholar]
- Dai S, Johansson K, Miginiac-Maslow M, Schürmann P, Eklund H (2004) Structural basis of redox signalling in photosynthesis: structure and function of ferredoxin:thioredoxin reductase and target enzymes. Photosynth Res 79: 233–248 [DOI] [PubMed] [Google Scholar]
- Dani DN, Sainis JK (2005) Isolation and characterization of a thylakoid membrane module showing partial light and dark reactions. Biochim Biophys Acta 1669: 43–52 [DOI] [PubMed] [Google Scholar]
- Emanuelsson O, Nielsen H, von Heijne G (1999) ChloroP, a neural network-based method for predicting chloroplast transit peptides and their cleavage sites. Protein Sci 8: 978–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figge RM, Schubert M, Brinkmann H, Cerff R (1999) Glyceraldehyde-3-phosphate dehydrogenase gene diversity in eubacteria and eukaryotes: evidence for intra- and inter-kingdom gene transfer. Mol Biol Evol 16: 429–440 [DOI] [PubMed] [Google Scholar]
- Gavin AC, Superti-Furga G (2003) Protein complexes and proteome organization from yeast to man. Curr Opin Chem Biol 7: 21–27 [DOI] [PubMed] [Google Scholar]
- Gontero B, Lebreton S, Graciet E (2002) Multienzyme complexes involved in the Benson-Calvin cycle and fatty acid metabolism. In MT McManus, W Laing, AC Allan, eds, Protein-Protein Interactions in Plant Biology. Annual Plant Reviews, Vol 7. Sheffield Academic Press, Sheffield, UK, pp120–150
- Goodsell DS (1991) Inside a living cell. Trends Biochem Sci 16: 203–206 [DOI] [PubMed] [Google Scholar]
- Graciet E, Gans P, Wedel N, Lebreton S, Camadro JM, Gontero B (2003. a) The small protein CP12: a protein linker for supramolecular complex assembly. Biochemistry 42: 8163–8170 [DOI] [PubMed] [Google Scholar]
- Graciet E, Lebreton S, Camadro JM, Gontero B (2003. b) Characterization of native and recombinant A4 glyceraldehyde 3-phosphate dehydrogenase. Kinetic evidence for conformation changes upon association with the small protein CP12. Eur J Biochem 270: 129–136 [DOI] [PubMed] [Google Scholar]
- Graciet E, Lebreton S, Gontero B (2004) Emergence of new regulatory mechanisms in the Benson-Calvin pathway via protein-protein interactions: a glyceraldehyde-3-phosphate dehydrogenase/CP12/phosphoribulokinase complex. J Exp Bot 55: 1245–1254 [DOI] [PubMed] [Google Scholar]
- Heineke D, Riens B, Grosse H, Hoferichter P, Heldt HW (1991) Redox transfer across the inner chloroplast envelope membrane. Plant Physiol 95: 1131–1137 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirasawa M, Ruelland E, Schepens I, Issakidis-Bourguet E, Miginiac-Maslow M, Knaff D (2000) Oxidation-reduction properties of the regulatory disulfides of sorghum chloroplast nicotinamide adenine dinucleotide phosphate-malate dehydrogenase. Biochemistry 39: 3344–3350 [DOI] [PubMed] [Google Scholar]
- Hirasawa M, Schürmann P, Jacquot J-P, Manieri W, Jacquot P, Keryer E, Hartman F, Knaff D (1999) Oxidation-reduction properties of chloroplast thioredoxins, ferredoxin:thioredoxin reductase, and thioredoxin f-regulated enzymes. Biochemistry 38: 5200–5205 [DOI] [PubMed] [Google Scholar]
- Hutchinson RS, Ort DR (1995) Measurement of equilibrium midpoint potentials of thiol/disulfide regulatory groups on thioredoxin-activated chloroplast enzymes. Methods Enzymol 252: 220–228 [DOI] [PubMed] [Google Scholar]
- Kobayashi D, Tamoi M, Iwaki T, Shigeoka S, Wadano A (2003) Molecular characterization and redox regulation of phosphoribulokinase from the cyanobacterium Synechococcus sp. PCC 7942. Plant Cell Physiol 44: 269–276 [DOI] [PubMed] [Google Scholar]
- Li AD, Anderson LE (1997) Expression and characterization of pea chloroplastic glyceraldehyde-3-phosphate dehydrogenase composed of only the B-subunit. Plant Physiol 115: 1201–1209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marri L, Sparla F, Pupillo P, Trost P (2005) Coordinated gene expression of photosynthetic glyceraldehyde-3-phosphate dehydrogenase, phosphoribulokinase and CP12 in Arabidopsis thaliana. J Exp Bot 56: 73–80 [DOI] [PubMed] [Google Scholar]
- Michels AK, Wedel N, Kroth PG (2005) Diatom plastids possess a phosphoribulokinase with an altered regulation and no oxidative pentose phosphate pathway. Plant Physiol 137: 911–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller B (1972) A labile CO2-fixing enzyme complex in spinach chloroplasts. Z Naturforsch B 27: 925–932 [Google Scholar]
- Muto S, Miyachi S, Usuda H, Edwards GE, Bassham JA (1980) Light-induced conversion of nicotinamide adenine dinucleotide to nicotinamide adenine dinucleotide phosphate in higher plant leaves. Plant Physiol 68: 324–328 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pohlmeyer K, Paap BK, Soll J, Wedel N (1996) CP12: a small nuclear-encoded chloroplast protein provides novel insights into higher-plant GAPDH evolution. Plant Mol Biol 32: 969–978 [DOI] [PubMed] [Google Scholar]
- Porter MA, Milanez S, Stringer CD, Hartman FC (1986) Purification and characterization of ribulose-5-phosphate kinase from spinach. Arch Biochem Biophys 245: 14–23 [DOI] [PubMed] [Google Scholar]
- Porter MA, Stringer CD, Hartman FC (1988) Characterization of the regulatory thioredoxin site of phosphoribulokinase. J Biol Chem 263: 123–129 [PubMed] [Google Scholar]
- Pupillo P, Faggiani R (1979) Subunit structure of three glyceraldehyde-3-phosphate dehydrogenases of some flowering plants. Arch Biochem Biophys 154: 475–482 [DOI] [PubMed] [Google Scholar]
- Pupillo P, Giuliani Piccari G (1975) The reversible depolymerization of spinach chloroplast glyceraldehyde-3-phosphate dehydrogenase. Interaction with nucleotides and dithiothreitol. Eur J Biochem 51: 475–482 [DOI] [PubMed] [Google Scholar]
- Rault M, Giudici-Orticon M-T, Gontero B, Ricard J (1993) Structural and functional properties of a multienzyme complex from spinach chloroplasts. I. Stoichiometry of the polypeptide chains. Eur J Biochem 217: 1065–1073 [DOI] [PubMed] [Google Scholar]
- Ruelland E, Miginiac-Maslow M (1999) Regulation of chloroplast enzyme activities by thioredoxin: activation or relief from inhibition? Trends Plant Sci 4: 136–141 [DOI] [PubMed] [Google Scholar]
- Scagliarini S, Trost P, Pupillo P (1998) The non-regulatory isoform of NAD(P)-glyceraldehyde-3-phosphate dehydrogenase from spinach chloroplasts. J Exp Bot 49: 1307–1315 [Google Scholar]
- Scheibe R, Wedel N, Vetter S, Emmerlich V, Sauermann SM (2002) Co-existence of two regulatory NADP-glyceraldehyde 3-P dehydrogenase complexes in higher plant chloroplasts. Eur J Biochem 269: 5617–5624 [DOI] [PubMed] [Google Scholar]
- Schürmann P, Jacquot JP (2000) Plant thioredoxin systems revisited. Annu Rev Plant Physiol Plant Mol Biol 51: 371–400 [DOI] [PubMed] [Google Scholar]
- Sparla F, Pupillo P, Trost P (2002) The C-terminal extension of glyceraldehyde-3-phosphate dehydrogenase subunit B acts as an autoinhibitory domain regulated by thioredoxins and nicotinamide adenine dinucleotide. J Biol Chem 277: 44946–44952 [DOI] [PubMed] [Google Scholar]
- Sparla F, Tedeschi G, Pupillo P, Trost P (1999) Cloning and heterologous expression of NAD(P)H:quinone reductase of Arabidopsis thaliana, a functional homologue of animal DT-diaphorase. FEBS Lett 463: 382–386 [DOI] [PubMed] [Google Scholar]
- Sparla F, Zaffagnini M, Wedel N, Scheibe R, Pupillo P, Trost P (2005) Regulation of photosynthetic GAPDH dissected by mutants. Plant Physiol 138: 2210–2219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Süss K-H, Arkona C, Manteuffel R, Adler K (1993) Calvin cycle multienzyme complexes are bound to chloroplast thylakoid membranes of higher plants in situ. Proc Natl Acad Sci USA 90: 5514–5518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamoi M, Myazaki T, Fukamizo T, Shigeoka S (2005) The Calvin cycle in cyanobacteria is regulated by CP12 via NAD(H)/NADP(H) ratio under light/dark conditions. Plant J 42: 504–513 [DOI] [PubMed] [Google Scholar]
- Trost P, Scagliarini S, Valenti V, Pupillo P (1993) Activation of spinach chloroplast glyceraldehyde-3-phosphate dehydrogenase: effect of glycerate 1,3-bisphosphate. Planta 190: 320–326 [Google Scholar]
- Wara-Aswapati O, Kemble RJ, Bradbeer JW (1980) Activation of glyceraldehyde-phosphate dehydrogenase (NADP) and phosphoribulokinase in Phaseolus vulgaris leaf extracts involves the dissociation of oligomers. Plant Physiol 66: 34–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel N, Soll J (1998) Evolutionary conserved light regulation of Calvin cycle activity by NAPDH-mediated reversible phosphoribulokinase/CP12/glyceraldehyde-3-phosphate-dehydrogenase complex dissociation. Proc Natl Acad Sci USA 95: 9699–9704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wedel N, Soll J, Paap BK (1997) CP12 provides a new mode of light regulation of Calvin cycle activity in higher plants. Proc Natl Acad Sci USA 94: 10479–10484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weigel D, Glazebrook J (2002) Arabidopsis: A Laboratory Manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Wolosiuk RA, Ballicora MA, Hagelin K (1993) The reductive pentose phosphate cycle for photosynthetic CO2 assimilation: enzyme modulation. FASEB J 7: 622–637 [DOI] [PubMed] [Google Scholar]
- Wolosiuk RA, Buchanan BB (1978) Activation of chloroplast NADP-linked glyceraldehyde-3-phosphate dehydrogenase by the ferredoxin/thioredoxin system. Plant Physiol 61: 669–671 [DOI] [PMC free article] [PubMed] [Google Scholar]