Abstract
In higher plants, ferredoxin (Fd):NADPH oxidoreductase (FNR) catalyzes reduction of NADP+ in the final step of linear photosynthetic electron transport and is also implicated in cyclic electron flow. We have identified three leaf FNR isoenzymes (LFNR1, LFNR2, and LFNR3) in maize (Zea mays) chloroplasts at approximately equivalent concentrations. Fractionation of chloroplasts showed that, while LFNR3 is an exclusively soluble enzyme, LFNR1 is only found at the thylakoid membrane and LFNR2 has a dual location. LFNR1 and LFNR2 were found to associate with the cytochrome b6f complex following its partial purification. We cloned LFNR3 and produced all three isoenzymes as stable, soluble proteins. Measurement of Fd reduction ability showed no significant differences between these recombinant enzymes. Column chromatography revealed variation between the interaction mechanisms of LFNR1 and LFNR2 with Fd, as detected by differential dependence on specific intermolecular salt bridges and variable sensitivity of interactions to changes in pH. A comparison of LFNR transcripts in leaves of plants grown on variable nitrogen regimes revealed that LFNR1 and LFNR2 transcripts are relatively more abundant under conditions of high demand for NADPH. These results are discussed in terms of the functional differentiation of maize LFNR isoenzymes.
Ferredoxin (Fd):NADPH oxidoreductase (FNR; EC 1.18.1.2) is a flavoenzyme that catalyzes reduction of NADP+ or oxidation of NADPH through electron transfer with Fd. In the final step of photosynthetic electron transport, FNR reduces NADP+, using Fd that has accepted electrons from PSI (Carrillo and Ceccarelli, 2003). In linear photosynthesis, the resulting NADPH fuels carbon fixation by the Calvin cycle and drives other reductive metabolism in the stroma or outside the chloroplast following export by the malate-oxaloacetate shuttle. In higher plant roots, genetically distinct, soluble, root-type FNR (RFNR) enzymes catalyze the NADPH-dependent reduction of Fd to provide reducing power to the many Fd-dependent enzymes involved in assimilation of nitrogen and sulfur, etc. (Neuhaus and Emes, 2000).
It has been reported that FNR in higher plant chloroplasts is localized peripherally on the stromal side of thylakoid membranes through association with an intrinsic protein (Vallejos et al., 1984; Matthijs et al., 1986), PSI (Andersen et al., 1992), cytochrome (cyt) b6f complex (Clark et al., 1984; Zhang et al., 2001), and NAD(P)H dehydrogenase complex (Quiles et al., 2000).
In addition to linear electron transport, a photosynthetic proton gradient may also be driven solely by PSI cycling electrons via Fd or NADPH back to the cyt b6f complex (Bendall and Manasse, 1995). This alternative electron flow results in ATP production without the generation of new reducing species and is essential for photosynthesis in higher plants (Munekage et al., 2004), although its exact physiological role remains unclear. The localization of FNR at the cyt b6f and/or NAD(P)H dehydrogenase complexes implies a role in cyclic electron transport, and it has been suggested that FNR could be involved in modulating the two photosynthetic energy flows (Bojko et al., 2003), although there is currently no direct evidence.
Higher plant FNRs are encoded by a small multiple gene family as shown by the recent whole-genome analysis of Arabidopsis (Arabidopsis thaliana; http://www.plantgdb.org/AtGDB) and rice (Oryza sativa; http://cdna01.dna.affrc.go.jp/cDNA), in both of which two leaf-type FNR (LFNR) isoenzymes and two RFNR isoenzymes are annotated. The presence of multiple isoenzymes at the protein level has been reported in several plant species (Green et al., 1991). In maize (Zea mays), two different cDNAs encoding the precursors of LFNR have been cloned (Onda et al., 2000).
The reductive assimilation of nitrogen requires abundant reducing equivalents and ATP, and some molecular species of FNRs and Fds are known to respond to changes in nitrogen status. In both Arabidopsis (Wang et al., 2003) and maize (Ritchie et al., 1994; Sakakibara, 2003), transfer of plants to high-nitrate growth media resulted in short-term induction of genes encoding RFNRs but not LFNRs. However, assimilation of nitrate in photosynthetic tissue requires abundant export of reductant to the cytosol, demanding that Fd is reduced to NADPH to power the malate shuttle, whereas assimilation of ammonium requires reduced Fd in the chloroplast. Reflecting this, long-term growth of Arabidopsis on nitrate resulted in higher LFNR transcript levels than under ammonium conditions (Hanke et al., 2005).
Here we report the identification at the cDNA and protein level of a new, third maize LFNR isoenzyme. Comparison of the subplastid location, expression pattern, and Fd interactions of native isoenzymes, combined with analysis of all three as recombinant enzymes following cloning and recombinant expression, indicates that multiple LFNR isoenzymes enable higher plants to respond rapidly to varied reductive demands.
RESULTS
There Are Three Leaf FNR Proteins in Maize Chloroplasts and They Vary in Subchloroplast Location
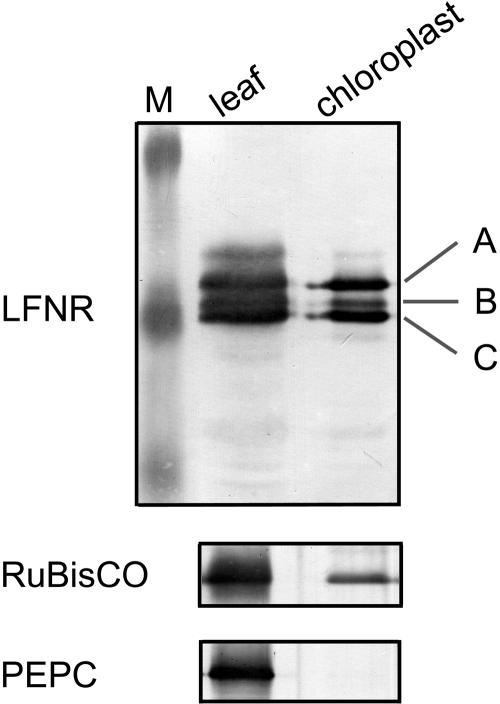
Antibodies raised against recombinant maize LFNR1 (Onda et al., 2000) were used to challenge crude extract and isolated chloroplast proteins from maize leaves by western blotting (Fig. 1). In both cases, three bands, labeled A, B, and C in order of mobility, were detected close to the 32.5-kD protein maker. A comparison of recovery with phosphoenolpyruvate carboxylase and Rubisco indicates that all three bands are chloroplast proteins.
Figure 1.
Immunodetection of LFNR polypeptides. Protein extracts from total maize leaves and isolated maize chloroplasts were separated by SDS-PAGE and challenged with antisera specific for maize LFNR. Samples applied to the gels were equivalent on a chlorophyll basis. Three bands detected with LFNR1 antibody are labeled A, B, and C in order of increasing mobility. The same samples were used for detection of phosphoenolpyruvate carboxylase as a cytosolic marker, and Rubisco large subunit as a chloroplast marker.
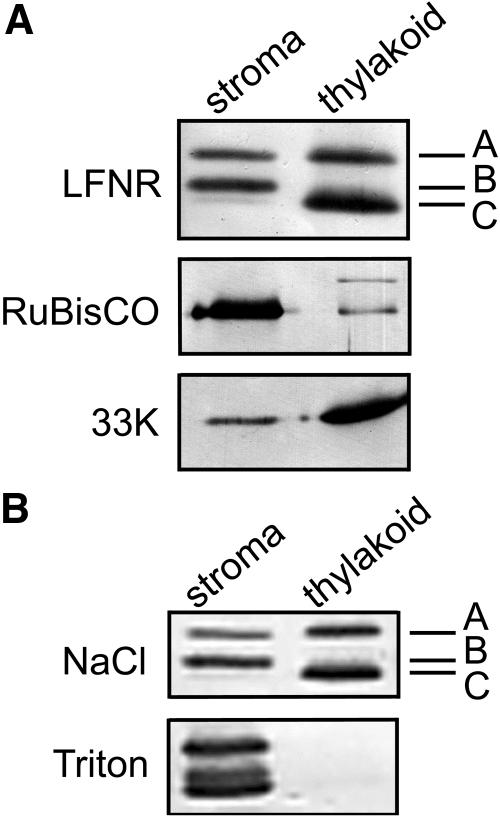
Subchloroplast distribution of putative LFNR isoenzymes was investigated by separating stroma and thylakoid membranes and western blotting (Fig. 2). Separation into predominantly stromal and thylakoid fractions was confirmed by distribution of the stromal enzyme Rubisco and the 33-kD PSII protein, which is intrinsic in the thylakoid membrane (Fig. 2A). The three LFNR bands were differentially distributed between the stroma and the thylakoid membranes: Band A is present in both fractions, band B is located only in the stroma, and band C is associated exclusively with the thylakoid membrane. Membrane association of bands A and C was unaffected by washing with 500 mm NaCl, but disrupted by 0.1% Triton X-100 (Fig. 2B).
Figure 2.
LFNR polypeptides vary in subchloroplast distribution. A, Isolated maize chloroplasts were separated into stromal and thylakoid fractions, equivalent volumes were subject to SDS-PAGE, and then western blotting using antisera against maize LFNR, the 33-kD oxygen-evolving protein of spinach PSII (as a marker for the thylakoid membrane), and maize Rubisco (as a stromal marker). B, Broken chloroplasts were treated with 0.5 m salt or 0.1% Triton X-100 prior to the separation of thylakoid and stromal fractions and western blotting.
Cloning of a New Maize LFNR
Two maize LFNR cDNAs (LFNR1 and LFNR2) had previously been identified (Onda et al., 2000) and so the detection of three polypeptides immunologically related to FNR encouraged an extensive database search of maize expressed sequence tag (EST) clones. This yielded cDNA fragments corresponding to another LFNR (Maize GDB; http://www.maizegdb.org, a long stretch of DNA formed by seven overlapping sequences, AW056238, BG316642, BG349441, BG349767, BG410251, BI233764, and AI001302). Several other EST sequences related to LFNR3, but with a few nucleotide substitutions presumably due to cultivar differences, were also identified.
During previous cloning of LFNR1 and LFNR2 cDNAs (Onda et al., 2000), 30 clones hybridizing to a LFNR1 probe were obtained from a maize cDNA library under weak stringency conditions. We further screened these clones and obtained a novel LFNR cDNA, which was named LFNR3 and perfectly matched the sequence constructed from the EST database. We confirmed the existence of the new LFNR in planta by reverse transcription (RT)-PCR amplification of mRNA using primers specific to LFNR3 cDNA (data not shown).
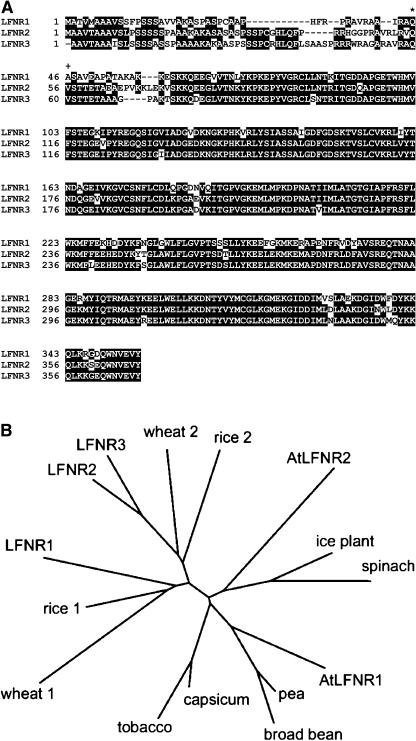
When aligned, the mature LFNRs have high amino acid sequence homology (LFNR1:LFNR2 = 83%, LFNR1:LFNR3 = 84%, LFNR2:LFNR3 = 92%; Fig. 3A). The phylogenetic tree in Figure 3B indicates that LFNR1 belongs to a genetically distinct group of isoenzymes from LFNR2 and LFNR3, and that these two groups are conserved among other cereals.
Figure 3.
Comparison of three maize LFNR isoenzymes. A, Alignment of amino acid sequences of the three maize LFNR isoenzymes using ClustalW 1.8 (http://searchlauncher.bcm.tmc.edu/multi-align/multi-align.html). Residues common to at least two sequences are shown as white on black. N-terminal protein sequencing (see later) determined the first amino acid of LFNR1 and LFNR2 (*) and LFNR3 (+). B, Phylogenetic tree of mature LFNRs from several higher plants. Where the N-terminal sequence was not known, it was estimated by comparison with known data. Sequences were aligned in ClustalW 1.8, and the tree drawn using the philodendron software (http://iubio.bio.indiana.edu/treeapp/treeprint-form.html).
The Three Putative LFNR Isoenzymes Correspond to LFNR1, LFNR2, and LFNR3
LFNRs were partially purified from stroma and thylakoid membranes by Fd affinity chromatography (Supplemental Fig. 1). The three putative LFNR bands were either blotted to polyvinylidene difluoride (PVDF) membrane and N-terminal sequenced, or excised, modified, digested, and analyzed by matrix-assisted laser-desorption ionization time-of-flight (MALDI-TOF) mass spectrometry (MS). This analysis demonstrated that band C corresponded to LFNR1, band B corresponded to LFNR3, and band A obtained from both stroma and thylakoid membranes corresponded to LFNR2. The mass of detected peptides is compared to that of predicted digestion products of LFNR1, LFNR2, and LFNR3 in Supplemental Table I. The sequence coverage of LFNR1, LFNR2, and LFNR3 by MS data was 40.3%, 43.9%, and 39.2%, respectively. N-terminal sequences of the mature enzymes are also shown in Supplemental Table I. The cleavage point of transit peptides, derived from this sequencing information, is indicated in the alignment in Figure 3A and reveals that the presequences of LFNR2 and LFNR3 are very similar (61%), and different from those of LFNR1 (22% and 23%, respectively). The molecular masses of mature LFNR1, LFNR2, and LFNR3 are 34.97, 35.57, and 34.7 kD, respectively, as predicted by the Compute pI/Mw tool program at ExPASy (http://ca.expasy.org/tools/pi_tool.html).
LFNR Isoenzymes at the Thylakoid Membrane Are Associated with the Cyt b6f Complex
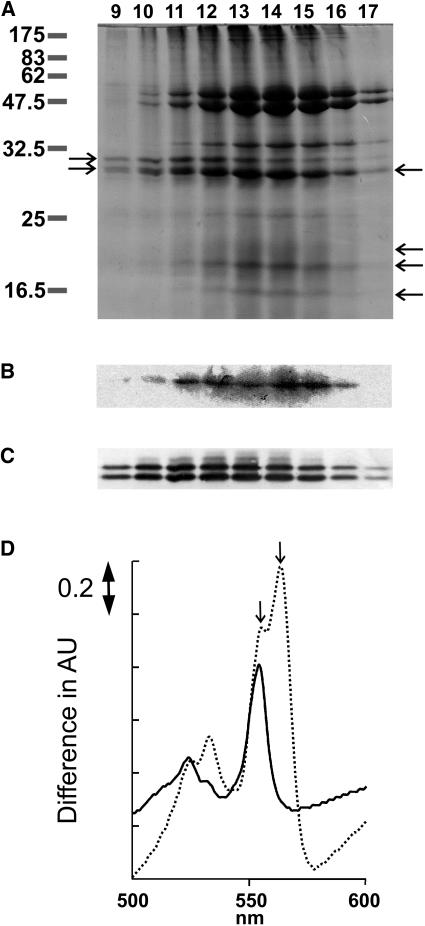
FNR is an α-subunit of the cyt b6f complex in spinach (Spinacia oleracea; Zhang et al., 2001), and so we performed a partial purification of cyt b6f to determine whether this association is isoenzyme specific in maize. Figure 4A shows SDS-PAGE separation of fractions from the final chromatography in partial purification of cyt b6f. Bands of equivalent size to subunits of the cyt b6f complex were present (Fig. 4A, arrows on the right), and the identity of cyt f was further confirmed by heme staining (Fig. 4B). Proteins of the same size as LFNR1 and LFNR2 in roughly equal quantities (Fig. 4A, left-hand arrows) were also detected and their identity was confirmed by western blotting (Fig. 4C). Differential spectra in Figure 4D were generated from fraction 13. Addition of ascorbate or dithionite resulted in differential absorption maxima at 555 and 564 nm, respectively, indicating the presence of cyt f and cyt b6. Elution of both LFNRs was very similar to that of cyt b6f.
Figure 4.
Copurification of LFNR isoenzymes with cyt b6f. Solubilized thylakoid proteins were fractionated over propyl agarose chromatography. A, SDS-PAGE separation of eluted fractions, stained with Coomassie Brilliant Blue. Numbers to the left indicate size in kilodaltons of molecular mass markers. Arrows to the left indicate the position of LFNR2 and LFNR1 from top to bottom. Arrows to the right of the gel indicate the position of cyt b6f subunits, from top to bottom; cyt f, cyt b6, Rieske iron sulfur protein, and subunit IV. B, Lanes as for A, but showing a heme stain of the band corresponding to cyt f. C, Lanes as for A, but showing a western blot using antiserum against maize LFNR to confirm the identity of the LFNR2 and LFNR1 proteins. D, Difference in absorbance spectra of the partially purified cyt b6f complex. A 1:3 dilution of fraction 13 into 50 mm Tris-HCl, pH 7.5, was reduced with sodium ascorbate (full line) or sodium dithionite (dashed line), and the differential spectra between the reduced and nonreduced forms are presented. The left-hand arrow indicates 555 nm and the right-hand arrow 564 nm.
Relative Abundance of LFNRs
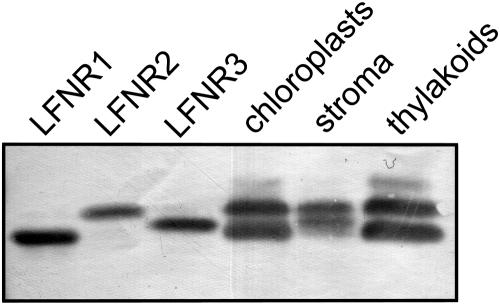
LFNR1 was recombinantly expressed in Escherichia coli previously (Onda et al., 2000) and the same principles were used to construct expression plasmids of LFNR2 and LFNR3. Cells transformed with the resulting plasmids (pQE-LFNR2 and pQE-LFNR3) had a yellow-green color. All enzymes were stable in solution and were purified to homogeneity (Supplemental Fig. 2). Both LFNR2 and LFNR3 showed absorption spectra typical of flavin-containing enzymes and similar to LFNR1 (data not shown) and migration of recombinant LFNR proteins through SDS-PAGE closely resembles that of the native chloroplast isoenzymes (Fig. 5). LFNR2 and LFNR3 have slightly lower reactivity to the antibody raised against LFNR1, and all isoenzymes are present at an approximately equivalent concentration in the leaf (15–30 μg/mg chlorophyll) based on western-blot comparisons with standard curves of recombinant proteins (Table I). A comparison of LFNR2 signal in western blots of equivalent thylakoid and stromal proteins from ruptured chloroplasts indicates that 58.4% of LFNR2 is located at the thylakoid (data not shown).
Figure 5.
Comparison of recombinant and native enzyme LFNRs. Recombinant LFNR isoenzymes and chloroplast fractions were separated by SDS-PAGE and western blotted with antisera specific for maize LFNR1.
Table I.
Quantitation of LFNR isoenzymes
Crude protein fractions from mature maize leaves were separated by SDS-PAGE and western blotted with an antibody raised against LFNR1. Reacting bands were compared densitometrically with standard curves of recombinant LFNR1, LFNR2, and LFNR3 between 5 and 25 ng. Concentrations were calculated using an estimate of chloroplast stromal volume of 66 μL/mg chlorophyll (Winter et al., 1994). Soluble concentration of LFNR2 was calculated as 41.6% of total LFNR2 protein, based on comparison of stromal and thylakoid proteins in western blots of lysed chloroplasts. Values are the mean of measurements from three separate leaves.
| μg LFNR/mg Chlorophyll | μm in Stroma | |
|---|---|---|
| LFNR1 | 16.1 | – |
| LFNR2 | 24.4 | 3.9 |
| LFNR3 | 27.3 | 11.8 |
Catalytic Activity and Differential Interaction with Fd
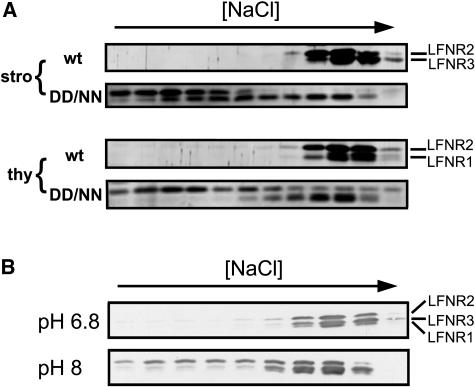
Fd reduction activity of recombinant LFNR isoenzymes was assayed using an NADPH-dependent cyt c reduction system, and kinetic parameters are shown in Table II. Activity of LFNR2 and LFNR3 was similar and higher than that of LFNR1, the Vmax value of LFNR1 being about two-thirds that of LFNR2 and LFNR3. The x-ray crystal structure of maize LFNR1 and Fd revealed that LFNR1 and Fd interact mainly through electrostatic forces (Kurisu et al., 2001). In particular, disruption of the salt bridges formed between Asp-65 and Asp-66 in Fd and Lys-88 and Lys-91 in LFNR1 was found to strongly disrupt electron transfer between FNR and Fd (Akashi et al., 1999). To establish whether the contribution of these salt bridges is consistent between LFNR isoenzymes, we used D65N/D66N Fd to prepare a mutant Fd-immobilized resin column. While the three LFNRs elute from the wild-type Fd column at an equivalent salt concentration, there is great variation between their elution from the mutant Fd column (Fig. 6A). LFNR1 elutes at a slightly lower salt concentration, LFNR3 binding is more perturbed, and LFNR2 interactions are greatly disrupted. The apparently biphasic elution profile of LFNR3 may be an artifact or due to as-yet unresolved posttranslational modification of the peptide. By contrast to column binding, ability to reduce the D65N/D66N Fd mutant did not vary dramatically between LFNR isoenzymes (Table II) as Km increased for all recombinant LFNRs. LFNR2 showed the greatest difference in elution profile among the three FNR isoenzymes and a slightly greater increase in Km for the mutant.
Table II.
Kinetic parameters of LFNR isoenzymes with wild-type and mutant Fd
Purified recombinant maize LFNR isoenzymes were used in cyt c-based Fd reduction assays over a 0 to 20 μm concentration range of the indicated Fds. Kinetic parameters were calculated using a least-squares fit of the Michaelis-Menten equation to experimental data.
| Wild-Type Fd
|
D65N/D66N
|
|||
|---|---|---|---|---|
| Km | Vmax | Km | Vmax | |
| μm | μmol cyt c min−1 | μm | μmol cyt c min−1 | |
| LFNR1 | 2.7 | 0.209 | 8.0 | 0.187 |
| LFNR2 | 3.4 | 0.346 | 10.4 | 0.355 |
| LFNR3 | 2.7 | 0.314 | 7.1 | 0.317 |
Figure 6.
Investigation of LFNR isoenzyme and Fd interactions by affinity chromatography. Maize LFNR isoenzyme bands are labeled on the right. A, Mutant Fd protein fractions from maize chloroplast stroma (stro) and thylakoids (thy) were eluted over a linear salt gradient from Fd affinity columns made from wild type (wt) and a D65N/D66N mutant (DD/NN). Fractions were western blotted with antiserum specific for maize LFNR. B, pH effects. Chloroplast extracts were eluted over a linear salt gradient from a wild-type Fd column at either pH 6.8 or pH 8. Eluted fractions were western blotted to detect LFNR isoenzymes as before.
Variable dependence of LFNRs on salt bridging for complex formation with Fd means that changes in pH may also have a variable effect on the formation of different FNR-Fd complexes, and we therefore compared the Fd binding of maize LFNRs at pH 8 and pH 6.8 (Fig. 6B). While LFNR1 eluted from the Fd column at approximately the same salt concentration, LFNR3-Fd interactions were slightly weakened at high pH and LFNR2 showed a dramatically faster elution.
Relative Expression of LFNR Isoenzymes Varies in Response to Nitrogen Status
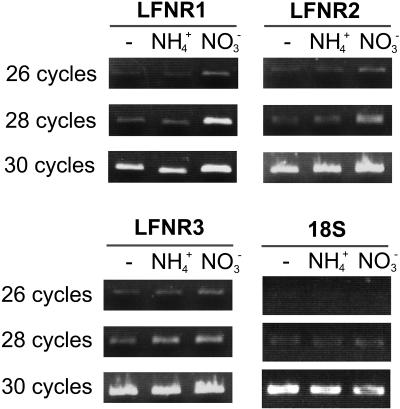
Assimilation of ammonium places very different reductive demands on the chloroplast than assimilation of nitrate, and different LFNR isoenzymes in Arabidopsis showed variation in relative expression when grown on variable nitrogen regimes (Hanke et al., 2005). Therefore, to investigate possible variable contribution of LFNR isoenzymes to redox metabolism, we compared their transcript levels in leaves of plants grown for 20 d on no nitrogen, ammonia, and nitrate using isoenzyme-specific primers (Fig. 7). This analysis revealed that transcripts of LFNR1 and LFNR2 were more abundant in nitrate conditions than ammonium or no-nitrogen conditions (this difference being greatest for LFNR1), while LFNR3 transcripts were equally abundant in nitrate- and ammonium-grown leaves, but reduced in no-nitrogen growth conditions.
Figure 7.
Response of LFNR isoenzyme transcripts to different nitrogen status. Purified mRNA isolated from maize seedlings grown on no-nitrogen source (-), ammonium (2 mm NH4+), and nitrate (10 mm NO3−) conditions was used as a template for cDNA synthesis. Specific PCR amplification of LFNR1, LFNR2, LFNR3, and 18S ribosome (as a control for loading) was performed for the indicated number of cycles, and the products separated on an agarose gel and visualized with ethidium bromide.
DISCUSSION
Variation in LFNR Isoenzyme Location
Following our identification of LFNR3 at cDNA and protein levels, we now know of three different LFNR isoenzymes present at roughly equal concentrations in maize chloroplasts. Our analysis indicates that a primary difference among them is their subplastid location; LFNR1 is restricted to the thylakoid membrane and LFNR3 is a soluble stromal enzyme, while LFNR2 is present in both fractions. Previous failure to detect LFNR3 is probably due to its stromal location, which is not consistent with the current dogma that LFNR principally catalyzes NADP+ photoreduction on the thylakoid membrane. At least one-third of all LFNR, in the form of the LFNR3 isoenzyme, is in fact in the stroma of maize chloroplasts (Table I), although it is not yet clear whether it acts in physiological reduction and/or oxidation of NADP(H).
The structural basis for differential location of LFNR isoforms remains unclear. The region of greatest variation between LFNR isoforms is at the N terminus, immediately following the transit peptide cleavage site (Fig. 3A). This is disordered in the crystal structure of maize LFNR1 (Kurisu et al., 2001), indicating flexibility, and could form the basis of thylakoid interactions, although a recent NMR study suggests it is involved in Fd binding (Maeda et al., 2005). Bruns and Karplus (1995) proposed that a hydrophobic pocket, far from the active site, on the crystal structure of spinach LFNR could be involved in membrane interaction. However, all side chains that contribute to this pocket (LFNR1 I207, A208, W241, F243, I288, M292, L299, W300, L303, T308, V310, M312, M318, I322, M326, A330, W337, W350, and V354) are conserved among maize LFNRs. In some cases, there is variation in adjacent amino acids (LFNR1 I206, G240, V327, S328, E331, and F338), and these may prove to be related to differential localization, but further work is required to address this question.
Approximately 40% of LFNR2 is present in the stroma, and it is unclear what determines this dual subplastid location and whether there is a dynamic equilibrium between the two locations. One possibility is that this difference may be due to posttranslational modification. There is an interesting difference between the composition of digestion products from stromal and thylakoid species of LFNR2, as detected by MALDI-TOF MS. Peptide279-290 was detected only in stromal LFNR2, whereas peptide193-212 was detected only in thylakoid-associated LFNR2 (Supplemental Table I), indicating that these regions may somehow be related to differential localization. As peptide coverage by MALDI-TOF MS was not complete, it is also possible that some kind of modification, located on an unmeasured peptide fragment, remained undetected.
Recombinant expression of mature LFNR1, LFNR2, and LFNR3 in E. coli generated stable, soluble, and functional proteins whose migration through SDS-PAGE was very similar to that of LFNR1, LFNR2, and LFNR3 extracted from maize leaves (Fig. 5). This suggests that membrane localization is not critical for protein assembly and that if any modification is associated with localizing LFNR at the thylakoid, it is relatively small. FNR interactions with the thylakoid are probably not electrostatic because they were not disrupted by a 2 m NaBr wash (data not shown) or EDTA (Zhang et al., 2001).
LFNR1 and LFNR2 were copurified with the cyt b6f complex, although both eluted a little faster than cyt b6f subunits during the final hydrophobic chromatography (Fig. 4). Previous reports from spinach (Zhang et al., 2001; Zhang and Cramer, 2004) in combination with these results indicate that thylakoid LFNRs are located at the cyt b6f complex, and that this interaction is not isoenzyme specific. It is probable that not all LFNR is associated with the cyt b6f complex. LFNR1 and LFNR2 may also form isoenzyme-specific or nonspecific associations with PSI and the NAD(P)H dehydrogenase complex, as described by Andersen et al. (1992) and Quiles et al. (2000), respectively. The presence of LFNR at the cyt b6f complex implies a role interconnecting the linear electron transport chain with Fd-dependent cyclic electron transport. Recent structural studies on the cyt b6f complex (Kurisu et al., 2003; Stroebel et al., 2003) revealed a novel heme on the stromal side of the complex, which could potentially support such cyclic electron transfer.
LFNR Isoenzyme-Dependent Fd Interactions
LFNR residues contributing intermolecular salt bridges to the complex with Fd (Kurisu et al., 2001) are conserved among all maize isoenzymes. However, chromatography with Fd mutated at Asp-65 and Asp-66 suggested that the relative contribution of LFNR Lys-88 and Lys-91 (the corresponding salt bridge pairs) to complex formation varies between isoenzymes. Interaction with LFNR1 was largely unaffected, while that with LFNR2 was particularly disrupted.
Despite this variation, the Fd reduction activity of all LFNR isoenzymes was disrupted to roughly the same extent by the D65N/D66N Fd mutation, although LFNR2 did show a slightly greater loss of affinity (Table II). The difference between the effect of the D65N/D66N Fd mutation on the column (a complex between oxidized Fd and LFNR) and during Fd reduction (when LFNR has been reduced by NADPH) indicates that there may be redox state-dependent changes in the mode of interaction between Fd and FNR. Additionally, there may be a discrepancy between the contribution of these salt bridges to formation of the kinetically competent complex (measured as Km), as compared to the initial, nonproductive complex, known to be dominated by ionic forces (Hurley et al., 1999).
Variation in Fd binding between LFNRs occurs despite conservation of all residues contributing salt bridges to the LFNR1-Fd complex. The structural basis of such differences remains to be elucidated, but their physiological implications are intriguing and lead us to investigate pH dependency. In high pH conditions, equivalent to those in the stroma of an actively photosynthesizing chloroplast, interaction of Fd with LFNR2 is dramatically weaker as compared to LFNR1, while interaction with LFNR3 is also slightly reduced. In Table I, the total LFNR in maize chloroplasts is 270 μg/mg chlorophyll (around 30 μm if all LFNR were soluble), which is of the same molar order as Fd as measured by Yonekura-Sakakibara et al. (2000), suggesting that there is competition for reduced Fd under photosynthetic conditions, even though the Km value for each LFNR is lower than the Fd concentration in the stroma. Considering the massive flux through the photosynthetic pathway, small changes in affinity could result in electron partitioning through different LFNR isoenzymes. Further experiments are required to resolve physiological differences between LFNR isoenzymes, which would clarify the metabolic result of any such electron partitioning.
Varied Physiological Responses of LFNR Isoenzymes
Continuous growth on different nitrogen regimes revealed isoenzyme-specific differences in LFNR transcripts (Fig. 7). This is consistent with our work on Arabidopsis (Hanke et al., 2005) and suggests that LFNR transcript levels respond indirectly to the differing demands of nitrate and ammonium assimilation, possibly through the redox state of the cell and chloroplast. Assimilation of nitrate rather than ammonium requires a greater supply of reducing power both inside and outside the chloroplast (Crawford, 1995; Stitt, 1999). Therefore, the higher abundance of LFNR1 (and to a lesser extent LFNR2) transcripts in nitrate-grown plants indicates a relatively greater contribution to linear electron flow and generation of NADPH to drive the malate shuttle and transfer reducing power to the cytosol. By contrast, LFNR3 transcripts are more abundant in both ammonium- and nitrate-grown leaves compared to no-nitrogen growth conditions. The reason for this is unclear, but one possible function of LFNR3 in the stroma could be to provide reduced Fd for ammonium and/or nitrite assimilation in the dark. LFNR protein concentrations were also compared by western blotting, but did not vary significantly between growth conditions (data not shown). This indicates a high turnover of enzymes, presumably to counter oxidative damage caused by increased electron transfer activity.
In this article, we describe the identification of a new, soluble LFNR isoenzyme in maize and present a thorough comparison of the subchloroplast location, physical properties, expression, and Fd interactions of all three maize LFNRs. This analysis leads us to propose that LFNR1 and LFNR2 are the predominantly active isoenzymes in NADP+ reduction during linear and cyclic electron transport, and that electron flow through LFNR2 may be dynamically regulated by the photosynthetic state of the chloroplast. The exact functional differentiation of the three LFNRs remains to be elucidated, in particular the physiological role of soluble LFNR.
MATERIALS AND METHODS
Plant Growth
Maize (Zea mays L. cv Golden Cross Bantam T51) seedlings were grown in vermiculite for 2 weeks with Hoagland nutrients (Arnon and Hoagland, 1940). The photoperiod was 10-h dark at 26°C/14-h light at 28°C, with a light intensity of approximately 700 μE m−2 s−1. Plants on varied nitrogen regimes were grown on washed sand as described by Suzuki et al. (2001). For large-scale preparation of thylakoid membranes, young maize plants (3 to 4 weeks old) were grown in the field during the summer season.
Electrophoresis and Immunoblotting
SDS-PAGE and western blotting using an antibody raised against maize LFNR1 was essentially as described by Onda et al. (2000). Protein staining of gels was with Coomassie Brilliant Blue or a Silver Stain MS kit (Wako Pure Chemical Industries).
Chloroplast Isolation and Subchloroplast Fractionation
Chloroplasts were isolated from maize leaves essentially as described by Jenkins and Boag (1984). Isolated chloroplasts were ruptured at 4°C by osmotic pressure in 10 mm Tris (hydroxymethyl)aminomethane (Tris)-HCl, pH 7.5, before adding NaCl to 100 mm. Stroma and thylakoids were separated by centrifugation at 12,000 rpm for 5 min in a benchtop centrifuge at 4°C. The pellet was resuspended in an equivalent volume of 10 mm Tris-HCl, pH 7.5, 100 mm NaCl to make the thylakoid membrane fraction.
Fd Affinity Chromatography
Fd affinity resin was prepared from recombinant maize wild-type Fd and D65N/D66N mutant Fd as described previously (Hase et al., 1991). Column chromatography was carried out essentially as described by Hanke et al. (2005) using an ÄKTA prime system (Amersham Biosciences). The Fd column was equilibrated with 50 mm Tris-HCl, pH 7.5, except for experiments conducted at pH 8.0 and pH 6.8.
N-Terminal Amino Acid Sequencing and MALDI-TOF MS
LFNRs separated by Fd affinity chromatography were reduced and carboxymethylated essentially as described by Crestfield et al. (1963). Modified samples were dialyzed against 50 mm NH4HCO3 and lyophilized, then dissolved in 2 mm NaOH and separated by SDS-PAGE. Proteins were either blotted to PVDF membrane (Millipore) for N-terminal protein sequencing (performed as described by Hanke et al., 2005), or stained with Coomassie Brilliant Blue or silver stain. Coomassie Brilliant Blue and silver-stained bands were manually excised from the gel and destained in 50 mm NH4HCO3, 50% methanol for 1 h at 40°C, or according to the manufacturer's notes, respectively. Gel pieces were ground, dried in a vacuum, and resuspended in 100 μL MilliQ water containing 1 milliunit activity Achromobacter lysyl endopeptidase (Wako) for digestion over 12 h at room temperature. Peptides were sequentially extracted from the gel in 0%, 100%, 50%, and 100% acetonitrile, all in 0.1% trifluoroacetic acid. From this point, mass spectrometry was performed as described by Hanke et al. (2005).
Molecular Cloning and Sequencing of cDNA for a New Leaf FNR Isoenzyme
Thirty candidate FNR clones isolated from a maize seedling cDNA library (Onda et al., 2000) were further screened by PCR amplification followed by restriction site mapping. One cDNA clone with a restriction digest pattern unique from LFNR1 and LFNR2 was sequenced by conventional methods (Hokkaido System Science) and named pSO1.
Partial Purification of Cyt b6f
Maize leaves (500 g) were homogenized in 1 L of 0.2 m Suc, 25 mm HEPES-NaOH, pH 7.5, 10 mm CaCl2, and 10 mm MgCl2 at 4°C and filtered through two layers of gauze and two layers of miracloth. The partial purification of cyt b6f from this homogenate was then essentially the same as the published protocol for spinach (Zhang and Cramer, 2004) until the step of membrane solubilization, when the protocol was further optimized for maize. All centrifugation steps were at 10,000 rpm in a GSA rotor at 4°C for 30 min unless otherwise stated. The thylakoid membrane pellet was resuspended in 30 mm Tris-HCl, pH 7.5, 50 mm NaCl, 5 mm KCl, 5 mm MgCl2, 1 mm EDTA, and 10% Suc. The solution was further diluted 1:1 with this buffer containing 50 mm n-octyl-β-d-glucoside (Anatrace) and 0.6% sodium cholate (Sigma), incubated for 15 min at room temperature, and centrifuged at 25,000 rpm for 30 min at 4°C. The cyt b6f complex was then separated by propyl-agarose chromatography as described by Zhang et al. (2001). Elution of the complex was followed by the differential spectrum of cyt f as measured following addition of ascorbate. Heme-containing proteins transferred to PVDF membrane were visualized essentially according to Thomas et al. (1976).
Construction of Plasmids and Recombinant Expression of LFNR2 and LFNR3
pYOLFNR2 was used as a template for LFNR2 and primer pairs were GGCGCCATGGTATCTACAACAGAAACCGCGGAGGCGGAGCCGGTCAAG and GTCGAAGTCTACTGACATGCGGATCCTTA. The pSO1 vector was used as a template for LFNR3 and primer pairs were GGCGCCATGGTATCTACAACAGAAACCGCGGCGGCGGGGCCGGCGAAG and GTCGAGGTCTACTGACATGCGGATCCTTA. Sense primers contained the initiation Met, codons of 13 amino acids from the mature protein N terminus and an NcoI site. Antisense primers contained a stop codon, codons of four amino acids from the C terminus, and a BamHI site. PCR was carried out for 35 cycles of 15 s at 94°C, 30 s at 55°C for LFNR2 or 65°C for LFNR3, and 60 s at 72°C using KOD-Plus Taq (Toyobo). Products were purified using a MinElute gel extraction kit (Qiagen) after electrophoresis on a 1% agarose gel and digested with NcoI and BamHI (Toyobo). The resulting fragments were inserted into a NcoI- and BamHI-digested pQE-60 vector (Qiagen) using DNA ligation kit version 2.1 (TaKaRa Bio). Plasmids were named pQE-LFNR2 and pQE-LFNR3 and, in essentially the same procedure described by Onda et al. (2000), used to transform Escherichia coli TG1 cells from which FNR proteins were purified following large-scale cultivation.
Cyt c Reduction Assays
Fd reduction activities of LFNRs were measured as described in Onda et al. (2000) using a cyt c reduction assay system in 0.6 mL containing 100 mm NaCl, 200 μm cyt c, 200 nm FNR, and 50 μm NADPH in the presence of a NADPH regenerating system. The reaction was initiated by the addition of maize FdI or the maize FdI mutant D65N/D66N at final concentrations from 1 to 20 μm and carried out at room temperature. Cyt c reduction was measured by monitoring the increase in A550.
RT-PCR
Total RNA was prepared from maize leaves using the RNeasy plant mini kit (Qiagen) and cDNA was synthesized using Omniscript kit (Qiagen). PCR was hot started at 94°C and carried out at 94°C for 15 s, 65°C for 30 s, and 72°C for 2 min using TaKaRa LA Taq (TaKaRa Bio). LFNR1 primers were AAGGCCAAGAAGGAGTCCAAGAAG and ACAACACAAAATGTCAGCTGCAAAA, LFNR2 primers were GAGCCGGTCAAGAAGCTGGAG and TTGCTTGAGCTGAACAATACAATGAA, and LFNR3 primers were CGGCGAAGACGTCCAAGAAG and AGTCGTCAACGGATGGATGGAT. PCR products were visualized by ethidium bromide staining after electrophoresis through a 1% agarose gel. An initial PCR with quantum RNA 18S standards (Ambion) was used to confirm equivalence of the cDNA template.
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession numbers AB035644, AB035644, At5g66190, At1g20020, AJ457979, AJ457980, D17790, XM_506676, X12446, U14956, M25528, X07981, AJ250378, and Y14032.
Supplementary Material
This work was supported by a grant-in-aid (no. 15GSO320) for Creative Scientific Research from the Ministry of Education, Culture, Sports, Science and Technology of Japan and a Joint Research Program between the Japan Society for the Promotion of Science and the Institut National de la Recherche Agronomique, France.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Toshiharu Hase (enzyme@protein.osaka-u.ac.jp).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.070813.
References
- Akashi T, Matsumura T, Ideguchi T, Iwakiri K, Kawakatsu T, Taniguchi I, Hase T (1999) Comparison of the electrostatic binding sites on the surface of ferredoxin for two ferredoxin-dependent enzymes, ferredoxin-NADP(+) reductase and sulfite reductase. J Biol Chem 274: 29399–29405 [DOI] [PubMed] [Google Scholar]
- Andersen B, Scheller HV, Moller BL (1992) The PSI E subunit of photosystem I binds ferredoxin:NADP+ oxidoreductase. FEBS Lett 311: 169–173 [DOI] [PubMed] [Google Scholar]
- Arnon DI, Hoagland DR (1940) Crop production in artificial solutions and soils with special reference to factors influencing yield and absorption of inorganic nutrient. Soil Sci 50: 463–471 [Google Scholar]
- Bendall DS, Manasse RS (1995) Cyclic photophosphorylation and electron transport. Biochim Biophys Acta 1229: 23–38 [Google Scholar]
- Bojko M, Kruk J, Wieckowski S (2003) Plastoquinones are effectively reduced by ferredoxin:NADP+ oxidoreductase in the presence of sodium cholate micelles: significance for cyclic electron transport and chlororespiration. Phytochemistry 64: 1055–1060 [DOI] [PubMed] [Google Scholar]
- Bruns CM, Karplus PA (1995) Refined crystal structure of spinach ferredoxin reductase at 1.7Å resolution: oxidized, reduced and 2′-phospho-5′-AMP bound states. J Mol Biol 247: 125–145 [DOI] [PubMed] [Google Scholar]
- Carrillo N, Ceccarelli EA (2003) Open questions in ferredoxin-NADP+ reductase catalytic mechanism. Eur J Biochem 270: 1900–1915 [DOI] [PubMed] [Google Scholar]
- Clark RD, Hawkesford MJ, Coughlan SJ, Bennett J, Hind G (1984) Association of ferredoxin-NADP+ oxidoreductase with the chloroplast cytochrome b-f complex. FEBS Lett 174: 137–142 [Google Scholar]
- Crawford NM (1995) Nitrate: nutrient and signal for plant growth. Plant Cell 7: 859–868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crestfield AM, Stanford M, Stein WH (1963) The preparation and enzymatic hydrolysis of reduced and S-carboxymethylated proteins. J Biol Chem 238: 622–627 [PubMed] [Google Scholar]
- Green LS, Yee BC, Buchanan BB, Kamide K, Sanada Y, Wada K (1991) Ferredoxin and ferredoxin-NADP reductase from photosynthetic and nonphotosynthetic tissues of tomato. Plant Physiol 96: 1207–1213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanke GT, Okutani S, Satomi Y, Takao T, Suzuki A, Hase T (2005) Multiple iso-proteins of FNR in Arabidopsis: evidence for different contributions to chloroplast function and nitrogen assimilation. Plant Cell Environ 28: 1146–1157 [Google Scholar]
- Hase T, Mizutani S, Mukohata Y (1991) Expression of maize ferredoxin cDNA in Escherichia coli: comparison of photosynthetic and nonphotosynthetic ferredoxin isoenzymes and their chimeric molecule. Plant Physiol 97: 1395–1401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurley JK, Hazzard JT, Martinez-Julvez M, Medina M, Gomez-Moreno C, Tollin G (1999) Electrostatic forces involved in orienting Anabaena ferredoxin during binding to Anabaena ferredoxin:NADP+ reductase: site-specific mutagenesis, transient kinetic measurements, and electrostatic surface potentials. Protein Sci 8: 1614–1622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkins CLD, Boag S (1984) Large scale, rapid preparation of functional mesophyll chloroplasts from Zea mays and other C4 species. Plant Sci Lett 39: 19–24 [Google Scholar]
- Kurisu G, Kusunoki M, Katoh E, Yamazaki T, Teshima K, Onda Y, Kimata-Ariga Y, Hase T (2001) Structure of electron transfer complex between ferredoxin and ferredoxin-NADP+ reductase. Nat Struct Biol 8: 117–121 [DOI] [PubMed] [Google Scholar]
- Kurisu G, Zhang H, Smith JL, Cramer WA (2003) Structure of the cytochrome b6f complex of oxygenic photosynthesis: tuning the cavity. Science 302: 1009–1014 [DOI] [PubMed] [Google Scholar]
- Maeda M, Lee YH, Ikegami T, Tamura K, Hoshino M, Yamazaki T, Nakayama M, Hase T, Goto Y (2005) Identification of the N- and C-terminal substrate binding segments of ferredoxin-NADP+ reductase by NMR. Biochemistry 44: 10644–10653 [DOI] [PubMed] [Google Scholar]
- Matthijs HCP, Coughlan SJ, Hind G (1986) Removal ferredoxin-NADP+ oxidoreductase from thylakoid membranes, rebinding to depleted membranes, and identification of the binding site. J Biol Chem 261: 12154–12158 [PubMed] [Google Scholar]
- Munekage Y, Hashimoto M, Miyake C, Tomizawa K, Endo T, Tasaka M, Shikanai T (2004) Cyclic electron flow around photosystem I is essential for photosynthesis. Nature 429: 579–582 [DOI] [PubMed] [Google Scholar]
- Neuhaus HE, Emes MJ (2000) Nonphotosynthetic metabolism in plastids. Annu Rev Plant Physiol Plant Mol Biol 51: 111–140 [DOI] [PubMed] [Google Scholar]
- Onda Y, Matsumura T, Kimata-Ariga Y, Sakakibara H, Sugiyama T, Hase T (2000) Differential interaction of maize root ferredoxin:NADP+ oxidoreductase with photosynthetic and nonphotosynthetic ferredoxin isoenzymes. Plant Physiol 123: 1037–1045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiles MJ, Garcia A, Cuello J (2000) Separation by blue-native PAGE and identification of the whole NAD(P)H dehydrogenase complex from barley stroma thylakoids. Plant Physiol Biochem 38: 225–232 [Google Scholar]
- Ritchie SW, Redinbaugh MG, Shiraishi N, Vrba JM, Campbell WH (1994) Identification of a maize root transcript expressed in the primary response to nitrate: characterization of a cDNA with homology to ferredoxin-NADP+ oxidoreductase. Plant Mol Biol 26: 679–690 [DOI] [PubMed] [Google Scholar]
- Sakakibara H (2003) Differential response of genes for ferredoxin and ferredoxin:NADP+ oxidoreductase to nitrate and light in maize leaves. J Plant Physiol 160: 65–70 [DOI] [PubMed] [Google Scholar]
- Stitt M (1999) Nitrate regulation of metabolism and growth. Curr Opin Plant Biol 2: 178–186 [DOI] [PubMed] [Google Scholar]
- Stroebel D, Choquet Y, Popot JL, Picot D (2003) An atypical haem in the cytochrome b(6)f complex. Nature 426: 413–418 [DOI] [PubMed] [Google Scholar]
- Suzuki A, Rioual S, Lemarchand S, Godfroy N, Roux Y, Boutin JP, Rothstein S (2001) Regulation by light and metabolites of ferredoxin-dependent glutamate synthase in maize. Physiol Plant 112: 524–530 [DOI] [PubMed] [Google Scholar]
- Thomas PE, Ryan D, Levin W (1976) An improved staining procedure for the detection of the peroxidase activity of cytochrome P-450 on sodium dodecyl sulfate polyacrylamide gels. Anal Biochem 75: 168–176 [DOI] [PubMed] [Google Scholar]
- Vallejos RH, Ceccarelli E, Chan R (1984) Evidence for the existence of a thylakoid intrinsic protein that binds ferredoxin-NADP+ oxidoreductase. J Biol Chem 259: 8048–8051 [PubMed] [Google Scholar]
- Wang R, Okamoto M, Xing X, Crawford NM (2003) Microarray analysis of the nitrate response in Arabidopsis roots and shoots reveals over 1,000 rapidly responding genes and new linkages to glucose, trehalose-6-phosphate, iron, and sulfate metabolism. Plant Physiol 132: 556–567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter H, Robinson DG, Heldt HW (1994) Subcellular volumes and metabolite concentrations in spinach leaves. Planta 193: 530–535 [Google Scholar]
- Yonekura-Sakakibara K, Onda Y, Ashikari T, Tanaka Y, Kusumi Y, Hase T (2000) Analysis of the reductant supply systems for ferredoxin-dependent sulfite reductase in photosynthetic and nonphotosynthetic organs in maize. Plant Physiol 122: 887–894 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Cramer WA (2004) Purification and crystallization of the cytochrome b6f complex in oxygenic photosynthesis. Methods Mol Biol 274: 67–78 [DOI] [PubMed] [Google Scholar]
- Zhang H, Whitelegge JP, Cramer WA (2001) Ferredoxin:NADP+ oxidoreductase is a subunit of the chloroplast cytochrome b6f complex. J Biol Chem 276: 38159–38165 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.