Abstract
Cryptochromes are widespread in higher plants but their physiological roles as blue-light photoreceptors have been examined in relatively few species. Screening in a phyA null mutant background has identified several blue-light response mutants in pea (Pisum sativum), including one that carries a substitution of a highly conserved glycine residue in the N-terminal photolyase-homologous domain of the pea CRY1 gene. Analyses of cry1, phyA, and phyB mutants show that all three photoreceptors contribute to seedling photomorphogenesis under high-irradiance blue light, whereas phyA is the main photoreceptor active under low irradiances. Triple phyA phyB cry1 mutants grown under high-irradiance blue light are indistinguishable from dark-grown wild-type plants in length and leaf expansion but show a small residual response to higher-irradiance white light. Monogenic cry1 mutants have little discernable phenotype at the seedling stage, but later in development are more elongated than wild-type plants. In addition, the loss of cry1 moderates the short-internode phenotype of older phyA mutants, suggesting an antagonism between phyA and cry1 under some conditions. Pea cry1 has a small inhibitory effect on flowering under long and short days. However, the phyA cry1 double mutant retains a clear promotion of flowering in response to blue-light photoperiod extensions, indicating a role for one or more additional blue-light photoreceptors in the control of flowering in pea.
Light plays a central role in plant growth and development. It supplies not only the energy required for photosynthesis but also information about time and place that is crucial for appropriate development. Plants monitor specific wavelengths of light using a number of photoreceptors, which include the red and far Pr (phy) photoreceptor family and the blue-light specific cryptochrome (cry) and phototropin photoreceptor families.
Roles for the phytochrome family of photoreceptors have been well characterized through the isolation of specific phytochrome-deficient mutants in a range of higher plant species (Takano et al., 2001; Weller et al., 2001a, 2001b; Fankhauser and Staiger, 2002). In contrast, the roles of specific blue-light receptors are less widely characterized. Blue light regulates many important processes in plant development, including seedling de-etiolation, stem elongation, entrainment of the circadian clock, and photoperiod-responsive flowering (Liscum et al., 2003). In Arabidopsis (Arabidopsis thaliana), many of these responses to blue light are now known to be mediated at least in part by members of the cryptochrome photoreceptor family. The cryptochromes are flavoproteins that share homology with DNA photolyases, but are distinguished by lack of photolyase activity and the presence of a C-terminal extension (Lin and Todo, 2005). Cryptochromes were first identified in Arabidopsis, and have subsequently been characterized in several plant species. They fall into two distinct types, CRY1 and CRY2, based largely on differences in the C-terminal extension (Lin and Shalitin, 2003). An additional more distantly related class of plant cryptochrome proteins (variously termed CRY3 or CRY-DASH) have recently been identified (Brudler et al., 2003), although it is not currently known if they function in a similar manner to CRY1 and CRY2. Cryptochromes are also found in animals, where they play an important role in circadian rhythms (Lin and Todo, 2005).
Cryptochromes have been most extensively studied in Arabidopsis, which contains one representative of each of the three cryptochrome subtypes (Lin and Todo, 2005). However, studies in other plant species are beginning to reveal variation in the size of the CRY gene family. Rice (Oryza sativa), barley (Hordeum vulgare), and tomato (Lycopersicon esculentum) each contain two CRY1-like genes (Perrotta et al., 2000; Matsumoto et al., 2003), whereas two distinct CRY2 genes are present in pea (Pisum sativum) and Medicago truncatula (Platten et al., 2005). CRY genes are also present in lower plants, and have been described for the fern Adiantum capillus-veneris, the moss Physcomitrella patens, and the alga Chlamydomonas reinhardtii, although these genes cluster separately from higher plant cryptochromes (Kanegae and Wada, 1998; Imaizumi et al., 2002; Platten et al., 2005). To date, cryptochrome mutants in higher plants have only been identified in Arabidopsis and tomato. Arabidopsis and tomato cry1 mutant seedlings exhibit reduced de-etiolation under relatively high-irradiance blue light (Ahmad and Cashmore, 1993; Weller et al., 2001a). CRY1 also plays a role in the development of adult plants, contributing to the promotion of flowering by long days in Arabidopsis (Jackson and Jenkins, 1995; Mockler et al., 1999) and influencing internode elongation and chlorophyll levels in leaves and fruit in tomato (Weller et al., 2001a). Arabidopsis plants containing mutations in the gene encoding the light-labile CRY2 protein have been identified on the basis of late flowering in long-day conditions (Guo et al., 1998) and independently by reduced de-etiolation under relatively low levels of blue light (Lin et al., 1998). In addition to these mutant studies, the phenotypes of transgenic rice and tomato plants with altered CRY expression have also been reported (Matsumoto et al., 2003; Giliberto et al., 2005).
Mutant studies in several species have shown that phytochromes also play an important role in blue-light responses. In tomato and Arabidopsis, both phyA and phyB photoreceptors contribute to the promotion of de-etiolation under blue light (Ahmad and Cashmore, 1997; Neff and Chory, 1998; Weller et al., 2001a). In pea, phyB promotes de-etiolation under high-irradiance blue light, while phyA appears to play a somewhat anomalous role, promoting most aspects of de-etiolation but slightly reducing the effectiveness of blue light for inhibition of stem elongation (Weller et al., 2001b). While phytochromes and cryptochromes may act independently in some blue-light responses, in other cases they show clear physiological interactions. For example, in Arabidopsis, the promotion of flowering by cry2 and phyA is achieved by antagonizing the inhibitory effects of phyB (Valverde et al., 2004). Also, the cry1 and phyA-dependent suppression of hypocotyl elongation by blue light in early seedling development is antagonized by phyB (Folta and Spalding, 2001). Finally, cry1-mediated blue-light effects on membrane polarization in Arabidopsis hypocotyl protoplasts are enhanced by prior activation of phyA or phyB (Wang and Iino, 1998). The molecular basis for these physiological interactions is not yet known. However, phytochromes and cryptochrome are known to interact physically with each other and with other proteins such as COP1 and ADO1/ZTL (Ahmad et al., 1998; Más et al., 2000; Jarillo et al., 2001; Wang et al., 2001), and both cry and phy photoreceptors contribute to the nuclear exclusion and inactivation of COP1 (Osterlund et al., 2000).
We previously reported that phyA phyB double mutants of pea show clear residual responses to blue light, indicating an important role for one or more blue-light-specific photoreceptors in pea development (Weller et al., 2001b). In this article, we report on the characterization of a pea cry1 mutant, which was identified on a phyA-deficient background on the basis of reduced de-etiolation under blue light. We have used this mutant to explore the role of cry1 in several aspects of pea development, including de-etiolation, adult vegetative development, and the timing of flowering.
RESULTS
Isolation of a Mutant with Reduced De-Etiolation under Blue Light
Studies in Arabidopsis and tomato have revealed a high degree of redundancy between phytochromes and cryptochromes in the control of seedling de-etiolation under blue light. One result of this functional overlap is that the effect of cryptochrome deficiency is much less in a wild-type background than observed on a phy-deficient (and particularly on a phyA-deficient) background (Neff and Chory, 1998; Mockler et al., 1999; Weller et al., 2001a). To exploit any similar functional overlap between phyA and cryptochromes in blue-light responses in pea, we screened for cry mutants in the phyA-1 null mutant background (Weller et al., 2001b). Progeny of ethyl methanesulfonate-mutagenized seed were screened in the M2 generation for de-etiolation defects under 4 μmol m−2 s−1 blue light, as studies in Arabidopsis and tomato indicate that this intermediate irradiance has the potential to expose the action of CRY1 and CRY2 (Lin et al., 1998; Weller et al., 2001a). M3 lines showing reduced de-etiolation under blue light were rescreened under red and far-red light to exclude mutants with more general defects in de-etiolation.
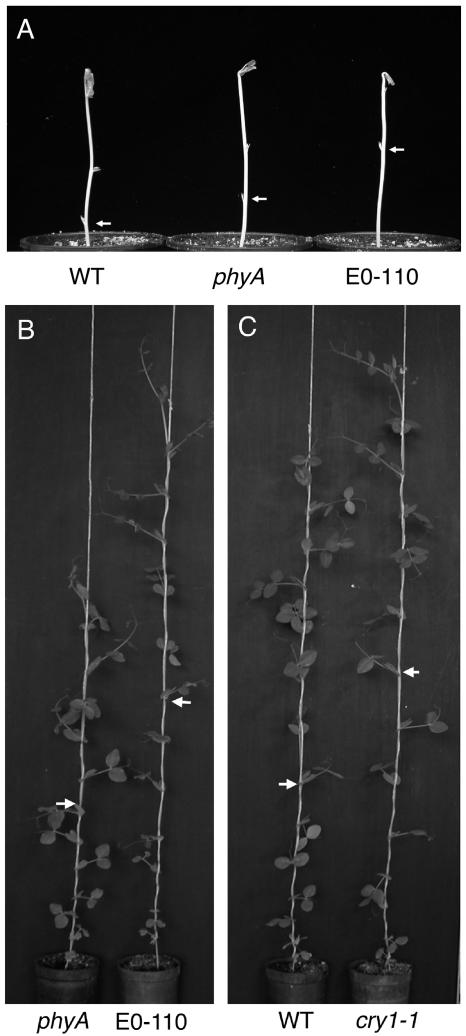
Several lines isolated from this screen showed substantially longer internodes than the phyA-1 parent line under blue light but not under red or far-red light. One line (E0-110) also exhibited reduced leaflet expansion, a paler stem, and a greater tendency to retain an apical hook compared to phyA-1 single mutant plants when grown under blue light (Fig. 1A). When grown under an 18-h photoperiod in the glasshouse, the E0-110 line also displayed longer internodes and smaller leaves than the phyA-1 parental line (Fig. 1B). In addition, the characteristic development of thickened internodes in older phyA-1 mutant plants (Weller et al., 2001b) was significantly reduced in the E0-110 line (diameter of internode 18; phyA-1: 4.62 ± 0.17 mm; E0-110: 3.91 ± 0.16 mm; P < 0.01). No marked difference in flowering node between E0-110 and phyA-1 was observed under these conditions (data not shown).
Figure 1.
Phenotype of a new pea mutant line with reduced responsiveness to blue light. A, Phenotype of wild-type, phyA-1, and line E0-110 seedlings grown for 8 d under continuous blue light (4 μmol m−2 s−1). Arrows indicate node 1. B and C, Phenotype of 35-d-old E0-110, phyA-1, wild-type, and cry1-1 plants grown under standard glasshouse conditions. Arrows indicate node 9.
The F2 progeny of an E0-110 × phyA-1 backcross segregated 39 phyA-like and 16 E0-110-like plants (χ12 = 0.491, P > 0.4) under blue light, indicating that the E0-110 selection phenotype is inherited in a monogenic recessive manner. Transfer of these seedlings to glasshouse conditions confirmed that this seedling trait cosegregated with the elongated adult plant phenotype (data not shown), consistent with both aspects of the E0-110 phenotype resulting from mutation at a single locus.
The E0-110 Line Contains a Mutation in the CRY1 Gene
The phenotype of the E0-110 line appeared generally similar to that of previously described cry mutants in Arabidopsis and tomato (Ahmad and Cashmore, 1993; Lin et al., 1998; Weller et al., 2001a). To determine if the E0-110 line contained a lesion in the CRY1 gene (Platten et al., 2005), we directly sequenced the CRY1 gene in wild type cv Torsdag and the E0-110 mutant. The E0-110 line contained a single nucleotide substitution in the CRY1 cDNA (G778A), directing a substitution of Gly with Glu at position 250 in the CRY1 protein. This Gly residue is perfectly conserved across a wide range of CRY and photolyase proteins from plants, animals, and bacteria (Fig. 2). The substitution of the corresponding Gly residue in the Arabidopsis CRY2 protein by an Arg residue has been reported in the fha-2 (cry2-2) mutant, which has a phenotype equivalent in severity to that of cry2 null mutants (Guo et al., 1998). A Csp45I restriction polymorphism introduced by the G778A mutation was converted to a cleaved amplified polymorphic sequence (CAPS) marker that showed perfect cosegregation with the E0-110 mutant phenotype in the F2 progeny of a phyA-1 × E0-110 backcross (n = 53).
Figure 2.
The cry1-1 mutation affects a highly conserved amino acid. Alignment of amino acid sequences for cryptochrome (CRY and CPH) and photolyase (PHR, 6-4) proteins are shown for the region surrounding the Gly (G) residue substituted in the E0-110 line (G250; marked with an asterisk). Ps, Pisum sativum; At, Arabidopsis; Os, Oryza sativa; Le, Lycopersicon esculentum; Ac, Adiantum capillus-veneris; Pp, Physcomitrella patens; Cr, Chlamydomonas reinhardtii; Mm, Mus musculus; and Dm, Drosophila melanogaster. The alignment was performed using ClustalX.
The E0-110 line thus shows a blue-light-specific de-etiolation defect that cosegregates with a mutation in the CRY1 gene predicted to substitute a highly conserved residue in the CRY1 protein. We therefore conclude that the E0-110 mutant phenotype is likely to result from this mutation, and have designated this mutation cry1-1. The original E0-110 line is subsequently referred to as the phyA-1 cry1-1 double mutant.
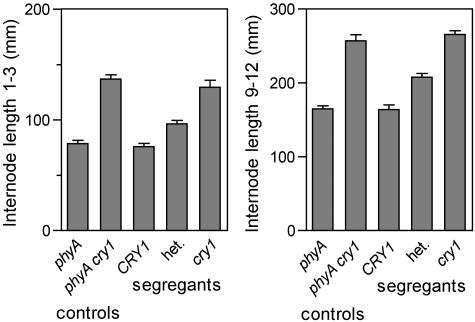
Analysis of the phyA-1 × phyA-1 cry1-1 backcross under blue light also enabled us to examine the dominance of the wild-type CRY1 allele on a phyA background. CRY1/cry1 heterozygotes were identified using the cry1-1 CAPS marker and displayed a phenotype intermediate between cry1/cry1 and CRY1/CRY1 homozygotes (Fig. 3). This incomplete dominance of the CRY1 wild-type allele on a phyA mutant background is consistent with the haploinsufficiency previously reported for CRY1 and various phytochrome genes in Arabidopsis and tomato (Koornneef et al., 1980; Whitelam et al., 1993; Weller et al., 2001a).
Figure 3.
Haploinsufficiency of CRY1. An F2 progeny from a cross between the phyA-1 mutant and the phyA-1 cry1-1 double mutant was grown for 14 d under blue light (10 μmol m−2 s−1) before transfer to standard glasshouse conditions. Segregants were genotyped at the CRY1 locus using molecular markers. Values represent mean ± se, n = 15 to 25.
The cry1-1 Mutation Influences Seedling De-Etiolation and Vegetative Development
We next selected a monogenic cry1-1 mutant line from F2 progeny of a cross between wild type cv Torsdag and the phyA-1 cry1-1 double mutant, using molecular markers for the cry1-1 and phyA-1 mutations. Under blue light at moderately high irradiance (20 μmol m−2 s−1), cry1-1 mutant plants exhibited a substantial reduction in leaflet expansion relative to wild-type seedlings, but did not show a significant difference in internode length (Fig. 4). The effect of the cry1-1 mutation on stem elongation was therefore less clear on a wild-type background than on a phyA-1 background, suggesting a functional overlap between cry1 and phyA in pea similar to that reported previously in Arabidopsis and tomato. However, cry1-1 mutants grown in the glasshouse under an 18-h photoperiod did show a clearly elongated phenotype later in development (Fig. 1C), regardless of the presence of phyA, although this phenotype was not as extreme as that of phyB mutants (Weller et al., 2001b). Under glasshouse conditions cry1-1 monogenic mutants flowered at a similar node to wild-type plants (data not shown).
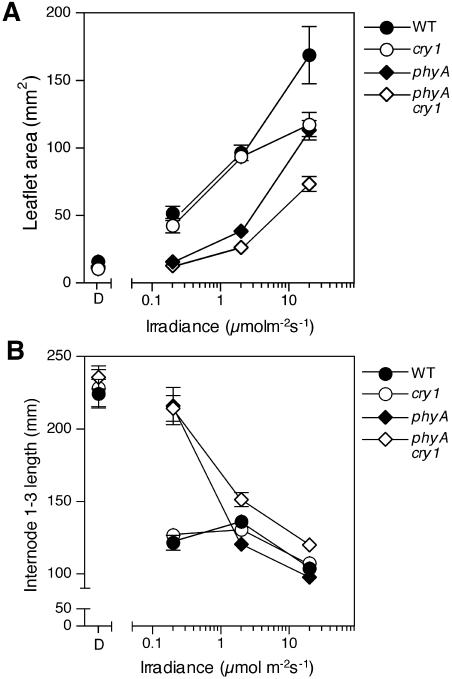
Figure 4.
Irradiance-response relationship for blue-light-induced de-etiolation in wild-type, phyA, cry1, and phyA cry1 seedlings. A, Leaflet area estimated as length × width of the larger leaflet from leaf 3. B, Length between nodes 1 and 3 (mm). Values represent means ± se, n = 8 to 12. Where not visible, error bars are smaller than plot symbols.
To examine the range of blue-light irradiances under which phyA and cry1 act to promote de-etiolation in pea, single and double mutant seedlings were grown under three different irradiances of blue light (Fig. 4). Under the highest irradiance used (20 μmol m−2 s−1), the cry1-1 mutant showed a substantial reduction in leaflet expansion relative to wild type but did not differ from wild type under the two lower irradiances (0.2 and 2 μmol m−2 s−1; Fig. 4A). However, internode elongation in cry1-1 mutants did not differ substantially from wild type across the range of blue-light irradiances tested (Fig. 4B). This contrasted with the phyA-1 mutant, which was substantially more elongated than both wild type and cry1 seedlings under the lowest irradiance (0.2 μmol m−2 s−1) and exhibited little if any response for either stem elongation or leaflet expansion. Compared to wild-type seedlings, phyA-1 mutant seedlings also showed reduced leaflet expansion under both the intermediate and high-irradiance conditions. The phyA cry1 double mutant did not differ significantly from the phyA single mutant in either stem elongation or leaflet expansion under the lowest irradiance, but was more etiolated than phyA under intermediate and high-irradiance blue light. These results show that in pea, phyA is the main photoreceptor controlling de-etiolation responses to low-irradiance blue light, whereas both phyA and cry1 play a role under high-irradiance blue light. Also, since no significant effect of low-irradiance blue light was observed for either the phyA or phyA cry1 mutant, there is no evidence for any substantial contribution from any other photoreceptor apart from phyA under these conditions. Finally, the results also suggest that the threshold irradiance for cry1 action may be close to the intermediate irradiance used, at around 2 μmol m−2 s−1.
PhyB Also Contributes to Seedling De-Etiolation under Blue Light
Although phyA-1 cry1-1 double mutant seedlings exhibit some reduction in de-etiolation under relatively high levels of blue light, these plants still display a substantial response to blue light compared to dark-grown plants (Fig. 4). To examine the contribution of phyB to this response, we selected phyB cry1 and phyA phyB cry1 mutants from the progeny of a cross between the phyA-1 cry1-1 double mutant and the phyB-5 null mutant, using a combination of phenotypic screening and molecular genotyping. Seedlings showing an almost completely etiolated appearance under white light were readily identified in this progeny, and were confirmed as phyA-1 phyB-5 cry1-1 triple mutant plants by molecular markers. These plants failed to survive until flowering and were consequently maintained through heterozygous lines. The phyB cry1 double mutants also yielded extremely few seed.
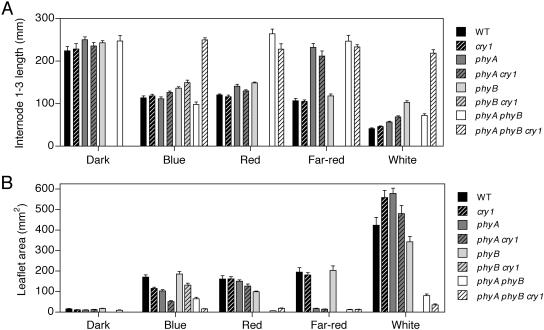
To gain a more complete picture of the interaction of phyA, phyB, and cry1 during de-etiolation in pea, various mutant combinations were grown under different monochromatic light conditions and internode length and leaflet area were measured (Fig. 5). Due to limited seed availability, the phyB cry1 and phyA phyB cry1 genotypes were only grown under selected light conditions. As previously observed, de-etiolation in response to red and far-red light was regulated exclusively by phyA and phyB, as phyA phyB double mutants are fully etiolated under both conditions (Fig. 5; Weller et al., 2001b). As expected, phyA phyB cry1 triple mutants were also not responsive to red or far-red light. However, the triple mutant also showed an essentially complete loss of responsiveness to blue light, indicating that the residual response to blue light seen in the phyA cry1 mutant is largely controlled by phyB. A small effect of phyB on internode elongation in blue light was also evident on a wild-type background. The striking difference in internode elongation under blue light between the triple mutant and all three double mutants (Fig. 5A) indicates a large degree of functional overlap among all three photoreceptors in the control of this response. Interestingly, this strong functional overlap was not as apparent in the control of leaflet expansion under blue light, as both phyA and cry1 appear to contribute to this process in an essentially additive manner (Fig. 5B). PhyB also contributes to this response, although this is only apparent in the absence of phyA.
Figure 5.
Interaction of phyA, phyB, and cry1 in pea seedling de-etiolation. Internode length (A) and leaflet area (B) estimated as length × width of the larger leaflet from leaf 3 for seedlings grown in complete darkness or under continuous blue (20 μmol m−2 s−1), red (20 μmol m−2 s−1), far-red (20 μmol m−2 s−1), or white light (110 μmol m−2 s−1). Values represent means ± se, n = 8 to 12.
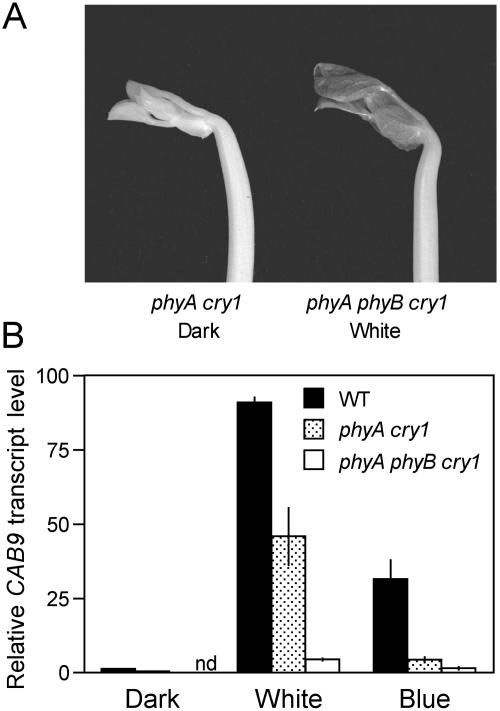
The only clear response to light in the triple mutant was a small increase in leaflet expansion under white light (Figs. 5B and 6A), a response also seen under natural daylight conditions. Compared to dark-grown seedlings of other genotypes, the phyA phyB cry1 triple mutant also retained a small induction of CAB gene expression in response to white light at 100 μmol m−2 s−1, but not to blue light at 20 μmol m−2 s−1 (Fig. 6B).
Figure 6.
Residual response to continuous white light in the phyA phyB cry1 triple mutant. A, Shoot apices of a phyA cry1 double mutant grown in complete darkness and a phyA phyB cry1 triple mutant grown under continuous white light (110 μmol m−2 s−1). Both seedlings are 14 d old. Double mutant phyA cry1 seedlings are visually indistinguishable from triple mutant phyA phyB cry1 siblings in segregating progenies grown in darkness. B, Levels of CAB9 transcript in shoot apices of 14-d-old wild-type, phyA cry1, and phyA phyB cry1 seedlings grown under continuous blue (20 μmol m−2 s−1) or white light (110 μmol m−2 s−1), or in complete darkness. Values are normalized for actin transcript level and are expressed as mean ± se, n = 3. nd, Not determined.
Photoreceptors Remaining in the phyA phyB cry1 Triple Mutant
The pea genome is known to contain two expressed CRY2 genes (Platten et al., 2005) and two homologs of the phototropin gene PHOT1 (Elliott et al., 2004), any of which might function in the residual responses observed in the phyA phyB cry1 mutant. Residual responses could potentially also be mediated by additional phytochrome photoreceptors. Among the angiosperm species examined so far, most possess at least three phytochromes, including phyA, phyB, and phyC. Many dicot species, including Arabidopsis and tomato, have at least one additional divergent phyB-like phytochrome, PHYE (Clack et al., 1994; Hauser et al., 1995; Mathews et al., 1995).
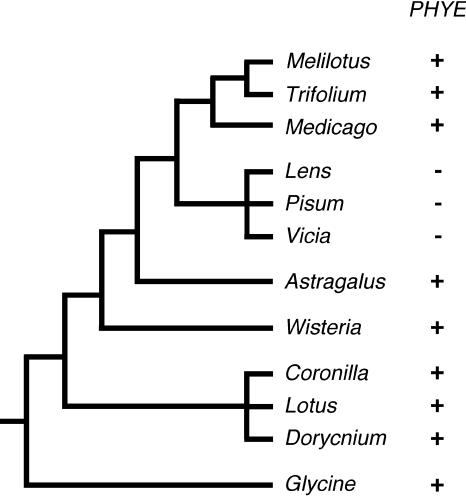
However, a number of lines of evidence suggest that in pea the phytochrome gene family may be limited to only two members, PHYA and PHYB. First, a broad PCR-based survey identified PHYA, PHYB, and PHYE sequences but found no evidence of PHYC-like genes in legumes (Lavin et al., 1998). Second, Medicago, soybean (Glycine max), and Lotus databases contain genomic and/or expressed sequence tag sequences for PHYA, PHYB, and PHYE but not PHYC (Fig. 7; Hecht et al., 2005). Third, we have used Medicago PHYE to search for PHYE sequences from pea and other related legumes by degenerate-primer PCR and genomic Southern blotting. These approaches confirmed the presence of PHYE in the Trifolieae clade (including Medicago and Trifolium), but failed to identify PHYE sequences in pea and other species in the sister tribe Vicieae, despite also identifying PHYE from species in the more distantly related Loteae clade (Fig. 7; data not shown). Finally, pea phyA phyB double mutants completely lack a shade-avoidance response to supplementary far-red light under conditions where both tomato phyA phyB1 phyB2 triple mutants and Arabidopsis phyA phyB phyD triple mutants show a strong response (Devlin et al., 1999; Weller et al., 2000, 2001a).
Figure 7.
Taxonomic distribution of legume PHYE sequences. Diagrammatic representation of the taxonomic relationships among legume genera examined for the presence of PHYE-related sequence. Redrawn from Choi et al. (2004) with additional genera placed according to Kajita et al. (2001). PHYE sequence from wisteria (U78061) was previously reported by Lavin et al. (1998). PHYE genomic sequences were identified from Medicago (AC158464) and Lotus (AP006687), and PHYE expressed sequence tags were identified from soybean (AW186474, BE822159) and Lotus (AU088958, BP055499). All other sequences were isolated by degenerate-primer PCR.
Taken together, these findings suggest that that pea and related species in the tribe Vicieae may have lost a PHYE-like gene that is present in Medicago and more broadly throughout the hologalegoid legumes. A similar situation may exist in black cottonwood (Populus trichocarpa), which has been reported to contain only PHYA and PHYB genes (Howe et al., 1998). In view of this evidence, it is most likely that residual de-etiolation responses to white light seen in the phyA phyB cry1 triple mutant are due to the action of one or more specific blue-light photoreceptors rather than an additional phytochrome.
PhyA and Cry1 Both Contribute to Blue-Light Regulation of CRY2b Gene Expression
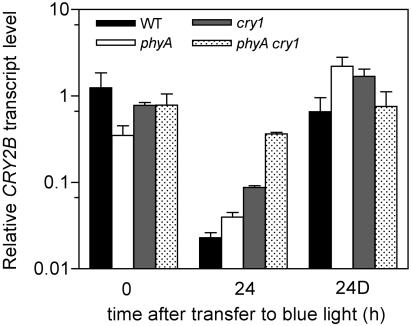
Of the two expressed CRY2 genes in pea, the expression of CRY2b in particular is strongly down-regulated by blue light (Platten et al., 2005). Light regulation of photoreceptor transcript abundance has previously been reported for the phytochromes where both phyA and phyB contribute to the down-regulation of PHYA (Cantón and Quail, 1999). However, similar interactions between blue-light photoreceptors at the level of transcriptional control have not been reported. We examined the role of cry1 in the regulation of CRY2b transcript accumulation in shoot tissue of wild-type and mutant plants transferred from darkness to continuous blue light. Figure 8 shows that the expression of CRY2b was strongly down-regulated in wild-type plants 24 h after transfer to blue light, and a smaller down-regulation occurred in phyA-1 and cry1-1 single mutant plants. In contrast, no significant difference in CRY2b expression was observed in phyA-1 cry1-1 double mutant plants 24 h after transfer to blue light, and these plants had significantly higher CRY2b expression than comparable wild-type, phyA-1, and cry1-1 seedlings (P < 0.001). This shows that phyA and cry1 act together to mediate the initial repression of CRY2b expression that follows exposure of etiolated seedlings to blue light.
Figure 8.
Role of phyA and cry1 in the regulation of CRY2b expression by blue light. Seven-day-old wild-type, phyA, cry1, and phyA cry1 seedlings were transferred from complete darkness to continuous blue light at 20 μmol m−2 s−1. Whole shoots were harvested at various times after transfer, and expression of CRY2b was monitored by quantitative reverse transcription-PCR. Expression was also monitored in plants kept in darkness for a further 24 h (24D). Values were normalized for actin transcript level and are expressed as mean ± se, n = 2 to 3.
Cry1 Has Only a Minor Effect on Flowering in Pea
Pea is a long-day plant and flowers earlier under long-day photoperiods (LD) than under short-day photoperiods (SD). Previous studies have shown that phyA acts to promote flowering in response to photoperiod extensions while phyB inhibits flowering under noninductive SD conditions (Weller and Reid, 1993; Weller et al., 1997). However, phyA mutant plants still retain a substantial response to photoperiod extensions with white light of high red:far-red ratio (Weller et al., 2001b). This response is unlikely to be mediated by phyB, as phyB activation clearly inhibits flowering (Weller et al., 2001b). Another possibility is that it may reflect a response to the blue-light component of the white light, mediated by one or more cryptochromes. In Arabidopsis, the cryptochrome cry2 is known to play an important role in the blue-light-specific promotion of flowering (Guo et al., 1998) and acts by preventing the phyB-mediated degradation of the flower-promoting CONSTANS protein (Valverde et al., 2004).
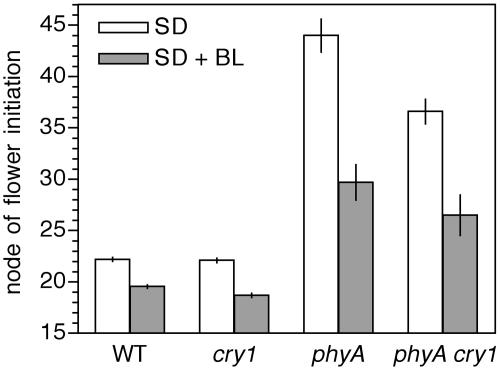
To test this possibility and to examine the possible involvement of cry1, we compared the flowering response of wild-type, cry1, phyA, and phyA cry1 double mutant plants to the extension of an 8-h SD with an additional 16 h of blue light (10 μmol m−2 s−1). Figure 9 shows that wild-type and cry1 lines responded similarly to the blue-light extension, both flowering at a significantly earlier node than when grown in SD (P < 0.05). Monogenic cry1 mutant plants grown in SD flowered at a similar node to wild-type plants, and under the extended-day treatment in fact flowered slightly but significantly earlier than wild-type plants (P < 0.05). As previously observed, phyA plants grown in SD flowered later than wild-type plants (Weller et al., 2001b) and retained a strong response to the blue-light extension. The phyA cry1 double mutant flowered significantly earlier than the phyA mutant under SD (P < 0.01), but retained a clear promotion of flowering in response to the extension. It therefore appears that in pea, cry1 has a small inhibitory effect on flowering especially in the absence of phyA, but plays little if any role in the blue-light-specific promotion of flowering.
Figure 9.
The effect of cry1 on the promotion of flowering in response to blue-light extensions. Node of flower initiation was recorded from plants grown in growth cabinets under an 8-h photoperiod of cool-white fluorescent light (SD; 150 μmol m−2 s−1) extended with 16 h of blue light (SD + BL; 10 μmol m−2 s−1). Values represent means ± se, n = 9 to 10.
DISCUSSION
To date, much of our understanding of the role of cryptochromes in plant development has been gained from mutant studies in Arabidopsis. In Arabidopsis, both cry1 and cry2 contribute to the control of deetiolation and photoperiod-responsive flowering, although cry1 plays a more dominant role in de-etiolation and cry2 in flowering (Liscum et al., 2003). While both cry1 and cry2 are likely to be ubiquitous in higher plants (Platten et al., 2005), relatively little is known about the roles of these photoreceptors in other plant species. Recent isolation of a tomato cry1 mutant has shown that cry1 in this species also contributes to other aspects of development, including apical dominance and chlorophyll accumulation (Weller et al., 2001a). Preliminary characterizations of the role of cry1 in rice and cry2 in tomato have also been undertaken using transgenic and silencing approaches (Matsumoto et al., 2003; Giliberto et al., 2005). We have previously characterized the roles of phyA and phyB in pea and have identified several roles for specific blue-light receptors (Weller et al., 2001b). More recently, we have demonstrated the presence of three expressed CRY genes in pea, including CRY1 and two distinct CRY2 genes (Platten et al., 2005).
In this study we have isolated a line carrying a single nucleotide substitution in the pea CRY1 gene. Several lines of evidence indicate that this mutation is likely to severely impair CRY1 function. First, the Gly residue affected by the mutation is perfectly conserved across a wide range of cryptochromes from plants and animals and is also conserved in the more distantly related DNA photolyases (Fig. 2). Second, this residue is located in an N-terminal region of the CRY1 protein that is known to bind the FAD chromophore in cryptochromes and photolyases (Fig. 2; Brudler et al., 2003; Brautigam et al., 2004) and might, therefore, be predicted to have an important role in the chromophore-apoprotein interaction. Third, the substitution of this same Gly residue in the Arabidopsis CRY2 gene results in complete loss of function, as determined by phenotypic comparisons with null deletion mutants (Guo et al., 1998).
Our analyses of the pea cry1 mutant show that cry1 contributes to seedling de-etiolation. In seedlings, cry1 acts together with phyA and phyB in a highly redundant manner to mediate the effects of high-irradiance blue and white light on stem elongation (Figs. 4 and 5). All three photoreceptors also contribute to leaflet expansion under high-irradiance blue light, but in a more independent manner (Figs. 4 and 5). PhyA, phyB, and cry1 also contribute to de-etiolation of tomato and Arabidopsis seedlings under high-irradiance blue light, in most cases acting in a partially redundant manner to regulate traits including anthocyanin accumulation, hypocotyl elongation, and cotyledon expansion (Ahmad and Cashmore, 1997; Neff and Chory, 1998; Poppe et al., 1998; Weller et al., 2001a). However, on a wild-type background the loss of cry1 appears to have a greater effect on seedling de-etiolation in tomato and Arabidopsis than in pea. The reason for this difference is not clear, but could reflect a greater contribution from phyA in pea.
Cry1 also plays a clear role in the later development of pea plants grown under white light. Mutant cry1 plants grown under glasshouse conditions have internodes that are 20% to 40% longer than wild type, and also have slightly smaller leaflets (Fig. 1C; data not shown). These effects of the cry1 mutation are clearly independent of phyA as they are also observed on a phyA background (Fig. 1B), and are consistent with reports of cry1 function in Arabidopsis and tomato (Jackson and Jenkins, 1995; Weller et al., 2001a).
An examination of the irradiance response curves for both stem elongation and leaflet expansion (Fig. 4) suggests that the threshold blue-light irradiance for cry1 action in pea is similar to that previously reported for Arabidopsis and tomato cry1 (Lin et al., 1998; Poppe et al., 1998; Weller et al., 2001a). Pea phyA mutant seedlings exhibit no substantial response to very low-irradiance blue light (Fig. 4), showing that in pea, as in Arabidopsis and tomato, phyA is the primary photoreceptor promoting de-etiolation under very low-irradiance blue light (Poppe et al., 1998; Weller et al., 2001a). The absence of both phyA and cry1 reveals a strong contribution of phyB to de-etiolation under high-irradiance blue light (Fig. 5) that is consistent with the role of phyB in other species (Neff and Chory, 1998; Poppe et al., 1998; Weller et al., 2001a).
Our preliminary analysis of triple mutants containing lesions in phyA, phyB, and cry1 suggests that these are the major photoreceptors regulating seedling de-etiolation in pea. At the level of gross morphology, phyA phyB cry1 triple mutant pea seedlings are effectively blind to blue light (20 μmol m−2 s−1) and under higher-irradiance white light (110 μmol m−2 s−1) show only a slight increase in leaflet expansion and CAB transcript accumulation relative to dark-grown control plants (Figs. 5 and 6). A similar phenotype has been reported for a phyA phyB1 phyB2 cry1 quadruple mutant of tomato (Weller et al., 2001a) and Arabidopsis phyA phyB cry1 mutants (Neff and Chory, 1998; Mazzella et al., 2001). Studies in Arabidopsis have shown that an additional cryptochrome, cry2, also plays a role in de-etiolation under some conditions, and it is possible that the small effect on leaflet expansion seen in the pea phyA phyB cry1 triple mutant could reflect the action of a cry2. In Arabidopsis, a cry2 mutant line was initially isolated on the basis of reduced de-etiolation under low-irradiance blue light (Lin et al., 1998) and cry2 has also been reported to act across a range of sunlight irradiances in the absence of cry1 (Mazzella and Casal, 2001). However, other experiments with phyA phyB cry1 triple and phyA phyB cry1 cry2 quadruple mutants have found that cry2 does not contribute to the inhibition of hypocotyl elongation under high-irradiance white light or full sunlight, and plays only a small role in cotyledon opening under these conditions, at least in the absence of phyA, phyB, and cry1 (Neff and Chory, 1998; Mazzella et al., 2001; Mazzella and Casal, 2001).
A number of physiological interactions between phytochrome and cryptochrome photoreceptors have been described in Arabidopsis (Casal, 2000) and tomato (Weller et al., 2000, 2001a). These include an antagonistic effect of phyA on phyB under red light (Cerdán et al., 1999; Weller et al., 2000), a dependence of cry1-mediated responses on phyA and/or phyB action (Wang and Iino, 1998; Casal, 2000), and an antagonism of phyA/cry1-mediated inhibition of elongation by phyB (Folta and Spalding, 2001). Many of these interactions have been identified under specific illumination regimes and for specific phases of growth. Our more general analyses of long-term growth responses of photoreceptor mutant phenotypes under continuous monochromatic light have not identified any of these interactions in de-etiolating pea seedlings. However, we did observe one clear example of photoreceptor interaction not previously observed in other species. In contrast to phyA and phyA phyB mutants in Arabidopsis (Neff and Chory, 1998) and tomato (Weller et al., 2001a), pea phyA and phyA phyB mutant seedlings exhibit reduced stem elongation compared to their isogenic PHYA lines when grown in high-irradiance blue light (Fig. 4; Weller et al., 2001b). This enhanced response to blue light in phyA mutants is clearly dependent on cry1 as it is not seen in either cry1 or phyB cry1 background, and could thus be understood as an antagonistic effect of phyA on cry1 action. In addition, when grown to maturity under white light, phyA and phyA phyB mutants develop short, thickened internodes (Fig. 1B; Weller et al., 2001b), a phenotype that also appears to require cry1, as internodes of phyA cry1 plants are significantly longer and thinner than phyA plants (Fig. 1B; data not shown). We are currently examining whether these two effects of phyA mutations are related, and testing the possibility that they may result from altered hormone status (Weller et al., 2001b).
We have shown previously that the expression of both of the pea CRY2 genes is down-regulated in young seedlings in response to blue light (Platten et al., 2005). For CRY2b at least, this down-regulation is particularly strong and appears to be dependent on both phyA and cry1 (Fig. 8). The functional importance of this regulation is at present not clear, although in general, it suggests that CRY2b levels may be elevated under conditions where cry1 and phyA are relatively inactive. However, because any role of cry2 photoreceptors in de-etiolation of pea seedlings is likely to be relatively minor (Figs. 4 and 5), it may be that they have a more important role in the control of other developmental responses.
In photoperiodic species such as pea and Arabidopsis, photoreceptors play important roles in regulating the transition from vegetative to floral development. In pea, the promotion of flowering by LD photoperiods is partially mediated by phyA. Mutant phyA plants grown under LD flower later than wild-type plants but still show a substantial promotion of flowering in response to photoperiod extensions with cool-white fluorescent light (Weller et al., 2001b). It now appears that much of this response may have been due to the blue component of the white light, because monochromatic blue-light extensions are equally as effective at promoting flowering in phyA plants (Fig. 9). It is also clear that cry1 does not mediate this response, because flowering was promoted to a similar extent in phyA and phyA cry1 double mutant plants exposed to blue-light extensions, and because the only consistent effect of cry1 on flowering that we observed was a slight delay.
One possible explanation for the residual blue-light promotion of flowering seen in the phyA mutant is the partial inactivation of phyB, which we have previously shown to inhibit flowering in pea (Weller et al., 2001b). Compared to SD conditions, which would establish a high red:far-red ratio at the end of the day and maintain phyB in its active inhibitory form during the night, the blue-light extension might be expected to establish an intermediate phyB photoequilibrium less inhibitory for flowering. However, a more likely explanation is that the blue-light response reflects an active promotion of flowering mediated by one or both of the two pea CRY2 genes (Platten et al., 2005). In Arabidopsis, the only other species in which the role of cryptochromes in flowering has been examined, cry2 plays a major role in promoting flowering under LD and in response to supplemental blue light (Mockler et al., 1999; Valverde et al., 2004). Although it might be possible to distinguish between these explanations photophysiologically, direct demonstration of the involvement of cry2a and/or cry2b obviously requires the isolation of corresponding mutants. As studies presented here have not identified a clear role for cry2 in de-etiolation, this flowering response suggests a basis for a mutant screen to search for cry2 mutants in pea. We have recently shown that CRY2b transcript levels exhibit strong diurnal regulation (Platten et al., 2005), and this evidence does seem consistent with a role for cry2b in photoperiodic flowering. Current phenotypic screening for late flowering mutants and molecular screening for cry2b mutants will help determine if this is so.
MATERIALS AND METHODS
Plant Material, Mutagenesis, and Growth Conditions
All pea (Pisum sativum) lines were derived from cv Torsdag. The phyA-1, phyB, and phyA phyB double mutant lines have been described previously (Weller et al., 1997, 2001b). For the flowering experiment shown in Figure 9, all lines also carried the le-3 mutation, which has no influence on flowering response (Reid et al., 1990). For mutagenesis, approximately 1,000 phyA-1 seed were allowed to imbibe in a 1% (v/v) ethyl methanesulfonate solution for 5 h at room temperature (approximately 18°C). M1 plants were grown to maturity under standard glasshouse conditions under an 18-h photoperiod, which consisted of a natural photoperiod extended at dawn and dusk with mixed white fluorescent tubes and white incandescent globes. M2 seeds from individual plants were planted in plastic tote boxes and grown for 10 d under 4 μmol m−2 s−1 fluorescent blue light supplied by 36W/15 blue fluorescent tubes (Philips TLD) wrapped in two layers of plastic film (cutting sheet 521C, Nakagawa Chemical Company).
All plants were grown in a 1:1 mixture of dolerite chips and vermiculite topped with potting mix, and if grown to maturity received nutrient solution weekly. Mutant screening, seedling de-etiolation experiments, and gene expression studies were carried out in growth cabinets at 20°C. Red and far-red monochromatic light sources are described by Reid et al. (2002), and blue-light sources are described by Platten et al. (2005). Fluence rates were measured at pot top. Plants grown for the flowering experiment in Figure 9 were also grown in growth cabinets at 20°C and received an 8-h photoperiod of 150 μmol m−2 s−1 white light supplied by cool-white fluorescent tubes (L40W/20S cool white) extended daily with either 16 h of darkness or 16 h of blue light at an irradiance of 10 μmol m−2 s−1. All other plants were grown under glasshouse conditions under an 18-h photoperiod.
Sequencing and Molecular Markers
The full PsCRY1 cDNA was amplified from the E0-110 line using the following primers: 5′-ACCCTTATTTTTCTTCTTGT-3′ (forward) and 5′-CATCCCACTTGGTGAGATAG-3′ (reverse). RNA was extracted from E0-110 plants using the RNeasy plant mini kit (Qiagen) and cDNA synthesized using Superscript III RNase H– reverse transcriptase kit (Invitrogen). PCR products were sequenced using a Beckman-Coulter CEQ 8000 genetic analysis system (Beckman-Coulter). The cry1-1 mutation introduced a novel Csp451 restriction site and was converted into a CAPS marker with the primers 5′-CGGCCAACCATTCACAAC-3′ (forward) and 5′-CAACCAGTAGCCCACAACTCT-3′ (reverse) and was used to follow the cosegregation of the molecular lesion with the cry1-1 phenotype. Both the phyA-1 and phyB-5 mutations introduce a novel MnlI restriction site and were converted to CAPS markers with the following primers: for phyA-1, PHYAF1 5′-TGATGGGGCTGCACTCTTTTAT-3′ and PHYAR1 5′-CACGCTTCTGGCTTTCACAACT-3′; and for phyB-5, PHYBF1 5′-TGGGGCTGCTTTGTATTATC-3′ and PHYBR1 5′-ACGGCTCTTCACCACTTCCTA-3.
PHYE sequences were isolated by PCR from genomic DNA using degenerate primers 5′-AAGAGTTGGCATATATACTGCAAGAGATSAAGAARCC-3′ and 5′-AGGATTTTCCTGGACATATATAGTCCTARWCCWTC-3′.
Gene Expression Studies
RNA was extracted as described above and cDNA was synthesized from 4.5 μg of RNA with Superscript II (Invitrogen) according to the manufacturer's instructions. cDNA was diluted and duplicate PCR reactions were carried out with Dynamo SYBR green master mix (Geneworks) in a Rotor Gene 2000 (Corbett). PCR was carried out with 100 to 200 pmol of each primer under the following conditions: 94°C 15 min, 60 cycles of 94°C 15 s, 58°C to 59°C 20 s, 72°C 30 s, and 75°C 15 s. Primers for CRY2b have been described by Platten et al. (2005). Quantification of CAB9 transcript employed specific primers (CAB9F2 5′-AACAGGAAAAGGACCAATAG-3′ and CAB9R 5′-TATATTCACACATTGTGG-3′) designed from sequence previously reported by White et al. (1992) and an annealing temperature of 52°C. Actin primer sequences and calculation of relative expression values are described by Foo et al. (2005).
Sequence data from this article can be found in the GenBank/EMBL data libraries under accession number AY508971.
Acknowledgments
We thank Ian Cummings, Tracey Winterbottom, Jackie Vanderschoor, and Jenny Smith for technical support.
This work was supported by the Australian Research Council through Large Grant A00105316 and Discovery Project DP044972 (to J.B.R. and J.L.W.), and an Australian Postgraduate Award (to J.D.P.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: James L. Weller (jim.weller@utas.edu.au).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.067462.
References
- Ahmad M, Cashmore AR (1993) HY4 gene in A. thaliana encodes a protein with characteristics of a blue-light photoreceptor. Nature 366: 162–166 [DOI] [PubMed] [Google Scholar]
- Ahmad M, Cashmore AR (1997) The blue-light receptor cryptochrome 1 shows functional dependence on phytochrome A or phytochrome B in Arabidopsis thaliana. Plant J 11: 421–427 [DOI] [PubMed] [Google Scholar]
- Ahmad M, Jarillo JA, Smirnova O, Cashmore AR (1998) The CRY1 blue light photoreceptor of Arabidopsis interacts with phytochrome A in vitro. Mol Cell 1: 939–948 [DOI] [PubMed] [Google Scholar]
- Brautigam CA, Smith BS, Ma Z, Palnitkar M, Tomchick DR, Machius M, Deisenhofer J (2004) Structure of the photolyase-like domain of cryptochrome 1 from Arabidopsis thaliana. Proc Natl Acad Sci USA 101: 12142–12147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brudler R, Hitomi K, Daiyasu H, Toh H, Kucho K, Ishiura M, Kanehisa M, Roberts VA, Todo T, Tainer JA, et al (2003) Identification of a new cryptochrome class: structure, function, and evolution. Mol Cell 11: 59–67 [DOI] [PubMed] [Google Scholar]
- Cantón FR, Quail PH (1999) Both phyA and phyB mediate light-imposed repression of PHYA gene expression in Arabidopsis. Plant Physiol 121: 1207–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casal JJ (2000) Phytochromes, cryptochromes, phototropin: photoreceptor interactions in plants. Photochem Photobiol 71: 1–11 [DOI] [PubMed] [Google Scholar]
- Cerdán PD, Yanovsky MJ, Reymundo FC, Nagatani A, Staneloni RJ, Whitelam GC, Casal JJ (1999) Regulation of PHYB signaling by phytochrome A and FHY1 in Arabidopsis thaliana. Plant J 18: 499–507 [DOI] [PubMed] [Google Scholar]
- Choi HK, Mun JH, Kim DJ, Zhu H, Baek JM, Mudge J, Roe B, Ellis N, Doyle J, Kiss GB, et al (2004) Estimating genome conservation between crop and model legume species. Proc Natl Acad Sci USA 101: 15289–15294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clack T, Mathews S, Sharrock RA (1994) The phytochrome apoprotein family in Arabidopsis is encoded by five genes; the sequences and expression of PHYD and PHYE. Plant Mol Biol 25: 413–427 [DOI] [PubMed] [Google Scholar]
- Devlin PF, Robson PRH, Patel SR, Goosey L, Sharrock RA, Whitelam GC (1999) Phytochrome D acts in the shade-avoidance syndrome by controlling elongation growth and flowering time. Plant Physiol 119: 909–915 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott RC, Platten JD, Watson JC, Reid JB (2004) Phytochrome regulation of pea phototropin. J Plant Physiol 161: 265–270 [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Staiger D (2002) Photoreceptors in Arabidopsis: light perception, signal transduction and entrainment of the endogenous clock. Planta 216: 1–16 [DOI] [PubMed] [Google Scholar]
- Folta KM, Spalding EP (2001) Opposing roles of phytochrome A and phytochrome B in early cryptochrome-mediated growth inhibition. Plant J 28: 333–340 [DOI] [PubMed] [Google Scholar]
- Foo E, Bullier E, Goussot M, Foucher F, Rameau C, Beveridge CA (2005) The branching gene RAMOSUS1 mediates interactions among two novel signals and auxin in pea. Plant Cell 17: 464–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giliberto L, Perrotta G, Pallara P, Weller JL, Fraser PD, Bramley PM, Fiore A, Tavazza M, Giuliano G (2005) Manipulation of the blue light photoreceptor cryptochrome 2 in tomato affects vegetative development, flowering time, and fruit antioxidant content. Plant Physiol 137: 199–208 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Yang H, Mockler TC, Lin C (1998) Regulation of flowering time by Arabidopsis photoreceptors. Science 279: 1360–1363 [DOI] [PubMed] [Google Scholar]
- Hauser BA, Cordonnier-Pratt MM, Daniel-Vedele F, Pratt LH (1995) The phytochrome gene family in tomato includes a novel subfamily. Plant Mol Biol 29: 1143–1155 [DOI] [PubMed] [Google Scholar]
- Hecht V, Foucher F, Ferrandíz C, Macknight R, Navarro C, Vardy ME, Ellis THN, Beltrán JP, Rameau C, Weller JL (2005) Conservation of Arabidopsis flowering genes in model legumes. Plant Physiol 137: 1420–1434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe GT, Bucciaglia PA, Hackett WP, Furnier GR, Cordonnier-Pratt M-M, Gardner G (1998) Evidence that the phytochrome gene family in black cottonwood has one PHYA locus and two PHYB loci but lacks members of the PHYC/F and PHYE subfamilies. Mol Biol Evol 15: 160–175 [DOI] [PubMed] [Google Scholar]
- Imaizumi T, Kadota A, Hasebe M, Wada M (2002) Cryptochrome light signals control development to suppress auxin sensitivity in the moss Physcomitrella patens. Plant Cell 14: 373–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson JA, Jenkins GI (1995) Extension-growth responses and expression of flavonoid biosynthesis genes in Arabidopsis hy4 mutant. Planta 197: 233–239 [DOI] [PubMed] [Google Scholar]
- Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, Ecker JR, Cashmore AR (2001) An Arabidopsis circadian clock component interacts with both CRY1 and PHYB. Nature 410: 487–490 [DOI] [PubMed] [Google Scholar]
- Kajita T, Ohashi H, Tateishi Y, Bailey CD, Doyle JJ (2001) rbcL and legume phylogeny, with particular reference to Phaseoleae, Millettieae, and allies. Syst Bot 26: 515–536 [Google Scholar]
- Kanegae T, Wada M (1998) Isolation and characterization of homologues of plant blue-light photoreceptor (cryptochrome) genes from the fern Adiantum capillus-veneris. Mol Gen Genet 259: 345–353 [DOI] [PubMed] [Google Scholar]
- Koornneef M, Rolff E, Spruit CJP (1980) Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol 100: 147–160 [Google Scholar]
- Lavin M, Eshbaugh E, Hu JM, Mathews S, Sharrock RA (1998) Monophyletic subgroups of the tribe Millettieae (Leguminosae) as revealed by phytochrome nucleotide sequence data. Am J Bot 85: 412–433 [Google Scholar]
- Lin C, Shalitin D (2003) Cryptochrome structure and signal transduction. Annu Rev Plant Biol 54: 469–496 [DOI] [PubMed] [Google Scholar]
- Lin C, Todo T (2005) The cryptochromes. Genome Biol 6: 220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin C, Yang H, Guo H, Mockler T, Chen J, Cashmore AR (1998) Enhancement of blue light sensitivity of Arabidopsis seedlings by a blue light receptor cryptochrome 2. Proc Natl Acad Sci USA 95: 2686–2690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liscum E, Hodgson DW, Campbell TJ (2003) Blue light signaling through the cryptochromes and phototropins: so that's what the blues is all about. Plant Physiol 133: 1429–1436 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Más P, Devlin PF, Panda S, Kay SA (2000) Functional interaction of phytochrome B and cryptochrome 2. Nature 408: 207–211 [DOI] [PubMed] [Google Scholar]
- Mathews S, Lavin M, Sharrock RA (1995) Evolution of the phytochrome gene family and its utility for phylogenetic analysis of angiosperms. Ann Mo Bot Gard 82: 296–321 [Google Scholar]
- Matsumoto N, Hirano T, Iwasaki T, Yamamoto N (2003) Functional analysis and intracellular localization of rice cryptochromes. Plant Physiol 133: 1494–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazzella MA, Casal JJ (2001) Interactive signalling by phytochromes and cryptochromes generates de-etiolation homeostasis in Arabidopsis thaliana. Plant Cell Environ 24: 155–161 [Google Scholar]
- Mazzella MA, Cerdán PD, Staneloni RJ, Casal JJ (2001) Hierarchical coupling of phytochromes and cryptochromes reconciles stability and light modulation of Arabidopsis development. Development 128: 2291–2299 [DOI] [PubMed] [Google Scholar]
- Mockler TC, Guo H, Yang H, Duong H, Lin C (1999) Antagonistic action of Arabidopsis cryptochromes and phytochrome B in the regulation of floral induction. Development 126: 2073–2082 [DOI] [PubMed] [Google Scholar]
- Neff MM, Chory J (1998) Genetic interactions between phytochrome A, phytochrome B, and cryptochrome 1 during Arabidopsis development. Plant Physiol 118: 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterlund MT, Hardtke CS, Wei N, Deng XW (2000) Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature 405: 462–466 [DOI] [PubMed] [Google Scholar]
- Perrotta G, Ninu L, Flamma F, Weller JL, Kendrick RE, Nebuloso E, Giuliano G (2000) Tomato contains homologues of Arabidopsis cryptochromes 1 and 2. Plant Mol Biol 42: 765–773 [DOI] [PubMed] [Google Scholar]
- Platten JD, Foo E, Foucher F, Hecht V, Reid JB, Weller JL (2005) The cryptochrome gene family in pea includes two differentially expressed CRY2 genes. Plant Mol Biol 59: (in press) [DOI] [PubMed]
- Poppe C, Sweere U, Drumm-Herrel H, Schäfer E (1998) The blue light receptor cryptochrome 1 can act independently of phytochrome A and B in Arabidopsis thaliana. Plant J 16: 465–471 [DOI] [PubMed] [Google Scholar]
- Reid JB, Botwright NA, Smith JJ, O'Neill DP, Kerckhoffs LHJ (2002) Control of gibberellin levels and gene expression during de-etiolation in pea. Plant Physiol 128: 734–741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid JB, Hasan O, Ross JJ (1990) Internode length in Pisum: gibberellins and the response to far-red-rich light. J Plant Physiol 137: 46–52 [Google Scholar]
- Takano M, Kanegae H, Shinomura T, Miyao A, Hirochika H, Furuya M (2001) Isolation and characterization of rice phytochrome A mutants. Plant Cell 13: 521–534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valverde F, Mouradov A, Soppe W, Ravenscroft D, Samach A, Coupland G (2004) Photoreceptor regulation of CONSTANS protein in photoperiodic flowering. Science 303: 1003–1006 [DOI] [PubMed] [Google Scholar]
- Wang HY, Ma LG, Li JM, Zhao HY, Deng XW (2001) Direct interaction of Arabidopsis cryptochromes with COP1 in light control development. Science 294: 154–158 [DOI] [PubMed] [Google Scholar]
- Wang X, Iino M (1998) Interaction of cryptochrome 1, phytochrome, and ion fluxes in blue-light-induced shrinking of Arabidopsis hypocotyl protoplasts. Plant Physiol 117: 1265–1279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Beauchamp N, Kerckhoffs LHJ, Platten JD, Reid JB (2001. b) Interaction of phytochrome A and B in the control of de-etiolation and flowering in pea. Plant J 26: 283–294 [DOI] [PubMed] [Google Scholar]
- Weller JL, Murfet IC, Reid JB (1997) Pea mutants with reduced sensitivity to far-red light define an important role for phytochrome A in day-length detection. Plant Physiol 114: 1225–1236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weller JL, Perrotta G, Schreuder MEL, van Tuinen A, Koornneef M, Giuliano G, Kendrick RE (2001. a) Genetic dissection of blue-light sensing in tomato using mutants deficient in cryptochrome 1 and phytochromes A, B1 and B2. Plant J 25: 427–440 [DOI] [PubMed] [Google Scholar]
- Weller JL, Reid JB (1993) Photoperiodism and photocontrol of stem elongation in two photomorphogenic mutants of Pisum sativum L. Planta 189: 15–23 [Google Scholar]
- Weller JL, Schreuder MEL, Koornneef M, Kendrick RE (2000) Physiological interactions of phytochromes A, B1 and B2 in the control of development in tomato. Plant J 24: 345–356 [DOI] [PubMed] [Google Scholar]
- White MJ, Fristensky BW, Falconet D, Childs LC, Watson JC, Alexander L, Roe BA, Thompson WF (1992) Expression of the chlorophyll-a/b-protein multigene family in pea (Pisum sativum L.): evidence for distinct developmental responses. Planta 188: 190–198 [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP (1993) Phytochrome A mutants of Arabidopsis display a wild-type phenotype in white light. Plant Cell 5: 757–768 [DOI] [PMC free article] [PubMed] [Google Scholar]