Abstract
Hahsp17.6G1 is the promoter of a small heat stress protein (sHSP) from sunflower (Helianthus annuus) that is activated during zygotic embryogenesis, but which does not respond to heat stress. We report here the cloning of a transcription factor (TF), sunflower drought-responsive element binding factor 2 (HaDREB2), by one-hybrid interaction with functional cis-elements in Hahsp17.6G1. We have analyzed the functional interaction between HaDREB2 and a second transcription factor, sunflower heat stress factor A9 (HaHSFA9), which was previously assigned to the regulation of Hahsp17.6G1. HaDREB2 and HaHSFA9 synergistically trans-activate the Hahsp17.6G1 promoter in bombarded sunflower embryos. This synergistic interaction is heat stress factor (HSF) specific and requires the binding of both factors to the promoter. The C-terminal region of HaHSFA9 is sufficient for the HSF specificity. Our results represent an example of a functional interaction between members of the Apetala 2 (HaDREB2) and HSF (HaHSFA9) families of transcription factors. We suggest new roles in zygotic embryogenesis for specific members of the AP2 transcription factor family.
The functional assignment of individual transcription factors (TFs) from multimember families is one of the major goals in postgenomic analyses of plants. We focus our attention on factors that are involved in the developmental induction of small heat stress protein (sHSP) genes during zygotic embryogenesis. In plants, as in other eukaryotes, HSPs of different molecular masses, including the sHSPs (17–30 kD), are expressed not only during the heat shock response but also during development in the absence of exogenous stresses (for review, see Sun et al., 2002).
The elucidation of the regulator genes involved in the developmental control of HSP expression has just begun. A crucial step in the elucidation is the precise identification of cis-regulatory elements; however, to date, few sHSP promoters have been characterized fully. The combination of point mutation and deletion analysis allowed the functional characterization of heat shock cis-elements (HSEs), and in the case of the Hahsp17.6G1 promoter, the detection and broad definition of additional upstream cis-element(s; Carranco et al., 1999). These latter additional elements are distinct from HSEs and thus are not directly involved in binding of heat stress factors (HSFs). In all plant sHSP promoters analyzed thus far, including Hahsp17.6G1, HSEs appear essential for developmental regulation, indicating the relevance of HSF(s). We recently characterized HSEs from sunflower (Helianthus annuus) promoters, which led to the cloning and functional identification of HaHSFA9 (Almoguera et al., 2002). HaHSFA9 is a peculiar HSF: Its expression patterns are mostly embryo specific, and it preferentially trans-activates promoters with imperfect HSEs (such as Hahsp17.7G4 and the seed-specific Hahsp17.6G1). Thus, we recognized HaHSFA9 as an activator of these two promoters during embryogenesis (Almoguera et al., 2002).
Analyses of the expression of sHSP chimeric genes in Arabidopsis (Arabidopsis thaliana) mutants resulted in the identification of FUS3, LEC1, and ABI3 as additional TFs that may cooperate with HSFs during embryogenesis (Wehmeyer and Vierling, 2000). FUS3, LEC1, and ABI3 have broad effects on late seed maturation programs but do not affect the heat shock response. Their direct binding to sHSP promoter(s) has not yet been demonstrated. Using the Hahsp17.7G4 promoter, we showed that ABI3 could function as a transcriptional coactivator that interacts with the HSFs bound to HSEs (Rojas et al., 1999). The role of additional accessory TFs appears significant as indicated by the phenotype of specific developmental mutants, such as abi3.6 in Arabidopsis (Wehmeyer and Vierling, 2000), and by the transcriptional effect of deletions in the Hahsp17.6G1 promoter (Carranco et al., 1999). However, knowledge of such factors is currently limited.
Here, we exploited the functional identification of HaHSFA9 (Almoguera et al., 2002). We showed that mutation of HSE eliminated developmental induction of the Hahsp17.6G1 promoter, which was similar to the effect of deletion of upstream elements, in the −126(G1):β-glucuronidase (GUS) gene (Carranco et al., 1999). From these observations, we infer that a synergistic interaction exists between HaHSFA9 and unknown additional TFs. We predict that the interaction requires the concurrent binding to the promoter of all involved factors. In this work we further define cis-elements in the Hahsp17.6G1 promoter, which allowed us to clone a second TF, sunflower drought-responsive element binding factor 2 (HaDREB2). HaDREB2 belongs to the Apetala 2 (AP2) gene family, which contains 145 different members in Arabidopsis (Sakuma et al., 2002). Using transient expression analyses, we demonstrate that a synergistic interaction exists between HaDREB2 and HaHSFA9 in sunflower embryos. Such interaction meets the requirements deduced from the analyses of the Hahsp17.6G1 promoter when stably integrated in transgenic plants. Thus, we propose that HaDREB2 cooperates with HaHSFA9 in the developmental regulation of the Hahsp17.6G1 promoter during embryogenesis in sunflower.
RESULTS
Identification of Positive cis-Acting Elements in the Hahsp17.6G1 Promoter
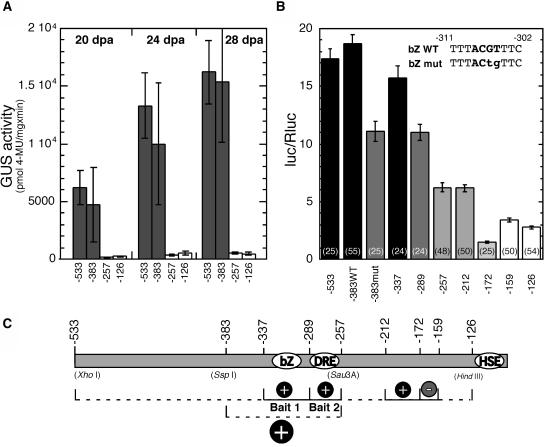
Our aim was to define short DNA fragments in the Hahsp17.6G1 promoter that could be used as bait for cloning novel TFs using the yeast one-hybrid technique (Meijer et al., 1998; Almoguera et al., 2002). Using transgenic tobacco (Nicotiana tabacum) and the GUS reporter gene, we have analyzed the effects of additional 5′ deletions in the Hahsp17.6G1 promoter. These deletion constructs, −383(G1):GUS and −257(G1):GUS, contain promoter 5′-flanking sequences located between those previously analyzed: −533(G1):GUS and −126(G1):GUS (Carranco et al., 1999). We compared the expression patterns of the four genes in tobacco seeds during zygotic embryogenesis, in the absence of exogenous stress (Fig. 1A). The −533(G1):GUS and −383(G1):GUS genes were activated during the desiccation stage, from 20 DPA, and reached maximal activity in mature seeds at 28 DPA. No significant statistical difference was observed between the reporter activities of −533(G1):GUS and −383(G1):GUS at any stage. Statistically similar reporter activities were also observed for −257(G1):GUS and −126(G1):GUS. However, in these cases the reporter activity was much lower than that obtained for −533(G1):GUS and −383(G1):GUS (Fig. 1A). This low activity was consistent with the expression levels observed for all chimeric genes before 20 DPA (data not shown; see Carranco et al., 1999). These results refined our location of the positive cis-acting elements (initially between positions −533 and −126 [Carranco et al., 1999]) to a shorter promoter region from −383 to −257 (Fig. 1A; see also the map in Fig. 1C).
Figure 1.
A, Fluorometric quantification of GUS activity in whole seeds during maturation in transgenic tobacco. Hahsp17.6G1:GUS chimeric genes (containing the 5′ end indicated in each case, see the map in C) were compared at 20, 24, and 28 DPA. The following numbers of independent transgenic lines were analyzed for each chimeric gene: −533(G1):GUS, 10; −383(G1):GUS, 5; −257(G1):GUS, 8; and −126(G1):GUS, 12. B, Transcriptional activation assays in bombarded sunflower embryos. Comparisons between different Hahsp17.6G1:LUC fusions are shown. The scheme in the top-right corner shows the bZIP-binding core sequences in wild-type (bZ WT) and mutated (bZ mut) forms. Reporter activity is given as the ratio between Photinus (luc) and Renilla (Rluc) luc activities. Numbers in parentheses show the number of replicates for each chimeric gene. In A and B, mean reporter activity values and se bars are represented. The same shading indicates statistically similar reporter activity. Different shading corresponds to statistically significant differences. C, Map indicating positions of the 5′ borders in the different chimeric genes and cis-element localization. Dashed lines indicate regions as originally determined (top) in transgenic tobacco (Carranco et al., 1999) and (bottom) after the subsequent analyses shown in A. Solid lines and other symbols indicate the positive (+) and negative (−) cis-elements defined by the results in A and B. The Bait 1 and Bait 2 promoter fragments used for one-hybrid cloning are shown, as well as the positions of the cis-elements HSE, DRE like (DRE), and bZIP core binding site (bZ). Reference restriction sites in the Hahsp17.6G1 promoter are given in parentheses.
Our previous studies indicated conservation between the developmental regulation of sHSP promoters in sunflower and transgenic tobacco (Almoguera et al., 1998; Carranco et al., 1999; Almoguera et al., 2002). However, further determination of cis-acting elements using tobacco was not practical because of time considerations. Therefore, we performed transient-expression assays using a dual-luciferase (luc) system in sunflower embryos (the homologous system; see details in “Materials and Methods” section). In bombarded sunflower embryos, we reproduced the effects of deletion of Hahsp17.6G1 promoter sequences that were observed in transgenic tobacco (compare Fig. 1, A and B). Thus, 5′ deletion of sequences up to positions −126 or −257 caused significant decreases in chimeric reporter gene expression (Fig. 1B). However, the reporter activities for the −533(G1):LUC and −383(G1):LUC genes were statistically similar.
Intermediate deletions (Fig. 1C) allowed us to further define the promoter region as an array of at least three positive and one negative cis-acting elements, ranging from 13 to 48 bp in length. Thus, we observed significant decreases in the reporter activity between the genes with deletion end points of −337 and −289, and between −289 and −257. This indicated the presence of at least two separate cis-acting elements that enhance transcription and that likely bind different trans-acting factors that are conserved between sunflower and tobacco. A significant decrease in reporter gene activity upon deletion of the fragment between −212 and −172 indicated the presence of a positive regulatory element. Conversely, an increase in expression after the deletion of sequences between −172 and −159 indicated the presence of a negative regulatory element. Other deletions did not show statistically significant effects. This would indicate the absence of additional elements. In summary, the results in Figure 1B (transient expression in sunflower embryos), mostly agree with those in Figure 1A (stable expression in transgenic tobacco). The main difference was an absence of functional effect of sequences downstream of position −257 in transgenic tobacco.
For further characterization, we selected the two promoter fragments that are labeled, respectively, Bait 1 and Bait 2 (Fig. 1C). We analyzed the DNA sequence by searching for potential binding sites using the nucleotide distribution matrices of the TRANSFAC database and the program MatInspector (Quandt et al., 1995). In Bait 1, we found a potential bZIP-binding site with an ACGT core between positions −311 and −302 (bZ, Fig. 1C). The effect of mutating two nucleotides within the core sequences of this site was analyzed in the context of the −383 gene (Fig. 1B, see diagram on top right). This change significantly reduced the activity of the chimeric gene to a level statistically similar to that of the −289 gene. We conclude that bZIP factor(s) are involved in the developmental activation of the Hahsp17.6G1 promoter together with the HSF(s) acting through the imperfect HSE (Carranco et al., 1999; Almoguera et al., 2002). Attempts to clone the sunflower embryo bZIP factor(s) using a reporter yeast (Saccharomyces cerevisiae) strain with a tetramerized form of Bait 1 were unsuccessful, as the HIS3 reporter was activated by yeast bZIPs. In Bait 2, we found possible binding sites for TFs with the AP2 domain (Sakuma et al., 2002). These include core sequences with high similarity, or identical to those in drought-responsive/C-repeat element binding-site matrices. Such sequences are located on the coding (−277 to −272) and noncoding (−263 to −258) DNA strands of the Hahsp17.6G1 promoter.
Yeast One-Hybrid Cloning and Characteristics of HaDREB2
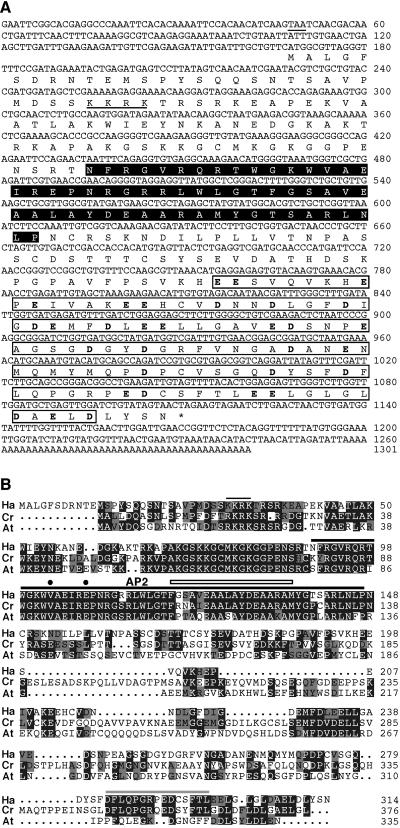
A monomer of the Bait 2 sequences (Fig. 1C) was used in yeast to screen a sunflower embryo cDNA library (Almoguera et al., 2002). We obtained 22 positive clones after screening approximately eight library equivalents (12.6 × 106 cDNAs). Of the initial clones, we selected the 14 that grew fastest on selective medium. These clones were independent cDNA isolates encoding a TF containing a single AP2 domain (Fig. 2). The AP2 domain is a DNA-binding domain that recognizes the drought-responsive/C-repeat element core sequences (Sakuma et al., 2002) in Bait 2. The nucleotide and deduced-amino acid sequences of the cDNA are shown in Figure 2A. In this figure, we also indicate the AP2 domain and other predicted protein regions with putative functions, including a C-terminal portion (of 114 amino acids) enriched in acidic residues, which could participate in transcriptional activation. Sequence characteristics indicate that the new factor, here cloned and named HaDREB2, belongs to Group A in the drought-responsive/C-repeat element binding factor subfamily of single-AP2-domain TFs. This group has 56 different members in Arabidopsis, all of which show Val at position 14 of the AP2 domain, and most of them (including HaDREB2) encode Glu at position 19 (Sakuma et al., 2002). Additional comparisons with homologous TFs further classify HaDREB2 as belonging to the A-2 subgroup defined by Sakuma et al. (2002). Thus, the sequence alignment in Figure 2B illustrates extensive sequence similarity with AtDREB2A and CrORCA1, both classified as subgroup A-2 factors that, respectively, are 40% or 42% identical to HaDREB2. Both factors show extensive (51%) amino acid similarity to HaDREB2. This similarity includes the AP2 domain and extends to the N terminus, with additional identical sequence segments located outside the AP2 domain. However, only in the case of CrORCA1, the sequence similarity also extended to sequences in the C-terminal region, including some of the mentioned acidic residues (Fig. 2B).
Figure 2.
A, The HaDREB2 cDNA and characteristics of its predicted protein. On the nucleotide sequence, a stop codon located 5′ upstream of the predicted protein is underlined. Below the nucleotide sequence of HaDREB2, we show the predicted amino acid sequence. An asterisk marks the stop codon. The AP2 and the acidic CTDs are boxed and shaded black or white, respectively, on background. The acidic residues in the CTD are indicated in boldface. Four basic amino acid residues that could function as a nuclear localization sequence are underlined. B, Alignment of the predicted HaDREB2 protein (Ha) with the subgroup A-2 DREB proteins CrORCA1 (Cr) and AtDREB2A (At; Sakuma et al., 2002). The putative nuclear localization sequence and the conserved AP2 domain are marked with black lines. Within the AP2 domain, the Valine-14 and Aspartic acid-19 residues characteristic of DREB proteins (Sakuma et al., 2002) are indicated by black dots. The 18-amino acid core region predicted to form an amphipathic α-helix in AP2 (Okamuro et al., 1997) is indicated by a thin rectangular box. In the less conserved C-terminal region, amino acid residues conserved only between HaDREB2 and CrORCA1 are indicated by a gray line.
The HaDREB2 cDNA was sufficient for function in yeast, as the GAL4 activation domain in the library plasmid was not in frame with the predicted HaDREB2 protein. We inferred that the HaDREB2 cDNA encoded a complete TF. This was further supported by sequence comparisons with homologous TFs in other plants (Fig. 2B) and by the functional characterization of HaDREB2 in sunflower embryos (see below). In addition, we indicate a stop codon located 5′ upstream and in frame with the predicted protein in the HaDREB2 cDNA (Fig. 2A).
Synergistic Transcriptional Activation of the Hahsp17.6G1 Promoter by HaDREB2 and HaHSFA9 in Sunflower Embryos: HSF Specificity
Our published analyses of the Hahsp17.6G1 promoter indicated a strong synergism between the HSE and other cis-acting elements located upstream of position −126 (Carranco et al., 1999). Therefore, we predicted that HSF(s) bound to the HSE would interact with different factor(s) bound upstream of the HSE to activate this promoter in embryos. We previously identified HaHSFA9 as an HSF involved in Hahsp17.6G1 activation (Almoguera et al., 2002). We could thus use HaHSFA9 as a tool for functional verification of the newly cloned TF. If HaDREB2 is involved in the developmental activation of the Hahsp17.6G1 promoter in sunflower embryos, it should synergistically interact with HaHSFA9. We also predict that this synergy should not extend to different HSFs.
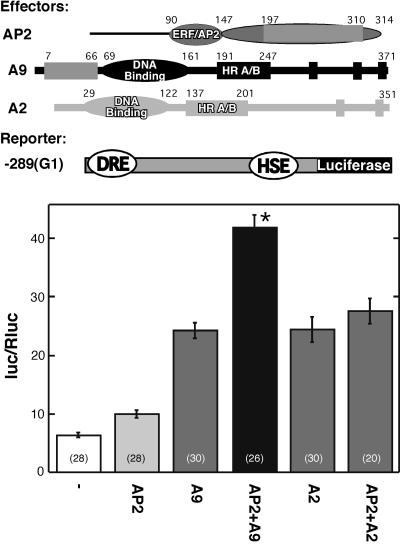
We constructed a HaDREB2 effector plasmid, which we tested by transient expression in micro projectile-bombarded embryos. We used the −289(G1):LUC reporter gene, which is driven by Hahsp17.6G1 promoter sequences including the HSE and Bait 2 (which contains the DRE-like sequences used to clone HaDREB2). We investigated whether HaDREB2 activated −289(G1):LUC, and the effect of cobombardment with the HaHSFA9 (Almoguera et al., 2002) or LpHSFA2 effector plasmids. LpHSFA2 is a similar (class A) HSF from tomato (Lycopersicon esculentum) that was previously shown to trans-activate the Hahsp17.6G1 promoter (Rojas et al., 2002). LpHSFA2 was selected as a negative control to investigate the specificity of the interaction between HaDREB2 and HaHSFA9.
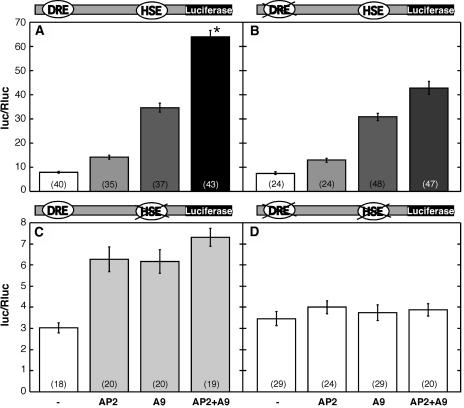
The results in Figure 3 show that HaDREB2 alone activated reporter gene expression above basal levels. Each HSF (HaHSFA9 or LpHSFA2) activated the reporter to statistically similar levels. However, when cobombarded with HaDREB2, only HaHSFA9, but not LpHSFA2, produced higher expression levels than observed with either HSF alone. Furthermore, the effect observed upon cobombardment of HaHSFA9 and HaDREB2 was synergistic, as the expression level attained was significantly higher than the sum of activation induced by either factor alone. We conclude that HaDREB2 is a functional TF that activates the Hahsp17.6G1 promoter in sunflower embryos. Furthermore, HaDREB2 meets the expected HSF specificity requirement for involvement in the developmental regulation of this promoter, as it was able to synergistically interact with HaHSFA9 but not with LpHSFA2.
Figure 3.
Transcriptional activation of the Hahsp17.6G1 promoter by HaDREB2 in sunflower embryos: specific synergistic interaction with HaHSFA9. The −289(G1):LUC reporter gene (−289[G1]) was bombarded without effector plasmid (−), or with the following effector plasmids: HaDREB2 (AP2), HaHSFA9 (A9), or LpHSFA2 (A2) alone, or in combination: HaDREB2 + HaHSFA9 (AP2 + A9) or HaDREB2 + LpHSFA2 (AP2 + A2). Construct maps are at the top of the figure. Data representing mean Photinus luc activity normalized with Renilla luc (Rluc) are plotted. The asterisk denotes significant synergism between HaDREB2 and HaHSFA9. Statistical significance, sample sizes, and remaining symbols are described in Figure 1. The results of statistical analyses are detailed in Supplemental Table I.
The Synergic Interaction between HaDREB2 and HaHSFA9 Requires the Binding of Both Factors to the Hahsp17.6G1 Promoter
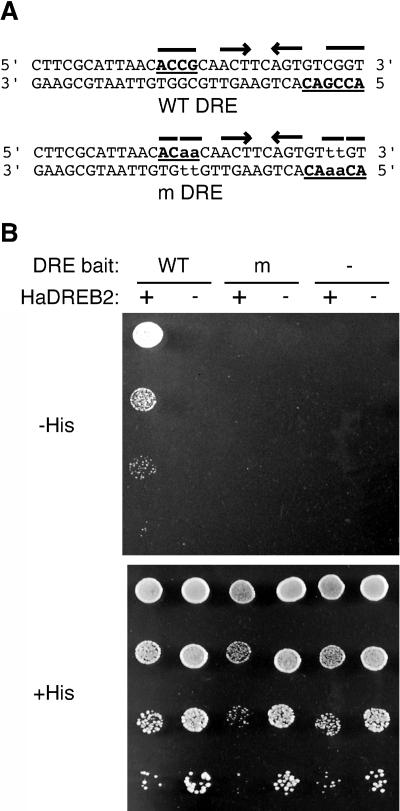
We investigated promoter DNA-binding requirements for the functional interaction between HaDREB2 and HaHSFA9. Mutations in each factor-binding site were incorporated in separate plasmid constructs or in the same construct. The mutant reporter genes were constructed in the context of −289(G1):LUC. The mutant HSE sequences that were assessed have been described previously (Carranco et al., 1999). In the case of HaDREB2, putative DRE sequences that were present in Bait 2 were mutated by introducing four nucleotide changes (Fig. 4A). These were identical to mutations that abolished DRE binding by similar DREB2 factors (Sakuma et al., 2002). In vitro DNA-binding assays were conducted using recombinant HaDREB2 proteins and a range of procedures, including gel retardation protocols used in the analysis of other DREB proteins (for example, Sakuma et al., 2002). Our attempts to detect binding to the wild-type DRE sequences were unsuccessful however, perhaps due to the requirement of other factors or protein modifications. Therefore, the effects of the mutant DRE were analyzed in yeast by examining the one-hybrid interaction of HaDREB2 with the wild-type and mutant Bait 2 sequences. HaDREB2 activated the HIS3 reporter in the plasmid with the wild-type sequence but not with the mutant DRE sequence, as determined by cell growth in selective medium (Fig. 4B). The HIS3 reporter appeared to be weakly activated by HaDREB2 in the mutant DRE plasmid in selective medium lacking 3-amino-1′,2′,4′-triazole (3-AT; data not shown), but the level of activation was insufficient to support cell growth in selective medium containing 10 mm 3-AT (Fig. 4B).
Figure 4.
Yeast one-hybrid DNA-binding assays. A, The Bait 2 sequence between positions −289 and −258 in the Hahsp17.6G1 promoter, showing the positions of wild-type (WT) and mutant (m) DRE sequences. Nucleotides that partially, or totally, match the DRE core consensus 5′-(A/G)CCGnc-3′ (Sakuma et al., 2002) are underlined. Mutated nucleotides are in lowercase. Arrows indicate palindromes present in the wild-type and mutant DRE sequences. B, One-hybrid assays with yeast strains containing different combinations of prey and bait plasmids. The DRE Bait 2 plasmid was wild type (WT), mutant (m), or absent (−). The prey plasmid expressing HaDREB2 was present (+) or absent (−). Yeast cultures were diluted (1:10 successive dilution series), spotted onto selective medium containing 10 mm 3-AT and no Histidine (−His), or onto nonselective medium with Histidine (+His), then incubated for 3 d at 30°C.
Cis-element mutants were analyzed by transient expression in sunflower embryos using the respective mutant −289(G1):LUC reporter plasmids (Fig. 5). Each mutant was compared to a positive control (the wild-type plasmid with the relevant combinations of effector plasmids), to compensate for variability among different batches of sunflower embryos. The separate mutation of each cis-element abolished the synergic activation (Fig. 5, compare sections B and C with A). This demonstrated that HaHSFA9 and HaDREB2 should bind DNA at their respective promoter cis-elements to attain a synergic effect. The simultaneous mutation of both elements also abolished the synergism. Only basal reporter activity levels were observed in the HSE/DRE double mutant with all the effector plasmid combinations (Fig. 5D). Therefore, transcriptional activation by HaHSFA9 or by HaDREB2 was also eliminated in this double mutant. The latter finding contrasts with results obtained for the activation, by each factor, of the individual HSE and DRE mutant reporter plasmids. Mutation of HSE drastically reduced transcriptional activation by HaHSFA9, but HaDREB2 was still able to activate this mutant reporter gene (Fig. 5C). Mutation of the DRE did not affect transcriptional activation by HaHSFA9 (Fig. 5B; A9, compared to levels for the wild-type reporter in Fig. 5A). Surprisingly, the DRE mutation also did not affect activation by HaDREB2 (Fig. 5B; AP2). The observation that mutation of DRE severely reduced the binding of HaDREB2 to DNA in yeast (Fig. 4) suggested that HaDREB2 might bind the promoter through HSF(s) already bound to HSE, at least in conditions of transient expression. Results from the double-mutation study (Fig. 5D) rule out the direct binding of HaDREB2 to plasmid DNA at spurious DRE sites. Therefore, HaDREB2 either directly binds the HSE with the same nucleotide requirements as an HSF, or it binds indirectly via HSFs already bound to HSE.
Figure 5.
Mutation of the HSE or DRE sequences eliminates synergism between HaHSFA9 and HaDREB2. Sunflower embryos were bombarded with the −289(G1):LUC reporter gene constructs indicated above each section and with the effector plasmids indicated at the bottom. Mutated cis-elements are crossed on the construct maps. Sample sizes, statistical comparisons, and symbols for effector plasmids are described in Figures 1 and 3. The results of statistical analyses are detailed in Supplemental Table I.
Physical Interaction between HaDREB2 and HaHSFA9 in Vitro
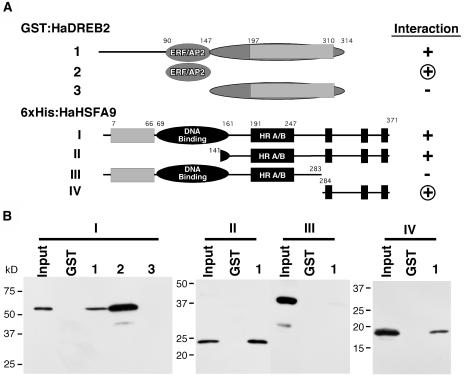
The complete HaDREB2 and HaHSFA9 proteins appeared to interact directly in glutathione S-transferase (GST) pull-down assays conducted under in vitro conditions (Fig. 6). However, this interaction was very inefficient. In further analysis of HaDREB2, the AP2 (amino acids 90–147; Fig. 6, A2), and C-terminal (amino acids 148–314; Fig. 6, A3) domains fused to GST were tested for interaction with the complete HaHSFA9 protein (Fig. 6, A1). The AP2 domain showed a more efficient interaction with HaHSFA9 than that observed for the full-length HaDREB2 protein. In contrast, the C-terminal domain (CTD) of HaDREB2 was not sufficient for interaction with HaHSFA9 (Fig. 6B). It has been postulated that an amphipathic α-helix in the AP2 domain is involved in protein-protein interactions (Okamuro et al., 1997; Fig. 2). Our results in Figure 6B provide support for this hypothesis.
Figure 6.
In vitro interaction between HaDREB2 and HaHSFA9: mapping of interacting domains using GST pull-down experiments. A, The negative control, GST alone (GST), and each of the GST:HaDREB2 fusion proteins (A, 1–3) were bound to glutathione affinity beads then incubated with protein extracts containing the 6×His:HaHSFA9 fusions shown in A, I to IV. B, Interacting proteins were eluted, separated by SDS-PAGE, and detected by western blotting using an anti-Xpress antibody. Input lanes contain aliquots comprising from left to right: 0.32%, 0.40%, 1.6%, and 0.32% of the total protein in the respective extracts (constructs I–IV). Negative (−) and positive (+) interactions as well as the smallest fragment of each TF that was sufficient for interaction (encircled +) are indicated in A.
N-terminal and C-terminal deletions of HaHSFA9 (II and III in Fig. 6) were also tested for their ability to interact with the complete HaDREB2 protein fused to GST (Fig. 6, AI). Only the C-terminal deletion (III in Fig. 6) severely reduced the interaction (Fig. 6B), suggesting a crucial role for the region. However, regardless of the combination analyzed, between only 0.1% and 1.5% of the input protein was specifically retained by the GST:DREB2 Sepharose beads. This indicates that, besides requiring the C-terminal and AP2 domains of HaHSFA9 and HaDREB2, respectively, the direct interaction between both proteins is intrinsically weak. The CTD of HaHSFA9 by itself (amino acids 284–371; Fig. 6, AIV) was sufficient for interaction with the complete HaDREB2 protein and interacted even more efficiently with a truncated HaDREB2 protein containing only the AP2 domain (construct 2, data not shown). All these results confirmed the crucial importance of both domains for direct physical interaction between HaDREB2 and HaHSFA9.
The identification of interacting domains explains results obtained in transient expression analyses (Fig. 5). In the case of HaHSFA9, the CTD sequences that appear critical for interaction with HaDREB2 contain the putative activation domain. The direct interaction with HaDREB2 could therefore compete for contacts between this CTD and components of the transcription machinery. This may explain why HaHSFA9 showed reduced transcriptional activation in the absence of a functional HSE, when forced to interact through promoter-bound HaDREB2 (Fig. 5C). Conversely, the opposite result obtained for the activation by HaDREB2 might be also explained: In the absence of a functional DRE site, HaDREB2 may interact through promoter-bound, endogenous HSFs using the AP2 domain. This is a DNA-binding domain (Okamuro et al., 1997; Sakuma et al., 2002) not directly involved in or interfering with transcriptional activation by DREB factors. The experimental conditions conducive to directing contact between HaHSFA9 and HaDREB2 abolished their transcriptional synergism (Fig. 5B). It is unlikely that these conditions are functionally relevant in planta. A definite explanation for the abolishment of synergism observed with the mutant genes is not necessary for our main conclusion from data in Figure 5: the necessity of both the DRE and the HSE for the synergism.
The apparent interaction between HaDREB2 and HaHSFA9 in bombarded embryos appears to be efficient (compare activation by HaDREB2 in Fig. 5, A and B). We performed electrophoretic mobility shift assays using the wild-type HSE fragment and yeast extracts with either HaHSFA9, HaDREB2, or with both TFs to further investigate the nature of this contact. Specific binding to DNA of HaHSFA9 was not detectable in the electrophoretic mobility shift assays, alone or in combination with HaDREB2 (data not shown). Therefore, we conducted yeast monohybrid binding assays using an HSE:LacZ reporter in the RSY4 strain with HaHSFA9 as the sole HSF (Almoguera et al., 2002). The reporter activity obtained in this strain was compared with that of the same strain transformed with a 2-μm plasmid expressing HaDREB2. Similar activity levels were observed (respectively, 333.97 ± 14 and 399.26 ± 12, n = 8), which strongly argues against efficient contact (direct or indirect) between the two TFs in yeast. In agreement with this, interaction between HaHSFA9 and HaDREB2 was not detected in multiple yeast two-hybrid assays (Supplemental Fig. 1). Therefore, if HaHSFA9 and HaDREB2 do interact in plant cells the contact is most likely indirect i.e. involving additional accessory proteins.
The C-Terminal Region of HaHSFA9 Is Sufficient for Synergistic Transcriptional Activation with HaDREB2
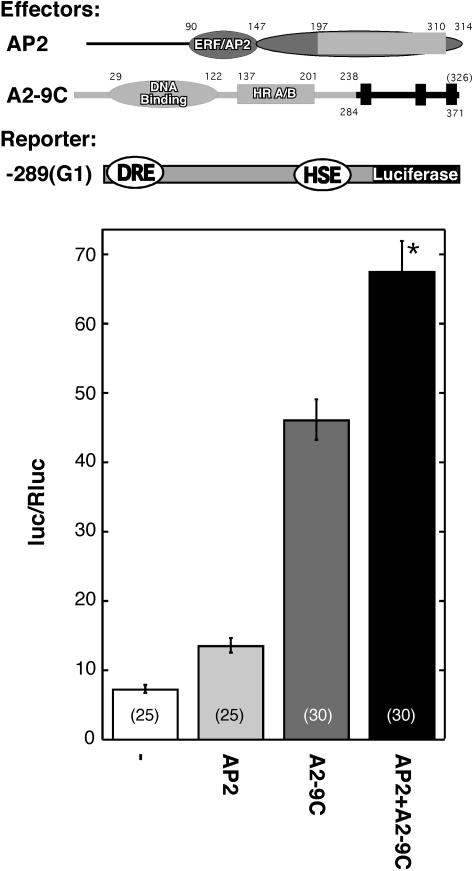
We investigated whether the C-terminal region of HaHSFA9 could explain the HSF specificity that was observed in the functional interaction with HaDREB2 (Fig. 3). A chimeric HSF, HSFA2-A9C, was constructed by fusion PCR according to methods described by Hobert (2002). To produce this construct we replaced the native C-terminal sequence of LpHSFA2 (from amino acid 239) with the C-terminal region of HaHSFA9 (amino acids 284–371; Fig. 7). We then assayed HSFA2-A9C via the same transient expression system used to analyze the specificity of functional interaction between HaDREB2 and other HSFs in sunflower embryos (Fig. 3).
Figure 7.
A chimeric HSF containing the C-terminal region of HaHSFA9 synergistically interacts with HaDREB2 in sunflower embryos. Top, Maps of the effector plasmids HaDREB2 (AP2) and HSFA2-A9C (A2-9C) and the −289(G1):LUC reporter gene (−289[G1]). In the case of the chimeric HSF (A2-9C), domains originating from LpHSFA2 (light gray) or from HaHSFA9 (dark gray) are indicated. Numbers represent amino acid positions. Narrow rectangles at the C terminus of A2-9C represent putative AHA motifs in the HaHSFA9 CTD. Bottom, Bombardment experiments using the indicated effector plasmid combinations. Symbols as in Figures 1 and 3. The results of statistical analyses are detailed in Supplemental Table I.
HSFA2-A9C activated transcription from the Hahsp17.6G1 promoter in the −289(G1):LUC gene (Fig. 7). The chimeric HSF showed synergistic functional interaction with HaDREB2. This was demonstrated by the difference observed between the activity level obtained upon cobombardment and the value obtained by adding the activation levels for each factor. We conclude that the C-terminal region of HaHSFA9 is sufficient for the specific synergistic interaction with HaDREB2 in sunflower embryos.
DISCUSSION
The assembly of unique complexes from similar sets of TFs on promoters provides the specificity for regulation of complex patterns of gene expression in eukaryotes. The DNA components of the regulatory specificity have been termed cis-regulatory modules. These modules are made up of unique combinations of cis-elements, the deletion or mutation of which affects gene regulation in the same process (Davidson et al., 2002). The results in Figure 1 define a cis-regulatory module in the Hahsp17.6G1 promoter (Carranco et al., 1997, 1999). This module contains various cis-elements (Fig. 1C) and is involved in developmental transcriptional activation during embryo desiccation. Mutation of the HSE (Carranco et al., 1999) and deletion of different upstream regions (Carranco et al., 1999; Fig. 1A) indicated that strong synergistic interactions exist between the TFs that bind to these elements. In sunflower embryos, we demonstrated a specific synergistic interaction between HaHSFA9 and HaDREB2 (Fig. 3). HaDREB2 is a new TF belonging to the AP2 family. It was cloned by one-hybrid DNA interaction with the cis-element contained within the Bait 2 sequence (Figs. 1, 2, and 4A). Binding of HaDREB2 and HaHSFA9 to the Hahsp17.6G1 promoter was required for their synergic interaction in sunflower embryos (Fig. 5). This observation was consistent with functional analyses of chimeric genes when stably expressed in transgenic plants (Fig. 1A; Carranco et al., 1999). HaDREB2 (this work) and HaHSFA9 (Almoguera et al., 2002) were functionally identified from among large TF families with 125 (single AP2 domain) and 21 (HSF) members in Arabidopsis (Nover et al., 2001; Sakuma et al., 2002).
The importance of combinatorial control of transcriptional regulation in plants has been reviewed (Singh, 1998). For most plant genes, knowledge of the structure/function relation within cis-regulatory modules is very scarce. In each promoter, unidentified interactions between TFs are integrated by different combinations of cis-elements. Functional analyses of promoters, while time consuming, may be the sole alternative for safely revealing and analyzing TF interactions. Promoter studies are ineluctable when these functional interactions depend on binding to DNA of more than two TFs (as, for example, deduced from the results in Fig. 1), on their steric disposition on the promoter (Carey, 1998; Singh, 1998), and even on protein conformation changes induced by TF interaction with DNA (Santoro et al., 1998).
The mechanism for the synergism between HaDREB2 and HaHSFA9 is not fully understood. The simplest explanation for the synergism would be the specific recruitment by each TF of different components of the transcription machinery, as specific coactivators, mediator or basal machinery subunits (TBP, TAFs), etc. This hypothesis would be consistent with the simultaneous requirement of the DRE and the HSE cis-elements for the synergism, as activation by recruitment requires that the involved TFs bind DNA (Ptashne and Gann, 2002). However, we cannot completely exclude the interaction between HaDREB2 and HaHSFA9 in planta (i.e. in transgenic tobacco or in sunflower plants) when bound to the intact promoter. Such an interaction would be possible only if in these circumstances the transcriptional interference observed with the mutant promoter (Fig. 5) do not take place. Furthermore, it is possible that the interaction between HaDREB2 and HaHSFA9 is indirect, i.e. involving a third factor. Indeed, contacts through a third plant protein have been observed for the functional interaction between the HSFs: LpHSFB1 and LpHSFA1 (Bharti et al., 2004). In addition, the functional interaction between LpHSFA1 and LpHSFA2 (observed using the Hahsp17.6G1 promoter in bombarded embryos) also appears to require plant-specific proteins (Rojas et al., 2002).
Class A HSFs, the family to which HaHSFA9 belongs, have acidic activation domains in their CTDs. These domains have sequence singularities that comprise AHA motifs. Each AHA motif contains a unique combination of aromatic, hydrophobic, and acidic-amino acid residues (Döring et al., 2000). Distinct plant HSFs would have a different specificity for interactions with transcriptional machinery (Döring et al., 2000; Bharti et al., 2004). For class A HSFs, the specificity may be conferred by different AHA motifs (Döring et al., 2000). HSFs have furthermore been shown to interact with different components of the transcription machinery. These components include TBP (Reindl and Schöffl, 1998), TFIIB (Yuan and Gurley, 2000), TAF130, SNF2, and ADA3 (Kotak et al., 2004) and p300/CBP-like proteins (Bharti et al., 2004). Only class B HSFs have been observed to specifically interact with such components (Bharti et al., 2004). For class A HSFs, interactions with plant-specific transcription machinery components have been proposed for the AHA1 motif of AtHSFA2 (Kotak et al., 2004). This was based on the observed differential effect of AHA mutation on transcription activation in yeast or in plant protoplasts. In addition, the CTDs of A4c, A7a, and A9 HSFs from Arabidopsis were active in tobacco protoplasts but not in yeast, further indicating plant-specific interactions. The results of Figure 7 suggest that the CTD of HaHSFA9 containing putative AHA motifs (Almoguera et al., 2002) confers to a chimeric HSF the capacity of synergizing with HaDREB2. The sequences in this CTD would be sufficient for the specific recruitment mediated by HaHSFA9.
We propose that HaDREB2 is involved in the developmental activation of the Hahsp17.6G1 promoter during zygotic embryogenesis. HaDREB2 appears to specifically interact with HaHSFA9, at least indirectly through different components of the transcription machinery. Our work suggests the involvement of an AP2 protein (HaDREB2) in transcriptional control during embryo desiccation. These findings broaden previous observations for similar TFs (AtDREB2A and B) involved in vegetative drought stress in Arabidopsis (Shinozaki et al., 2003). The developmental activation of the Hahsp17.6G1 promoter requires the concurrent involvement of additional TFs such as bZIPs (Fig. 1B). These factors would bind to the cis-elements functionally defined in this work (Fig. 1). The strategy we have used here to functionally verify HaDREB2 might be applied to the additional factors. HaHSFA9, HaDREB2, and other verified TFs could be used alone or in combination as tools for genetic modifications (in gain-/loss-of-function approaches), which could verify the proposed functions for the proteins encoded by target HSP genes in embryos (Sun et al., 2002).
MATERIALS AND METHODS
Stable Expression Analyses in Transgenic Tobacco
The chimeric genes −383(G1):GUS and −257(G1):GUS were cloned into the SmaI site of vector pBI101.2 to produce translational fusions. These genes contained the indicated Hahsp17.6G1 5′-flanking sequences (see map in Fig. 1C) and coding sequences to position +115 to allow comparison with the previously analyzed −533(G1):GUS and −126(G1):GUS genes (Carranco et al., 1999). We generated primary (T0) tobacco (Nicotiana tabacum) transgenic plants using published procedures (Almoguera et al., 1998). We compared the developmental expression patterns of independent transformants (between 4 and 10 per construct) with a similar number (1–3) of integration events. Fluorometric GUS assays during seed maturation of the T0 plants and statistical analyses were conducted as previously reported (Almoguera et al., 1998; Rojas et al., 2002). Statistical significance was considered for values of P < 0.05. The same applied to the analysis of results of transient expression (see Supplemental Table I).
One-Hybrid Cloning and Methods for Yeast Assays
General methods for one-hybrid cloning and experimental manipulations in yeast (Saccharomyces cerevisiae) were as described by Meijer et al. (1998). The sunflower (Helianthus annuus) embryo cDNA library has been previously described (Almoguera et al., 2002). The reporter plasmid used for library screening was integrated into yeast strain Y187 (Clontech). The plasmid was constructed by insertion between the SmaI and EcoRI sites of pHIS3NX:pINT1 (Meijer et al., 1998) of the DNA fragment generated by hybridization of the following complementary oligonucleotides: 5′-CTTCGCATTAACACCGCAACTTCAGTGTCGGTG-3′ (top strand) and 5′-aattCACCGACACTGAAGTTGCGGTGTTAATGCGAAG-3′ (bottom strand). The insert contained an EcoRI overhang (lowercase) and the 32-bp sequence from the Hahsp17.6G1 promoter defined by Bait 2 (Figs. 1C and 4A). The reporter yeast was transformed with DNA from the library and grown on synthetic dextrose (SD)-His-Leu-selective medium supplemented with 10 mm 3-AT. The initial positive clones were selected after 3 to 4 d growth at 30°C. Further selection was performed after subsequent growth in selective medium supplemented with 100 mm 3-AT. The HaDREB2 cDNA was subcloned into pBluescript SK+ (Stratagene) as an EcoRI-BglII fragment and the two strands of DNA sequenced with an ABI PRISM 3700 DNA analyzer.
For yeast one-hybrid DNA-binding assays, two additional strains were constructed, one with the pHIS3NX:pINT1 reporter plasmid lacking inserted bait sequences and a second strain with inserted mutant Bait 2 sequences. The mutant sequences were generated by hybridization of the following complementary oligonucleotides: 5′-CTTCGCATTAACACaaCAACTTCAGTGTttGTG-3′ and 5′-aattCACaaACACTGAAGTTGttGTGTTAATGCGAAG-3′. Nucleotide mutations and EcoRI overhang are indicated in lowercase. The double-stranded mutant sequence was cloned into pHIS3NX:pINT1 as described above for the wild-type Bait 2 sequence. DNA-binding assays were performed after transformation of each yeast reporter strain with the empty vector (pGAD424) or with the HaDREB2 library plasmid (pGAD424:HaDREB2). Yeast cells were grown for 3 d at 30°C in SD-Ura-Leu medium (nonselective). Cultures were then diluted to 0.4 optical density at 600 nm and then further diluted with three 1/10-dilution steps. Approximately 3 μL of each dilution were spotted onto nonselective medium and onto selective medium (SD-Ura-Leu-His) with and without 10 mm 3-AT. DNA binding to the DRE was assessed by observing cell growth on selective medium at 30°C.
Transient Expression
Conditions for transient expression assays of chimeric genes after bombardment of sunflower embryos were essentially as previously described (Almoguera et al., 2002; Rojas et al., 2002; Fig. 1B), except a Dual-Luciferase reporter assay was used (Promega). For the experiments in Figures 3, 5, and 7, the total amount of plasmid DNA used for bombardment and the relative amounts of effector, reporter, and reference plasmids were optimized to ablate systematic variations in the reporter activity of the reference plasmid. The optimized plasmid amounts per DNA precipitate (5 shots) were 2.5 μg for the reporter plasmids, 1.0 μg for the reference plasmid, 0.5 μg each for the HaHSFA9 and HSFA2-A9C effector plasmids, 3 μg for the LpHSFA2 effector plasmid, and 1 μg for the HaDREB2 effector plasmid. The total amount of plasmid was adjusted to 20 μg with pBI221 DNA.
The different reporter plasmids were constructed in a vector derived from pDR102 (Riggs and Chrispeels, 1987). The Photinus pyralis luc sequences in pDR102 were substituted for the optimized LUC+ gene in pSP-LUC+ (Promega). The T3 promoter and polylinker sequences of pBluescript SK+ (between SacI and SalI) were also included 5′ upstream of LUC+. The reporter plasmids contained different translational fusions between position +49 in the coding sequence of Hahsp17.6G1 and LUC+. First, a −1486(G1):LUC plasmid was constructed by inserting the 1,547-bp fragment obtained by partial HindIII digestion of plasmid −1486(G1):GUS in the appropriate orientation in the pDR102-derived vector (Carranco et al., 1999). The −533(G1):LUC plasmid is a XhoI, 5′-deletion derivative of −1486(G1):LUC. The 5′ end points of the other deletion-derived fusions are indicated in the map of Figure 1C. The different PCR-amplified DNA fragments were digested with SalI and inserted between the EcoRV and SalI sites of the pDR102-derived vector. A common 3′-LUC+ primer, 5′-GCGGTTCCATCTTCCAGCGG-3′, and different Hahsp17.6G1 primers were used for PCR amplification from the larger (−533) reporter plasmid. The reporter plasmid with the mutant HSE was constructed by replacing the wild-type sequences in the −289(G1):LUC gene with a 175-bp HindIII fragment containing the HSE mutations described by Carranco et al., (1999). For construction of the −289(G1):LUC mutant DRE plasmid, the mutagenic primer was 5′-CTTCGCATTAACACaaCAACTTCAGTGTttGTGATCGCTGACAGTAG-3′ (nucleotide substitutions indicated in lowercase). In this case, the mutant DNA fragment was amplified from the wild-type −289(G1):LUC plasmid. The reporter plasmid, with mutant HSE and DRE elements, was constructed from the mutant HSE −289(G1):LUC plasmid after PCR amplification using the mutant DRE oligonucleotide described above. The reference plasmid contained the luc reporter gene from Renilla reniformis (Rluc, obtained from plasmid pRL-CMV, Promega). This gene was cloned between the promoter and terminator sequences of plasmid pBI221. More details on restriction sites and/or primers for PCR amplification used in each construct are available upon request.
We have previously described the effector plasmids for HaHSFA9 (Almoguera et al., 2002) and LpHSFA2 (Rojas et al., 2002). The effector plasmid for the complete HaDREB2 protein was constructed by replacing the GUS sequences in pBI221 (removed as a SmaI-SacI fragment) with the HaDREB2 cDNA sequence, which was taken as an EcoRV-SacI fragment from the pBluescript SK-HaDREB2 plasmid described above. The effector plasmid for the chimeric HSFA2-A9C protein (see map in Fig. 7) was constructed using a PCR fusion-based approach, which was performed essentially as described by Hobert (2002). The LpHSFA2 sequences in HSFA2-A9C were PCR amplified from plasmid pRT-LpHSFA2 (Rojas et al., 2002). We used (in PCR-1) the oligonucleotides 5′-ACTATCCTTCGCAAGACCCTTCC-3′ (OliA, 35S promoter sequences from position −46) and 5′-cttgaaaacgatcgagatcatcTGTAACACTGGGGGTCATCGTTAG-3′ (OliB). Uppercase sequences in OliB encode LpHSFA2 amino acids ending at position 238. The lowercase sequences are a 22-nucleotide overhang encoding HaHSFA9 amino acids from position 284. The HaHSFA9 sequences in HSFA2-A9C were PCR amplified from the HaHSFA9 effector plasmid (Almoguera et al., 2002). We used (in PCR-2) the oligonucleotides 5′-GATGATCTCGATCGTTTTCAAG-3′ (OliC) and 5′-CAGGAAACAGCTATGAC-3′ (OliD, M13-reverse). The OliC sequences are the reverse complement of the 22-nucleotide overhang present in OliB. The fusion PCR was performed with approximately 10 ng (each) of the unpurified PCR-1 and PCR-2 products, using the nested primers 5′-AAGGAAGTTCATTTCATTTGGAG-3′ (OliA′, 35S sequences eight nucleotides downstream of OliA) and 5′-TCGGAATTAACCCTCACTAAAG-3′ (OliD′, 17 nucleotides downstream of OliD). The fusion PCR product was digested with XhoI and XbaI, and the resultant HSFA2-A9C DNA-fragment of 1,110 nucleotides was cloned into the corresponding sites of the pRT101 vector. Annealing temperatures were as follows: 57°C (PCR-1), 51°C (PCR-2), and 54°C (Fusion PCR). The nucleotide sequences of the effector plasmids, which had been amplified with Pwo-DNA polymerase (Roche), were verified before performing functional tests.
Dual-Luciferase assays were performed with the Promega system according to the manufacturer's instructions. The bombarded embryo samples were homogenized in 150 to 250 μL of Passive Lysis buffer (Promega). After centrifugation for 7 min and 17,000g at 4°C, we assayed 2 μL of each extract in 40 μL of Luciferase Assay Reagent (LAR II, Promega). For the Rluc assays, we added 40 μL of 1× Stop & Glo solution (Promega). Luc and Rluc activities were sequentially measured in a TD-20/20 luminometer (Turner Designs). In most cases, each plasmid combination (reporter ± effector[s] + reference) was bombarded at least 25 times (five replicates in five independent experiments).
Production of Recombinant Proteins and GST Pull Down
The complete HaDREB2 coding sequence (Fig. 6, A1) and two deletion derivatives, (Fig. 6A, 2 and 3), were fused in frame with GST in plasmid pGEX4T-1 (Amersham Biosciences, construct 1) or pGEX4T-3 (Amersham Biosciences, constructs 2 and 3). In the case of the complete protein (construct 1), a XhoI site was introduced 12 nucleotides upstream of the start codon using the oligonucleotide primer 5′-aaaagagctcgagttcccCATGGCGTTAG-3′ (synthetic sequences in lowercase).
We cloned in frame with the 6×His tag of pRSET vectors A to C (Invitrogen) the complete HaHSFA9 protein (I); the N-terminal (II), and C-terminal (III) deletions, as well as the C-terminal amino acids of HaHSFA9 (IV) indicated in the corresponding maps of Figure 6A. pRSET-A was used for I and III; pRSET-B was used for II; and pRSET-C was used for IV. Construct I was obtained by insertion of a 1,195-bp XhoI-PstI fragment, which was obtained by PCR amplification from the pSKHSFA9-F plasmid (Almoguera et al., 2002). The XhoI site was introduced three nucleotides upstream from the start codon using the oligonucleotide primer 5′-GATTGTTTTTGATcTCGAgTTCATG-3′ (mutated sequences in lowercase). The PstI end is a natural restriction site at position 1,247 within the HaHSFA9 cDNA (Almoguera et al., 2002). The insert in construct II was a 783-bp EcoRI fragment that was obtained from construct I and cloned in the appropriate orientation. Construct III was obtained from construct I by substituting the 564-bp BglII-PstI fragment for a shorter sequence (259 bp). The insert in construct IV was a 342-bp fragment that was obtained after PCR amplification from construct I, followed by digestion with PstI. In this PCR, we used the oliC (see above) and pRSET3 (5′-TCAGCAAAAAACCCCTC-3′) oligonucleotide primers. Additional details of plasmid construction are available upon request.
Recombinant plasmids and empty vectors (negative controls) were transformed into Escherichia coli BL21 cells (Invitrogen). The expression of respective proteins was induced, cells were sonicated, and GST fusion proteins were isolated from cell lysate by immobilization on Glutathione-Sepharose 4B beads (Amersham Biosciences). All cultures were grown in liquid medium at 37°C, and the following specific induction conditions were used: GST protein, 0.5 mm isopropyl β-D-thiogalactopyranoside (IPTG) for 2 h; GST-HaDREB2 fusions (1, 2, and 3), 1 mm IPTG for 5 h; and HaHSFA9 fusions, 0.5 mm IPTG for 4 h. Lysis buffer contained 10 mm Tris-HCl (pH 8.0), 0.2 m NaCl, 5% (v/v) glycerol, 1% (v/v) Triton X-100, 0.1 mm phenylmethylsulfonylfluoride, and 1× complete protease inhibitor cocktail (Roche). Aliquots of the soluble protein extracts were stored at −80°C. The Sepharose beads were washed four times using lysis buffer without Triton X-100 and stored at 4°C as 50% (v/v) slurry until use.
Different amounts of each soluble extract and Sepharose beads with GST, or the tested GST:HaDREB2 fusion, were incubated for 1 h at 4°C in an orbital shaker. We normalized the amount of total protein for each combination using Coomassie-Blue-R-stained gels (data not shown) or immunodetection. When the 6×His deletion fusion was expressed at a much higher level than the complete fusion protein (i.e. Fig. 6, AII), the amount of GST fusion protein used (1) was reduced proportionally. The final amounts of protein used were: I, 750 μg; II, 42 μg; III, 950 μg; and IV, 8,250 μg. We used 135 μg of proteins 1, 2, and 3, except when 6 μg of 1 were incubated with the II extracts. Incubations were in a total volume of 750 μL in lysis buffer without Triton X-100 and supplemented with 1% (v/v) Nonidet NP-40. Beads were washed five times, each time with 1 mL of buffer. After the last wash, proteins immobilized on the sedimented beads (400 g, 1 min) were released by boiling in 20 μL of 2× Laemmli sample buffer. Proteins were resolved by SDS-PAGE (with 10% [w/v] or 12.5% [w/v] acrylamide), transferred to Hybond-P membranes (Amersham), and analyzed on western blots. Conditions for western-blot analyses were essentially as described by Almoguera et al. (2002). The primary antibody, Anti-XpressTm-HRP (Invitrogen) was used at 1/7,000 dilution for detection of the HaHSFA9 fusions. The secondary antibody, goat anti-mouse IgG (Calbiochem), was used at 1/30,000 dilution.
The nucleotide sequence and conceptual translation of HaDREB2 (Ha, H. annuus) have been deposited in the GenBank/EMBL data libraries under accession number AY508007.
Supplementary Material
Acknowledgments
We thank Drs. Pieter B.F. Ouwerkerk and Annemarie H. Meijer for plasmids pHIS3NX, pINT1, and for experimental advice for one-hybrid cloning. We are thankful to Anabel Rojas and to Sebastián Chavez for helpful manuscript comments, and Victoria Palau for efficient technical assistance.
This work was supported by the Spanish Ministry of Education and Science (grant no. BIO02–01463) and Junta de Andalucía (grant no. CVI148). We also received support from Ph.D. fellowships awarded to J.D.-M. and J.M.E.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Juan Jordano (fraga@cica.es).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.069963.
References
- Almoguera C, Prieto-Dapena P, Jordano J (1998) Dual regulation of a heat shock promoter during embryogenesis: stage-dependent role of heat shock elements. Plant J 13: 437–446 [DOI] [PubMed] [Google Scholar]
- Almoguera C, Rojas A, Díaz-Martín J, Prieto-Dapena P, Carranco R, Jordano J (2002) A seed-specific heat-shock transcription factor involved in developmental regulation during embryogenesis in sunflower. J Biol Chem 277: 43866–43872 [DOI] [PubMed] [Google Scholar]
- Bharti K, von Koskull-Döring P, Bharti S, Kumar P, Tintschl-Körbitzer A, Treuter E, Nover L (2004) Tomato heat stress transcription factor HsfB1 represents a novel type of general transcription coactivator with a histone-like motif interacting with the plant CREB binding protein ortholog HAC1. Plant Cell 16: 1521–1535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carey M (1998) The enhanceosome and transcriptional synergy. Cell 92: 5–8 [DOI] [PubMed] [Google Scholar]
- Carranco R, Almoguera C, Jordano J (1997) A plant small heat shock protein gene expressed during zygotic embryogenesis but noninducible by heat stress. J Biol Chem 272: 27470–27475 [DOI] [PubMed] [Google Scholar]
- Carranco R, Almoguera C, Jordano J (1999) An imperfect heat shock element and different upstream sequences are required for the seed-specific expression of a small heat shock protein gene. Plant Physiol 121: 723–730 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson EH, Rast JP, Oliveri P, Ransick A, Calestani C, Yuh CH, Minokawa T, Amore G, Hinman V, Arenas-Mena C, et al (2002) A genomic regulatory network for development. Science 295: 1669–1678 [DOI] [PubMed] [Google Scholar]
- Döring P, Treuter E, Kistner C, Lyck R, Chen A, Nover L (2000) The role of AHA motifs in the activator function of tomato heat stress transcription factors HsfA1 and HsfA2. Plant Cell 12: 265–278 [PMC free article] [PubMed] [Google Scholar]
- Hobert O (2002) PCR fusion-based approach to create reporter gene constructs for expression analysis in transgenic C. elegans. Biotechniques 32: 728–730 [DOI] [PubMed] [Google Scholar]
- Kotak S, Port M, Ganguli A, Bicker F, von Koskull-Döring P (2004) Characterization of C-terminal domains of Arabidopsis heat stress transcription factors (Hsfs) and identification of a new signature combination of plant class A Hsfs with AHA and NES motifs essential for activator function and intracellular localization. Plant J 39: 98–112 [DOI] [PubMed] [Google Scholar]
- Meijer AH, Ouwerkerk PBF, Hoge JHC (1998) Vectors for transcription factor cloning and target site identification by means of genetic selection in yeast. Yeast 14: 1407–1415 [DOI] [PubMed] [Google Scholar]
- Nover L, Bharti K, Döring P, Mishra SK, Ganguli A, Scharf KD (2001) Arabidopsis and the heat stress transcription factor world: how many heat stress transcription factors do we need? Cell Stress Chaperones 6: 177–189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okamuro JK, Caster B, Villarroel R, Van Montagu M, Jofuku KD (1997) The AP2 domain of APETALA2 defines a large new family of DNA binding proteins in Arabidopsis. Proc Natl Acad Sci USA 94: 7076–7081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M, Gann A (2002) Genes and Signals. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
- Quandt K, Frech K, Karas H, Wingender E, Werner T (1995) MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res 23: 4878–4884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reindl A, Schöffl F (1998) Interaction between the Arabidopsis thaliana heat shock transcription factor HSF1 and the TATA binding protein TBP. FEBS Lett 436: 318–322 [DOI] [PubMed] [Google Scholar]
- Riggs CD, Chrispeels MJ (1987) Luciferase reporter gene cassettes for plant gene expression studies. Nucleic Acids Res 15: 8115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A, Almoguera C, Carranco R, Scharf KD, Jordano J (2002) Selective activation of the developmentally regulated Hahsp17.6G1 promoter by heat stress transcription factors. Plant Physiol 129: 1207–1215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rojas A, Almoguera C, Jordano J (1999) Transcriptional activation of a heat shock gene promoter in sunflower embryos: synergism between ABI3 and heat shock factors. Plant J 20: 601–610 [DOI] [PubMed] [Google Scholar]
- Sakuma Y, Liu Q, Dubouzet JG, Abe H, Shinozaki K, Yamaguchi-Shinozaki K (2002) DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem Biophys Res Commun 290: 998–1009 [DOI] [PubMed] [Google Scholar]
- Santoro N, Johansson N, Thiele DJ (1998) Heat shock element architecture is an important determinant in the temperature and transactivation domain requirements for heat shock transcription factor. Mol Cell Biol 18: 6340–6352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinozaki K, Yamaguchi-Shinozaki K, Seki M (2003) Regulatory network of gene expression in the drought and cold stress responses. Curr Opin Plant Biol 6: 410–417 [DOI] [PubMed] [Google Scholar]
- Singh KB (1998) Transcriptional regulation in plants: the importance of combinatorial control. Plant Physiol 118: 1111–1120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, van Montagu M, Verbruggen N (2002) Small heat shock proteins and stress tolerance in plants. Biochim Biophys Acta 1577: 1–9 [DOI] [PubMed] [Google Scholar]
- Wehmeyer N, Vierling E (2000) The expression of small heat shock proteins in seeds responds to discrete developmental signals and suggests a general protective role in desiccation tolerance. Plant Physiol 122: 1099–1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan CX, Gurley WB (2000) Potential targets for HSF1 within the preinitiation complex. Cell Stress Chaperones 5: 229–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.