Abstract
The CYP74B2 gene in Arabidopsis (Arabidopsis thaliana) ecotype Columbia (Col) contains a 10-nucleotide deletion in its first exon that causes it to code for a truncated protein not containing the P450 signature typical of other CYP74B subfamily members. Compared to CYP74B2 transcripts in the Landsberg erecta (Ler) ecotype that code for full-length hydroperoxide lyase (HPL) protein, CYP74B2 transcripts in the Col ecotype accumulate at substantially reduced levels. Consistent with the nonfunctional HPL open reading frame in the Col ecotype, in vitro HPL activity analyses using either linoleic acid hydroperoxide or linolenic acid hydroperoxide as substrates show undetectable HPL activity in the Col ecotype and C6 volatile analyses using leaf homogenates show substantially reduced amounts of hexanal and no detectable trans-2-hexenal generated in the Col ecotype. P450-specific microarrays and full-genome oligoarrays have been used to identify the range of other transcripts expressed at different levels in these two ecotypes potentially as a result of these variations in HPL activity. Among the transcripts expressed at significantly lower levels in Col leaves are those coding for enzymes involved in the synthesis of C6 volatiles (LOX2, LOX3), jasmonates (OPR3, AOC), and aliphatic glucosinolates (CYP83A1, CYP79F1, AOP3). Two of the three transcripts coding for aliphatic glucosinolates (CYP83A1, AOP3) are also expressed at significantly lower levels in Col flowers.
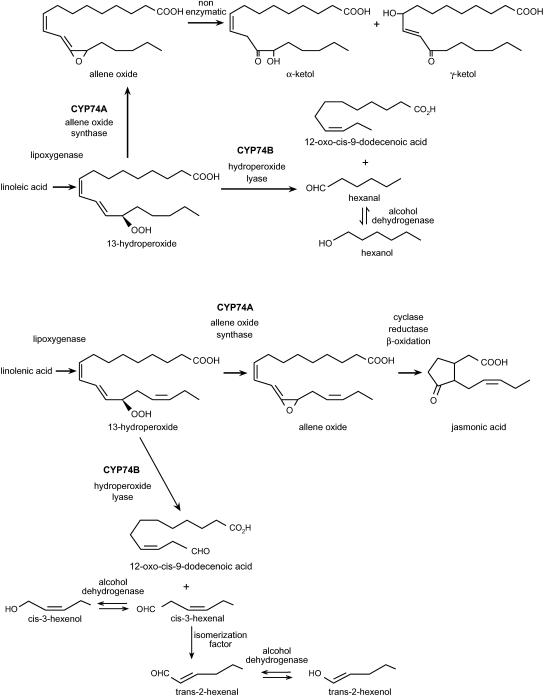
Among the many cytochrome P450 monooxygenase (P450s) genes existing in Arabidopsis (Arabidopsis thaliana; Paquette et al., 2000; Werck-Reichhart et al., 2002; http://arabidopsis-P450.biotec.uiuc.edu; http://www.p450.kvl.dk//p450.shtml), there are two members designated CYP74A1 and CYP74B2 within different branches (subfamilies) of the very small CYP74 family. P450 proteins within this family are unique when compared to most other P450 proteins in that they do not require molecular oxygen or utilize NADPH for dioxygen activation (Song and Brash, 1991; Lau et al., 1993; Shibata et al., 1995), a feature that has often caused them to be described as nonclassical P450 proteins. Also, in contrast to the endoplasmic reticulum localization of most other P450s, gene comparisons have revealed the existence of an N-terminal transit peptide appropriate for chloroplast targeting (Song et al., 1993; Laudert et al., 1996; Bate et al., 1998b). Subcellular fractionations in tomato (Lycopersicon esculentum) have indicated that the CYP74A member of this family in tomato is localized on the inner chloroplast membrane facing the stroma and the CYP74B member of this family is localized in the outer chloroplast membrane facing the intermembrane space (Froehlich et al., 2001); other studies simply localize members of this P450 family to the chloroplast without differentiating between its various subcompartments. Biochemically, each of these enzymes has a distinct function with CYP74A (allene oxide synthase [AOS]) members mediating the synthesis of 12-oxo-phytodienoic acid (12-OPDA), jasmonic acid (JA), and its methyl ester (MeJA) from fatty acid hydroperoxide (HP) in the lipoxygenase (LOX) pathway of oxylipin synthesis (Laudert et al., 1996; Laudert and Weiler, 1998; Kubigsteltig et al., 1999) and CYP74B (hydroperoxide lyase [HPL]) members mediating the breakdown of HP to C6 volatiles and octadecanoic acid (Bate et al., 1998b; Matsui et al., 1999; Fig. 1). As a pivotal enzyme in this LOX pathway, HPL directs the synthesis of a group of C6 volatiles (also called green leaf volatiles [GLV]) that include trans-2-hexenal, trans-2-hexenol, cis-3-hexenal, hexanal, and hexanol, depending on the available HP substrate (Fig. 1), thereby reducing the pools of HP available for production of jasmonates (JAs).
Figure 1.
Oxylipin pathway. In the oxylipin pathway, 13-HPOD is catabolized by CYP74B (HPL) to produce hexanal and 12-oxo-cis-9-dodecenoic acid, and 13-HPOT is catabolized by either CYP74B (HPL) to form cis-3-hexenal and 12-oxo-cis-9-dodecenoic acid or by CYP74A (AOS) to form JA and its derivatives. cis-3-Hexenal is further converted to trans-2-hexenal by an isomerization factor and all C6 aldehydes are interconverted to their alcohol forms by ADHs.
This oxylipin pathway (Farmer et al., 1998; Blee, 2002; Creelman and Mulpuri, 2002) is involved in many developmental and defense pathways, especially those involved in herbivore resistance (Baldwin, 1999), disease resistance (Dong, 1998), and general stress responses (Howe and Schilmiller, 2002). Most of the work contributing to our knowledge of genes regulated by the oxylipin pathway has focused on the AOS-derived JA-signaling branch of this pathway (Wasternack and Parthier, 1997; Liechti and Farmer, 2002; Devoto and Turner, 2003), regardless of the fact that HPL-derived C6 volatiles are well known to exist (Hatanake, 1993) and have been suggested to have roles in the signaling of plant defense processes (Bate and Rothstein, 1998). Evidence implicating this group of volatiles as activators of defense signaling includes the fact that synthetic GLVs elicit the accumulation of phytoalexins in cotton (Gossypium hirsutum; Zeringue, 1992), production of anthocyanins in Arabidopsis (Bate and Rothstein, 1998), and accumulation of the systemin precursor in tomato (Sivasankar et al., 2000), and that terpenoid volatile organic compounds are released from wounded tomato (Farag and Paré, 2002) potentially as a mechanism for interplant signaling. At a molecular level, GLV treatment has been shown to increase transcript levels for a number of genes involved in phenylpropanoid and oxylipin synthesis in Arabidopsis (Bate and Rothstein, 1998) and bean and citrus (Arimura et al., 2000; Gomi et al., 2003), not all of which overlap those induced by JA. This has led to the proposition that C6 volatiles signal the activation of wound-related pathways different from some of those induced by JA and its structural analogs (Bate and Rothstein, 1998). The varying downstream effects of separately silencing the AOS and HPL branches of this pathway support this notion and effectively demonstrate that signaling molecules derived from both branches of this pathway cross-talk in the process of defense signaling (Halitschke et al., 2004).
Studies in Arabidopsis addressing the role of JAs and C6 volatiles in defense responses, especially to bacterial pathogens and insects, have dealt with two intensively studied accessions, Columbia (Col) and Landsberg erecta (Ler), as well as a number of naturally occurring accessions (ecotypes) present in different habitats. Considerable variation that, in fact, exists at the DNA sequence level results in an extensive range of phenotypic variations between these ecotypes in biochemical and morphological characteristics associated with defense responses. Examples of this variation include differences in disease resistance (Kunkel, 1996), leaf trichome density (Larkin et al., 1996), glucosinolate content (Kliebenstein et al., 2001a; Raybould and Moyes, 2001), epicuticular wax composition (Rashotte et al., 1997), and insect resistance (Jander et al., 2001). While complicating molecular analysis of this plant species, these natural variations have provided an imported genetic resource for analyzing gene functions. As evidence of this, we describe here our characterization of a natural, possibly recent, deletion in the coding region of the CYP74B2 gene in the Col ecotype that effectively prevents it from coding for HPL and limits production of C6 volatiles. Comparison of the transcripts expressed in the Col and Ler ecotypes from individual loci in the oxylipin pathway (via reverse transcription [RT]-PCR analysis) and across the entire genome (via microarray and oligoarray analyses) have tied the effects of silencing the CYP74B2 locus to effects on a number of defense genes that are potentially downstream targets for GLV-signaling pathways.
RESULTS
Sequence Polymorphism of CYP74B2 in Col and Ler Ecotypes
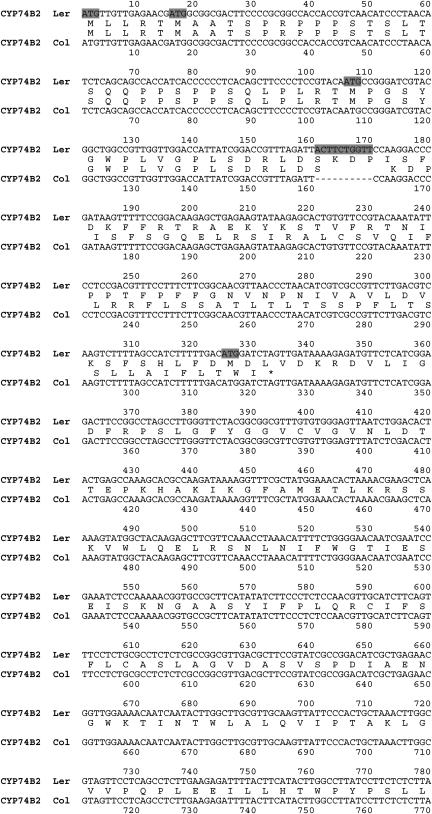
Database analyses have indicated that there are six full-length CYP74B2 cDNA records having GenBank accession numbers AF087932 (Bate et al., 1998b), NM_117633 (reference sequence derived from an annotated genomic sequence NC_003075), BX827042, BX826974, BX826954, and BX826430; because the last four records are individual clones from a single cDNA library, they are considered as one source. Although records from all three sources report that they are derived from the Col ecotype, the last two sets of sequences (NM_117633 and four BX82XXXX) differ in the deletion of 10 nucleotides (nts; gray box in Fig. 2) from the AY087932 coding sequence approximately 53 codons downstream from the translation initiation site. Comparisons with the genomic DNA sequences for the Col ecotype available at The Arabidopsis Information Resource (TAIR) and The Institute for Genomic Research (TIGR; (http://www.arabidopsis.org/; http://www.tigr.org) indicated that these 10 nts are also absent in the sequences of the CYP74B2 gene in the Col ecotype. To further confirm this polymorphism in the Col ecotype, we cloned the CYP74B2 transcripts and genomic DNAs from two of these Arabidopsis ecotypes (Col and Ler) using RT-PCR and PCR amplification strategies with primers directed against the full-length mRNA sequences and genomic DNA from the Wassilewskija (Ws) ecotype using PCR amplification strategies. Sequencing of these clones confirmed the presence of these 10 nts in the Ler and Ws ecotypes, but not in the Col ecotype. The existence of these sequences in the AF087932 cDNA reported to be from the Col ecotype is most likely explained by contamination of Col seed stocks with Ler or Ws seeds or recent deletion of these nucleotides from existing stocks of the Col ecotype.
Figure 2.
CYP74B2 cDNAs in the Col and Ler ecotypes. Alignment of CYP74B2 cDNA sequences from the Col and Ler ecotypes with deduced amino acid sequences shown in between the DNA sequences. Use of the first translation start site in the CYP74B2 transcript from the Col ecotype is predicted to generate a truncated protein of only 110 amino acids due to the 10-nt deletion region that is shaded in gray; the asterisk denotes the stop codon. The four translation start sites preceding the 385-amino acid open reading frame containing a P450 signature sequence are shaded in gray; current TAIR annotation for this locus in the Col genome uses the fourth one in its gene model.
Because of this 10-nt polymorphism, two distinct gene models exist for the Col and Ler CYP74B2 genes. As shown in Figure 2, the first gene model used for the Ler sequence utilizes the first ATG in the mRNA (5′ untranslated region [UTR]; Fig. 2, data not shown) to code for a full-length 492-amino acid protein containing an in-frame P450 signature characteristic of other members of the CYP74 family. Because of the 10-nt deletion existing in the Col ecotype, translation initiation at this ATG would result in production of a C-terminal truncated protein containing only 110 amino acids and lacking a P450 signature sequence. The second gene model currently assigned to the Col sequence by TAIR utilizes the fourth ATG in the mRNA (Fig. 2, gray box) to code for an N-terminal truncated protein containing 385 amino acids and an in-frame P450 signature sequence. Deleted from this second predicted protein, which is substantially shorter than all other known P450 proteins, is the entire transit sequence needed for chloroplast targeting (Bate et al., 1998b) as well as some N-terminal residues found within HPLs of different plant species (Froehlich et al., 2001).
CYP74B2 Expression Levels in Different Ecotypes
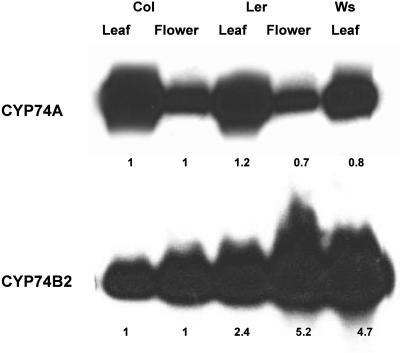
Since the 10-nt deletion in the CYP74B2 cDNA of the Col ecotype predicted that it might generate a short, truncated nonfunctional P450 protein or, less likely, because of multiple upstream translation starts, a longer P450 derivative not capable of targeting to the chloroplast, we speculated that nonsense-mediated decay mechanisms might reduce CYP74B2 transcript levels in the Col ecotype compared to either of the other ecotypes. Because tissue profile data of Arabidopsis P450s (S. Ali, H. Duan, Y. Ferhatoglu, A. Hehn, S. Goepfer, M. Band, D. Werck-Reichhart, and M.A. Schuler, unpublished data) indicated detectable levels of this transcript in leaf and flower tissues of 1-month-old plants of the Col ecotype, expression levels of CYP74B2 transcripts were compared in leaves and flowers of the Col, Ler, and Ws ecotypes using semiquantitative RT-PCR blot assays. As shown in Figure 3, accumulation of CYP74B2 transcripts in Col leaves and flowers are 2.4- to 5.2-fold lower than in Ler leaves and flowers and 4.7-fold lower than in Ws leaves. Consistent with this, P450-specific microarrays and full-genome Arabidopsis oligoarrays (described below) indicate that CYP74B2 transcripts accumulate to higher levels in leaf and flower tissues of the Ler ecotype (Table II; Supplemental Table I).
Figure 3.
CYP74A1 and CYP74B2 transcript levels. Total RNAs isolated from 1-month-old Col leaf (lane 1), Col flower (lane 2), Ler leaf (lane 3), Ler flower (lane 4), and Ws leaf (lane 5) were RT-PCR amplified as outlined in “Materials and Methods” using gene-specific primers for CYP74A1 and CYP74B2 transcripts. The RT-PCR products were electrophoresed on 1.0% agarose gels, blotted to Hybond-n nylon membranes, and probed with 32P-labeled gene-specific fragments. The expression levels for each transcript calculated after normalization to the amount of EF-1α transcript (data not shown) are recorded below each lane relative to the transcript level in corresponding Col tissues.
Table II.
Comparison of transcripts in Col and Ler ecotype using P450 microarrays
| ID | Locus | Normalized Ratio (Col/Ler) | Sampling Sizea | Annotation |
|---|---|---|---|---|
| Normalized ratios for P450 and marker transcripts expressed greater than 2-fold higher in the Col ecotype | ||||
| Flower | ||||
| CYP71A24 | At3g48290 | 25.59 | 8 | Cytochrome P450 |
| CYP71B3 | At3g26220 | 5.24 | 8 | Cytochrome P450 |
| CYP71B4 | At3g26280 | 4.12 | 6 | Cytochrome P450 |
| CYP71B7 | At1g13110 | 2.91 | 7 | Cytochrome P450 |
| CYP71B10 | At5g57260 | 3.32 | 4 | Cytochrome P450 |
| CYP71B26 | At3g26290 | 16.73 | 8 | Cytochrome P450 |
| CYP71B27-1 | At1g13070 | 2.56 | 4 | Cytochrome P450 |
| CYP71B27-2 | At1g13070 | 2.07 | 4 | Cytochrome P450 |
| CYP71B34 | At3g26300 | 2.07 | 8 | Cytochrome P450 |
| CYP72A10-1 | At3g14640 | 9.52 | 4 | Cytochrome P450 |
| CYP72A11-1 | At3g14650 | 4.26 | 8 | Cytochrome P450 |
| CYP72A11-2 | At3g14650 | 5.41 | 8 | Cytochrome P450 |
| CYP72A12P | At3g14640 | 4.29 | 8 | Cytochrome P450; pseudogene |
| CYP77A5P | N/A | 12.46 | 8 | Cytochrome P450; pseudogene |
| CYP77A9-2 | At5g04630 | 4.37 | 4 | Cytochrome P450 |
| CYP81D10-A | At1g66540 | 18.72 | 8 | Cytochrome P450 |
| CYP81D10-C | At1g66540 | 7.11 | 6 | Cytochrome P450 |
| CYP81D5 | At4g37320 | 5.65 | 7 | Cytochrome P450 |
| CYP81F1 | At4g37430 | 2.16 | 8 | Cytochrome P450 |
| CYP81H1 | At4g37310 | 22.41 | 8 | Cytochrome P450 |
| CYP81K1 | At5g10610 | 7.11 | 8 | Cytochrome P450 |
| CYP88A3 | At1g05160 | 4.31 | 8 | Cytochrome P450; multifunctional ent-kaurenoic acid oxidase in GA synthesis |
| CYP89A5 | At1g64950 | 4.50 | 7 | Cytochrome P450 |
| CYP94C1 | At2g27690 | 2.14 | 6 | Cytochrome P450 |
| CYP96A10-A | At4g39490 | 2.35 | 7 | Cytochrome P450 |
| CYP97B3 | At4g15110 | 4.96 | 8 | Cytochrome P450 |
| CYP701A3 | At5g25900 | 2.67 | 8 | Cytochrome P450; multifunctional ent-kaurene oxidase in GA synthesis |
| CYP706A1 | At4g22690 | 3.97 | 8 | Cytochrome P450 |
| CYP707A2 | At2g29090 | 2.32 | 4 | Cytochrome P450; 8′-hydroxylase for ABA in ABA degradation |
| CYP710A1 | At2g34500 | 2.91 | 4 | Cytochrome P450 |
| 4CL-like1-A | At4g19010 | 3.62 | 8 | Putative 4-CL |
| AC12E03 | At5g24780 | 2.61 | 8 | Vegetative storage protein (Vsp1) |
| AF111711 | N/A | 2.66 | 8 | Transcription factor-pan1 |
| ATR3 | At3g02280 | 2.93 | 5 | NADPH-ferrihemoprotein reductase-like |
| CHI1 | At1g53520 | 2.37 | 8 | Putative CHI |
| EST AA651310 | At5g28350 | 2.12 | 8 | Putative protein |
| MSU015H01 | At5g24770 | 2.11 | 6 | Vegetative storage protein (Vsp2) |
| MSU015H07 | At1g58807 | 2.15 | 6 | Putative disease-resistant protein (CC-NBS-LRR class) |
| MSU042C12 | At5g26220 | 2.05 | 8 | Putative protein |
| MSU043A08 | At1g10130 | 2.12 | 8 | Calcium-transporting ATPase 3, endoplasmic reticulum type |
| MSU096C08 | At2g16980 | 2.15 | 8 | Putative tetracycline transporter protein |
| Leaf | ||||
| CYP71B2 | At1g13080 | 2.31 | 5 | Cytochrome P450 |
| CYP71B3 | At3g26220 | 4.94 | 7 | Cytochrome P450 |
| CYP71B4 | At3g26280 | 3.01 | 7 | Cytochrome P450 |
| CYP71B7 | At1g13110 | 20.39 | 8 | Cytochrome P450 |
| CYP71B24 | At3g26230 | 27.93 | 4 | Cytochrome P450 |
| CYP71B26 | At3g26290 | 9.62 | 4 | Cytochrome P450 |
| CYP71B34 | At3g26300 | 2.38 | 6 | Cytochrome P450 |
| CYP72A11-1 | At3g14650 | 7.42 | 8 | Cytochrome P450 |
| CYP72A11-2 | At3g14650 | 4.73 | 6 | Cytochrome P450 |
| CYP72A12P | At3g14640 | 4.30 | 8 | Cytochrome P450; pseudogene |
| CYP72A13 | At3g14660 | 2.13 | 8 | Cytochrome P450 |
| CYP76C1 | At2g45560 | 8.65 | 4 | Cytochrome P450 |
| CYP83B1-1 | At4g31500 | 2.13 | 8 | Cytochrome P450; oxidation of indole-3-acetyldoxime in indole glucosinolate synthesis |
| CYP83B1-2 | At4g31500 | 2.37 | 7 | Cytochrome P450; oxidation of indole-3-acetyldoxime in indole glucosinolate synthesis |
| CYP89A4 | At2g12190 | 2.34 | 4 | Cytochrome P450 |
| CYP89A5 | At1g64950 | 4.28 | 4 | Cytochrome P450 |
| CYP96A10-B | At4g39490 | 2.40 | 7 | Cytochrome P450 |
| CYP96A12 | At4g39510 | 2.26 | 4 | Cytochrome P450 |
| CYP97B3 | At4g15110 | 2.74 | 4 | Cytochrome P450 |
| CYP98A9 | At1g74550 | 3.47 | 8 | Cytochrome P450 |
| CYP701A3 | At5g25900 | 4.77 | 4 | Cytochrome P450; multifunctional ent-kaurene oxidase in GA synthesis |
| CYP706A1 | At4g22690 | 2.10 | 6 | Cytochrome P450 |
| CYP708A3 | At1g78490 | 2.84 | 7 | Cytochrome P450 |
| CYP734A1 | At2g26710 | 6.47 | 4 | Cytochrome P450; 26-hydroxylase for brassinolide and castasterone in brassinolide degradation |
| 4CL-like1-A | At4g19010 | 2.15 | 4 | Putative 4-CL |
| AA02E09 | At2g33340 | 2.01 | 8 | PRP19-like splicing factor |
| AA06G01 | At3g26650 | 2.20 | 8 | Glyceraldehyde 3-P dehydrogenase A subunit (GapA) |
| AA08B05 | At5g41800 | 2.13 | 8 | Amino acid permease-like protein; Pro transporter-like protein |
| AA08E09 | AT2G36880 | 4.06 | 8 | S-adenosylmethionine synthetase |
| AA11D03 | At5g12860 | 2.48 | 8 | 2-Oxoglutarate/malate translocator precursor-like protein |
| AA12A11 | At4g05420 | 2.02 | 8 | UV-damaged DNA-binding factor-like protein |
| AA12B12 | At4g05070 | 2.10 | 8 | cDNA T44741 |
| AA17E07 | At1g08830 | 3.18 | 8 | Copper/zinc superoxidase dismutase (CSD1) |
| AA18E09 | At1g75210 | 4.44 | 8 | Putative cytosolic IMP-GMP specific 5′-nucleotidase |
| AA18H07 | At2g26430 | 4.08 | 8 | Putative cyclin |
| AB07D05 | At1g02930 | 3.74 | 8 | Putative glutathione transferase |
| AB07E06 | At3g47340 | 13.92 | 6 | Glutamine-dependent Asn synthetase |
| AB07F05 | N/A | 2.07 | 8 | N/A |
| AC01H11 | At1g20010 | 2.52 | 8 | Putative β-tubulin 1 |
| AC08F11 | At3g46970 | 3.33 | 8 | Starch phosphorylase |
| AC12D07 | At1g12090 | 4.14 | 8 | pEARLI 1-like protein |
| AF06H11 | At5g65310 | 2.55 | 8 | Homeobox-Leu zipper protein (ATHB-5) |
| AF07C12 | At5g11110 | 2.48 | 7 | Suc-P synthase-like protein |
| AF111711 | N/A | 5.44 | 4 | Transcription factor-pan1 |
| ATR3 | At3g02280 | 3.30 | 4 | NADPH-ferrihemoprotein reductase-like |
| EST AA651310 | At5g28350 | 4.34 | 4 | putative protein |
| MSU035F12 | At1g02840 | 3.28 | 8 | SRp34/SR1 splicing factor |
| MSU045G04 | At1g61740 | 2.37 | 8 | Unknown protein |
| MSU045G08 | At2g27710 | 2.94 | 8 | 60S Ribosomal protein P2 |
| MSU056C10 | At2g33210 | 2.51 | 8 | Mitochondrial chaperonin (HSP60) |
| MSU058D10 | At5g42250 | 2.48 | 7 | ADH |
| MSU067C01 | At1g09830 | 2.26 | 7 | Putative phosphoribosylglycinamide synthetase |
| MSU071D04 | At5g47560 | 2.11 | 6 | Sodium-dicarboxylate cotransporter-like |
| TGA3 | At1g22070 | 2.97 | 6 | Transcription factor |
| TGA5/OBF5 | At5g06960 | 2.60 | 6 | Transcription factor HBP-1b |
| Normalized ratios for P450 and marker transcripts expressed greater than 2-fold higher in the Ler ecotype | ||||
| Flower | ||||
| CYP71B11-B | At5g25120 | 0.46 | 5 | Cytochrome P450 |
| CYP71B12 | At5g25130 | 0.34 | 4 | Cytochrome P450 |
| CYP71B13 | At5g25140 | 0.38 | 8 | Cytochrome P450 |
| CYP71B31 | At3g53300 | 0.32 | 6 | Cytochrome P450 |
| CYP71B38 | At3g44250 | 0.39 | 6 | Cytochrome P450 |
| CYP74B2 | At4g15440 | 0.26 | 8 | Cytochrome P450; HPL1 |
| CYP83A1 | At4g13770 | 0.12 | 8 | Cytochrome P450; oxidation of Met-derived oximes in aliphatic glucosinolates |
| CYP86C4 | At1g13150 | 0.38 | 8 | Cytochrome P450 |
| CYP703A2 | At1g01280 | 0.46 | 6 | Cytochrome P450 |
| CYP707A4 | At3g19270 | 0.37 | 7 | Cytochrome P450; 8′-hydroxylase for ABA in ABA degradation |
| CYP709B2 | At2g46950 | 0.49 | 8 | Cytochrome P450 |
| CYP714A1 | At5g24910 | 0.35 | 5 | Cytochrome P450 |
| CYTB5-1 | At2g46650 | 0.24 | 7 | Putative cytochrome b5 protein |
| Leaf | ||||
| CYP71B17 | At3g26160 | 0.29 | 4 | Cytochrome P450 |
| CYP74B2 | At4g15440 | 0.30 | 5 | Cytochrome P450; HPL1 |
| CYP78A6 | At2g46660 | 0.47 | 4 | Cytochrome P450 |
| CYP78A9 | At3g61880 | 0.47 | 4 | Cytochrome P450 |
| CYP81F2 | At5g57220 | 0.34 | 4 | Cytochrome P450 |
| CYP81G1 | At5g67310 | 0.38 | 8 | Cytochrome P450 |
| CYP83A1 | At4g13770 | 0.43 | 8 | Cytochrome P450; oxidation of Met-derived oximes in aliphatic glucosinolates |
| CYP96A15 | At1g57750 | 0.11 | 4 | Cytochrome P450 |
| CYP706A2 | At4g22710 | 0.47 | 4 | Cytochrome P450 |
| CYP706A5 | At4g12330 | 0.39 | 4 | Cytochrome P450 |
| CYP707A3 | At5g45340 | 0.36 | 4 | Cytochrome P450; 8′-hydroxylase for ABA in ABA degradation |
| AA03H10 | At2g41410 | 0.41 | 8 | Calmodulin-like protein |
| AF02E02 | At2g34600 | 0.35 | 8 | Hypothetical protein |
| PR1-similar A | At2g19990 | 0.42 | 5 | PR protein |
Number of spots out of eight contributing to the normalized ratio; microarray signals derived from less than four of eight normalized ratios are not statistically significant and have been excluded from these datasets.
CYP74B2 Enzyme Activities and C6 Volatile Production in Different Ecotypes
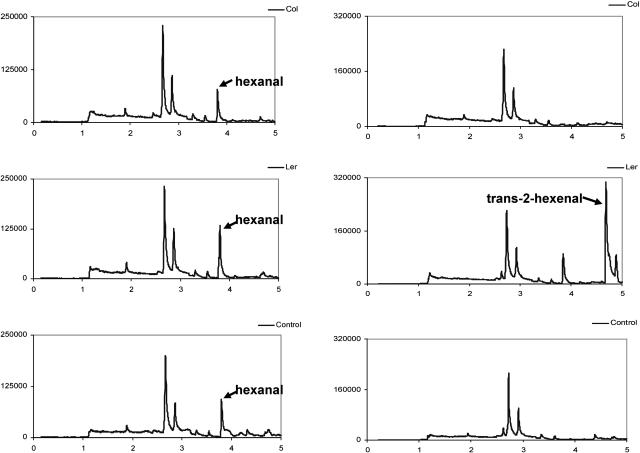
To determine whether the differences in coding and transcript levels potentially reduce the level of functional HPL due to the disruption of the single-copy CYP74B2 gene in the Col ecotype, HPL activities and C6 volatile emissions (representing products of the HPL pathway) were measured in leaf extracts of 1-month-old plants of both ecotypes. In the first of two methods used for assaying HPL activity in this study, indirect coupled-enzyme assays that distinguish between HPL and AOS activities by monitoring for the oxidation of NADH to NAD at A340 with linoleic acid HP (13-HPOD) and linolenic acid HP (13-HPOT) substrates (Vick, 1991) detect significant HPL activity in the Ler ecotype (4.85 and 35.35 with units presented as 10−2 times the change in A340 min−1 mg−1 protein for HPOD and HPOT, respectively) and none in the Col ecotype (Table I). In the second of two methods, direct enzyme assays that measure aldehydes generated by leaf homogenates supplemented with HP substrates detect significant HPL activity (9.6 μg min−1 mg−1 protein) in the Ler ecotype and no detectable activity in the Col ecotype when HPOT was used as substrate, and trans-2-hexenal production is monitored by headspace gas chromatography (GC). However, when HPOD was used as substrate, hexanal production was detected in the Col ecotype (2.6 μg min−1 mg−1 protein) at approximately one-half that in the Ler ecotype (4.3 μg min−1 mg−1 protein). Control reactions containing HPOD alone without leaf homogenates indicate that the lower levels of hexanal produced in assays conducted with the Col ecotype are due to the autooxidation/instability of the HPOD substrate in our assay system (Table II; Fig. 4). In agreement with this conclusion, previous studies in which HPOD was incubated alone (Grosch, 1987) or in Escherichia coli extracts (Bate et al., 1998b) have also indicated that this compound readily autooxidizes to form hexanal.
Table I.
Indirect assay for HPL activities in Col and Ler ecotypes
HPL activity values are the averages derived from four independent assays on leaves from 1-month-old individual plants of the specified ecotype. Activities are presented as 10−2 times the change in A340 min−1 mg−1 total protein as in Vick (1991). UD, Undetectable.
| Extract
|
Volatiles Produced (μg min−1 mg−1 Protein)
|
|
|---|---|---|
| Hexanal (HPOD) | trans-2-Hexenal (HPOT) | |
| Col | 4.3 ± 0.3 | UD |
| Ler | 2.6 ± 0.3 | 9.6 ± 1.1 |
| Control | 2.8 ± 0.2 | UD |
Figure 4.
Representative chromatograms of volatile compounds from direct assay of HPL activity. Volatile compounds from the headspace of tissue extracts (Col or Ler ecotypes) or buffer (control) incubated with either 13-HPOD (left) or 13-HPOT (right) substrates were analyzed by GC as outlined in “Materials and Methods.” Labeled peaks were identified on the basis of retention time by comparison with authentic standards and by MS.
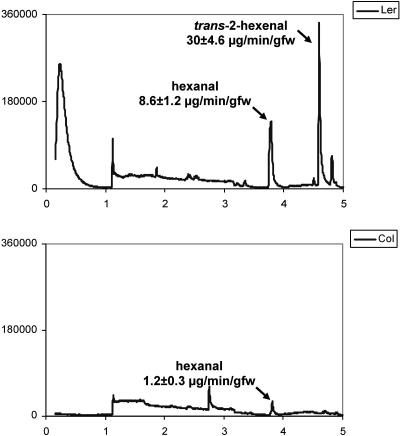
To determine the in vivo HPL activities in Col and Ler ecotypes, endogenous volatile production in 4-week-old leaves was also monitored with hexanal production used as a diagnostic of HPL activity for the endogenous HPOD substrate and trans-2-hexenal production used as a diagnostic of HPL activity for the endogenous HPOT substrate. In both this in vivo assay and the in vitro assay described before, the direct product of HPL metabolism of the HPOT substrate, cis-3-hexenal, is not directly detected in our system because it is isomerized readily to the more stable trans-2-hexenal. As shown in Figure 5, trans-2-hexenal is produced at a rate of 30 μg min−1 g−1 fresh weight in the Ler ecotype and is undetectable in the Col ecotype. Contrasting with this, hexanal is produced at 8.6 μg min−1 g−1 fresh weight and 1.2 μg min−1 g−1 fresh weight in the Ler and Col ecotypes, respectively. Based on the in vitro assays described above, the low levels of hexanal production in the Col ecotype are best explained by the autooxidation of endogenous HPOD, leading us to conclude that volatile production is significantly compromised in the Col ecotype.
Figure 5.
Volatile production in Col and Ler ecotypes. Volatile compounds from leaf tissue of Col (bottom) and Ler (top) 4-week-old plants. The production levels (in units of μg min−1 g −1 fresh weight) averaged from four individual plants are listed under the name of each labeled compound. Labeled peaks were identified on the basis of retention time by comparison with authentic standards and by MS. The quantities of volatiles generated are recorded relative to known quantities of standard compounds.
Transcript Profiling in Col and Ler Ecotypes
Because expression of HPL derived from the CYP74B2 locus is essential for production of C6 volatile compounds that serve as signaling molecules for insect defense and pathogen resistance, we further investigated variations in the levels of other transcripts that potentially arise from depletion of the CYP74B2 transcript and HPL activity in the Col ecotype. Using a P450 gene-specific microarray containing short (400 nt) elements for 265 of 272 Arabidopsis P450 loci (genes and pseudogenes) and 43 biochemical pathway marker loci and longer expressed sequence tag elements for 322 physiological function marker loci (S. Ali, H. Duan, Y. Ferhatoglu, A. Hehn, S. Goepfer, M. Band, D. Werck-Reichhart, and M.A. Schuler, unpublished data), we compared the gene expression profile in flower and leaf tissues of Col and Ler ecotypes. Table II shows that 37 P450 transcripts were expressed higher (2-fold cutoff) in the Col ecotype and 21 P450 transcripts were expressed higher in the Ler ecotype. Among 37 P450 transcripts expressed higher in Col, 27 were expressed higher in Col flower tissue and 22 were expressed higher in Col leaf tissue, with 12 from these two datasets overlapping (five CYP71B subfamily members, CYP72A11, and the CYP72A12P pseudogene in its 3′ UTR, CYP89A5, CYP97B3, CYP701A3, and CYP706A1). Of these 37 P450 transcripts, five code for defined biochemical functions with CYP88A3 and CYP701A3 being multifunctional kaurenoic and kaurene oxidases in GA synthesis (Helliwell et al., 1998, 1999, 2001), CYP83B1 mediating oxidations of indole-3-acetyldoxime in indole glucosinolate synthesis (Bak and Feyereisen, 2001; Bak et al., 2001; Naur et al., 2003), CYP707A2 mediating 8′-abscisic acid (ABA) hydroxylation in ABA degradation (Kushiro et al., 2004; Saito et al., 2004), and CYP734A1 mediating 26-hydroxylations on brassinolide and castasterone in brassinolide degradation (Neff et al., 1999; Turk et al., 2003). As to the 21 P450 transcripts expressed higher in the Ler ecotype, 12 were from flower tissue and 11 were from leaf tissue with two from these datasets overlapping (CYP74B2 and CYP83A1). The inclusion of CYP74B2 in the sets of transcripts expressed at higher levels in both types of Ler tissue corroborates the RT-PCR results shown in Figure 3. In addition to CYP74B2, three of the P450 transcripts expressed at higher levels in the Ler ecotype code for defined biochemical functions with CYP83A1, which overlaps in the flower and leaf datasets, mediating oxidations on Met-derived oximes in aliphatic glucosinolate synthesis (Bak and Feyereisen, 2001; Hemm et al., 2003; Naur et al., 2003) and CYP707A3 and CYP707A4 mediating 8′ ABA hydroxylation in ABA degradation (Kushiro et al., 2004; Saito et al., 2004). Both ecotypes overexpress different ranges of P450s with unknown function which, interestingly, contains 15 members of the 37-member CYP71B subfamily (nine overexpressed in Col, six overexpressed in Ler) and seven members of the 18-member CYP81 family (five overexpressed in Col and two overexpressed in Ler).
Among the biochemical pathway and physiological function marker loci represented on our microarrays, transcripts coding for 4-coumarate-CoA ligase (4-CL), chalcone isomerase (CHI), NADPH-dependent P450 reductase (ATR3), glyceraldehyde 3-P dehydrogenase A (GapA), copper/zinc superoxidase dismutase (CSD1), vegetative storage protein (Vsp1), Asn synthetase (ASN1), and a number of other loci were expressed higher in the Col ecotype. In contrast, only transcripts coding for cytochrome b5, a pathogenesis-related 1 (PR1)-similar protein, and two other unassigned proteins were expressed higher in the Ler ecotype; the PR1-similar transcript is related to the plant defense protein expressed in response to pathogen infection and treatment with resistance-inducing compounds (Laird et al., 2004).
To gain a broader perspective on the differences between the Col and Ler transcript profiles, we compared the gene expression profile in flower and leaf tissues using the Arabidopsis oligomer array containing 26,101 elements for 23,668 nonredundant loci (http://www.ag.arizona.edu/microarray). Supplemental Table I shows that, of 151 loci expressed at higher levels in the Col ecotype, 54 were expressed higher in Col flower tissue and 103 loci were expressed higher in Col leaf tissue, with six from these two datasets overlapping. Supplemental Table I shows that, of 469 loci expressed at higher levels in the Ler ecotype, 59 loci (60 elements) were expressed higher in flower tissue and 430 loci (441 elements) were expressed higher in leaf tissue, with 20 from these two datasets overlapping. Among these loci recorded by oligoarray analysis as varying between the two ecotypes, there are just seven P450s including CYP71B23 (which is expressed higher in the Col ecotype) and CYP74B2, CYP79F1, CYP83A1, CYP89A6, CYP94B3, and CYP704B1 (which are expressed higher in the Ler ecotype). Among these, the expression variations of the CYP74B2, CYP83A1, and CYP94B3 transcripts are consistent with results from our P450 microarray array. Variations in the CYP79F1 transcript agree with the trend of overexpression in Ler leaves evident in our P450 microarray analyses but, because its magnitude (1.7 higher in Ler leaves derived from five of eight spots) is below our 2-fold cutoff, this locus has not been included as an overexpressed Ler locus in Table II. Variations in the CYP71B23, CYP89A6, and CYP704B1 transcripts derived from oligoarray analysis are not consistent with P450 microarray lists either because their oligoarray element (i.e. CYP89A6) has potential for cross-hybridizing with several other loci in multimember P450 subfamilies (designated by the -m extension on the CYP89A6-m element name), because their normalized ratios fall just below the 2-fold cutoffs for our P450 microarray analysis (i.e. CYP71B23), or because the low signal levels for particular P450 transcripts (i.e. CYP704B1) did not meet our criteria for normalized ratios derived from at least four of eight microarray replicates.
The other loci showing variations in the two ecotypes on the full-genome oligoarrays are organized in Supplemental Table I based on their functional categories. Among 54 and 59 loci expressed at higher levels in Col and Ler flower tissues, respectively, many code for functions in carbon metabolism, cell defense, protein folding and processing, secondary metabolism, and transcription. Significantly more differences are found in leaf tissues with 103 and 430 loci expressed at higher levels in Col and Ler leaves, respectively, coding for functions in carbon metabolism, cell communication, cell defense, development, hormone regulation, lipid metabolism, protein folding and processing, secondary metabolism, transcription factor, and transport. Most interesting among the set overexpressed in the Ler ecotype are several loci in both branches of the oxylipin pathway. These include three LOXs (LOX2, LOX3, and At1g72520), which generate substrates for both the HPL and AOS branches of oxylipin synthesis, and allene oxide cyclase (AOC) and 12-oxophytodienoate reductase (OPR3), which exist in the AOS branch. Together with the higher expression levels of CYP74B2 (HPL) transcripts and volatiles produced, these data suggest that the levels of JAs are also elevated in Ler leaves compared to Col leaves. Also interesting among the set more highly expressed in the Ler ecotype are several loci in aliphatic glucosinolate synthesis, including CYP79F1, which exists upstream of CYP83A1 (Hansen et al., 2001; Reintanz et al., 2001; Chen et al., 2003), CYP83A1 itself, and 2-oxoglutarate-dependent dioxygenase (AOP3), which exists downstream of CYP83A1 (Kliebenstein et al., 2001b); Trp synthetase (TSB2; Last et al., 1991) also potentially impacts aliphatic glucosinolate synthesis because it is postulated to feed substrates into the aliphatic glucosinolate pathway via CYP79B2 and CYP79B3 (Naur et al., 2003). Contrasting with these glucosinolate synthetic enzymes that are more highly expressed in the Ler ecotype, one locus more highly expressed in the Col ecotype codes for thioglycosyl hydrolase (TGG2, myrosinase), which degrades glucosinolates to release toxic derivatives (Xue et al., 1995).
Transcript Profiling by RT-PCR Analysis
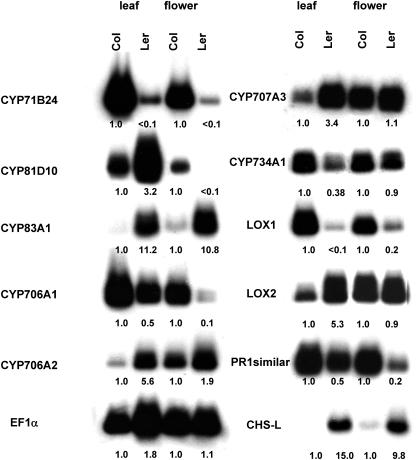
To further clarify the extent of ecotype variation in some of these loci, polyadenylated transcripts in flower and leaf tissues of the Col and Ler ecotypes were RT-PCR amplified using 5′ gene-specific primers and a 3′ oligo(dT) primer complementary to the poly(A) tract present on mature mRNAs using PCR cycle numbers determined to quantitatively amplify each transcript. RT-PCR gel blots hybridized with probes corresponding to the gene-specific microarray elements and normalized against the level of constitutive elongation factor (EF)-1α transcript in each sample provided comparisons between tissues and ecotypes. Following up on our P450 microarray, we analyzed CYP71B24, CYP706A1, CYP81D10, and CYP734A1, which were loci expressed at higher levels in the Col ecotype, as well as CYP83A1, CYP706A2, CYP707A3, and chalcone synthase-like (CHS-L), which were loci expressed at higher levels in the Ler ecotype. We also analyzed expression of the PR1-similar locus, the LOX2 locus that codes for a chloroplast 13-LOX providing substrates for HPL, and the LOX1 locus that codes for a 9-LOX not generating substrates for HPL (Royo et al., 1996; Blee, 2002). As shown in Figure 6, the expression patterns from RT-PCR analyses are generally in agreement with the P450 microarray results: Nearly all genes shown by microarray analysis to be expressed higher in one ecotype were also expressed higher in that ecotype by RT-PCR analysis. But, contrasting with P450 microarray analysis that indicated higher expression in only one tissue of an ecotype, RT-PCR analysis of several of these genes actually indicated high expression in both tissues of a particular ecotype. This is particularly apparent for CYP71B24 transcripts, which were recorded on microarray analysis as expressed at higher levels only in Col leaf tissue but documented in RT-PCR analysis as also expressed at higher levels in Col flower tissue, for CYP81D10 transcripts, which were recorded by microarray analysis as expressed at higher levels only in Col flower tissue but documented in RT-PCR analysis as also expressed at higher levels in Ler leaf tissue (just below our 2-fold cutoff), and CHS-L transcripts, which were not on the P450 microarray list because their sampling size was below our replicate cutoff. We attribute these variations to the greater sensitivity of the RT-PCR amplification assays and the relatively high stringencies that we have used to filter our microarray datasets.
Figure 6.
RT-PCR analysis of some loci differentially expressed in the Col and Ler ecotypes. Total RNAs isolated from 1-month-old Col leaf (lane 1), Col flower (lane 2), Ler leaf (lane 3), and Ler flower (lane 4) were RT-PCR amplified using gene-specific primers for CYP71B24, CYP81D10, CYP83A1, CYP706A1, CYP706A2, CYP734A1, LOX1, LOX2, PR1-similar, CHS-L, and EF-1α transcripts as outlined in “Materials and Methods.” The RT-PCR products were electrophoresed on 1.0% agarose gels, blotted to Hybond-N nylon membranes, and probed with 32P-labeled gene-specific fragments. The expression levels for each transcript calculated after normalization to the amount of EF-1α transcript are recorded below each lane relative to the transcript level in corresponding Col tissues.
For the LOX loci that were not present on our P450 microarray but were present on the oligoarray, LOX2 was expressed at higher levels in leaves of the Ler ecotype in agreement with oligoarray analysis and LOX1 was expressed at higher levels in flowers and leaves of the Col ecotype contrasting with oligoarray analysis, which records little difference between these ecotypes because of P values greater than 0.05. The contrasting expression of these LOX transcripts in these two ecotypes is especially interesting in that it shows reduced expression of LOX2 transcripts in leaves of the Col ecotype that, because of their HPL gene disruption, have no ability to metabolize the 13-hydroperoxides that are the products of the LOX2 protein and other 13-LOXs. Accommodating this deficiency, LOX1 transcripts are enhanced in the leaves of the Col ecotype potentially increasing metabolism of 9-HPs in a compensatory manner.
DISCUSSION
It is now clear that both branches of the oxylipin synthetic pathway mediate plant defense responses to diverse biotic and abiotic stresses (Blee, 2002; Creelman and Mulpuri, 2002; Howe and Schilmiller, 2002) and have potential for cross-talk (Halitschke et al., 2004). We have now demonstrated that the CYP74B2 locus encoding HPL needed for production of C6 volatiles is differentially expressed in the Arabidopsis Col and Ler ecotypes due to a 10-nt polymorphism within the coding sequence of this gene. Eliminating all ability to synthesize full-length HPL protein, this polymorphism does not prevent expression of aberrant HPL transcripts in the Col ecotype. Coupled with the position of this deletion and the presence of a premature stop codon more than 50 nt upstream from the sole splice junction in this transcript, the levels of aberrant HPL transcripts are reduced in the Col ecotype relative to the levels of wild-type HPL transcripts in the Ler and Ws ecotypes, suggesting to us that defective HPL transcripts are destabilized in the Col ecotype by a nonsense-mediated decay pathway (Frischmeyer and Dietz, 1999). Other of our P450 microarray analyses examining the MeJA-induced expression patterns of 7-d-old seedlings of the Col ecotypes indicate that CYP74B2 transcripts, presumably aberrant, accumulate specifically in response to this stress-signaling molecule and not in response to any other chemicals or environmental conditions tested (S. Ali, H. Duan, and M.A. Schuler, unpublished data). Together, these data indicate that basal and MeJA response elements in this promoter are still functional despite inactivation of the CYP74B2 coding sequence and also that the HPL branch of oxylipin synthesis is inducible by products of the AOS branch of oxylipin synthesis.
Our biochemical analyses of HPL activity and volatile production have confirmed the absence of functional HPL activity in the Col ecotype, indicating that this HPL activity is dispensable for normal growth and development and not replaced by an alternate enzyme capable of cleaving 13-HPOD. Branching from a common point in the oxylipin pathway, CYP74B2 and CYP74A1 utilize the same substrate for the production of different types of volatile signaling molecules. Previous work has shown that the relationship between these branches is far beyond that of two similar enzymes competing for the same substrate and suggestive of transcriptional cross-talk between activators of these branched pathways (Halitschke et al., 2004). Initiated with antisense depletion strategies in transgenic potato and tobacco plants that provided the first information on the physiological effects of reducing CYP74B2 expression (Vancanneyt et al., 2001; Halitschke et al., 2004), our discovery of a natural frame-shift deletion in the CYP74B2 locus extends this depletion analysis to analysis of plants completely eliminating functional CYP74B2 activity. We find no evidence for compensation between the HPL and AOS branches of this pathway; when HPL activity is not expressed in the Col ecotype, there is no significant increase in AOS or any of four AOC transcripts needed for synthesis of JA.
Using microarray technology, we have revealed a significant number of differences in transcript levels between the Col and Ler ecotypes, some of which potentially correlate with reduced levels of HPL in the Col ecotype and its consequent effects on the production of C6 volatiles. It is among the loci expressed at higher levels in the Ler ecotype that those regulated by C6 volatiles exist. Comparing the gene expression profiles of the Col and Ler ecotypes, there are clearly more plant defense-related genes expressed at higher levels in the Ler ecotype. The first examples of these are genes involved in the core of the oxylipin pathway and include CYP74B2 itself, At3g45140 (LOX2), At1g17420 (LOX3), At1g72520 (LOX family), and 12-oxophytodienoate reductase (OPR3). Representing three of eight LOX loci in the Arabidopsis genome, these data suggest the LOX proteins encoded by these three genes generate substrates for the HPL branch cleaving 13-HPs and/or that they are activated by the products of a functional HPL pathway. Other LOX loci whose expression patterns do not vary between these ecotypes would be predicted to code for proteins generating substrates for the AOS branch of this pathway and/or metabolize other fatty acid HPs (León et al., 2002; Halitschke and Baldwin, 2003). Underexpression of the LOX2 locus in the Col ecotype, which is induced by both insect damage and JA treatment in other ecotypes (Reymond et al., 2004) and implicated in generating precursors for both branches of the oxylipin pathway, is an especially important example of the autoregulatory cycles operating in this signaling pathway.
The second examples of genes differentially expressed in these ecotypes are cytochrome P450 genes, such as CYP83A1 and CYP79F1, which are clearly involved in the synthesis of aliphatic glucosinolates. The consistency of the low CYP83A1 transcript levels in the Col ecotype in both types of arrays as well as RT-PCR analysis confirm the prediction by Hemm et al. (2003) that allelic variation at the CYP83A1 locus exists among Arabidopsis ecotypes. The contrasting higher transcript levels in the Col ecotype (2.3-fold) of CYP83B1, which mediates synthesis of indole glucosinolates (Bak and Feyereisen, 2001; Bak et al., 2001; Hansen et al., 2001), suggests that compensatory interplay exists between the CYP83A1 and CYP83B1 loci as previously seen in the studies of Hemm et al. (2003) and Naur et al. (2003). The differential expression of CYP83A1 in these ecotypes partially explains the different basal glucosinolate profiles in the Col and Ler ecotypes (Kliebenstein et al., 2001a) and the higher overall glucosinolate content of the Ler ecotype (Jander et al., 2001). Presumably these different basal levels of glucosinolates contribute to variations in ecotype resistance to insect predators as myrosinases release toxic glucosinolate derivatives (Rask et al., 2000). But, in the long run, it is the induced levels of glucosinolates that provide the most effective, sustained protection against insect damage. Consistent with this need, CYP83B1 transcripts were shown to be induced in the Col ecotype in response to herbivore attack and JA application, and, inconsistently, CYP83A1 transcripts showed no response to these effectors (Reymond et al., 2004). In showing that the mutant CYP74B2 protein potentially expressed in the Col ecotype is incapable of generating C6 volatiles and other signaling derivatives even if it was induced, our results provide an explanation for the differential induction of the CYP83A1 and CYP83B1 loci in this earlier study. In this explanation, we postulate that, in some ecotypes such as Ler, the CYP83A1 locus is normally activated by HPL derivatives and the CYP83B1 locus is activated by AOS derivatives synthesized after insect damage. In other ecotypes containing nonfunctional CYP74B2 loci such as the Col ecotype described here, the CYP83A1 locus is incapable of being activated due to the absence of functional HPL and the consequent absence of some C6 volatiles.
Oligoarray analysis has indicated that a number of other defense-related genes are also expressed at higher levels in the Ler ecotype. These include 1-aminocyclopropane-1-carboxylate oxidase (At1g12010), which is a protein involved in ethylene synthesis, and cationic peroxidase (At1g30870) and dehydroascorbate reductase (At1g19570), which are both proteins involved in antioxidant defense. The fact that transcripts from these loci are also induced in herbivore-damaged plants (Reymond et al., 2004) and depleted in the Col ecotype suggests that they also exist downstream of the HPL pathway activated in plant-insect interactions. The immediate effects on insect resistance of inactivating the HPL pathway are complexed by the fact that GLVs have the ability to affect herbivore performance both negatively (by reducing feeding rate, repelling oviposition, or attracting predators; Hildebrand et al., 1993; DeMoraes et al., 2001; Kessler and Baldwin, 2001; Vancanneyt et al., 2001) and positively (by serving as feeding stimulants; Halitschke et al., 2004). Further comparison of the differences in the responses of the Col and Ler ecotypes to herbivore damage will provide more insight into the necessity of having a functional HPL branch in the oxylipin pathway.
MATERIALS AND METHODS
Chemicals
Linoleic acid, linolenic acid, NADH, DEAE-cellulose, soybean lipoxygenase, yeast alcohol dehydrogenase (ADH), hexanal, and trans-2-hexenal were purchased from Sigma.
Plant Materials and Growth Conditions
Arabidopsis (Arabidopsis thaliana) ecotype Col and Ler seeds were surface sterilized with 70% ethanol for 30 s, 12% commercial bleach for 20 min, and washed four times with sterile water prior to plating on one-half-strength Murashige and Skoog agar media (Murashige and Skoog salts plus B5 vitamins [Sigma]), pH 5.7, supplemented with 1% Suc. One-month-old plants were grown on these plates for 1 week at a temperature of 21°C with a 16-h-light/ 8-h-dark cycle, transferred to soil, and grown for an additional 3 weeks under the same temperature and light conditions. For RNA profiling, leaf and flower tissues were harvested from 1-month-old plants, frozen in liquid nitrogen, and stored at −80°C. Total RNA was isolated using TRIzol reagent (Invitrogen) from each of these tissues as described by Duan and Schuler (2005).
Microarray Hybridization and Data Analysis
P450 microarrays containing gene-specific elements for 265 of 272 annotated P450 sequences in the Arabidopsis genome, 48 biochemical pathway markers, and 322 physiological function markers printed at the University of Illinois were hybridized with Cy3- or Cy5-labeled probes (as described by S. Ali, H. Duan, Y. Ferhatoglu, A. Hehn, S. Goepfer, M. Band, D. Werck-Reichhart, and M.A. Schuler, unpublished data). RNA for each ecotype-tissue comparison was analyzed on four microarrays (each containing duplicate spots for each locus), with two technical replicates and two biological replicates. Technical replicates used RNA collected from tissues of different ecotypes grown and harvested at the same time point with dye labeling reversed to avoid incorporation biases and/or differences in recording fluorescence signals. Biological replicates used RNA from tissues grown in independent experiments to minimize biological variations. Oligoarrays containing 70-mer elements for >25,000 nonredundant Arabidopsis loci were printed at the University of Arizona (http://www.ag.arizona.edu/microarray; Galbraith, 2003). With these oligoarrays, RNA for each ecotype-tissue comparison was analyzed on three oligoarrays (each containing a single spot for each locus) with two technical (dye-swapped) replicates for the first biological sample and one sample for the second biological sample. Microarrays were scanned using a Genepix 4000B scanner (Axon Instruments) with the intensities of both Cy5 and Cy3 channels quantified in Genepix 4.0 and data filtering and background cutoffs were done in GeneSpring 7 (Silicon Genetics). Prior to analysis in GeneSpring, the median intensities of each spot were adjusted by subtracting the median intensities of local background and the average intensity of 58 buffer spots present on each microarray (as described by S. Ali, H. Duan, Y. Ferhatoglu, A. Hehn, S. Goepfer, M. Band, D. Werck-Reichhart, and M.A. Schuler, unpublished data). For both types of arrays, the expression ratios for each element were calculated by dividing the average intensities of the Col sample by that for each Ler sample, with Lowess normalization procedures used for chip normalization. In analysis of the P450 arrays, genes were classified as significantly different when the expression ratio calculated from at least four of eight spots was at least 2.0 or at most 0.5 (±2-fold); expression ratios calculated from three or fewer spots were eliminated from further consideration because they were not statistically significant. In the P450 microarrays, multiple elements for the same locus were distinguished with locus extensions of -1, -2, etc., and duplicate sets of the same locus element are distinguished with locus extensions of -A, -B, -C, etc. In analysis of the oligoarrays, genes were classified as significantly different when the expression ratio was at least 2.0 or at most 0.5 with t test P values less than 0.05 calculated from three arrays. In the process of analyzing these oligoarrays, all oligoarray elements were annotated to match locus annotations in TAIR as of December 2004. Multiple elements for the same locus were distinguished with locus extensions of -1, -2, etc., and elements having significant risk of hybridizing with multiple loci (>95% identity across the 70-nt oligomer) were marked with a locus extension of -m.
RNA Quantitation and Genomic DNA Sequencing
Semiquantitative RT-PCR gel-blot analysis of individual P450 transcripts was carried according to Duan and Schuler (2005). Briefly, approximately 0.1 μg total RNA isolated from different tissues was used for one-step RT-PCR amplifications with 21 PCR cycles used for all P450 and marker transcripts and 18 PCR cycles used for EF-Lα transcripts; these cycle numbers were determined to be within the linear amplification range for each of these transcripts. PCR products were fractionated on 1.0% agarose-Tris-borate/EDTA gels, transferred to Hybond-n (Amersham-Pharmacia Biotech), and probed with random hexamer 32P-labeled probes corresponding to the coding sequence of Arabidopsis EF-1α, CYP74A1, and CYP74B2 cDNAs or the 3′ UTR of other P450 genes. After hybridization, the blots were scanned by a Typhoon 8600 variable model imager (Amersham-Pharmacia Biotech) and quantified by ImageQuant 5.1 software. Comparisons between tissue samples were done after normalization to the level of the EF-1α RT-PCR product in each sample. The gene-specific primers used in this analysis were as follows: CYP74A1 5′, 5′-atggcttctatttcaaccccttttcc-3′; CYP74A1 3′, 5′-ctaaaagctagctttccttaacgacgagaa-3′; CYP74B2 5′, 5′-aaccgcatagaaatagaaggc-3′; CYP74B2 3′, 5′-aaactgaagatgcaacgttggagag-3′; CYP71B24 5′, 5′-aaacttgtcccagttcttca-3′; CYP81D10 5′, 5′-gtgcctttgttggaaaaat-3′; CYP83A1 5′, 5′-atggatgtcatgactggtct-3′; CYP706A1 5′, 5′-gaggttgaagagaagtttgg-3′; CYP706A2 5′, 5′-cacttgtggctattcctgtt-3′; CYP707A3 5′, 5′-ggtaatttgcagatggtcaa-3′; CYP734A1 5′, 5′-tcctcaacatggtgcaccaa-3′; CHS-L 5′, 5′-agaaaacgagtggggtctga-3′; PR1 similar 5′, 5′-ggttcgacttggttgtgc-3′; LOX1 5′, 5′-gaagaccatcacagctca-3′; LOX2 5′, 5′-tagtgatggtcacgttgg-3′; oligo(dT), 5′-cggaattcttttttttttttttttt-3′; EF-1α 5′, 5′-accaccaagtactactgcac-3′; and EF-1α 3′, 5′-gacctctcaatcatgttgtc-3′. Probe sequences for each of the P450 and marker transcripts are as follows: CYP74A1, full-length cDNA from the start to stop codon of the At5g42650 locus; CYP74B2, 66 nt upstream to 612 nt downstream from the start codon of the At4g15440 locus; CYP71B24, 61 nt upstream to 351 nt downstream from the stop codon of the At3g26230 locus; CYP81D10, 35 nt upstream to 395 nt downstream from the stop codon of the At1g66540 locus; CYP83A1, 75 nt upstream to 379 nt downstream from the stop codon of the At4g13770 locus; CYP706A1, 96 nt upstream to 380 nt downstream from the stop codon of the At4g22690 locus; CYP706A2, 125 nt upstream to 394 nt downstream from the stop codon of the At4g22710 locus; CYP707A3, 103 nt upstream to 385 nt downstream from the stop codon of the At5g45340 locus; CYP734A1, 52 nt upstream to 395 nt downstream from the stop codon of the At2g26710 locus; CHS-L, 79 nt upstream to 398 nt downstream from the stop codon of the At4g34850 locus; PR1 similar, 103 nt upstream to 304 nt downstream from the stop codon of the At2g19990 locus; LOX1, 310 nt upstream to 26 nt downstream from the stop codon of the At1g55020 locus; and LOX2, 278 nt upstream to 93 nt downstream from the stop codon of the At3g45140 locus.
Genomic CYP74B2 DNAs from Col, Ler, and Ws ecotypes were PCR amplified using the gene-specific primers listed above, cloned into pGEMT-easy vector (Promega), and sequenced with vector primers.
Volatile Production
Volatile compounds derived from leaf tissue of 4-week-old Arabidopsis plants were measured by GC/mass spectrometry (MS) analyses. Sample homogenates were prepared by homogenizing 2 g of diced Arabidopsis leaves in 2 mL of sample buffer (150 mm sodium phosphate, 250 mm sorbitol, 10 mm EGTA, 10 mm magnesium chloride, 1% [v/v] glycerol; Bate et al., 1998a) with mortar and pestle. The homogenate from each sample was transferred to a 5-mL tube and headspace volatiles were sampled with a polydimethylsiloxane SPEM fiber (Supelco) for 20 min at 50°C with constant stirring. Samples were injected into a gas chromatograph (model 6890; Hewlett-Packard) fitted with a mass selective detector (model 5973; Hewlett-Packard). Compounds were separated using a fused silica capillary column (30 m × 0.25 mm × 0.25 μm; Supelco) that was maintained at 40°C for the first minute of the run and programmed to shift 10°C/min up to 120°C, 20°C/min up to 300°C, and then maintained at 300°C for 5 min. Interesting products were identified using internal mass spectra libraries of the detector and authentic standards.
HPL Enzyme Assays
For fatty acid HPL assays, proteins were partially purified from Col and Ler leaves according to Vick (1991), with minor modifications. In brief, 100 mg of fresh leaf tissue of 1-month-old plants were ground in 200 μL of extraction buffer (100 mm potassium phosphate, pH 6.5, 0.1% Triton X-100) in a microfuge tube using a micropestle. The extract was centrifuged at 12,000g for 10 min and the supernatant was applied to a small column of DEAE-cellulose equilibrated with the same extraction buffer. Proteins were eluted with 200 μL of extraction buffer and used immediately in HPL assays. The 13-HPOD or 13-HPOT were prepared in a 45-mL solution containing 87,000 units/mL soybean LOX and 1.3 mm fatty acid substrates with continuous oxygen bubbling for 20 min at room temperature. Both indirect and direct assays were used to determine HPL activity. Indirect HPL assays were run in 1-mL reactions containing 100 mm potassium phosphate, pH 6.5, 0.1 mm NADH, 50 units/mL yeast ADH, 0.1 mm 13-HP of fatty acid, and 50 μL protein eluted from DEAE-cellulose, and the oxidation of NADH was monitored by following the decrease in A340 as described by Vick (1991). For the actual HPL assays, the background oxidation of NADH was measured in reactions lacking the HP substrate using a Cary 100 UV-visible spectrophotometer and reactions were initiated by the addition of substrate and recorded for 5 min. Direct HPL assays were run in 2-mL reactions containing 100 mm potassium phosphate (pH 6.5), 0.1 mm 13-HP substrate (HPOD or HPOT), and 50 μl of enzyme extract in a 5-mL vial and the aldehydes generated were measured by headspace GC/MS analysis as described above. HPL assays for each ecotype were replicated four independent times with tissues derived from individual plants.
Supplementary Material
Acknowledgments
The authors gratefully acknowledge Dr. Shahjahan Ali, Dr. Mark Band, Dr. Daniele Werck-Reichhart, and Mr. Alan Bari for their assistance in construction and analyses of the P450-specific microarrays used in this study. We also acknowledge Dr. Jyothi Thimmapuram, Dr. Dheepa Balasubramanian, and Mr. Dmitri Novikov for revisions in the Arabidopsis oligoarray annotation and statistical analyses of microarray data, Mr. Emerson Lacey for technical assistance in GC/MS analyses, Dr. Pinghua Li for training in oligoarray hybridizations and analyses, Dr. Minsoo Yoon for sequencing of CYP74B2 genomic DNAs, and Ms. Kara Sandfort for help with RT-PCR analyses.
This work was supported by the National Science Foundation (NSF 2010 grant no. MCB 0115068 to M.A.S.).
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Mary A. Schuler (maryschu@uiuc.edu).
The online version of this article contains Web-only data.
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.067249.
References
- Arimura G, Tashiro K, Kuhara S, Nishioka T, Ozawa R, Takabayashi J (2000) Gene responses in bean leaves induced by herbivory and by herbivore-induced volatiles. Biochem Biophys Res Commun 277: 305–310 [DOI] [PubMed] [Google Scholar]
- Bak S, Feyereisen R (2001) The involvement of two P450 enzymes, CYP83B1 and CYP83A1, in auxin homeostasis and glucosinolate biosynthesis. Plant Physiol 127: 108–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bak S, Tax FE, Feldmann KA, Galbraith DW, Feyereisen R (2001) CYP83B1, a cytochrome P450 at the metabolic branch point in auxin and indole glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 101–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baldwin IT (1999) The jasmonate cascade and the complexity of induced defense against herbivore attack. In M Wink, ed, The Role of Plant Secondary Metabolites and Their Utilization in Biotechnology. Sheffield Academic Press, Sheffield, UK, pp 155–186
- Bate NJ, Riley J, Thompson JE, Rothstein SJ (1998. a) Quantitative and qualitative differences in C6-volatile production from the lipoxygenase pathway in an alcohol dehydrogenase mutant of Arabidopsis thaliana. Physiol Plant 104: 97–104 [Google Scholar]
- Bate NJ, Rothstein SJ (1998) C6-volatiles derived from the lipoxygenase pathway induce a subset of defense-related genes. Plant J 16: 561–569 [DOI] [PubMed] [Google Scholar]
- Bate NJ, Sivasankar S, Moxon C, Riley JMC, Thompson JE, Rothstein SJ (1998. b) Molecular characterization of an Arabidopsis gene encoding hydroperoxide lyase, a cytochrome P-450 that is wound inducible. Plant Physiol 117: 1393–1400 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blee E (2002) Impact of phyto-oxylipins in plant defense. Trends Plant Sci 7: 315–322 [DOI] [PubMed] [Google Scholar]
- Chen S, Glawischnig E, Jørgensen K, Naur P, Jørgensen B, Olsen CE, Hansen CH, Rasmussen H, Pickett JA, Halkier BA (2003) CYP79F1 and CYP79F2 have distinct functions in the biosynthesis of aliphatic glucosinolates in Arabidopsis. Plant J 33: 923–937 [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mulpuri R (2002) The oxylipin pathway in Arabidopsis. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD [DOI] [PMC free article] [PubMed]
- DeMoraes CM, Mescher MC, Tunlinson JH (2001) Caterpillar-induced nocturnal plant volatiles repel conspecific females. Nature 410: 577–580 [DOI] [PubMed] [Google Scholar]
- Devoto A, Turner JG (2003) Regulation of jasmonate-mediated plant responses in Arabidopsis. Ann Bot (Lond) 92: 329–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong X (1998) SA, JA, ethylene, and disease resistance in plants. Curr Opin Plant Biol 1: 316–323 [DOI] [PubMed] [Google Scholar]
- Duan H, Schuler MA (2005) Differential expression and evolution of Arabidopsis CYP86A subfamily. Plant Physiol 137: 1067–1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farag MA, Paré PW (2002) C6-green leaf volatiles trigger local and systemic VOC emissions in tomato. Phytochemistry 61: 545–554 [DOI] [PubMed] [Google Scholar]
- Farmer EE, Weber H, Vollenweider S (1998) Fatty acid signaling in Arabidopsis. Planta 206: 167–174 [DOI] [PubMed] [Google Scholar]
- Frischmeyer PA, Dietz HC (1999) Nonsense-mediated mRNA decay in health and disease. Hum Mol Genet 8: 1893–1900 [DOI] [PubMed] [Google Scholar]
- Froehlich JE, Itoh A, Howe GA (2001) Tomato allene oxide synthase and fatty acid hydroperoxide lyase, two cytochrome P450s involved in oxylipin metabolism, are targeted to different membranes of chloroplast envelope. Plant Physiol 125: 306–317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galbraith DW (2003) Global analysis of cell type-specific gene expression. Comp Funct Genomics 4: 208–215 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomi K, Yamasaki Y, Yamamoto H, Akimitsu K (2003) Characterization of a hydroperoxide lyase gene and effect of C6-volatiles on expression of genes of the oxylipin metabolism in citrus. J Plant Physiol 160: 1219–1231 [DOI] [PubMed] [Google Scholar]
- Grosch W (1987) Reactions of hydroperoxide products of low molecular weight. In HWS Chan, ed, Autoxidation of Unsaturated Lipids. Academic Press, New York, pp 95–139
- Halitschke R, Baldwin IT (2003) Antisense LOX expression increases herbivore performance by decreasing defense responses and inhibiting growth-related transcriptional reorganization in Nicotiana attenuate. Plant J 36: 794–807 [DOI] [PubMed] [Google Scholar]
- Halitschke R, Ziegler J, Keinänen M, Baldwin IT (2004) Silencing of hydroperoxide lyase and allene oxide synthase reveals substrate and defense signaling crosstalk in Nicotiana attenuate. Plant J 40: 35–46 [DOI] [PubMed] [Google Scholar]
- Hansen CH, Wittstock U, Olsen CE, Hick AJ, Pickett JA, Halkier BA (2001) Cytochrome P450 CYP79F1 from Arabidopsis catalyzes the conversion of dihomomethionine and trihomomethionine to the corresponding aldoximes in the biosynthesis of aliphatic glucosinolates. J Biol Chem 276: 11078–11085 [DOI] [PubMed] [Google Scholar]
- Hatanake A (1993) The biogeneration of green odor by green leaves. Phytochemistry 34: 1201–1218 [Google Scholar]
- Helliwell CA, Chandler PM, Poole A, Dennis ES, Peacock WJ (2001) The CYP88A cytochrome P450, ent-kaurenoic acid oxidase, catalyzes three steps of the gibberellin biosynthesis pathway. Proc Natl Acad Sci USA 98: 2065–2070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Poole A, Peacock WA, Dennis ES (1999) Arabidopsis ent-kaurene oxidase catalyzes three steps of gibberellin biosynthesis. Plant Physiol 119: 507–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helliwell CA, Sheldon CC, Olive MR, Walker AR, Zeevaart JA, Peacock WJ, Dennis ES (1998) Cloning of the Arabidopsis ent-kaurene oxidase gene GA3. Proc Natl Acad Sci USA 95: 9019–9024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemm MR, Ruegger MO, Chapple C (2003) The Arabidopsis ref2 mutant is defective in the gene encoding CYP83A1 and shows both phenylpropanoid and glucosinolate phenotypes. Plant Cell 15: 179–194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrand DF, Brown GC, Jackson DM, Hamilton-Kemp TR (1993) Effects of some leaf-emitted volatile compounds on aphid population increase. J Chem Ecol 19: 1875–1887 [DOI] [PubMed] [Google Scholar]
- Howe GA, Schilmiller AL (2002) Oxylipin metabolism in response to stress. Curr Opin Plant Biol 5: 230–236 [DOI] [PubMed] [Google Scholar]
- Jander G, Cui J, Nhan B, Pierce NE, Ausubel FM (2001) The TASTY locus on chromosome 1 of Arabidopsis affects feeding of the insect herbivore Trichoplusia ni. Plant Physiol 126: 890–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kessler A, Baldwin IT (2001) Defensive function of herbivore-induced plant volatile emissions in nature. Science 291: 2141–2144 [DOI] [PubMed] [Google Scholar]
- Kliebenstein DJ, Kroymann J, Brown P, Figuth A, Pedersen D, Gershenzon J, Mitchell-Olds T (2001. a) Genetic control of natural variation in Arabidopsis glucosinolate accumulation. Plant Physiol 126: 811–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliebenstein DJ, Lambrix VM, Reichelt M, Gershenzon J, Mitchell-Olds T (2001. b) Gene duplication in the diversification of secondary metabolism: tandem 2-oxoglutarate-dependent dioxygenases control glucosinolate biosynthesis in Arabidopsis. Plant Cell 13: 681–693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubigsteltig I, Laudert D, Weiler EW (1999) Structure and regulation of the Arabidopsis thaliana allene oxide synthase gene. Planta 208: 463–471 [DOI] [PubMed] [Google Scholar]
- Kunkel BN (1996) A useful weed put to work: genetic analysis of disease resistance in Arabidopsis thaliana. Trends Genet 12: 63–69 [DOI] [PubMed] [Google Scholar]
- Kushiro T, Okamoto M, Nakabayashi K, Yamagishi K, Kitamura S, Asami T, Hirai N, Koshiba T, Kamiya Y, Nambara E (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8′-hydroxylases: key enzymes in ABA catabolism. EMBO J 23: 1647–1656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laird J, Armengaud P, Giuntini P, Laval V, Milner JJ (2004) Inappropriate annotation of a key defence marker in Arabidopsis: Will the real PR-1 please stand up? Planta 219: 1089–1092 [DOI] [PubMed] [Google Scholar]
- Larkin JC, Young N, Prigge M, Marks MD (1996) The control of trichome spacing and number in Arabidopsis. Development 122: 997–1005 [DOI] [PubMed] [Google Scholar]
- Last RL, Bissinger PH, Mahoney DJ, Radwanski ER, Fink GR (1991) Tryptophan mutants in Arabidopsis: the consequences of duplicated tryptophan synthase β genes. Plant Cell 3: 345–358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lau SMC, Harder PA, O'Keefe DP (1993) Low carbon monoxide affinity allene oxide synthase is the predominant cytochrome P450 in many plant tissues. Biochemistry 32: 1945–1950 [DOI] [PubMed] [Google Scholar]
- Laudert D, Pfannschmidt U, Lottspeich F, Holländer-Czytko H, Weiler EW (1996) Cloning, molecular and functional characterization of Arabidopsis thaliana allene oxide synthase (CYP 74), the first enzyme of the octadecanoid pathway to jasmonates. Plant Mol Biol 31: 323–335 [DOI] [PubMed] [Google Scholar]
- Laudert D, Weiler EW (1998) Allene oxide synthase: a major control point in Arabidopsis thaliana octadecanoid signalling. Plant J 15: 675–684 [DOI] [PubMed] [Google Scholar]
- León J, Royo J, Vancanneyt G, Sanz C, Silkowski H, Griffiths G, Sánchez-Serrano JJ (2002) Lipoxygenase H1 gene silencing reveals a specific role in supplying fatty acid hydroperoxides for aliphatic aldehyde production. J Biol Chem 277: 416–423 [DOI] [PubMed] [Google Scholar]
- Liechti R, Farmer EE (2002) The jasmonate pathway. Science 296: 1649–1650 [DOI] [PubMed] [Google Scholar]
- Matsui K, Wilkinson J, Hiatt B, Knauf V, Kajiwara T (1999) Molecular cloning and expression of Arabidopsis fatty acid hydroperoxide lyase. Plant Cell Physiol 40: 477–481 [DOI] [PubMed] [Google Scholar]
- Naur P, Petersen BL, Mikkelsen MD, Bak S, Rasmussen H, Olsen CE, Halkier BA (2003) CYP83A1 and CYP83B1, two nonredundant cytochrome P450 enzymes metabolizing oximes in the biosynthesis of glucosinolates in Arabidopsis. Plant Physiol 133: 63–72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neff MM, Nguyen SM, Malancharuvil EJ, Fujioka S, Noguchi T, Seto H, Tsubuki M, Honda T, Takatsuto S, Yoshida S, et al (1999) BAS1: a gene regulation brassinosteroid levels and light responsiveness in Arabidopsis. Proc Natl Acad Sci USA 96: 15316–15323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paquette SM, Bak S, Feyereisen R (2000) Intron-exon organization and phylogeny in a large superfamily, the paralogous cytochrome P450 genes of Arabidopsis thaliana. DNA Cell Biol 19: 307–317 [DOI] [PubMed] [Google Scholar]
- Rashotte AM, Jenks MA, Nguyen TD, Feldman KA (1997) Epicuticular wax variation in ecotypes of Arabidopsis thaliana. Phytochemistry 45: 251–255 [DOI] [PubMed] [Google Scholar]
- Rask L, Andreasson E, Ekbom B, Eriksson S, Pontoppidan B, Meijer J (2000) Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol Biol 42: 93–113 [PubMed] [Google Scholar]
- Raybould AF, Moyes CL (2001) The ecological genetics of aliphatic glucosinolates. Heredity 87: 383–391 [DOI] [PubMed] [Google Scholar]
- Reintanz B, Lehnen M, Reichelt M, Gershenzon J, Kowalczyk M, Sandberg G, Godde M, Uhl R, Palme K (2001) bus, a bushy Arabidopsis CYP79F1 knockout mutant with abolished synthesis of short-chain aliphatic glucosinolates. Plant Cell 13: 351–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Bodenhausen N, Van Poecke RMP, Krishnamurthy V, Dicke M, Farmer EE (2004) A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16: 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royo J, Vancanneyt G, Perez AG, Sanz C, Stormann K, Rosahl S, Sanchez-Serrano JJ (1996) Characterization of three potato lipoxygenases with distinct enzymatic activities and different organ-specific and wound-regulated expression patterns. J Biol Chem 271: 21012–21019 [DOI] [PubMed] [Google Scholar]
- Saito S, Hirai N, Matsumoto C, Ohigashi H, Ohta D, Sakata K, Mizutani M (2004) Arabidopsis CYP707As encode (+)-abscisic acid 8′-hydroxylase, a key enzyme in the oxidative catabolism of abscisic acid. Plant Physiol 134: 1439–1449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shibata Y, Matsui K, Kajiwara T, Hatanaka A (1995) Purification and properties of fatty acid hydroperoxide lyase from green bell pepper fruits. Plant Cell Physiol 36: 147–156 [Google Scholar]
- Sivasankar S, Sheldrick B, Rothstein SJ (2000) Expression of allene oxide synthase determines defense gene activation in tomato. Plant Physiol 122: 1335–1342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song WC, Brash AR (1991) Purification of an allene oxide synthase and identification of the enzyme as a cytochrome P-450. Science 253: 781–784 [DOI] [PubMed] [Google Scholar]
- Song WC, Funk CD, Brash AR (1993) Molecular cloning of an allene oxide synthase: a cytochrome P450 specialized for the metabolism of fatty acid hydroperoxides. Proc Natl Acad Sci USA 90: 8519–8523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turk EM, Fujioka S, Seto H, Shimada Y, Takatsuto S, Yoshida S, Denzel MA, Torres QI, Neff MM (2003) CYP72B1 inactivates brassinosteroid hormones: an intersection between photomorphogenesis and plant steroid signal transduction. Plant Physiol 133: 1643–1653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vancanneyt G, Sanz C, Farmaki T, Paneque M, Ortego F, Castañera P, Sánchez-Serrano JJ (2001) Hydroperoxide lyase depletion in transgenic potato plants lead to an increase in aphid performance. Proc Natl Acad Sci USA 98: 8139–8144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick BA (1991) A spectrophotometric assay for hydroperoxide lyase. Lipids 26: 315–320 [Google Scholar]
- Wasternack C, Parthier B (1997) Jasmonate-signalled plant gene expression. Trends Plant Sci 2: 302–307 [Google Scholar]
- Werck-Reichhart D, Bak S, Paquette S (2002) Cytochrome P450. In CR Somerville, EM Meyerowitz, eds, The Arabidopsis Book. American Society of Plant Biologists, Rockville, MD
- Xue J, Jorgensen M, Pihlgren U, Rask L (1995) The myrosinase gene family in Arabidopsis thaliana: gene organization, expression and evolution. Plant Mol Biol 27: 911–922 [DOI] [PubMed] [Google Scholar]
- Zeringue HJ Jr (1992) Effects of C6-C10 alkenals and alkanals on eliciting a defence response in the developing cotton boll. Phytochemistry 31: 2305–2308 [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.