Abstract
Indirect evidence previously suggested that Arabidopsis (Arabidopsis thaliana) vegetative storage protein (VSP) could play a role in defense against herbivorous insects. To test this hypothesis, other AtVSP-like sequences in Arabidopsis were identified through a Basic Local Alignment Search Tool search, and their transcriptional profiles were investigated. In response to methyl jasmonate application or phosphate starvation, AtVSP and AtVSP-like genes exhibited differential expression patterns, suggesting distinct roles played by each member. Arabidopsis VSP2 (AtVSP2), a gene induced by wounding, methyl jasmonate, insect feeding, and phosphate deprivation, was selected for bacterial expression and functional characterization. The recombinant protein exhibited a divalent cation-dependent phosphatase activity in the acid pH range. When incorporated into the diets of three coleopteran and dipteran insects that have acidic gut lumen, recombinant AtVSP2 significantly delayed development of the insects and increased their mortality. To further determine the biochemical basis of the anti-insect activity of the protein, the nucleophilic aspartic acid-119 residue at the conserved DXDXT signature motif was substituted by glutamic acid via site-directed mutagenesis. This single-amino acid alteration did not compromise the protein's secondary or tertiary structure, but resulted in complete loss of its acid phosphatase activity as well as its anti-insect activity. Collectively, we conclude that AtVSP2 is an anti-insect protein and that its defense function is correlated with its acid phosphatase activity.
Vegetative storage proteins (VSP) are proteinaceous storage reserves that have been identified from numerous plants, such as soybean (Glycine max; Wittenbach, 1983), potato (Solanum tuberosum; Mignery et al., 1984, 1988), sweet potato (Ipomoea batatas; Maeshima et al., 1985), white clover (Trifolium repens; Goulas et al., 2003), alfalfa (Medicago sativa; Meuriot et al., 2004b), and in the bark of deciduous trees such as poplar (Populus deltoides; Coleman et al., 1991) and elderberry (Sambucus nigra; Van Damme et al., 1997). These proteins can accumulate to a high abundance, up to 50% of the total soluble proteins, in various vegetative storage organs. They act as temporary storage of amino acids that can buffer the availability of nitrogen and other nutrients, and biosynthesis and degradation of VSPs are thought to be regulated by temporary storage needs (Staswick, 1994).
The best-characterized VSPs are soybean VSPα and VSPβ (Wittenbach, 1983). They accumulate in leaves of sink-deprived soybean plants and exist in seedling hypocotyls, developing leaves, stems, flowers, and pods of mature plants as well. They are located primarily in vacuoles of the paraveinal mesophyll cells (Franceschi et al., 1983). VSPα/β do not accumulate in seeds and share little sequence similarity with seed storage proteins (Staswick, 1988). Two Arabidopsis (Arabidopsis thaliana) VSPs (approximately 40% identical to the soybean VSP sequences) cross-reacted with anti-soybean VSP antibody and were found in flowers and buds in an abundance similar to the levels of soybean VSP found in young leaves (Berger et al., 1995). Currently, VSPs are thought to serve as a transient reserve that sequesters unused amino acids during plant development. Once new seed production begins, stored VSPs presumably make nitrogen and other nutrients immediately available for seed development. This hypothesis, however, was not supported by transgenic soybean expressing a soybean antisense VSP gene. In this experiment, VSP expression was abolished, but, contrary to what was expected, seed production was unaffected even under nitrogen-depriving conditions (Staswick et al., 2001). Although roles of VSPs during seed development could be compensated for by other storage proteins, such results raised a possibility that VSPs may serve other functions beyond source-sink interaction or plant productivity.
Arabidopsis VSP transcripts are induced by mechanical wounding, jasmonic acid (JA), insect herbivory, and osmotic and nutritional stresses (Mason and Mullet, 1990; Berger et al., 1995, 2002; McConn et al., 1997; Utsugi et al., 1998; Xie et al., 1998; Stotz et al., 2000; Gong et al., 2001; Reymond et al., 2004), a common response shared by many genes encoding anti-insect proteins. Positive correlation between Arabidopsis VSP gene expression and plant resistance to insects has been observed. For instance, the Arabidopsis fad3-2 fad7-2 fad8 triple mutant with abolished Arabidopsis VSP and other wound-regulated gene expression is highly susceptible to insect attack (McConn et al., 1997), while ethylene-insensitive mutants (ein2, ein3, and etr1) that showed higher insect resistance also exhibited enhanced Arabidopsis VSP accumulation (Rojo et al., 1999; Stotz et al., 2000). The Arabidopsis cev1 mutant that constitutively expresses AtVSP1 and AtVSP2 had higher disease resistance (Ellis and Turner, 2001). Although these observations are only correlative, it is tempting to hypothesize that Arabidopsis VSPs may serve as defense proteins.
Despite a previous lack of biochemical evidence, VSPs from Arabidopsis are classified as acid phosphatases of the haloacid dehalogenase superfamily, based on sequence motif analysis (Thaller et al., 1998; Selengut, 2001). Phosphatases are phosphohydrolases that catalyze dephosphorylation of a wide variety of substrates (Duff et al., 1994). Some members of the haloacid dehalogenase superfamily contain the signature motif DXDXT, and this motif is conserved in many acid phosphatases from bacteria and other eukaryotes (Thaller et al., 1998; Collet et al., 1999). Plant acid phosphatases do not normally have high substrate specificity, but are important in the hydrolysis, transport, and recycling of phosphate, as well as energy transfer and metabolic regulation in the plant cell (Duff et al., 1994). Some plant acid phosphatases have been shown to be associated with disease resistance. Expression of Hra28, a putative acid phosphatase gene from Phaseolus vulgaris, is specifically induced by Pseudomonas syringae, a bacterial plant pathogen (Jakobek and Lindgren, 2002). Barley (Hordeum vulgare) acid phosphatase expression was increased in response to powdery mildew (Blumeria graminis) infection (Beßer et al., 2000).
Many plant proteins have dual or multiple roles, such as class I β-glucanases in development and defense (Reymond and Farmer, 1998). Indeed, storage proteins often possess enzymatic as well as other activities. For instance, the legume storage protein vicilins conferred insect resistance (Yunes et al., 1998). Patatin, the storage protein from potato tubers, is a lipid acyl hydrolase (Andrews et al., 1988), whereas sporamin belongs to the superfamily of trypsin inhibitors (Yeh et al., 1997). The 32-kD VSP from alfalfa is a chitinase (Meuriot et al., 2004b). Soybean VSPs have weak acid phosphatase activity (De Wald et al., 1992). The major VSP proteins in the bark of the black mulberry tree (Morus nigra) are jacalin-like lectins (Van Damme et al., 2002). The relationship of the above activities to a storage function is unclear. Notably, however, some of these protein activities have been shown to contribute to plant defense against herbivore insects and plant diseases (Peumans and Van Damme, 1995; Radhamani et al., 1995; Yeh et al., 1997; Yunes et al., 1998; Van Damme et al., 2002; Meuriot et al., 2004b).
Despite the identification of acid phosphatase signature motifs in Arabidopsis VSPs and abundant indirect evidence implying their defense functionality, direct evidence for biochemical and anti-insect activities of the Arabidopsis protein is lacking. In this study, we measured acid phosphatase activity of recombinant AtVSP2 and evaluated its effect on three insect species by incorporating the protein into their diets. Bioassays of the recombinant protein with a site-specific mutation at the DXDXT motif indicated that the anti-insect activity of AtVSP was correlated with its acid phosphatase activity. To our knowledge, this is the first report that unambiguously links an Arabidopsis VSP and its phosphatase activity to anti-insect functionality.
RESULTS
Differential Expression of AtVSP-Like Genes in Response to Methyl Jasmonate and Phosphate Starvation
To define the AtVSP-like gene family in the Arabidopsis genome and to determine their potential functions, a BLASTP search was performed in The Arabidopsis Information Resource (TAIR) database using the AtVSP2 coding region as the query. Nine additional sequences were identified with E values less than 10−8 (Table I). Among the 10 sequences, seven were annotated as acid phosphatases and three as VSPs. Eight of them have been confirmed to be expressed genes (Asamizu et al., 2000; Seki et al., 2002). AtVSP1 and AtVSP2 shared the highest sequence identity, while all contained the DXDXT signature sequence motif.
Table I.
AtVSP-like proteins revealed through BLASTP search
| Gene ID | Identity to AtVSP2 | Annotation | Signal Peptide | cDNA/EST Support | The DXDXT Signature Motif |
|---|---|---|---|---|---|
| At5g24770 | – | AtVSP2, VSP | Yes | Yes | Yes |
| At5g24780 | 82% | AtVSP1, VSP | Yes | Yes | Yes |
| At4g25150 | 41% | Acid phosphatase | Yes | Yes | Yes |
| At5g51260 | 38% | Acid phosphatase | Yes | Yes | Yes |
| At4g29260 | 38% | Acid phosphatase | Yes | Yes | Yes |
| At4g29270 | 36% | Acid phosphatase | Yes | Yes | Yes |
| At2g38600 | 36% | Acid phosphatase | Yes | None | Yes |
| At1g04040 | 38% | Acid phosphatase | Yes | Yes | Yes |
| At5g44020 | 36% | VSP | Yes | Yes | Yes |
| At2g39920 | 23% | Acid phosphatase | None | None | Yes |
Kyte-Doolittle hydropathy plots (data not shown) displayed a hydrophobic region in each of the sequences evaluated that corresponds to the predicted signal peptide, with the exception of At2g39920. Thus, the encoded proteins likely enter the secretory pathway. Although the subcellular location of AtVSP is unknown, soybean VSPs that share similarities in sequence as well as expression pattern with Arabidopsis VSP accumulate in vacuoles (Franceschi et al., 1983), as do many other VSPs (such as plant lectins) that have protective functions against herbivorous insects (Chrispeels and Raikhel, 1991). At2g39920 did not appear to have a signal peptide, suggesting a possible cytoplasmic localization.
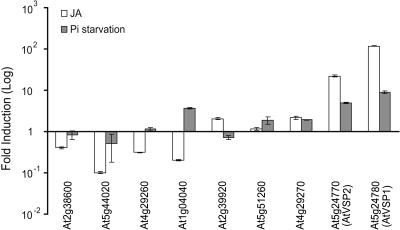
To determine methyl jasmonate (MeJA)- and phosphate deprivation-induced changes in AtVSP-like transcript profiles, real-time reverse transcription (RT)-PCR was carried out after Arabidopsis seedlings underwent MeJA application and phosphate starvation, respectively. Differential responses were detected among the genes tested (Fig. 1). Not all AtVSP-like genes were up-regulated by MeJA and, likewise, not all responded to phosphate starvation. The potential binding site of the PHOSPHATE STARVATION RESPONSE 1 (PHR1) transcription factor has been identified in the AtVSP2 promoter region (Franco-Zorrilla et al., 2004). However, the presence of this cis-element sequence in the promoter regions of AtVSP-like genes did not appear to be correlated with the gene expression patterns we observed. Further, presence of other motifs conserved among phosphate starvation-regulated genes (Mukatira et al., 2001) also was not correlated with regulation by phosphate starvation in our study (data not shown). Such gene expression profiles suggest diverse functionality exerted by these gene products. Alternatively, they may be subject to developmental or tissue-specific regulation. No PCR amplification was detected for At4g25150. One possibility is that this gene is expressed specifically in developing seeds (White et al., 2000). AtVSP1 and AtVSP2 exhibited dramatically higher induction in response to both MeJA and phosphate starvation than the rest, implying a unique function in plant stress responses.
Figure 1.
Differential mRNA expression of AtVSP-like genes. Total RNA was extracted from untreated Arabidopsis seedlings, as well as seedlings treated with MeJA and phosphate starvation, respectively. Gene-specific primers were used in real-time RT-PCR. At4g25150 was excluded from the graph as its primers failed to result in PCR product. Relative transcript abundance was calculated according to Salzman et al. (2005). Transcript levels in control seedlings were arbitrarily set at 1. Error bars indicate sd of the mean induction fold (n = 2).
Expression of Recombinant AtVSP2
Although mounting indirect evidence suggests that Arabidopsis VSPs may act as defense proteins against herbivore insects, direct support for this has not yet been demonstrated. In addition, the ascription of Arabidopsis VSPs as plant counterparts of the bacterial nonspecific acid phosphatases was solely based on the existence of common signature motifs (Thaller et al., 1998). Indeed, many plant acid phostphatases playing a role in phosphate production, transport, and recycling retained this DXDXT signature sequence (Table II). AtVSPs seemed likely to possess this enzymatic activity. To illustrate the biochemical properties and functional roles of AtVSP-like proteins, we selected AtVSP2 as a representative gene for further characterization. This gene was among those with the highest expression change under treatment conditions (Fig. 1) and has repeatedly been reported to respond to biotic and abiotic stresses (Berger et al., 1995, 2002; McConn et al., 1997; Utsugi et al., 1998; Xie et al., 1998; Stotz et al., 2000; Gong et al., 2001; Reymond et al., 2004).
Table II.
Signature motifs and flanking sequences conserved among selected bacterial and plant acid phosphatases and VSPs
| Namea | Organism | Accession | Conserved Motif 1b | Conserved Motif 2 | Reference |
|---|---|---|---|---|---|
| AtVSP2 | Arabidopsis | BAA22096 | 115-WIFDLDDTLLSSIPYY--102--IVGNIGDQWADL-20 | Utsugi et al. (1996) | |
| sAPase | Soybean | CAA11075 | 111-WVFDIDETTLSNLPYY--105--IIGNIGDQWSDL-20 | Penheiter et al. (1998) | |
| AtAPase | Arabidopsis | At4g25150 | 109-WIFDIDETLLSNLPYY--102--IRGNSGDQWSDL-20 | White et al. (2000) | |
| tAPase | Tomato | CAA39370 | 105-WIFDVDETLLSNLPYY--102--IVGNSGDQWSDL-20 | Williamson and Colwell (1991) | |
| BCI3 | Barley | AJ250282 | 122-WVFDIDETTLSNLPYY--101--IVGNIGDQWSDI-20 | Beßer et al. (2000) | |
| SIH5 | Soybean | AB083030 | 83-WILDVDDTCISNIDYY--103--IRGNVGDQWSDL-20 | Hagihara et al. (2004) | |
| Hra28 | Kidney bean | AY055218 | 113-WILDVDDTCISNVSYY--103--IWGNVGDQWSDL-20 | Jakobek and Lindgren (2002) | |
| Leps2 | Tomato | CAD30862 | 5-VVFDFDKTIIEVDSDN--147--RMIYLGDGIGDF-89 | Stenzel et al. (2003) | |
| B-NSAP | Bacteria | P32697 | 65-VGFDIDDTVLFSSPGF--104--IRIFYGDSDNDI-40 | Thaller et al. (1997) | |
| C-NSAP | Bacteria | O05471 | 95-IVLDIDETVLDNSPYQ--97--LIMLFGDNLVDF-65 | Gase et al. (1997) | |
| VSP-α | Soybean | P15490 | 102-FVFSIDGTVLSNIPYY--103--IVGIIGDQWSDL-21 | Mason et al. (1988) | |
| VSP-β | Soybean | P10743 | 103-FIFGIDNTVLSNIPYY--102--IVGIIGDQWSDL-21 | Mason et al. (1988) | |
APase, Acid phosphatase; B-NSAP, bacterial class B nonspecific acid phosphatase; C-NSAP, bacterial class C nonspecific acid phosphatase; BCI3, SIH5, Hra28, and Leps2 are pathogen defense-related acid phosphatases.
The conserved residues are shown in bold. Numbers indicate lengths of amino acid residues.
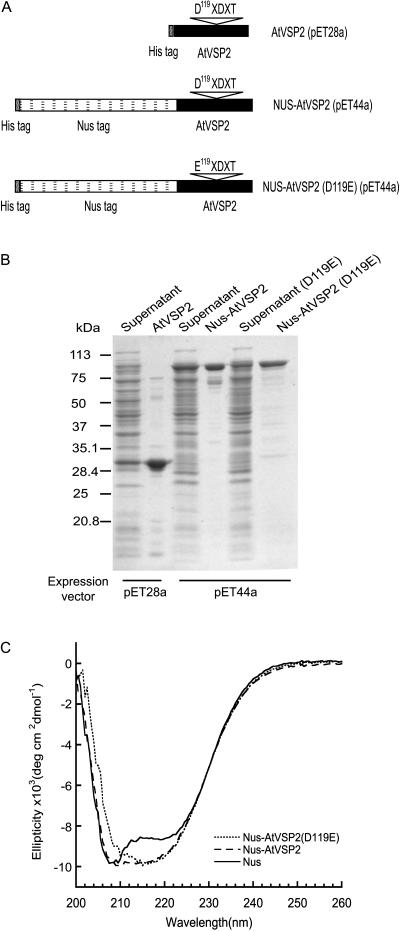
An active soybean nodule acid phosphatase has recently been expressed in the bacterial glutathione S-transferase system (Leelapon et al., 2004), suggesting the feasibility of characterizing this type of proteins using bacterial expression. An AtVSP2 cDNA devoid of the signal peptide was subcloned into a pET28a bacterial expression system for protein expression (Fig. 2A). We were able to detect the recombinant protein in the supernatant of the bacterial extract and subsequently to purify it via Ni2+ affinity chromatography (Fig. 2B). Removal of the signal peptide apparently is critical for protein expression because no recombinant protein was detected when the signal peptide was attached (data not shown).
Figure 2.
Production of recombinant proteins in E. coli. A, Constructs for expression of AtVSP2, Nus-AtVSP2, and Nus-AtVSP2(D119E). While AtVSP2 expressed via pET28a vector was soluble, mutated AtVSP2(D119E) could only be detected in the insoluble fraction. Thus, both cDNAs were cloned into the pET44a vector and proteins were expressed as fusion protein with Nus. B, SDS-PAGE analysis of expression and purification of recombinant proteins. The supernatant of bacterial cell extract and Ni2+ chelate affinity-purified recombinant proteins were subjected to SDS-PAGE and stained with Coomassie Blue. C, Site-specific mutation did not disrupt AtVSP2 structure. Shown are far-UV circular dichroism spectra of Nus-AtVSP2, Nus-AtVSP2(D119E), and Nus.
AtVSP2 Is an Acid Phosphatase
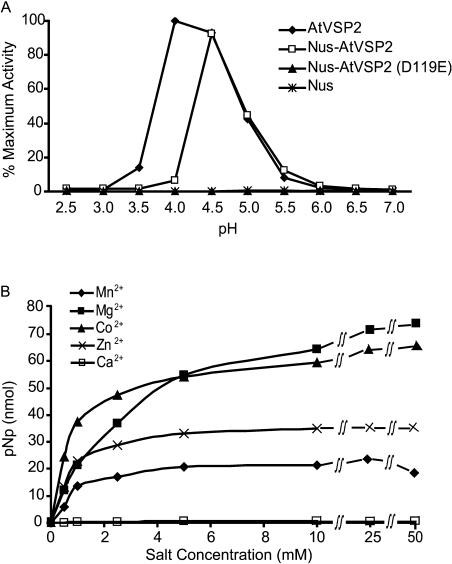
Expression of soluble AtVSP2 recombinant protein enabled biochemical and biological analyses of AtVSP2. Hydrolysis of p-nitrophenyl phosphate (pNPP) indicated that AtVSP2 is indeed an acid phosphatase. Activity kinetics for this substrate were determined: kcat = 22.0 s−1, Km = 14.3 mm, and kcat/Km = 1.5 × 103 m−1 s−1. It appears that the specific activity of AtVSP2 was similar to that of the soybean nodule acid phosphatase expressed in bacteria, and that both activities were significantly higher than soybean VSP activity (Leelapon et al., 2004). As expected, the pH optimum of AtVSP2 was acidic (Fig. 3A).
Figure 3.
AtVSP2 is a pH- and divalent cation-dependent acid phosphatase. A, Reactions were buffered by 50 mm citric acetate (pH 2.5–4.0), sodium acetate (pH 3.5–5.5), and Tris-acetate buffers (pH 5.5–7.5). B, Divalent cations Mg2+, Co2+, Zn2+, Ca2+, or Mn2+ at levels specified were evaluated for effects on phosphatase activity of AtVSP2 in 50 mm sodium acetate (pH 4.5). pNPP at 10 mm was used as substrate and incubated with purified recombinant proteins for 30 min at 37°C. Enzymatic activity was measured by release of p-nitrophenol according to a standard curve.
Metal ions are known to impact acid phosphatase activity (Duff et al., 1994). Here, effectiveness of Mg2+, Co2+, Zn2+, Mn2+, and Ca2+ as cofactors was evaluated at various doses (Fig. 3B). Apparently, enzymatic activity of AtVSP2 is dependent on the presence of divalent cations. They likely function to coordinate substrate-enzyme association as illustrated in phosphoserine phosphatase (Wang et al., 2002). While Mg2+, Co2+, Zn2+, and Mn2+ activated acid phosphatase activity, Ca2+ was ineffective (Fig. 3B), as were monovalent cations K+ and Na+ (data not shown).
AtVSP Has Anti-Insect Activity
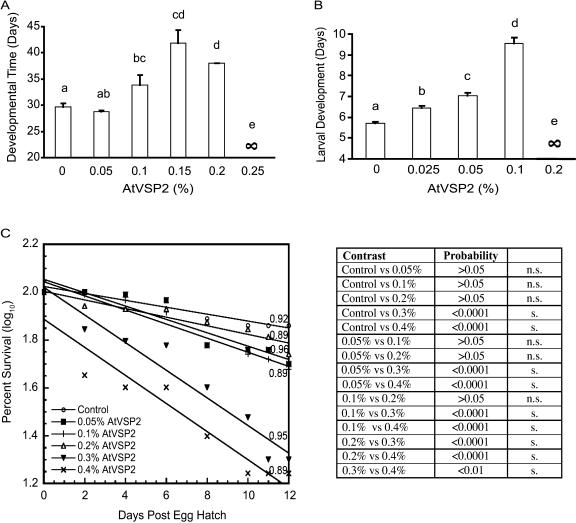
To determine whether AtVSP2 has negative effects when ingested by insects, we incorporated the recombinant protein into their diets. Insect species were selected based on the pH values of their digestive tracts. Many coleopteran, hemipteran, and dipteran insects have an acidic gut pH and use as their major digestive enzymes certain Cys and/or aspartic proteases that function optimally in the acidic pH range. Most lepidopteran insects, on the other hand, display very alkaline midgut contents and use Ser proteases (basic pH optimum) for food protein degradation (Murdock et al., 1987; Terra and Ferreira, 1994). We chose two coleopterans (southern corn rootworm [Diabrotica undecimpunctata howardi] and cowpea [Vigna unguiculata] bruchid [Callosobruchus maculatus]) and one dipteran (Drosophila) for our study (Fig. 4) because the major parts of the digestive tracts of these insects are acidic (Edmonds et al., 1996; Brenner and Atkinson, 1997; Bown et al., 2004), providing an environment that could promote AtVSP phosphatase activity.
Figure 4.
AtVSP2 is an anti-insect protein. A, Artificial seeds containing various doses of AtVSPs were infested with viable cowpea bruchid eggs. Within-seed developmental time was recorded and analyzed by one-way ANOVA. Fisher's protected lsd test (P = 0.05) was used for mean separation. Error bars indicate standard error. Means followed by the same letter are not significantly different at P = 0.05. ∞ indicates 100% mortality under a specified dose. B, Neonate Drosophila melanogaster were reared on AtVSP2 and AtVSP2-free diets. The development time from neonate larvae to pupae were recorded. The effect of AtVSP was analyzed by one-way ANOVA and Bonferroni multiple means comparison tests. Error bars indicated standard error. Means followed by the same letter were not significantly different at P = 0.05. ∞ indicates 100% mortality under a specified dose. C, Newly hatched southern corn rootworm larvae were reared in a diet containing AtVSP2 at doses indicated. Survival data were recorded as insects developed and were log10 transformed. The regression analysis was used to calculate regression equations and for each treatment; control, y = −0.01541x + 2.01737 (P < 0.0001); 0.05% AtVSP2, y = −0.02963x + 2.04037 (P < 0.0001); 0.1% AtVSP2, y = −0.03128x + 2.02984 (P < 0.0001); 0.2% AtVSP2, y = −0.01877x + 1.99290 (P < 0.0001); 0.3% AtVSP2, y = −0.06101x + 1.98955 (P < 0.0001); 0.4% AtVSP2, y = −0.06072x + 1.84539 (P < 0.0001). Linear regression R2 values were indicated in the graph. Comparative analysis was performed between each treatment. A probability of 0.05 was used to determine statistical significance. s, Significant; n.s., nonsignificant.
Developmental time is a useful measure of growth inhibition in cowpea bruchid (egg to adult) and Drosophila (neonate to pupa). Results from testing of both organisms showed that developmental time was directly correlated to AtVSP2 dose (Fig. 4, A and B). For southern corn rootworm, however, the assay was not applicable because larvae usually became infected by a fungus after rearing on an artificial diet for 2 weeks, leading to total death prior to reaching pupal or adult stages. Therefore, a bioassay method we previously developed (Liu et al., 2004) was followed for this insect (Fig. 4C). Retarded insect development and increased insect mortality was observed in all three insect species, indicating AtVSP2 was indeed inhibitory to their growth and development. Compared to a cystatin from soybean and a legume lectin from Griffonia simplicifolia that we previously studied in depth regarding their defense functions (Koiwa et al., 1998; Zhu-Salzman et al., 1998; Liu et al., 2004), AtVSP2 displayed more than 10-fold higher insecticidal activity on a molar basis. AtVSP2 thus could potentially be useful for pest control via a biotechnological approach.
Introduction of Single-Amino Acid Alteration Did Not Compromise the Protein Structural Integrity of AtVSP2
It was suggested that the DXDXT motif serves as an intermediate phosphoryl acceptor and that the Asp nucleophile (the first Asp-119 residue of the signature motif) was critical for catalysis (Collet et al., 1998; Selengut, 2001). To determine whether pNPP hydrolysis was due to this nucleophile, the Asp-119 residue was replaced with Glu. This also allowed us to subsequently determine whether there was any correlation between biochemical and biological activities of AtVSP2. Although unmutated recombinant AtVSP2 was soluble, the mutated protein AtVSP2(D119E) expressed in the pET28a vector was insoluble. We thus transferred cDNA inserts [both AtVSP2 and AtVSP2(D119E)] into the pET44a vector, where our proteins of interest were expressed as fusion proteins with the Nus tag, a solubility-enhancing protein from Escherichia coli (Fig. 2, A and B). Attempts to remove the Nus tag by thrombin excision were unsuccessful. Therefore, we decided to evaluate biochemical and biological changes associated with this site-specific mutation using fusion proteins. It should be noted that the Nus tag did not interfere with AtVSP2 protein characteristics. Nus-AtVSP2 expressed in the pET44a vector exhibited comparable enzymatic activity (kcat = 20.7 s−1, Km = 7.9 mm) to AtVSP2 produced from pET28a, despite a slight shift of the optimal pH, due most likely to the presence of a Nus domain in the fusion form (Fig. 3A). Therefore, while facilitating the solubility of the recombinant protein, the Nus tag did not interfere with the protein structure or enzymatic function.
To ensure that the amino acid substitution did not result in dramatic protein structural change, circular dichroism spectra were obtained for mutated and nonmutated proteins (Fig. 2C). Results did not reveal significant differences between Nus-AtVSP2 and Nus-AtVSP2(D119E). The characteristic pattern indicative of random protein structures was not detected. The potential for structural alteration was also examined by fluorescence spectra, which is based on measurements of Tyr fluorescence (Lackowicz, 1983). If substantial conformational changes occurred as the result of the mutation, such that the Tyr environments were altered, a shift toward a longer wavelength of max intensity would be expected. However, such a shift was not detected (data not shown), indicating the Tyr residues in the mutant protein molecule were in relatively hydrophobic environments; thus, major structural collapse was unlikely. Such a result was expected because replacing Asp with Glu is a rather conservative change.
Anti-Insect Activity of AtVSP2 Is Correlated with Acid Phosphatase Activity
The fact that the Nus domain did not interfere with AtVSP2 enzymatic activity permitted us to evaluate mutated protein with the nucleophilic Asp-119 replaced while fused with Nus protein. Although the protein structure remained intact, the single-amino acid alteration from Asp-119 to Glu voided all acid phosphatase activity (Fig. 3A). Complete elimination of enzymatic activity with conservative changes at this position also occurred in eukaryotic Mg2+-dependent acid phosphatases and phosphoserine phosphatases (Collet et al., 1998; Selengut, 2001), suggesting an essential role for this invariant residue in the signature motif.
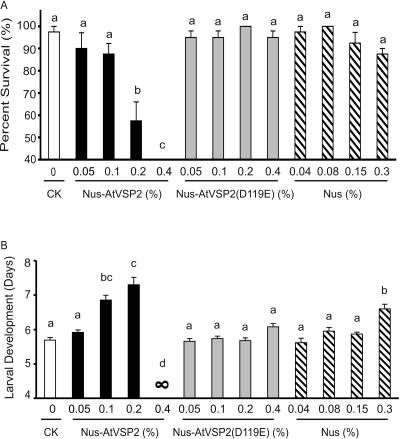
To understand the AtVSP2 anti-insect mechanism, we compared effects of Nus-AtVSP2 and Nus-AtVSP2(D119E) on Drosophila. Dietary Nus-AtVSP2 impacted insect mortality and development in a dose-dependent manner (Fig. 5), comparable on a molar basis to AtVSP2 expressed in pET28a (Fig. 4B). The anti-insect activity was revoked in the acid phosphatase-null Nus-AtVSP2(D119E) mutant protein, suggesting that the enzymatic activity is the basis for the biological function.
Figure 5.
Anti-insect activity of AtVSP2 is correlated with its acid phosphatase activity. The neonate Drosophila larvae were reared on a diet containing Nus-AtVSP2, Nus-AtVSP2(D119E), and Nus recombinant proteins. Surviving larvae were recorded daily until pupation, and the development time from neonate to pupae was calculated. The effect of Nus-AtVSP2, Nus-AtVSP2(D119E), and Nus on the survival (A) and development time (B) was analyzed by one-way ANOVA and Bonferroni multiple means comparison tests. Error bars indicate standard error. Means followed by the same letter are not significantly different at P = 0.05. CK, Control.
DISCUSSION
Temporary Nutrient Reservoir or Phosphate Metabolism?
Extensive work has been done on soybean VSPs as storage proteins. Soybean VSPs accumulate to nearly 50% of the total soluble proteins in leaves of depodded soybean, but decline to 1% during seed fill (Wittenbach, 1982; Staswick, 1994). Such a specific function in temporary protein storage has not been clearly established for Arabidopsis VSPs. Indeed, a significant difference in expression levels and tissue specificity has been recognized between the so-named VSPs from the two plant sources (Utsugi et al., 1996, 1998). Arabidopsis VSP genes were initially identified by random sequencing of Arabidopsis cDNAs, and the translated region of a partial cDNA sequence showed sequence similarity to soybean VSPs (Hofte et al., 1993). Full-length AtVSP1 and AtVSP2 cDNAs were later cloned from an Arabidopsis flower bud cDNA library (Utsugi et al., 1996), and their transcripts were shown to accumulate preferentially in flowers (Berger et al., 1995; Utsugi et al., 1996).
Classifying proteins based on short sequence motifs has received wide application as large quantities of sequence information have become available. Motif searches are particularly useful in assigning functions to proteins encoded by sequences with no significant matches in BLAST searches (Falquet et al., 2002). Undoubtedly, such motifs arise in the course of evolution due to the need to maintain protein structure in limited regions of functional importance. A new phosphatase family, the DDDD superfamily of phosphohydrolase, was recently defined by the occurrence of two conserved motifs that are separated by highly variable sequence regions (Thaller et al., 1998). Plant acid phosphatases were shown to be closely related to the bacterial nonspecific acid phosphatases by this classification criterion. Accordingly, Arabidopsis VSPs fall into this superfamily, suggesting their enzymatic functionality. Successful bacterial expression of active AtVSP2 has here allowed confirmation of this biochemical property (Figs. 2 and 3). Soybean VSPs, on the other hand, were excluded from this family due to lack of the nucleophilic Asp in the DXDXT motif domain, consistent with their proposed storage rather than enzymatic functionality.
Partial sequence homology with soybean VSPs led to assignment of Arabidopsis VSPs as VSPs. Illustration of weak phosphatase activity of the soybean VSPs further complicated the classification of the Arabidopsis proteins. However, the finding that soybean VSPs accounted for less than 0.1% of the total acid phosphatase activity in depodded plants while they comprised one-half of the total soluble proteins (Staswick, 1994) challenged the relevance of the enzymatic activity. Site-directed mutagenesis that restored the nucleophilic Asp in the DXDXT motif also greatly increased the acid phosphatase activity. Thus, most likely it is of evolutionary necessity in soybean for the phosphatases to inactivate their catalytic activity to serve as a storage protein (Leelapon et al., 2004). Retention of the signature sequences may have supported the acid phosphatase nature of Arabidopsis VSPs, distinguishing them functionally from the soybean VSPs. Substantially higher acid phosphatase activity in AtVSP2 than the residual phosphatase activity of the recombinant soybean VSP appears to support this view, although little is known of potential activities of these proteins on natural substrates in planta. Further, when the entire open reading frames were compared, AtVSP2 showed 40.5%, 36.0%, and 36.0% identity with soybean root nodule, barley, and tomato (Lycopersicon esculentum) acid phosphatases, respectively (data not shown). These are comparable to the homologies to soybean VSP sequences; 40.2% identical to soybean VSPα and 38.5% to VSPβ. Partial sequence alignment and lack of sufficient acid phosphatase sequence information when Arabidopsis VSP was initially identified may have biased its identification toward VSPs (Hofte et al., 1993). Taken together, acid phosphatases could be a better description for the Arabidopsis proteins initially named as VSPs.
Plant acid phosphatases are involved in phosphate acquisition and utilization and their expression is subject to developmental and environmental regulation. Phosphate starvation induces de novo synthesis of extra- and intracellular acid phosphatases, which is thought to be one of the strategies plants have evolved to adapt to phosphate-limiting conditions (Duff et al., 1994). In Arabidopsis, the purple acid phosphatase has been well studied for its response to phosphorus starvation. Induction at the mRNA and protein levels in roots as well as leaves under phosphate deficiency implies that it may function in scavenging phosphate from the soil as well as recycling it within the plant (del Pozo et al., 1999). A common cis-element motif in the promoter regions recognized by the PHR1 transcription factor is shared by AtVSP2 and the purple acid phosphatase, as well as other phosphate starvation-induced genes (Franco-Zorrilla et al., 2004). In contrast, the PHR1-binding motif is lacking in the soybean VSP promoter; rather, its response to phosphate deprivation is regulated by homeodomain Leu zipper proteins (Tang et al., 2001). On the other hand, the response of AtVSP to abscisic acid, JA, salt, water deficit, and wounding stresses is more similar to the soybean VSP than the Arabidopsis purple acid phosphatase (Berger et al., 1995; del Pozo et al., 1999; Gong et al., 2001; Franco-Zorrilla et al., 2004). The role of Arabidopsis VSPs in phosphate regulation thus needs to be further investigated.
Defense: One Possible Biological Function in Planta for AtVSP
Despite the overall sequence similarity between Arabidopsis and soybean VSPs, differences in tissue-specific expression are prominent. Arabidopsis expresses VSPs predominantly in the flower and very little in leaves (Utsugi et al., 1998). These contrasting expression patterns can be interpreted by the assumption that Arabidopsis VSPs may have other functions beyond a temporary protein reserve. A number of plant acid phosphatases have been implicated in resistance to pathogens and nematodes (Williamson and Colwell, 1991; Beßer et al., 2000; Jakobek and Lindgren, 2002; Petters et al., 2002; Stenzel et al., 2003). They appear to be induced by different regulatory pathways; barley acid phosphatase was induced by the salicylic acid mimic 2,6-dichloroisonicotinic acid and JA, while potato acid phosphatase responded only to intact bacterial organisms. Induction of gene expression in response to insect herbivory and correlation between increased Arabidopsis VSP expression and enhanced plant resistance to Egyptian cotton worm (Sponoptera littoralis; Rojo et al., 1999; Stotz et al., 2000; Berger et al., 2002) suggested a possible role in defense against herbivore insects. This concept has been confirmed here by feeding experiments that clearly illustrated AtVSP2 had deleterious effects on insects.
High expression of VSP proteins in flowers most likely represents a mechanism Arabidopsis uses to protect its reproductive structures. As sink tissues of the plant reproductive phase, floral organs and developing seeds draw significant amounts of sugars, amino acids, and ions from source tissues (Meuriot et al., 2004a) and are thus natural targets for herbivorous insects. Flowers, while being attractive to pollinators, may also be visual signals for herbivore insects. Therefore, it is not surprising that plants devote resources to synthesize defensive compounds to guard their reproductive organs and developing seeds, and these compounds are sometimes multifunctional. One illustration is the floral UV pigment flavonoids and dearomatized isoprenylated phloroglucinols from Hypericum calycinum (Gronquist et al., 2001). These UV pigments are responsible for forming UV patterns on flowers visible to insects to increase attractiveness to pollinators. Meanwhile, they are also toxic compounds to caterpillars. Therefore, they appear to play attractive and protective dual functions in the plant. Interestingly, they accumulate to very high concentrations in anthers and the ovary wall of the flower and are expressed at low levels in leaves (Gronquist et al., 2001). Tomato protease inhibitor 2 (Pin2), well established for its defense function, is also expressed at high levels in flowers (Creelman and Mullet, 1997). It is intriguing that a pattern of tissue specificity very similar to UV pigments and Pin2 also occurred in Arabidopsis VSP expression (Utsugi et al., 1998).
Like Pin2, Arabidopsis VSP expression can be rapidly induced by wounding, jasmonate, or insect feeding in leaves (Berger et al., 1995, 2002; Utsugi et al., 1998; Stotz et al., 2000; Reymond et al., 2004). AtVSP2 also responded to osmotic stresses exerted by drought and salt treatments (Gong et al., 2001). Since water deficit stress in plants tends to cause increased infestation by insects (Flint et al., 1996), activation of VSP may also strengthen plant defense aside from a possible role in source-sink regulation. Developmental regulation in a tissue-specific manner and in response to wounding and insect herbivory in the otherwise low-VSP-expressing leaf tissue suggested that VSP proteins belong to the defense arsenal that plants rely on to ensure protection of offspring.
Although the anti-insect activity appears to be related to acid phosphatase activity, it is unclear whether AtVSPs target specific substrates in the insects. Acid phosphatases are generally considered to lack substrate specificity, but specificity has been shown in a number of intracellular acid phosphates that have distinct metabolic functions, such as phytase, 3-phosphoglycerate phosphatase, and phosphoenolpyruvate phosphatase (Duff et al., 1994). Screening insect mutant lines that have altered susceptibility to AtVSP2 may help reveal insecticidal mechanisms of plant acid phosphatases such as Arabidopsis VSPs. Differential transcriptional regulation of AtVSP-like genes upon MeJA application and phosphate limitation implies distinct roles each member may play. Detailed characterization of individual genes will be necessary to reveal whether they all are associated with dephosphorylation and engaged in defense.
MATERIALS AND METHODS
Transcriptional Regulation of AtVSP-Like Genes
When TAIR database was queried with the AtVSP2 amino acid sequence, nine sequences (including AtVSP1) with E values lower than 10−8 were identified. These AtVSP-like sequences were functionally annotated as acid phosphatases and VSPs. The presence and location of signal peptide cleavage sites of these genes were determined by the SignalP 3.0 software (http://www.cbs.dtu.dk/services/SignalP), and their hydropathy plots were generated following Kyte and Doolittle (1982). The promoter regions of these genes were screened for phosphate starvation-responsive cis-elements [GNATATNC, CACGTG/C, TATCA(/T)A(/T), and CAT(/G)A(/C)TG] that were previously reported (Mukatira et al., 2001; Franco-Zorrilla et al., 2004).
To profile transcriptional regulation of AtVSP-like genes under MeJA treatment or phosphate deprivation, sterilized Arabidopsis (Arabidopsis thaliana) seeds were sown on 1× Murashige and Skoog solid medium, supplemented with 1% Suc, and cold treated at 4°C for 3 d, followed by a 7-d incubation period at 26°C. Seedlings sprayed with 0.2 mm MeJA were harvested 6 h after application. For the phosphate starvation treatment, cold-treated seeds were grown on nylon mesh placed on 0.2× Murashige and Skoog solid medium containing 3% Suc and 1.25 mm potassium phosphate for 5 d, then the nylon mesh was transferred to medium containing no phosphate and further incubated for 2 d prior to harvest.
Total RNA was extracted using Trizol reagent (Invitrogen) from control and treated plants and used for RT as described in Salzman et al. (2005). The mRNA levels of the above AtVSP-like sequences were compared between treated and control samples by quantitative real-time PCR using the ABI Prism 7900HT sequence detection system (Applied Biosystems). Gene-specific primers were synthesized using the ABI Prism Primer Express software (Applied Biosystems). PCR reactions (95°C for 15 s, 60°C for 1 min, 47 cycles), following an initial incubation of 50°C for 2 min and 95°C for 10 min, were performed in 2× SYBR Green master mix (Applied Biosystems). PCR amplification of 18S rRNA was used for normalization of input cDNA between samples. Each PCR reaction was run in duplicate.
Expression of AtVSP2 in Escherichia coli
The coding region of AtVSP2 cDNA was obtained by RT-PCR from MeJA-treated seedlings and the PCR product subjected to DNA sequencing to confirm its identity. The putative signal peptide was determined by the signal peptide-predicting software SignalP 3.0, as well as by sequence alignment with soybean VSPs, the N-terminal sequence of which is known. The cDNA fragment, excluding the putative signal peptide, was then cloned in frame into the bacterial expression vector pET28a (Novagen) at the BamHI and XhoI sites. The construct was transferred to the E. coli BL21(DE3) strain (Novagen) and AtVSP2 protein expression was induced by addition of isopropylthio-β-galactoside. The recombinant protein was purified through a Ni2+ chelate affinity column (Amersham-Pharmacia Biotech).
Site-Directed Mutagenesis of AtVSP2 and Expression of the Mutant Protein
Using the pET28a-AtVSP2 construct as template, we replaced the first Asp-119 residue in the D119XDXT motif with Glu via an inverse PCR approach (Ochman et al., 1988). Briefly, an antisense primer (5′-GGT ATC ATC TAG CTC AAA GAT CCA AAC-3′) was synthesized with the altered residue codon (underlined) located at the center. The sense primer (5′-CTC CTC TCT AGT ATT CCC TAC TAC GCA-3′) covered the immediate downstream sequence. Both primers were 5′ phosphorylated. The PCR reaction was done as follows: 94°C for 30 s, 51°C for 1 min, and 68°C for 6 min, for 35 cycles. The PCR product was gel purified, diluted to 0.5 μg/mL, self-ligated, and transformed into DH5α cells. The sequence was examined to confirm the site-specific mutation. Subsequent protein expression showed that the mutated protein was insoluble. To overcome this problem, cDNA inserts (sequence altered and unaltered) were cloned into pET44a so that mutant and native AtVSP2 could be expressed as a fusion protein with NusA. This Nus tag is an E. coli carrier protein used to improve the solubility of the expressed proteins (Novagen). Both constructs were transformed into Rosetta-gami host cells for protein expression (Novagen). The fusion proteins were purified through a Ni2+ chelate affinity column (Amersham-Pharmacia Biotech) and visualized by standard SDS-PAGE. Purified proteins were dialyzed against distilled water and lyophilized.
Acid Phosphatase Activity, pH Optimum, and Effects of Divalent Metals
Acid phosphatase activity of AtVSP2 purified from pET28, as well as Nus-AtVSP2 and mutant Nus-AtVSP2(D119E) fusion proteins obtained from pET44, were evaluated using pNPP (Sigma) as substrate following Hausmann and Shuman (2002). To determine the pH optimum, activity was measured in 50 mm citric acetate (pH 2.5–4.0), 50 mm sodium acetate (pH 3.5–5.5), and 50 mm Tris-acetate buffers (pH 5.5–7.5), respectively. Data from overlapping pH ranges of different buffers were averaged. The reaction mixtures [100 μL, containing 10 mm MgCl2, 10 mm pNPP, and approximately 0.03 μm purified AtVSP2 and Nus-AtVSP2, and 0.06 μm Nus and Nus-AtVSP2(D119E) in respective buffers] were incubated at 37°C for 30 min and quenched by addition of 900 μL of 1.0 m Na2CO3. Released p-nitrophenol was detected by measuring OD410 and the quantity calculated according to a standard curve.
A series of concentrations (from 0.5 to 50 mm) of MgCl2, CoCl2, ZnSO4, CaCl2, and MnCl2 were examined for their effects on acid phosphatase activity of AtVSP2 (0.05 μm). The aforementioned reactions were conducted under optimal pH of the enzyme, previously determined to be 4.5. Absorptions contributed by metals alone (i.e. reaction mixture without AtVSP2) were subtracted from the sample readings. Measurements were done in triplicate and plotted using Microsoft Excel.
Determination of Km and kcat
To determine apparent Km, Vmax, and kcat values of AtVSP2 and Nus-AtVSP2 for pNPP, the rate of dephosphorylation by 0.06 μm AtVSP was measured at substrate concentrations from 1 to 100 mm. Assays were performed in 50 mm sodium acetate (pH 4.5) and 10 mm MgCl2 at 37°C. Initial velocity for each substrate concentration was calculated. Data at each concentration were collected in triplicate and were fit to the Michaelis-Menten equation (v0 = Vmax [S]/Km + [S]) using the nonlinear least-squares-fitting analysis of KALEIDA-GRAPH software (Synergy).
Circular Dichroism and Fluorescence Spectra
Far-UV circular dichroism spectra of Nus-AtVSP2, Nus-AtVSP2(D119E), and Nus alone were obtained on an AVIV 62DS circular dichroism spectrometer (Aviv Associates) at 25°C. The instrument scanned from 200 to 260 nm with 10 scans for each protein sample [1.1 μm for Nus-AtVSP2 and Nus-AtVSP2(D119E) and 1.5 μm for Nus]. The path length was 0.5 cm. The solution for baseline spectra was 10 mm phosphate buffer, pH 7.0, the buffer in which proteins were dissolved.
Fluorescence spectra were acquired on a SLM 8100 spectrofluorometer in a 1.0-cm cuvette at 25°C. Protein samples (0.21 μm for Nus-AtVSP2 fusion proteins and 0.53 μm for Nus) were excited at 280 nm, and fluorescence emission was scanned from 300 to 400 nm. The fluorescence contribution from the buffer was subtracted from that of the samples.
Insect Feeding Assays
To analyze the effect of AtVSP2 on insect mortality and development, artificial diet/seeds incorporated with the recombinant protein were prepared and infested in the following manner. For southern corn rootworm (Diabrotica undecimpunctata howardi) assay, the artificial diet was prepared as instructed by the manufacturer (Bio-serv) using sterile techniques. Tetracycline and carbenicillin were added to prevent bacterial contamination. AtVSP was incorporated into the diet at various doses, dispensed into 24-well microtiter plates, and covered by parafilm to prevent drying. Nondiapausing southern corn rootworm egg masses, purchased from French Agricultural Research were incubated at 28°C and 60% relative humidity in the dark. For each AtVSP2 concentration, a total of 40 neonate larvae, in four replicates (10 larvae per replicate), were placed in the diet. Insects were transferred every other day to new plates with fresh diets and mortality recorded. Mean percent survival data were log10 transformed (Sokal and Rohlf, 1995). Linear regression analysis was performed to compare dose-mortality effects using PROC REG and user-defined contrasts in SAS 9.00 (SAS Institute).
For cowpea (Vigna unguiculata) bruchid (Callosobruchus maculatus), feeding procedures developed by Shade et al. (1986) were used with modifications. Decorticated cowpea seeds (California Blackeye No. 5) were milled into flour, into which AtVSP2 solutions were incorporated. Master pellets were made by injecting cowpea flour paste into the precooled Teflon mold, followed by immediate freezing in liquid nitrogen. After lyophilization, the master pellets were then ground into very fine flour, from which 10 cylindrical pellets (28 mg, 4.5-mm diameter × 1.6-mm high) for each treatment were made using a mechanical hand-operated press. Each pressed pellet was then coated with gelatin by dipping the pellet in an 8% gelatin solution and infested with one viable egg. Control artificial pellets without test proteins were made and infested in the same manner. Within-seed developmental time was recorded, as we have previously established that it is the most reliable variable for measuring the impact of compounds on cowpea bruchids. A one-way ANOVA test was used to analyze the data, with each infested pellet considered a replicate. Fisher's protected lsd test (P = 0.05) was used for mean separation.
Drosophila melanogaster strain Canton-S was maintained on a standard medium (1.5% agar, 10% corn meal, 4.1% yeast [Saccharomyces cerevisiae], 10% molasses, 0.8% propionic acid, and 0.2% Tegosept) and transferred onto an egg-collecting medium (40.5% apple juice, 5.3% Glc, 2.6% Suc, and 2% agar). Effects of AtVSP2, Nus-AtVSP2, and Nus-AtVSP2(D119E) on Drosophila were evaluated using Kankel/White Drososphila medium (White and Kankel, 1978). Briefly, diet mix containing 5% Glc, 5% yeast extract, 2% yeast, and 0.8% agar was boiled and cooled to 50°C, when respective protein solutions were incorporated. To prevent fungus contamination, a 1:1 (v/v) mixture of 83.6% propionic acid and 8.3% phosphoric acid was added to the diet at a final concentration of 0.5% (v/v). For each protein concentration, 40 newly hatched neonate larvae, in four replicates, were transferred to the AtVSP-containing diet and reared at 25°C. Fresh diet was supplied daily. Developmental time (from egg to pupa) was recorded. Statistical analysis was carried out using SPSS for Windows 11.0. Where necessary, log transformations were conducted to normalize distributions (Sokal and Rohlf, 1995). One-way ANOVA and Bonferroni multiple means comparison tests were performed to analyze treatment effects (Sokal and Rohlf, 1995).
Acknowledgments
We would like to thank Dr. J. Marty Scholtz and Dr. Abbas Razvi at the Texas A&M Department of Medical Biochemistry and Genetics for the use of laboratory equipment for circular dichroism and fluorescent spectra work. We appreciate thoughtful discussions with Dr. John Mullet and generous assistance in the enzymatic kinetics study provided by Dr. Jason Quinlan at the Department of Biochemistry and Biophysics. We thank Professor Richard Shade and Ms. Susan Balfe at the Department of Entomology, Purdue University, for their help in cowpea bruchid bioassays, and Dr. Tanya Pankiw at Texas A&M for help with statistical analysis.
The author responsible for distribution of materials integral to the findings presented in this article in accordance with the policy described in the Instructions for Authors (www.plantphysiol.org) is: Keyan Zhu-Salzman (ksalzman@tamu.edu).
Article, publication date, and citation information can be found at www.plantphysiol.org/cgi/doi/10.1104/pp.105.066837.
References
- Andrews DL, Beames B, Summers MD, Park WD (1988) Characterization of the lipid acyl hydrolase activity of the major potato (Solanum tuberosum) tuber protein, patatin, by cloning and abundant expression in a baculovirus vector. Biochem J 252: 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Asamizu E, Nakamura Y, Sato S, Tabata S (2000) A large scale analysis of cDNA in Arabidopsis thaliana: generation of 12,028 non-redundant expressed sequence tags from normalized and size-selected cDNA libraries. DNA Res 7: 175–180 [DOI] [PubMed] [Google Scholar]
- Berger S, Bell E, Sadka A, Mullet JE (1995) Arabidopsis thaliana AtVsp is homologous to soybean Vspa and Vspb, genes encoding vegetative storage protein acid-phosphatases, and is regulated similarly by methyl jasmonate, wounding, sugars, light and phosphate. Plant Mol Biol 27: 933–942 [DOI] [PubMed] [Google Scholar]
- Berger S, Mitchell-Olds T, Stotz HU (2002) Local and differential control of vegetative storage protein expression in response to herbivore damage in Arabidopsis thaliana. Physiol Plant 114: 85–91 [DOI] [PubMed] [Google Scholar]
- Beßer K, Jarosch B, Langen G, Kogel KH (2000) Expression analysis of genes induced in barley after chemical activation reveals distinct disease resistance pathways. Mol Plant Pathol 1: 277–286 [DOI] [PubMed] [Google Scholar]
- Bown DP, Wilkinson HS, Jongsma MA, Gatehouse JA (2004) Characterisation of cysteine proteinases responsible for digestive proteolysis in guts of larval western corn rootworm (Diabrotica virgifera) by expression in the yeast Pichia pastoris. Insect Biochem Mol Biol 34: 305–320 [DOI] [PubMed] [Google Scholar]
- Brenner R, Atkinson NS (1997) Calcium-activated potassium channel gene expression in the midgut of Drosophila. Comp Biochem Physiol B Biochem Mol Biol 118: 411–420 [DOI] [PubMed] [Google Scholar]
- Chrispeels MJ, Raikhel NV (1991) Lectins, lectin genes, and their role in plant defense. Plant Cell 3: 1–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman GD, Chen THH, Ernst SG, Fuchigami L (1991) Photoperiod control of poplar bark storage protein accumulation. Plant Physiol 96: 686–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collet JF, Stroobant V, Pirard M, Delpierre G, Van Schaftingen E (1998) A new class of phosphotransferases phosphorylated on an aspartate residue in an amino-terminal DXDX(T/V) motif. J Biol Chem 273: 14107–14112 [DOI] [PubMed] [Google Scholar]
- Collet JF, Stroobant V, Van Schaftingen E (1999) Mechanistic studies of phosphoserine phosphatase, an enzyme related to P-type ATPases. J Biol Chem 274: 33985–33990 [DOI] [PubMed] [Google Scholar]
- Creelman RA, Mullet JE (1997) Biosynthesis and action of jasmonates in plants. Annu Rev Plant Physiol Plant Mol Biol 48: 355–381 [DOI] [PubMed] [Google Scholar]
- del Pozo JC, Allona I, Rubio V, Leyva A, de la Pena A, Aragoncillo C, Paz-Ares J (1999) A type 5 acid phosphatase gene from Arabidopsis thaliana is induced by phosphate starvation and by some other types of phosphate mobilising/oxidative stress conditions. Plant J 19: 579–589 [DOI] [PubMed] [Google Scholar]
- De Wald DB, Mason HS, Mullet JE (1992) The soybean vegetative storage proteins VSP-alpha and VSP-beta are acid-phosphatases active on polyphosphates. J Biol Chem 267: 15958–15964 [PubMed] [Google Scholar]
- Duff SMG, Sarath G, Plaxton WC (1994) The role of acid-phosphatases in plant phosphorus-metabolism. Physiol Plant 90: 791–800 [Google Scholar]
- Edmonds HS, Gatehouse LN, Hilder VA, Gatehouse JA (1996) The inhibitory effects of the cysteine protease inhibitor, oryzacystatin, on digestive proteases and on larval survival and development of the southern corn rootworm (Diabrotica undecimpunctata howardi). Entomol Exp Appl 78: 83–94 [Google Scholar]
- Ellis C, Turner JG (2001) The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13: 1025–1033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falquet L, Pagni M, Bucher P, Hulo N, Sigrist CJA, Hofmann K, Bairoch A (2002) The PROSITE database, its status in 2002. Nucleic Acids Res 30: 235–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint HM, Naranjo SE, Leggett JE, Henneberry TJ (1996) Cotton water stress, arthropod dynamics, and management of Bemisia tabaci (Homoptera: Aleyrodidae). J Econ Entomol 89: 1288–1300 [Google Scholar]
- Franceschi VR, Wittenbach VA, Giaquinta RT (1983) Paraveinal mesophyll of soybean leaves in relation to assimilate transfer and compartmentation. 3. Immunohistochemical localization of specific glycopeptides in the vacuole after depodding. Plant Physiol 72: 586–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franco-Zorrilla JM, Gonzalez E, Bustos R, Linhares F, Leyva A, Paz-Ares J (2004) The transcriptional control of plant responses to phosphate limitation. J Exp Bot 55: 285–293 [DOI] [PubMed] [Google Scholar]
- Gase K, Liu GW, Bruckmann A, Steiner K, Ozegowski J, Malke H (1997) The LppC gene of Streptococcus equisimilis encodes a lipoprotein that is homologous to the e(P4) outer membrane protein from Haemophilus influenzae. Med Microbiol Immunol (Berl) 186: 63–73 [DOI] [PubMed] [Google Scholar]
- Gong ZZ, Koiwa H, Cushman MA, Ray A, Bufford D, Kore-eda S, Matsumoto TK, Zhu JH, Cushman JC, Bressan RA, et al (2001) Genes that are uniquely stress regulated in salt overly sensitive (sos) mutants. Plant Physiol 126: 363–375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulas E, Le Dily F, Ozouf J, Ourry A (2003) Effects of a cold treatment of the root system on white clover (Trifolium repens L.) morphogenesis and nitrogen reserve accumulation. J Plant Physiol 160: 893–902 [DOI] [PubMed] [Google Scholar]
- Gronquist M, Bezzerides A, Attygalle A, Meinwald J, Eisner M, Eisner T (2001) Attractive and defensive functions of the ultraviolet pigments of a flower (Hypericum calycinum). Proc Natl Acad Sci USA 98: 13745–13750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hagihara T, Hashi M, Takeuchi Y, Yamaoka N (2004) Cloning of soybean genes induced during hypersensitive cell death caused by syringolide elicitor. Planta 218: 606–614 [DOI] [PubMed] [Google Scholar]
- Hausmann S, Shuman S (2002) Characterization of the CTD phosphatase Fcp1 from fission yeast—preferential dephosphorylation of serine 2 versus serine 5. J Biol Chem 277: 21213–21220 [DOI] [PubMed] [Google Scholar]
- Hofte H, Desprez T, Amselem J, Chiapello H, Caboche M, Moisan A, Jourjon MF, Charpenteau JL, Berthomieu P, Guerrier D, et al (1993) An inventory of 1152 expressed sequence tags obtained by partial sequencing of cDNAs from Arabidopsis thaliana. Plant J 4: 1051–1061 [DOI] [PubMed] [Google Scholar]
- Jakobek JL, Lindgren PB (2002) Expression of a bean acid phosphatase cDNA is correlated with disease resistance. J Exp Bot 53: 387–389 [DOI] [PubMed] [Google Scholar]
- Koiwa H, Shade RE, Zhu-Salzman K, Subramanian L, Murdock LL, Nielsen SS, Bressan RA, Hasegawa PM (1998) Phage display selection can differentiate insecticidal activity of soybean cystatins. Plant J 14: 371–379 [DOI] [PubMed] [Google Scholar]
- Kyte J, Doolittle R (1982) A simple method for displaying the hydropathic character of a protein. J Mol Biol 157: 105–132 [DOI] [PubMed] [Google Scholar]
- Lackowicz JR (1983) Principles of Fluorescence Spectroscopy. Plenum Press, New York
- Leelapon O, Sarath G, Staswick PE (2004) A single amino acid substitution in soybean VSP alpha increases its acid phosphatase activity nearly 20-fold. Planta 219: 1071–1079 [DOI] [PubMed] [Google Scholar]
- Liu YL, Salzman RA, Pankiw T, Zhu-Salzman K (2004) Transcriptional regulation in southern corn rootworm larvae challenged by soyacystatin N. Insect Biochem Mol Biol 34: 1069–1077 [DOI] [PubMed] [Google Scholar]
- Maeshima M, Sasaki T, Asahi T (1985) Characterization of major proteins in sweet-potato tuberous roots. Phytochemistry 24: 1899–1902 [Google Scholar]
- Mason HS, Guerrero FD, Boyer JS, Mullet JE (1988) Proteins homologous to leaf glycoproteins are abundant in stems of dark-grown soybean seedlings—analysis of proteins and cDNAs. Plant Mol Biol 11: 845–856 [DOI] [PubMed] [Google Scholar]
- Mason HS, Mullet JE (1990) Expression of two soybean vegetative storage protein genes during development and in response to water deficit, wounding, and jasmonic acid. Plant Cell 2: 569–579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn M, Creelman RA, Bell E, Mullet JE, Browse J (1997) Jasmonate is essential for insect defense in Arabidopsis. Proc Natl Acad Sci USA 94: 5473–5477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuriot F, Avice JC, Simon JC, Laine P, Decau ML, Ourry A (2004. a) Influence of initial organic N reserves and residual leaf area on growth, N uptake, N partitioning and N storage in alfalfa (Medicago sativa) during post-cutting regrowth. Ann Bot (Lond) 94: 311–321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuriot F, Noquet C, Avice JC, Volenec JJ, Cunningham SM, Sors TG, Caillot S, Ourry A (2004. b) Methyl jasmonate alters N partitioning, N reserves accumulation and induces gene expression of a 32-kDa vegetative storage protein that possesses chitinase activity in Medicago sativa taproots. Physiol Plant 120: 113–123 [DOI] [PubMed] [Google Scholar]
- Mignery GA, Pikaard CS, Hannapel DJ, Park WD (1984) Isolation and sequence analysis of cDNAs for the major potato tuber protein, patatin. Nucleic Acids Res 12: 7987–8000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mignery GA, Pikaard CS, Park WD (1988) Molecular characterization of the patatin multigene family of potato. Gene 62: 27–44 [DOI] [PubMed] [Google Scholar]
- Mukatira UT, Liu CM, Varadarajan DK, Raghothama KG (2001) Negative regulation of phosphate starvation-induced genes. Plant Physiol 127: 1854–1862 [PMC free article] [PubMed] [Google Scholar]
- Murdock LL, Brookhart G, Dunn PE, Foard DE, Kelley S, Kitch L, Shade RE, Shukle RH, Wolfson JL (1987) Cysteine digestive proteinases in Coleoptera. Comp Biochem Physiol Biochem Mol Biol 87: 783–787 [Google Scholar]
- Ochman H, Gerber AS, Hartl DL (1988) Genetic applications of an inverse polymerase chain-reaction. Genetics 120: 621–623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penheiter AR, Klucas RV, Sarath G (1998) Purification and characterization of a soybean root nodule phosphatase expressed in Pichia pastoris. Protein Expr Purif 14: 125–130 [DOI] [PubMed] [Google Scholar]
- Petters J, Gobel C, Scheel D, Rosahl S (2002) A pathogen-responsive cDNA from potato encodes a protein with homology to a phosphate starvation-induced phosphatase. Plant Cell Physiol 43: 1049–1053 [DOI] [PubMed] [Google Scholar]
- Peumans WJ, Van Damme EJM (1995) The role of lectins in plant defense. Histochem J 27: 253–271 [DOI] [PubMed] [Google Scholar]
- Radhamani TR, Sudarshana L, Krishnan R (1995) Defense and carnivory: dual role of bracts in Passiflora foetida. J Biosci 20: 657–664 [Google Scholar]
- Reymond P, Bodenhausen N, Van Poecke RMP, Krishnamurthy V, Dicke M, Farmer EE (2004) A conserved transcript pattern in response to a specialist and a generalist herbivore. Plant Cell 16: 3132–3147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reymond P, Farmer EE (1998) Jasmonate and salicylate as global signals for defense gene expression. Curr Opin Plant Biol 1: 404–411 [DOI] [PubMed] [Google Scholar]
- Rojo E, Leon J, Sanchez-Serrano JJ (1999) Cross-talk between wound signalling pathways determines local versus systemic gene expression in Arabidopsis thaliana. Plant J 20: 135–142 [DOI] [PubMed] [Google Scholar]
- Salzman RA, Brady JA, Finlayson SA, Buchanan CD, Summer EJ, Sun F, Klein PE, Klein RR, Pratt LH, Cordonnier-Pratt MM, et al (2005) Transcriptional profiling of sorghum induced by methyl jasmonate, salicylic acid, and aminocyclopropane carboxylic acid reveals cooperative regulation and novel gene responses. Plant Physiol 138: 352–368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seki M, Narusaka M, Kamiya A, Ishida J, Satou M, Sakurai T, Nakajima M, Enju A, Akiyama K, Oono Y, et al (2002) Functional annotation of a full-length Arabidopsis cDNA collection. Science 296: 141–145 [DOI] [PubMed] [Google Scholar]
- Selengut JD (2001) MDP-1 is a new and distinct member of the haloacid dehalogenase family of aspartate-dependent phosphohydrolases. Biochemistry 40: 12704–12711 [DOI] [PubMed] [Google Scholar]
- Shade RE, Murdock LL, Foard DE, Pomeroy MA (1986) Artificial seed system for bioassay of cowpea weevil (Coleoptera, Bruchidae) growth and development. Environ Entomol 15: 1286–1291 [Google Scholar]
- Sokal RR, Rohlf FJ (1995) Biometry. The Principles and Practice of Statistics in Biological Research, Ed 3. W.H. Freeman and Company, New York
- Staswick PE (1988) Soybean vegetative storage protein-structure and gene-expression. Plant Physiol 87: 250–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick PE (1994) Storage proteins of vegetative plant-tissue. Annu Rev Plant Physiol Plant Mol Biol 45: 303–322 [Google Scholar]
- Staswick PE, Zhang Z, Clemente TE, Specht JE (2001) Efficient down-regulation of the major vegetative storage protein genes in transgenic soybean does not compromise plant productivity. Plant Physiol 127: 1819–1826 [PMC free article] [PubMed] [Google Scholar]
- Stenzel I, Ziethe K, Schurath J, Hertel SC, Bosse D, Kock M (2003) Differential expression of the LePS2 phosphatase gene family in response to phosphate availability, pathogen infection and during development. Physiol Plant 118: 138–146 [DOI] [PubMed] [Google Scholar]
- Stotz HU, Pittendrigh BR, Kroymann J, Weniger K, Fritsche J, Bauke A, Mitchell-Olds T (2000) Induced plant defense responses against chewing insects. Ethylene signaling reduces resistance of Arabidopsis against Egyptian cotton worm but not diamondback moth. Plant Physiol 124: 1007–1017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang ZJ, Sadka A, Morishige DT, Mullet JE (2001) Homeodomain leucine zipper proteins bind to the phosphate response domain of the soybean VspB tripartite promoter. Plant Physiol 125: 797–809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terra WR, Ferreira C (1994) Insect digestive enzymes—properties, compartmentalization and function. Comp Biochem Physiol B Biochem Mol Biol 109: 1–62 [Google Scholar]
- Thaller MC, Schippa S, Bonci A, Cresti S, Rossolini GM (1997) Identification of the gene (aphA) encoding the class B acid phosphatase/phosphotransferase of Escherichia coli MG1655 and characterization of its product. FEMS Microbiol Lett 146: 191–198 [DOI] [PubMed] [Google Scholar]
- Thaller MC, Schippa S, Rossolini GM (1998) Conserved sequence motifs among bacterial, eukaryotic, and archaeal phosphatases that define a new phosphohydrolase superfamily. Protein Sci 7: 1647–1652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Utsugi S, Sakamoto W, Murata M, Motoyoshi F (1998) Arabidopsis thaliana vegetative storage protein (VSP) genes: gene organization and tissue-specific expression. Plant Mol Biol 38: 565–576 [DOI] [PubMed] [Google Scholar]
- Utsugi S, Sakamoto W, Ogura Y, Murata M, Motoyoshi F (1996) Isolation and characterization of cDNA clones corresponding to the genes expressed preferentially in floral organs of Arabidopsis thaliana. Plant Mol Biol 32: 759–765 [DOI] [PubMed] [Google Scholar]
- Van Damme EJM, Hause B, Hu JL, Barre A, Rouge P, Proost P, Peumans WJ (2002) Two distinct jacalin-related lectins with a different specificity and subcellular location are major vegetative storage proteins in the bark of the black mulberry tree. Plant Physiol 130: 757–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme EJM, Roy S, Barre A, Citores L, Mostafapous K, Rouge P, VanLeuven F, Girbes T, Goldstein IJ, Peumans WJ (1997) Elderberry (Sambucus nigra) bark contains two structurally different Neu5Ac(alpha 2,6)Gal/GalNAc-binding type 2 ribosome-inactivating proteins. Eur J Biochem 245: 648–655 [DOI] [PubMed] [Google Scholar]
- Wang WR, Cho HS, Kim R, Jancarik J, Yokota H, Nguyen HH, Grigoriev IV, Wemmer DE, Kim SH (2002) Structural characterization of the reaction pathway in phosphoserine phosphatase: crystallographic “snapshots” of intermediate states. J Mol Biol 319: 421–431 [DOI] [PubMed] [Google Scholar]
- White JA, Todd T, Newman T, Focks N, Girke T, de Ilarduya OM, Jaworski JG, Ohlrogge JB, Benning C (2000) A new set of Arabidopsis expressed sequence tags from developing seeds: the metabolic pathway from carbohydrates to seed oil. Plant Physiol 124: 1582–1594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- White K, Kankel DR (1978) Patterns of cell-division and cell movement in formation of imaginal nervous-system in Drosophila-melanogaster. Dev Biol 65: 296–321 [DOI] [PubMed] [Google Scholar]
- Williamson VM, Colwell G (1991) Acid phosphatase-1 from nematode resistant tomato—isolation and characterization of its gene. Plant Physiol 97: 139–146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenbach VA (1982) Effect of pod removal on leaf senescence in soybeans. Plant Physiol 70: 1544–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wittenbach VA (1983) Purification and characterization of a soybean leaf storage glycoprotein. Plant Physiol 73: 125–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie DX, Feys BF, James S, Nieto-Rostro M, Turner JG (1998) COI1: an Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280: 1091–1094 [DOI] [PubMed] [Google Scholar]
- Yeh KW, Chen JC, Lin MI, Chen YM, Lin CY (1997) Functional activity of sporamin from sweet potato (Ipomoea batatas Lam): a tuber storage protein with trypsin inhibitory activity. Plant Mol Biol 33: 565–570 [DOI] [PubMed] [Google Scholar]
- Yunes ANA, de Andrade MT, Sales MP, Morais RA, Fernandes KVS, Gomes VM, Xavier-Filho J (1998) Legume seed vicilins (7S storage proteins) interfere with the development of the cowpea weevil (Callosobruchus maculatus (F)). J Sci Food Agric 76: 111–116 [Google Scholar]
- Zhu-Salzman K, Shade RE, Koiwa H, Salzman RA, Narasimhan M, Bressan RA, Hasegawa PM, Murdock LL (1998) Carbohydrate binding and resistance to proteolysis control insecticidal activity of Griffonia simplicifolia lectin II. Proc Natl Acad Sci USA 95: 15123–15128 [DOI] [PMC free article] [PubMed] [Google Scholar]