Abstract
Microevolution is regarded as changes in the frequencies of genes in populations over time. Ancient DNA technology now provides an opportunity to demonstrate evolution over a geological time frame and to possibly identify the causal factors in any such evolutionary event. Using nine nuclear microsatellite DNA loci, we genotyped an ancient population of Adélie penguins (Pygoscelis adeliae) aged ≈6,000 years B.P. Subfossil bones from this population were excavated by using an accurate stratigraphic method that allowed the identification of individuals even within the same layer. We compared the allele frequencies in the ancient population with those recorded from the modern population at the same site in Antarctica. We report significant changes in the frequencies of alleles between these two time points, hence demonstrating microevolutionary change. This study demonstrates a nuclear gene-frequency change over such a geological time frame. We discuss the possible causes of such a change, including the role of mutation, genetic drift, and the effects of gene mixing among different penguin populations. The latter is likely to be precipitated by mega-icebergs that act to promote migration among penguin colonies that typically show strong natal return.
Keywords: microsatellites, Adélie penguins, ancient DNA
For at least half a century, microevolution has meant changes in gene frequencies in natural populations over time (1). Most attempts to demonstrate such change have been limited to examining populations over contemporary time scales. These range from the historically important example of microevolutionary changes in the frequency of different morphs of the peppered moth consequent with changes in levels of air pollution (2) to more recent studies that show gene-frequency changes in small isolated salmon populations (3). Other studies have demonstrated microsatellite gene-frequency shifts among avian populations known to have gone through severe population bottle-necks (4, 5). However, the detection of microevolutionary changes over longer time frames is likely to be more difficult. Ancient DNA technology enables us to examine populations over longer time frames (6, 7), thereby increasing the probability of detecting any microevolutionary change and providing the opportunity to identify the causal factors responsible for such change. Nevertheless, this approach would require sufficient ancient DNA deposits from a narrow time period, together with samples from modern populations of the same species.
Any evolutionary changes that are detected are likely to result from environmental change mediated by natural selection or stochastically through the process of genetic drift (8). Alternatively, for some classes of DNA such as microsatellite DNA repeat sequences, mutational processes may play an important role. Microsatellite DNA loci have become the most common nuclear DNA marker for population genetic studies. This dominance is because these loci are biparentally inherited, show moderate levels of polymorphism, can be detected by using PCR, and are abundant in almost all eukaryotes examined to date. Microsatellite loci have been used predominantly to determine degrees of relatedness and levels of population structure. Nuclear markers are generally difficult to recover from ancient DNA samples, compared with mitochondrial DNA sequences (9, 10). This difficulty is because, in contrast with mitochondrial DNA, nuclear loci are typically present in only two copies per cell. Moreover, ancient DNA samples often are degraded and, consequently, the number of intact copies available for amplification is much lower, compared with modern samples. We have previously suggested that Adélie penguin subfossil bone deposits in Antarctica represent the highest quality ancient DNA yet discovered (7). Hence, the expectation is that there will be a relatively large number of intact DNA copies from which to amplify. This abundance of copies implies that single-copy microsatellite DNA changes could be detected from this ancient penguin material. It has recently been demonstrated that the amplification of other single-copy nuclear genes from large numbers of ancient avian bones is feasible (11). This finding, therefore, raises the possibility of a comparison of microsatellite changes between ancient and modern Adélie penguin populations.
Adélie penguins breed in high-density colonies at a number of sites around the margins of the Antarctic continent during the summer months. Colonies range in size from 100 to 170,000 breeding pairs. The ecology and population dynamics of Adélie penguins are well understood (12, 13). Individuals typically show strong natal return over their reproductive lifetime, and birds spend much of the winter on pack ice around the Antarctic continent. In early summer, breeding pairs arrive at nesting sites in coastal ice-free areas. Mating occurs in the colony, and pair bonds are quickly established. Females usually lay two eggs, and ≈23% of hatched chicks fail to fledge (12). This high level of chick mortality, in combination with adult mortality during the breeding season, has resulted in the accumulation of extensive subfossil remains beneath many colonies. Subfossil bones of Adélie penguins are preserved within ornithogenic soils that develop below penguin nests in presently occupied and relict rookeries. Droppings, feathers, egg fragments, and other penguin remains, mixed with sand, gravel, and pebbles, constitute distinct soil horizons. We have previously shown that this subfossil record is typically highly stratified, showing a consistent relationship between the ages of bones as determined by radiocarbon dating and the depth from which they are recovered (7). Given the strong natal return shown by Adélie penguins, these subfossil remains typically represent individuals ancestral to the present colony. The unique combination of ancient and modern populations, together with well understood life-history characteristics, suggests that a study of Adélie penguins would provide an ideal opportunity to detect microevolutionary changes. However, although in the Antarctic there are very large numbers of Adélie penguins and subfossil bones, it is rare to be able to collect a significant number of these from a single stratum (i.e., from approximately the same time point) and at one location.
Despite this difficulty, we were able to sample Adélie penguin bones from a single stratum on Inexpressible Island, Antarctica. Bones from this population were ≈6,000 years old. We isolated eight highly variable microsatellite DNA loci from an Adélie penguin genomic library. We used these loci, together with one heterologous locus, to genotype individuals and to compare the allelic composition of the modern and ancient populations. Factors likely to affect gene frequency shifts over time, such as the presence of mega-icebergs, were examined, as were mutational processes likely to affect microsatellite DNA (14).
Materials and Methods
Sample Collection and Extraction. Adélie penguin subfossil bones were collected from Inexpressible Island, Terra Nova Bay (74°53′S, 163°45′E). Approval to collect subfossil bones from specially protected areas and sites of special scientific interest was granted by Antarctica New Zealand (permit nos. 99/9 and 01/7). Subfossil bones were collected and stored in a dedicated ancient DNA laboratory. Forty-eight blood samples were collected from the modern Inexpressible Island Adélie penguin population. Genomic DNA was extracted from whole blood by using the InstaGene kit (Bio-Rad) according to the manufacturer's instructions.
Stratigraphy and 14C Dating of Subfossil Bones. We collected bone samples from abandoned nesting sites in the vicinity of the presently occupied rookery at Inexpressible Island (Fig. 4, which is published as supporting information on the PNAS web site). We dug for subfossil bones in ornithogenic soils by using an accurate stratigraphic method of excavation. Pits from 1-2 m2 to 6 m2 were excavated layer-by-layer, cleaning the entire surface from the top. In this way, we could identify and separate different individual remains even within the same layer. This method prevents contamination from the top and enabled us to identify wedges of sediment reworked by periglacial processes. A total of 15 ancient Adélie penguin individuals were collected from one distinct layer. Two of these bones were dated by using accelerator mass spectrometry at the Institute of Geological and Nuclear Sciences (Lower Hutt, New Zealand). 14C dates from the remains of Adélie penguin individuals that lived or fed in the sea are affected by an error known as the “reservoir effect” (15), induced by the depletion of 14C in seawater. In the Antarctic Ocean, this effect is elevated and is due to the upwelling of deep, old oceanic water and depleted glacial meltwaters (15-17). As a result, Antarctic samples appear 1,000-1,300 years older than their true age. Given that this error varies with time, we used the calibration program calib, Version 5.0 (18), to calibrate radiocarbon dates. Consequently, a DR value of 688 ± 55 was applied as a correction factor appropriate for our Adélie penguin samples. The parameter DR represents the constant difference in reservoir age of a regional part of the ocean.
PCR and Microsatellite Genotyping. The PCR primers used to amplify the microsatellite DNA loci TP500, RM6, AM13, AM3, and HrU2 were as detailed in ref. 19. Additional microsatellite DNA primer sequences (5′ to 3′) used are as follows: XVC11, F-GGTTCAACCAGAAGCAAAGC and R-GTGTAACCGGCCCTGATG; XXG12a, F-GGTCCTGGATTCCCTTGACT and R-GCCAACTGCTGGAGAGACC; XIXB3, F-TGATGATTCCCATTGCCATA and R-TCAAGTCAGGAGTATTGCCATTT; and XVIIIB2, F-AGTAGCCAGCTCCTCCAGGT and R-CTGGTGAGTAGTTGGGACCAG. In five of the nine loci, the sequenced clones contained a pure repeat: TP500, (CA)14; RM6, (CA)10; XVC11, (CA)13; XVIIIB2, (CA)16; and XIXB3, (CA)11. The remaining four loci were composed of interrupted or compound repeats: XXG12a, (CA)6CAAA(CA)2CG(CA)4; AM13, A11N33(GT)9(GC)4(GT)5; AM3, A8N5(TA)4; and HrU2. In relation to the latter locus, which was originally derived from swallows (20), a 124-bp allele from Adélie penguins had the following organization: (AG)2(GT)6A(GT)2(GA)5 N9(GA)3G5N3(GA)3(GT)3. Primers for the nine loci were labeled with 6-FAM (HrU2, TP500, AM3, and XVC11) or HEX (RM6 and AM13) from Sigma Genosys or PET (XIXB3, XXGI2a) or VIC (XVIIIB2) phosphoramidite dye from Applied Biosystems. The microsatellite DNA loci were amplified from the subfossil bone samples in a dedicated ancient DNA laboratory. PCR amplification conditions for HrU2, TP500, AM3, RM6, and AM13 were as described in ref. 19, except that 4 μg/μl BSA was added per reaction and all loci were amplified for 40 PCR cycles. Single-locus amplification products were pooled for genotyping when samples had high band intensity on an agarose gel. Loci XIXB3, XXG12a, XVIIIB2, and XVCII were isolated from an enriched genomic library from Adélie penguins by using a method similar to that of Armour et al. (21). These loci were independently amplified from the ancient samples by using a PCR mix containing 1 unit of Taq DNA polymerase (Roche), 200 μM each dNTP, 0.3 μM each primer, 1 M betaine (Sigma Genosys), 1.6 μg/μl BSA, 2 mM MgCl2, and 1.5× PCR buffer (Roche) in a final volume of 10 μl. XXGI2a and XVIIIB2 were amplified in the modern samples by using a 10-μl PCR mix containing 1 unit of Taq DNA polymerase, 200 μM each dNTP, 0.25 μM (XVIIIB2) or 0.3 μM (XXG12a) each primer, 1 M betaine, 0.4 μg/μl BSA, 1.5 mM (XVIIIB2) or 3 mM (XXG12a) MgCl2, and 1.5× (XXG12a) or 2× (XVIIIB2) PCR buffer. Loci XVC11 and XIXB3 were multiplexed in the modern samples by PCR-amplifying in 10-μl volumes containing 0.9 unit of Taq DNA polymerase, 300 μM each dNTP, 0.4 μg/μl BSA, 1 M betaine, 0.4 μM each XVC11 primer, 0.25 μM each XIXB3 primer, 2 mM MgCl2, and 2× PCR buffer. PCR amplification for these four loci was performed in a Bio-Rad iCycler. Thermal cycling conditions were denaturation at 94°C for 2 min, followed by 30 (for modern samples) or 40 (for ancient samples) cycles of 94°C for 20 s, 58°C for 30 s, and 72°C for 30 s, followed by 1 cycle of 78°C for 5 min. Samples were genotyped by using a Prism 377 sequencer (Applied Biosystems), and genotyping data were analyzed by using genescan analysis (Version 3.1, Applied Biosystems).
Ancient DNA Methods. Ancient DNA methods were generally as used by Lambert et al. (7). Mock DNA extractions and PCR blanks were continually screened, and contamination was not typically seen. When there was evidence of contamination, the resulting samples were not used. All controls yielded results that were consistent with authentic ancient DNA. As an additional authentication step, five ancient DNA samples also were reextracted and amplified at the University of Auckland. The AM13 locus was amplified for all five samples, and the TP500 locus was amplified for two samples. The genotypes for all samples were identical to those that had been previously determined. This replication represents 5% of our ancient samples and is comparable to the levels of replication in other studies (6% as recorded in ref. 7).
Statistical Analysis of Microsatellite DNA Data. A test of conformity with Hardy-Weinberg equilibrium predictions at each locus and Fisher's exact test of genic differentiation between temporally spaced samples were performed as described in ref. 19. The extent of differentiation was investigated by calculating FST and RST estimates, and the significance of these values was tested by permutating alleles among populations (10,000 permutations for both FST and RST). The estimation and testing of these statistics was calculated by using fstat [ref. 22; Version 2.9.3 (2001) is available at www2.unil.ch/popgen/softwares/fstat.htm] for FST and rstlcalc (23) for RST. An assignment test was used to classify individuals as belonging to ancient or modern populations (24).
Quantification of Penguin Philopatry and Migration. During 1961-1969, 5,000 near-to-fledging penguins were banded annually at Cape Crozier, Ross Island (Fig. 4). In subsequent years, searches were made for these birds, mainly at Cape Crozier but also among neighboring colonies (25). During 1994 and 1996-2001, 1,000 fledglings were banded annually at both Cape Crozier and Cape Bird, and 400 were banded at Cape Royds, all on Ross Island. Beginning in 1999, 400 birds also were marked at nearby Beaufort Island each year. Extensive searches at each of these colonies have been made annually for banded recruits. Bands were applied to the bird's left flipper. Imprinted numerals on these bands were legible through binoculars, thus avoiding the need to recapture and handle the birds a second time.
Results
Change in Microsatellite DNA Allele Frequencies Over Time. Using nine microsatellite DNA loci, we genotyped 48 samples from modern penguins from Inexpressible Island, together with 15 subfossil bone samples collected from one stratigraphic layer at the same site. The quality of the ancient DNA in Adélie penguins is evident because we were able to determine 119 of the 135 possible genotypes. An estimate of the age of the layer from which these samples came was determined from two bones that were carbon-dated to 6,082 ± 55 and 6,092 ± 60 years B.P. This site has been continuously occupied over this time period (26), and given that natal philopatry is characteristic of Adélie penguins (12), we suggest that these modern and ancient populations have been genealogically connected for >6,000 years.
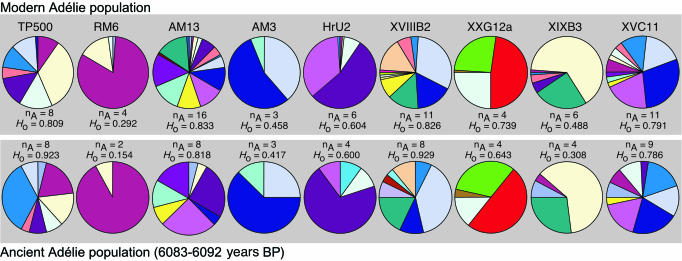
We present gene-frequency data for the nine loci from these temporally separated populations (Fig. 1 and Table 3, which is published as supporting information on the PNAS web site). Both populations showed genotypes in expected Hardy-Weinberg proportions (ancient populations, P = 0.682; living populations, P = 0.375). This result indicates that our ancient DNA methods detected genotypes accurately, in contrast to several other studies (27, 28), and that the modern and ancient samples are each derived from randomly mating populations. We performed several analyses to detect any gene-frequency shifts over this time period. Fisher's exact test (29) indicated a significant level of genetic differentiation between the two populations (Table 1). Additionally, we tested the null hypothesis that the ancient and modern samples represent subsets of the same population by generating 1,000 random samples from the modern data set. Each of these replicates comprised the same number of samples genotyped from the ancient population and had the same number of missing values. We then examined the level of genetic differentiation (as measured by Fisher's exact test) among each of the 1,000 random samples and the total modern data set (Fig. 5, which is published as supporting information on the PNAS web site). Only 0.7% of replicates resulted in values that were less than those observed for the ancient-vs.-modern comparison. Additional tests of genetic divergence between modern and ancient samples also were performed. FST (22) and RST (23) values, and tests of their significance are presented in Table 1. These values are indicative of a low but significant level of genetic structure. In addition, an assignment test (24) correctly allocated 60% of ancient and 85% of modern samples to their actual population, with 79% correctly identified overall (Table 2). This result, statistically better than random, again indicates gene-frequency differences over the time period examined.
Fig. 1.
Microsatellite DNA allele frequencies for nine loci, number of alleles (nA) and heterozygosity values (Ho) for modern and ancient populations at Inexpressible Island.
Table 1. Genetic divergence between modern and ancient populations of Adélie penguins from Inexpressible Island.
| Locus
|
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Test | TP500 | RM6 | AM13 | AM3 | HrU2 | XVIIIB2 | XXG12a | XIXB3 | XVCII | Overall |
| Fisher's exact test P value | 0.060 | 0.702 | 0.298 | 0.308 | 0.004 | 0.096 | 0.402 | 0.168 | 0.317 | 0.010 |
| FST Θ | 0.037 | 0.001 | 0.011 | −0.006 | 0.065 | −0.002 | −0.008 | −0.018 | −0.012 | 0.009 (P = 0.005) |
| RST Θ | −0.017 | 0.017 | 0.048 | 0.027 | 0.138 | 0.055 | 0.022 | 0.086 | 0.037 | 0.050 (P = 0.003) |
Table 2. Assignment test showing the classification of samples in relation to their source.
| Classification
|
||
|---|---|---|
| Source | Modern | Ancient |
| Modern | 41 | 7 |
| Ancient | 6 | 9 |
χ2 = 24.366. P = 6.75 × 10−7.
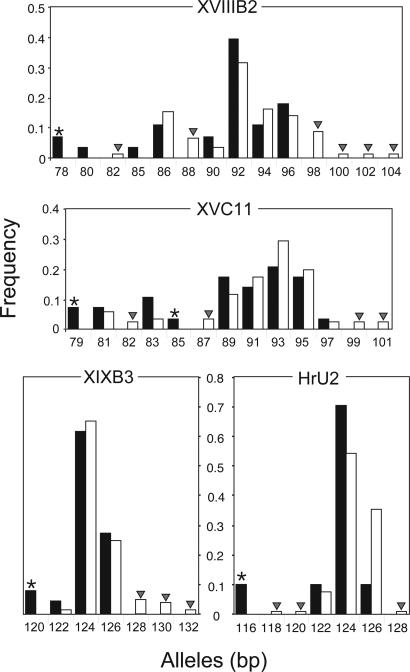
Microsatellite DNA Allele Lengths. A comparison of alleles from ancient and modern populations revealed that, at four of the nine loci, there was a significant increase in the average allele lengths recorded from the modern population, compared with the 123.2 bp, and the average modern allele size was 124.5 bp (two-tailed t test, P < 0.05) (Fig. 2). In addition, of the remaining five loci, three showed an increase in allele size from ancient to modern populations, whereas two showed the reverse trend. Also, for all loci, the ancient population was shown to be characterized by a total of eight private alleles, of which seven were present at loci that showed a significant increase in allele length over time (Fig. 2, denoted by an asterisk). Interestingly, five of these alleles were the shortest alleles recorded at each of five loci. The three remaining private alleles were among the shortest alleles at the XVIIIB2 and XVC11 loci. In addition, there were 27 private alleles recorded from the modern population. This larger number of private alleles, compared with the ancient population, is likely to be due, at least in part, to the larger sample size from the modern population. The private alleles from the modern population were generally among the longest alleles at each locus. Fig. 2 shows the private alleles for each of the four significant loci (denoted by an arrowhead).
Fig. 2.
The distribution of microsatellite DNA allele sizes for the four loci that show a significant increase in average allele size. Ancient and modern alleles are shown by black and white bars, respectively. Alleles are denoted by their size in base pairs. Private alleles for the ancient population are denoted by an asterisk, and those alleles for the modern population are represented by an arrowhead.
The detection of nuclear microsatellite alleles might be subject to biased amplification of shorter alleles. To test this hypothesis, we examined the distribution of missing genotypes in relation to the loci examined. Only 4 of the 16 missing genotypes were from the four loci that showed a significant difference between allele lengths in ancient and modern populations; the expected number of missing genotypes for the four loci is 7.1. Hence, there cannot be a significant excess of missing alleles at these loci. In fact, the missing genotypes derive proportionately more from the non-significant loci. We expected to find 8.5 missing genotypes from these loci and, in fact, recorded 12.
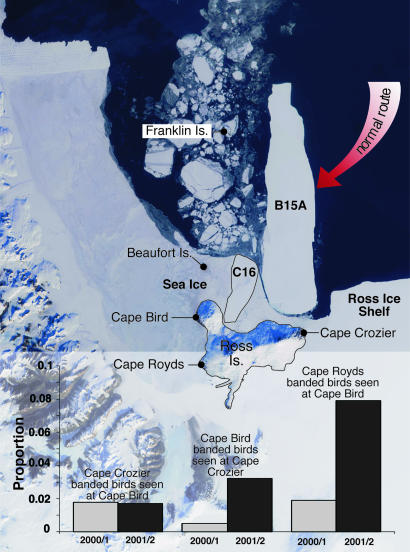
Penguin Movement During a Recent Period of Environmental Upheaval. We detected a significant alternation in Adélie penguin visitation patterns (Fig. 3) caused by the arrival and grounding of a 180-km iceberg, B15A, and a smaller iceberg, C16, which forced penguins to change their usual migration routes. Our data are based on resightings of young recruits that were banded as chicks at Cape Crozier, Cape Bird, and Cape Royds on Ross Island (n = 9,556). Specifically, among Cape Royds-banded penguins, there was a significant increase in the subsequent appearance of these individuals at Cape Bird (G = 8.58, df = 1, P < 0.01). This increase was a result of the extensive sea ice along the route normally taken by migrating Cape Royds' penguins. This sea ice was blocked from leaving the area as a result of the presence of B15A. As a result, the Cape Royds penguins more frequently stopped en route at the Cape Bird rookery. In addition, chicks that were banded at Cape Bird showed higher than expected visitation rates at Cape Crozier (G = 8.98, df = 1, P < 0.01). In this case, it was not extensive sea ice but rather blockage of the normal migration route by the B15A itself that forced the change.
Fig. 3.
Southern Ross Sea showing penguin colonies (black dots) and the position of mega-icebergs C16 and B15A in relation to Ross Island. The large arrow shows the usual migration route of Adélie penguins during spring (October). Upon reaching B15A, penguins attempting to reach Cape Bird or Cape Royds had to turn in either direction to round B15A. Graphs show intercolony visitation levels before (2000/2001) and after (2001/2002) the icebergs lodged in the position shown. This natural-color satellite image from the National Aeronautics and Space Administration's Multiangle Imaging SpectroRadiometer was acquired during two polar overpasses of the Terra satellite on Dec. 9, 2001. Details of the satellite image are available at www-misr.jpl.nasa.gov.
During the austral summer of 2000-2001 (pre-B15A), of 385 Cape Crozier-banded individuals (2- to 4-year-olds), 7 visited the Cape Bird and 2 visited Cape Royds. Of 348 Cape Bird-banded individuals, 2 visited Cape Crozier and 4 visited Cape Royds. From the 154 Cape Royds-banded birds, 2 and 3 visited Cape Crozier and Cape Bird, respectively. The next summer, with B15A now blocking the way, sightings of 344 Cape Crozier-banded (2- to 4-year-olds) were as follows: 8 visited Cape Bird (33% more than the in previous year) and 3 visited Cape Royds (40% more than in the previous year). These rates were much lower than expected (annual increases not statistically significant), because they should have reflected larger cohorts from enhanced production and survival at all colonies than in the previous few years. In contrast, of 160 Cape Bird-banded penguins seen, at least 7 visited Cape Crozier (700% more than in the previous year; G = 8.98, df = 1, P < 0.01) and thus appear to have been diverted from their usual route (Fig. 3). In subsequent years, with the iceberg remaining in place, similar altered patterns of movement have resulted. Moreover, individuals that were banded on Ross Island and Beaufort Island have been resighted at more distant colonies at Franklin Island, Edmonson Point, and Coulman Island, despite infrequent visits by observers to these locations (D.G.A., G.B., K. M. Dugger, and P.R.W., unpublished data).
Discussion
The microsatellite DNA data presented here reveal a pattern of significant change between the ancient and the modern populations of Adélie penguins from Inexpressible Island. Despite evidence for the genetic equilibrium of both populations, genic heterozygosity tests, measures of population subdivision (FST and RST), and assignment tests all indicate significant changes in allele frequencies over the >6,000-year time frame (Tables 1 and 2). Our study represents a detailed population genetic comparison of a modern and an ancient population. In addition, to our knowledge, no population genetic analyses have previously illustrated a significant difference in the genetic composition of populations separated by a geological time period.
Earlier comparisons of temporally separated populations, using microsatellite DNA loci, have been restricted to studies employing a small number of loci (e.g., refs. 1-4). Moreover, these studies have been difficult to conduct because of a range of technical obstacles, e.g., the accurate amplification of alleles (27). Some recent studies have been conducted over short time frames, for example, within 50-200 years (e.g., ref. 30). The general focus of these studies has been to detect any changes in the overall levels of genetic variation between temporarily separated populations that were severely bottlenecked as a result of human exploitation. Perhaps not surprisingly, a loss of genetic variation over time has been demonstrated in a number of cases (30, 31). In contrast, Limburg and Weider (32) examined potential changes in an undisturbed natural population. They examined four microsatellite DNA loci from the egg banks of natural populations of a number of species of Daphnia that were ≈200 years old. The authors compared changes in allele frequencies between these historical and contemporary Daphnia egg banks. They recorded a pattern of substantial genetic differentiation between these time-separated populations. However, as Limburg and Weider explain, this result could be a consequence of changes in the composition of Daphnia species over time, because different species cannot easily be distinguished morphologically. Other studies have compared both human (28) and animal (33) populations over longer time periods. These studies have typically recorded no microevolutionary shifts over time. For example, Zierdt et al. (28) compared allele frequencies at one microsatellite DNA locus in an ancient (1,200-1,500 years old) and a modern human population from Weingarten, Germany. These authors recorded “... a remarkable similarity of the allele frequencies between past and modern populations” (28). One early study of 5,000-year-old human teeth even concluded that a microsatellite DNA analysis of such material was not reliable enough to provide useful results (27). Therefore, it is clear that the demonstration of microevolutionary change among temporarily separated populations of Adélie penguins, as estimated by nine microsatellite DNA loci, is highly significant. We suggest that the success of our study is attributable, in part, to the high quality of the ancient DNA of Adélie penguins in the extremely cold and dry Antarctic environment.
The potential causes of this microevolutionary change are now open to investigation. Natural selection is unlikely to represent a significant force, because most microsatellite DNA loci are noncoding. Because of the large number of excavations carried out in the Adélie penguin colony at Inexpressible Island, it appears that this population has been large over the time period examined in this study (C.B., unpublished data). However, we cannot exclude the possibility that, at some point(s) over the >6,000-year period, there was a population bottleneck that resulted in gene-frequency changes. Unlike other genetic markers that typically mutate by single-nucleotide changes, microsatellite DNA loci, because of their internal repeat structure have recoverable histories that can be interpreted in terms of the mutational mechanisms that generate them. Changes in the length of repeat tracts are thought to be the predominant type of mutation for such loci. The principal mechanisms responsible for such length variation are slippage during replication (34) and recombination through unequal exchange (35) and/or gene conversion (36). Certainly, it is widely recognized that microsatellite DNA alleles tend to increase in length over time (14), at least when alleles are short and phylogenetically young. Consequently, we compared the length of alleles in ancient and modern populations. We show that the ancient population had significantly shorter microsatellite DNA alleles than does the modern population at four of the nine loci examined (Fig. 2). Although not statistically significant, three of the remaining loci showed longer alleles in the modern population, compared with the ancient population. In addition, the ancient population is characterized by short private alleles, and the modern population has a larger number of private alleles that are generally among the longest variants at each locus. This result suggests that mutational processes may have contributed to the microevolutionary event detected in this study.
Our present finding is in contrast with our previous demonstration of a lack of differentiation among modern Adélie penguin populations from around Antarctica (19). This geographic homogeneity is likely to be a result of the large population sizes of most colonies and/or gene flow among colonies (19). Both of these factors will contribute to the equilibration of gene frequencies and the genetic homogeneity that we now observe among the modern colonies (19). However, until recently, Adélie penguins have been thought to exhibit low levels of emigration and pronounced natal philopatry (12). This philopatric behavior has been observed during periods of relatively stable environmental and climatic conditions. Strong natal philopatry would be expected to promote genetic divergence among populations. However, in recent years, evidence is emerging of high levels of episodic gene flow among colonies.
In 2001, a mega-iceberg broke from the Ross Ice Shelf and drifted into an area where there was a high concentration of penguin colonies. This iceberg movement resulted in the blockage of the normal penguin migration routes whereby adults return to natal colonies. Both the presence of mega-icebergs and the consequent increased expanse of sea ice made it difficult for breeding penguins to reach natal colonies and, consequently, had negative effects on their foraging behavior. This situation resulted in the almost complete breeding failure of some colonies on Ross Island (D.G.A., G.B., K. M. Dugger, and P.R.W., unpublished data). Although the samples on which this study was based were collected before B15A broke from the Ross Ice Shelf, the appearance of such mega-icebergs is unlikely to be an unusual occurrence over long time periods. In fact, on average, it has been estimated that during the Holocene, 20 mega-icebergs per 1,000 years have broken from the Ross Ice Shelf (37, 38). Moreover, a large proportion of Adélie penguin colonies, Antarctica-wide, are located near ice shelves and glacier tongues (39), both of which produce mega-icebergs. In addition, it is known that these icebergs typically remain close to shore (near colonies) for much of their existence (40). Hence, altered rates of emigration, precipitated by changing ice conditions, are likely to represent a longstanding evolutionary force resulting in the homogenization of gene frequencies among colonies.
This study demonstrates the feasibility of applying population genetic methods and analyses to natural populations at different “slices” in time. Our comparison of a >6,000-year-old Adélie penguin population with its modern descendants is significant because it demonstrates the power of ancient DNA technology to now directly study microevolution. Importantly, this approach also has provided evidence for the causes of the microevolutionary change that we detected.
Supplementary Material
Acknowledgments
We thank J. Adams, S. Allen, M. Hester, B. Karl, H. Nevins, P. Ritchie, and S. Webb for field assistance; Argos Inc. (Largo, MD) for tracking the penguins; I. Anderson, J. Robins, and T. White for laboratory assistance; K. Newman and V. Ward for graphics; and D. MacAyeal for information about iceberg behavior. This work was supported by grants from the New Zealand Marsden Fund; Massey University; the University of Pisa; the Italian Antarctic Research Program; National Science Foundation Grants OPP 9526865, OPP 9814882, and OPP 0125608; Antarctica New Zealand; the U.S. Antarctic Program; the U.S. Coast Guard; and New Zealand Foundation for Research, Science, and Technology Grant C09527. This is PRBO contribution no. 1293 from the Point Reyes Bird Observatory.
Author contributions: C.D.M., G.B., D.G.A., P.R.W., and D.M.L. designed research; L.D.S., G.B., D.G.A., P.R.W., G.D.H., and C.B. performed research; C.D.M., G.B., D.G.A., and D.M.L. contributed new reagents/analytic tools; L.D.S., G.B., D.G.A., G.D.H., C.B., and D.M.L. analyzed data; and L.D.S., C.D.M., G.B., D.G.A., P.R.W., and D.M.L. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Dobzhansky, T. (1951) Genetics and the Origin of Species (Columbia Univ. Press, New York).
- 2.Majerus, M. E. M. (1998) Melanism: Evolution in Action (Oxford Univ. Press, Oxford).
- 3.Koskinen, M., Haugen, T. & Primmer, C. (2001) Nature 419, 826-830. [DOI] [PubMed] [Google Scholar]
- 4.Mundy, N., Winchell, C., Bur, T. & Woodruff, D. (1997) Proc. R. Soc. London Ser. B 264, 869-875. [Google Scholar]
- 5.Lambert, D. M., King, T., Shepherd, L. D., Livingston, A., Anderson, S. & Craig, J. L. (2005) Conserv. Genet. 6, 1-14. [Google Scholar]
- 6.Villablanca, F. X. (1990) in Ancient DNA, eds. Herrmann, B. & Hummel, S. (Springer, New York), pp. 31-58.
- 7.Lambert, D. M., Ritchie, P. A., Millar, C. D., Holland, B., Drummond, A. J. & Baroni, C. (2002) Science 295, 2270-2273. [DOI] [PubMed] [Google Scholar]
- 8.Freeman, S. & Herron, J. (2001) Evolutionary Analysis (Prentice-Hall, Engle-wood Cliffs, NJ).
- 9.Zhang, D.-X. & Hewitt, G. M. (2003) Mol. Ecol. 12, 563-584. [DOI] [PubMed] [Google Scholar]
- 10.Pääbo, S., Poinar, H., Serre, D., Jaenicke-Després, V., Hebler, J., Rohland, N., Kuch, M., Krause, J., Vigilant, L. & Hofreiter, M. (2004) Annu. Rev. Genet. 38, 645-679. [DOI] [PubMed] [Google Scholar]
- 11.Huynen, L., Millar, C. D., Scofield, R. P. & Lambert, D. M. (2003) Nature 425, 175-178. [DOI] [PubMed] [Google Scholar]
- 12.Ainley, D. G. (2002) The Adélie Penguin: Bellwether of Climate Change (Columbia Univ. Press, New York).
- 13.Ainley, D. G., LeResche, R. E. & Sladen, W. J. L. (1983) Breeding Biology of Adélie Penguins (Univ. Calif. Press, Los Angeles).
- 14.Goldstein, D. B. & Schlotterer, C., eds. (1999) Microsatellites: Evolution and Applications (Oxford Univ. Press, Oxford).
- 15.Stuiver, M., Pearson, G. & Baziunas, T. (1986) Radiocarbon 28, 980-1021. [Google Scholar]
- 16.Harkness, D. D. (1979) Brit. Antarc. Surv. Bull. 47, 43-59. [Google Scholar]
- 17.Omoto, K. (1983) in Antarctic Earth Science, eds. Oliver, R. L., James, P. R. & Jago, J. B. (Cambridge Univ. Press, London), pp. 450-452.
- 18.Stuiver, M. & Reimer, P. J. (1993) Radiocarbon 35, 215-230. [Google Scholar]
- 19.Roeder, A., Marshall, R. K., Mitchelson, A. J., Visagathilagar, T., Ritchie, P. A., Love, D. R., Pakai, T. J., McPartlan, H. C., Murray, N. D., Robinson, N. A., et al. (2001) Mol. Ecol. 10, 1645-1656. [DOI] [PubMed] [Google Scholar]
- 20.Primmer, C. R., Moller, A. P. & Ellegren, H. (1996) Mol. Ecol. 5, 365-378. [DOI] [PubMed] [Google Scholar]
- 21.Armour, J. A. L., Neumann, R., Gorbert, S. & Jeffreys, A. J. (1994) Hum. Mol. Genet. 3, 599-605. [DOI] [PubMed] [Google Scholar]
- 22.Goudet, J. (1995) J. Hered. 86, 485-486. [Google Scholar]
- 23.Goodman, S. (1997) Mol. Ecol. 6, 881-885. [Google Scholar]
- 24.Paetkau, D., Calvert, W., Sterling, I. & Strobeck, C. (1995) Mol. Ecol. 4, 347-354. [DOI] [PubMed] [Google Scholar]
- 25.Ainley, D. G. & DeMaster, D. (1980) Ecology 61, 522-530. [Google Scholar]
- 26.Baroni, C. & Orombelli, G. (1994) Geology 22, 23-26. [Google Scholar]
- 27.Ramos, M., Lalueza, C., Girbau, E., Perez-Perez, A., Quevedo, S., Turbon, D. & Estivill, X. (1995) Hum. Genet. 96, 205-212. [DOI] [PubMed] [Google Scholar]
- 28.Zierdt, H., Hummel, S. & Herrmann, B. (1996) Hum. Biol. 68, 185-199. [PubMed] [Google Scholar]
- 29.Raymond, M. & Rousset, F. (1995) J. Hered. 86, 248-249. [Google Scholar]
- 30.Larson, S. L., Jameson, R. J., Etnier, M., Fleming, M. & Bentzen, P. (2002) Mol. Ecol. 11, 1899-1903. [DOI] [PubMed] [Google Scholar]
- 31.Hutchinson, W. F., van Oosterhour, C., Rogers, S. & Carvalho, G. R. (2003) Proc. R. Soc. London Ser. B 270, 2125-2132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Limburg, P. A. & Weider, L. J. (2002) Proc. R. Soc. London Ser. B 269, 281-287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards, C. J., Connellan, J., Wallace, P. F., Park, S. D., McCormick, F. M., Olsaker, I., Eythorsdottir, E., MacHugh, D. E., Bailey, J. F. & Bradley, D. G. (2003) Anim. Genet. 34, 410-416. [DOI] [PubMed] [Google Scholar]
- 34.Levinson, G. & Gutman, G. (1987) Mol. Biol. Evol. 4, 203-221. [DOI] [PubMed] [Google Scholar]
- 35.Harding, R. M., Boyce, A. J. & Clegg, J. B. (1992) Genetics 132, 847-859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jakupciak, J. P. & Wells, R. D. (2000) J. Biol. Chem. 275, 4003-4013. [DOI] [PubMed] [Google Scholar]
- 37.Jacobs, S., MacAyeal, D. & Ardai, J. (1986) J. Glaciol. 32, 464-474. [Google Scholar]
- 38.Lazzara, M., Jezek, K., Scambos, T., MacAyeal, D. & Veen, C. V. D. (1999) Polar Geogr. 23, 201-212. [Google Scholar]
- 39.Woehler, E. (1993) The Distribution and Abundance of Antarctic and Subantarctic Penguins (Scott Polar Research Institute, Cambridge, U.K.).
- 40.Anderson, J. (1999) Antarctic Marine Geology (Cambridge Univ. Press, Cambridge, U.K.).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.