Abstract
Persistence of the opportunistic bacterial pathogen Vibrio cholerae in aquatic environments is the principal cause for seasonal occurrence of cholera epidemics. This causality has been explained by postulating that V. cholerae forms biofilms in association with animate and inanimate surfaces. Alternatively, it has been proposed that bacterial pathogens are an integral part of the natural microbial food web and thus their survival is constrained by protozoan predation. Here, we report that both explanations are interrelated. Our data show that biofilms are the protective agent enabling V. cholerae to survive protozoan grazing while their planktonic counterparts are eliminated. Grazing on planktonic V. cholerae was found to select for the biofilm-enhancing rugose phase variant, which is adapted to the surface-associated niche by the production of exopolymers. Interestingly, grazing resistance in V. cholerae biofilms was not attained by exopolymer production alone but was accomplished by the secretion of an antiprotozoal factor that inhibits protozoan feeding activity. We identified that the cell density-dependent regulator hapR controls the production of this factor in biofilms. The inhibitory effect of V. cholerae biofilms was found to be widespread among toxigenic and nontoxigenic isolates. Our results provide a mechanistic explanation for the adaptive advantage of surface-associated growth in the environmental persistence of V. cholerae and suggest an important contribution of protozoan predation in the selective enrichment of biofilm-forming strains in the out-of-host environment.
Keywords: grazing, resistance, protozoa, quorum sensing
Epidemics of cholera, an acute intestinal infection caused by toxigenic strains of the facultative pathogen Vibrio cholerae, are a major public health problem in developing countries around the globe. Both toxigenic and nontoxigenic strains of V. cholerae are natural inhabitants of a wide range of aquatic ecosystems, including estuarine and coastal waters, that provide the environmental reservoir of virulent V. cholerae strains (1). The fact that many environmental nontoxigenic strains carry virulence genes (2, 3) and that the occurrence of epidemics coincides with the increased prevalence of the causative V. cholerae strain in the aquatic environment (4, 5) supports the notion of an environmental origin of toxigenic V. cholerae clones. This view has led to the hypothesis that cholera epidemics are triggered by environmental factors and selective forces governing aquatic microbial communities. In recent years, studies on the ecology of V. cholerae have considerably increased our understanding of physical and biological parameters that influence the persistence of V. cholerae in the environment and hold the potential to predict the outbreak of cholera epidemics (6).
As a member of the natural bacterioplankton community, V. cholerae is an integral part of the pelagic microbial food web and is thus constrained in its growth and survival by the predatory action of bacterivorous protists, so-called protozoa. Grazing by small heterotrophic flagellates is recognized as the major mortality factor in bacterioplankton communities (7, 8) and as such has been suggested to tightly control cell numbers of Vibrio spp. in these systems (9). The fact that the high elimination rates frequently observed for bacterial pathogens in aquatic environments can be assigned to the predatory activity of protozoa (10-12) has nourished the concept that defensive strategies to survive protozoan grazing are an essential prerequisite for the environmental persistence of bacterial pathogens, such as V. cholerae (13).
An increasing number of studies have demonstrated that surface-associated growth on zooplankton, phytoplankton, and other suspended particulates is an important strategy in the life cycle of V. cholerae for its persistence and accumulation in natural aquatic habitats (14-16). Biofilms generally have been proposed to constitute an environmental refuge for a number of bacterial pathogens and to provide pathogens with an adaptive advantage promoting their environmental persistence (17, 18). The significant reduction of cholera incidence by water filtration procedures illustrates that particle-associated growth of V. cholerae is a key mechanism for concentrating bacterial numbers to the minimum infectious dose and to facilitate transmission to humans (19). Consequently, the central role of biofilm formation in the persistence of V. cholerae in natural habitats raises the question how and by what mechanisms environmental factors select for biofilm-associated phenotypes.
The aim of our study was to elucidate the role of protozoan predation in the persistence and accumulation of V. cholerae and to characterize the underlying mechanisms. We hypothesized that biofilms formed by V. cholerae exhibit a higher antipredator fitness than their planktonic counterparts, providing a surface-associated refuge from a main mortality factor. To test for planktonic versus biofilm fitness, we designed a model system employing two niche-specific predatory flagellates typically found in marine plankton communities (20), the surface feeder Rhynchomonas nasuta and the suspension feeder Cafeteria roenbergensis. The two flagellates are among the 20 most commonly reported species of heterotrophic flagellates (21) and feature similar cell sizes and feeding rates despite the contrasting niches they occupy (22). This simple model system allowed us to identify effective mechanisms of V. cholerae resistance to protozoan grazing, which has implications for understanding how V. cholerae persists and diversifies in the environment and how toxigenic strains reach the minimum infectious dose.
Materials and Methods
Strains and Culture Conditions. The V. cholerae strains used were the smooth and rugose phase variants of A1552 (wild-type, El Tor); the vpsR, vpsA, vpsL, vpsT, hapR, and luxT mutants of the rugose phase variant; and a collection of eight toxigenic and eight nontoxigenic V. cholerae isolates. All V. cholerae strains were routinely grown on LB plates and in 2M medium (23). C. roenbergensis (Bicosoecida) was isolated from the Baltic Sea by A. P. Mylnikov, and Rhynchomonas nasuta (Kinetoplastida) from the Atlantic Ocean by H. Arndt and M. Weitere. Both flagellates were routinely grown on heat-killed Pseudomonas aeruginosa PAO1 (final concentration 106 cells per ml-1) in 50% NSS medium (24). To eradicate indigenous bacteria, flagellate cultures were subject to a combination of antibiotic treatment (50 μg·ml-1 ampicillin, 30 μg·ml-1 gentamycin, 30 μg·ml-1 kanamycin, 60 μg·ml-1 neomyxin, 30 μg·ml-1 polymyxin, 30 μg·ml-1 streptomycin, and 50 μg·ml-1 tobramycin) and serial microdrop dilution over >1,000 generations. Before the experiments, growth tests on 2M liquid medium and agar plates were used to rule out bacterial contamination.
Grazing Experiments. Experiments testing the survival of V. cholerae in the presence of the two bacterivorous flagellates were performed in 24-well tissue culture plates. Overnight cultures of the V. cholerae strains were diluted to 105 cells per ml-1 in carbon-reduced 2M medium (0.08% glucose), transferred into tissue culture plates and incubated at 20°C with shaking (75 rpm). For the comparison of plankton vs. biofilm persistence, planktonic bacteria were separated from surface-associated cells after 24 h incubation by transferring the planktonic phase of each well to a new plate. Subsequently, the suspension-feeder C. roenbergensis was added to the planktonic subpopulation while the surface-feeder R. nasuta was introduced to the biofilm subpopulation (both at a final concentration of 1 × 103 ml-1). Numbers of flagellates and planktonic bacteria and the biofilm biomass were followed over 4 days. Generally, each treatment was run in replicate wells of four.
Enumeration of Bacteria and Flagellates. Numbers of planktonic V. cholerae cells and frequencies of rugose and smooth-phase variants were assessed by plating serial dilutions on LB agar. Biofilms were sampled mechanically from the well bottom and moderately sonicated before being plated on LB agar. Growth rates of the surface-associated flagellate R. nasuta were calculated from direct cell counts by means of inverted microscopy over 4 days. Cell numbers of C. roenbergensis were determined from formalin-fixed (2%) plankton samples by means of epifluorescence microscopy by using the DNA-stain DAPI.
Quantification of Biofilm Formation. Biomass of V. cholerae biofilms was quantified by a crystal violet (CV) staining assay (25). Briefly, the ambient aqueous phase was removed from each well, and planktonic cells were thoroughly washed off the surface with 50% NSS. CV was added to each well, and plates were incubated for 20 min and rinsed repeatedly before CV-stained biofilms were solubilized in 95% ethanol and the absorbance was determined with a plate reader at 490 nm (Wallac, Gaithersburg, MD). Values were corrected by blank readings and readings of grazer-only control treatments. Grazing-mediated changes in biofilm structure were evaluated by staining the biofilm with SYTO 9 (Molecular Probes) and by employing confocal laser scanning microscopy (IMT-2, Olympus, Melville, NY).
Supernatant Toxicity Assay. Cell-free supernatants from V. cholerae biofilms grown in microtiter plates were collected by centrifugation and 0.22-μm filtration. Toxicity to flagellate grazers was evaluated by adding R. nasuta (at a final concentration of 2 × 103 ml-1) to biofilm supernatants supplemented with heat-killed P. aeruginosa prey and by determining the number of active flagellate cells by direct inspection with an inverted light microscope (IMT-2, ×200 magnification, Olympus) on 3 consecutive days.
Results
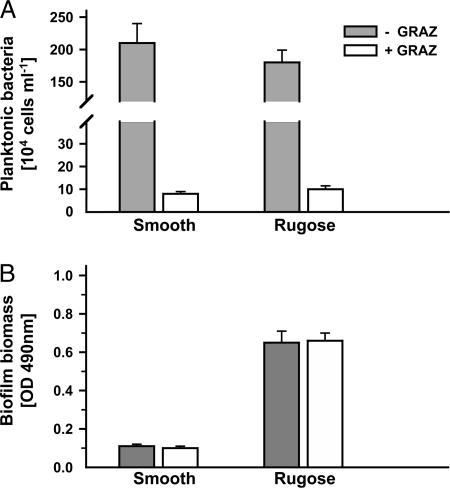
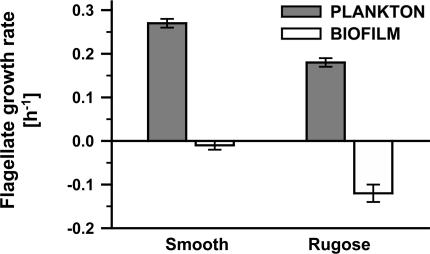
Planktonic V. cholerae Cells Are Eliminated by Grazing Whereas Biofilms Persist. Planktonic cells of V. cholerae readily colonize submersed surfaces and develop biofilms. To assess whether surface-associated bacteria reveal higher antipredator fitness than their planktonic counterparts, suspended V. cholerae cells were separated from biofilms, and both populations were subjected to niche-specific flagellate grazing. The addition of the suspension-feeding flagellate C. roenbergensis resulted in a drastic reduction of planktonic V. cholerae cells by >94% within 72 h for both smooth and rugose V. cholerae A1552 (Fig. 1A, P < 0.001). Furthermore, at a 100-fold higher initial concentration (2 × 108 bacteria ml-1) of planktonic V. cholerae, cells were reduced to the same extent (data not shown). In contrast, biofilms of both smooth and rugose variants were unaffected by the presence of the surface-feeding flagellate R. nasuta, so that biofilm biomass remained stable (Fig. 1B). Growth rates of C. roenbergensis reached maximum values by feeding on planktonic V. cholerae (0.27 h-1 and 0.18 h-1, respectively, Fig. 2) whereas the assessment of R. nasuta numbers revealed that flagellate growth was impaired by feeding on biofilms of both V. cholerae variants (-0.01 h-1 and -0.12 h-1, respectively). Biofilm-mediated inhibition of flagellate growth was significantly more pronounced in biofilms of the rugose variant (P < 0.001).
Fig. 1.
Persistence of V. cholerae biofilms as opposed to planktonic cells during flagellate grazing. Planktonic (A) and biofilm (B) subpopulations of the smooth and rugose phase variant of V. cholerae A1552 were examined in the absence (-GRAZ) and in the presence (+GRAZ) of flagellate grazers. Planktonic bacteria were exposed to the suspension feeder C. roenbergensis and biofilms to the surface feeder R. nasuta for 72 h. Error bars indicate standard deviations of four replicates.
Fig. 2.
Inhibition of flagellate growth by V. cholerae biofilms. Growth rates were determined for the suspension feeder C. roenbergensis on planktonic cells (A) and for the surface-feeder R. nasuta on biofilms (B) of V. cholerae A1552 over 4 days. Error bars indicate standard deviations of four replicates.
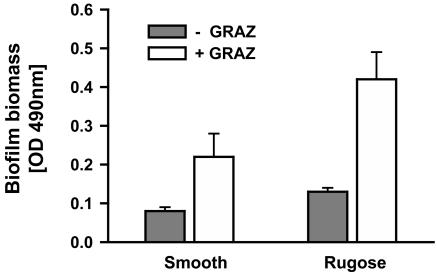
Planktonic Grazing Selects for Biofilm Formation and Rugose Phase Variants. Given the significant differences in grazing mortality of planktonic and biofilm populations, we tested whether grazing on unprotected planktonic V. cholerae stimulates the formation of grazing-resistant biofilms. Therefore, we followed the formation of biofilms from planktonic V. cholerae populations in the presence and the absence of the suspension-feeder C. roenbergensis. The grazing activity of C. roenbergensis on planktonic V. cholerae cells resulted in significantly enhanced biofilm formation of both smooth and rugose variants (Fig. 3, both P < 0.001). Grazing-stimulated increase of biofilm biomass was highest for the rugose strain (>3-fold).
Fig. 3.
Stimulation of biofilm formation by planktonic grazers. Planktonic subpopulations of the smooth and rugose phase variant of V. cholerae A1552 were cultivated without (-GRAZ) and with (+GRAZ) the suspension feeder C. roenbergensis. Biofilm formation was quantified by a crystal violet staining assay. Error bars indicate standard deviations of four replicates.
Throughout our experiments, the rugose phase variant showed enhanced biofilm formation compared with the smooth variant. To examine whether predation selects specifically for the rugose phenotype of V. cholerae, we followed the frequencies of colony morphotypes in the presence and the absence of the suspension-feeder C. roenbergensis. In the absence of the flagellate, the rugose variant exhibited high switching frequencies to the smooth colony variant 24 h after inoculation (Table 1). Specifically, the planktonic environment selected for the smooth variant constituting >96% of colonies. During flagellate grazing, however, the prevalence of the smooth variant in suspension was gradually reduced. The significant increase of rugose colony type frequencies from 1.6% to 43% demonstrates the high antipredator fitness of the rugose relative to the smooth variant. Furthermore, plating assays revealed the high affinity of the rugose phenotype to the biofilm life-style because 67-97% of the biofilm-derived colonies comprised the rugose colony type. Frequencies of biofilm-derived colonies were unaffected by planktonic grazing.
Table 1. Niche-specific frequencies of rugose colony variant of V. cholerae A1552 without (−GRAZ) and with (+GRAZ) the suspension-feeding flagellate C. roenbergensis.
| Frequency of rugose colonies, %
|
||||
|---|---|---|---|---|
| Plankton
|
Biofilm
|
|||
| Time, h | −GRAZ | +GRAZ | −GRAZ | +GRAZ |
| 24 | 3.4 ± 1.0 | 1.6 ± 1.2 | 67.6 ± 4.7 | 70.1 ± 2.3 |
| 48 | 3.7 ± 1.2 | 17.5 ± 2.6*** | 92.4 ± 1.8 | 89.4 ± 2.0 |
| 72 | 3.3 ± 0.2 | 43.3 ± 8.2*** | 97.0 ± 4.3 | 96.7 ± 1.9 |
All values are means ± SD (n = 4) derived from plate inspection after 2 days. Statistical analyses were performed by using arcsine square root-transformed data. ***, P < 0.001.
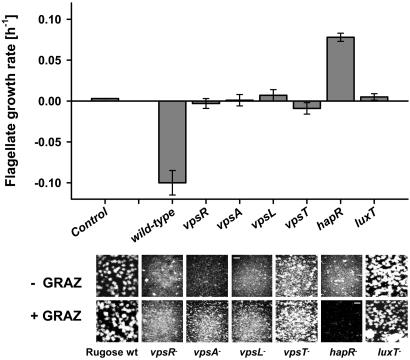
Grazing Inhibition in Biofilms Is Controlled by Quorum Sensing. Hallmarks characteristic of biofilm development and phase variation in V. cholerae are the production of the Vibrio exopolysaccharide (VPS) and gene regulation in a cell density-dependent fashion (i.e., quorum sensing) (26-28). To elucidate the underlying mechanisms of grazing resistance in V. cholerae biofilms, we exposed biofilms of mutants defective in genes of the VPS operon and in the regulatory genes hapR and luxT to the surface-feeding flagellate R. nasuta. In contrast to the negative growth rates on the rugose wild-type biofilm, mutants defective in genes required for VPS synthesis (vpsR, vpsA, vpsL, and vpsT) caused no reduction of flagellate numbers but did not restore flagellate growth (Fig. 4). The only mutant that provided strong flagellate growth (μ = 0.08 h-1) was the hapR mutant. As a consequence of the relatively high flagellate growth rate, hapR mutant biofilms were eliminated by R. nasuta after 5 days, whereas biofilms of the wild-type and the other mutants remained unaffected.
Fig. 4.
Inhibition of protozoan grazers in V. cholerae biofilms is regulated by hapR. Growth of the surface-feeding flagellate R. nasuta was evaluated on biofilms of the rugose wild-type A1552 and the isogenic mutants vpsR, vpsA, vpsL, vpsT, hapR, and luxT. Biofilms without (-GRAZ) and with (+GRAZ) grazer were examined by staining with SYTO 9 and by using confocal laser scanning microscopy. (Magnification, ×200; scale bar, 50 μm.) Note the elimination of the hapR- biofilm. Error bars indicate standard deviations of four replicates.
hapR (Quorum Sensing) Controls the Secretion of an Antiprotozoal Factor. We collected biofilm supernatants of the wild-type and the hapR mutant of both rugose and smooth V. cholerae A1552 and tested for their inhibitory effect on the activity of R. nasuta. Compared with the high number of active flagellates in the control treatment (>95%), flagellate activity dropped dramatically to ≈20% after adding supernatants of wild-type biofilms (Table 2). Supernatants of hapR mutant biofilms of both phase variants, however, had a significantly lower inhibitory effect on flagellate activity (P < 0.001), which indicates that the secretion of the antiprotozoal factor is controlled by quorum sensing.
Table 2. Activity of the surface-feeding flagellate R. nasuta in response to biofilm supernatants of V. cholerae A1552.
| % of active flagellates
|
||
|---|---|---|
| Strain | Rugose variant | Smooth variant |
| Wild-type | 18.5 ± 3.2 | 23.6 ± 4.4 |
| hapR− | 89.3 ± 11.1 | 84.4 ± 5.3 |
All values are means ± SD (n = 4) derived from microscopical inspection after 2 days. Statistical analyses were performed by using arcsine square root-transformed data.
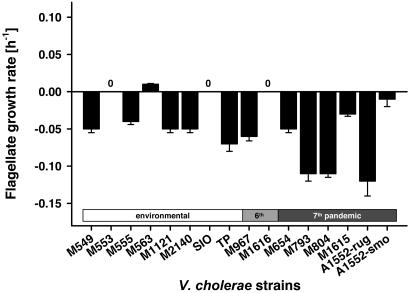
Biofilm-Mediated Grazing Resistance Is Widespread Among Toxigenic and Nontoxigenic V. cholerae Isolates. Based on our findings of biofilm-mediated grazing resistance in V. cholerae A1552, we extended our examinations to suspensions and biofilms of eight environmental nontoxigenic and eight toxigenic V. cholerae strains (two isolates from the sixth pandemic and six from the seventh pandemic). Flagellate grazing on V. cholerae suspensions revealed that all of the strains used provided high growth rates of C. roenbergensis, resulting in the rapid elimination of planktonic V. cholerae cells (data not shown). Biofilms of both environmental and toxigenic isolates, however, inhibited growth of R. nasuta efficiently, leading to zero or negative flagellate growth (Fig. 5). High flagellate mortality rates were observed more often with isolates from the seventh pandemic (three of six strains with flagellate loss rates >0.1 h-1), indicating a more drastic mechanism of grazing resistance of these isolates.
Fig. 5.
Widespread resistance of V. cholerae biofilms to protozoan grazing among toxigenic and nontoxigenic isolates. Growth of the surface-feeding flagellate R. nasuta was followed on biofilms of eight environmental, two sixth pandemic, and six seventh pandemic strains over 4 days. Error bars indicate standard deviations of three replicates.
Discussion
Recent attention has focused on the abiotic and biotic factors controlling the persistence of V. cholerae in aquatic ecosystems and the inherent selection pressures for the enrichment of toxigenic strains. The present study elucidates the role of a major mortality factor in natural microbial communities, predation by protozoans. The high elimination rates found for V. cholerae in planktonic ecosystems (29-32) suggest a tight control of V. cholerae growth and survival by protozoan predation. These observations raise the question how long-term persistence and seasonal accumulation of V. cholerae are possible in aquatic environments despite the predatory control by protozoans. The present study sheds light on this conundrum by directly comparing the antipredator fitness of planktonic versus biofilm V. cholerae cells. Our data reveal that V. cholerae biomass in biofilms remains stable in the presence of surface-feeding protozoa whereas planktonic V. cholerae cells are rapidly eliminated (Fig. 1), which illustrates the importance of surface-associated growth as a protective niche for the environmental persistence of V. cholerae. Intriguingly, our laboratory findings correspond well with a recent field study that reports the successful suppression of V. cholerae planktonic growth by protozoan grazing but a relative increase of V. cholerae cells associated with phytoplankton detritus (32). The protective nature of biofilms relative to planktonic cells has been reported for anthropogenic chemical stressors commonly used in infection control of bacterial pathogens (e.g., antibiotics, chlorine, and hydrogen peroxide; refs. 33 and 34) which has led to the assumption that biofilms in natural environments generally function as protective life strategy for bacteria (35). The present study on V. cholerae directly demonstrates the persistence advantage for biofilm bacteria over planktonic cells in the presence of an environmentally relevant stressor such as predation.
Interestingly, we found that flagellate grazing not only eliminates planktonic V. cholerae populations but also that the selective reduction of planktonic bacteria stimulates the formation of a grazing resistant biofilm population (Fig. 3). Apparently, biofilm populations are favored by the protective nature of cell consortia, the selective elimination of planktonic “competitor” cells, and grazing-mediated nutrient recycling, all of which can be expected to boost biofilm growth. Our findings specify a dual role for protozoan grazing as both a powerful mortality factor and an active driving force for biofilm formation and growth of V. cholerae in a protected niche. Therefore, the benefits of particle-associated growth are not limited to the availability of nutrients as previously thought but are extendable to effective grazing protection, which may jointly render biofilms the preferred ecological niche for V. cholerae.
The formation of biofilms is enhanced by the excessive production of exopolymers as commonly described for the rugose variants of V. cholerae (34, 36). Rugose V. cholerae strains have been isolated from cholera patients and the environment (28, 37) and were found to be equally as pathogenic as smooth strains (38, 39). The observed high-frequency switching between the smooth and rugose phenotype (40) has raised the question as to how phase variation may contribute to the environmental survival of V. cholerae. Our findings of the niche-specific separation into a mobile planktonic population consisting of the smooth-phase variant and a sessile biofilm population with a prevailing rugose phenotype (Table 1) illustrate the ecological consequences of the phenotypic and genetic differences of smooth and rugose variants reported earlier (28). Cells of the rugose variant are characterized by the production of excessive amounts of VPS due to the up-regulation of vps operons I and II and extracellular protein secretion (eps) genes and by a reduced transcription of flagellar (fla) and some of the chemotaxis genes (28). The marked differences in grazing mortality between planktonic and biofilm bacteria found in the present study specify the fitness conflicts of the “explorer strategy” of the smooth phenotype versus the “persister strategy” of rugose biofilm cells. Similar to the observations made on mucoid and nonmucoid variants of Pseudomonas fluorescens (41), protozoan grazing selects for the persister phenotype in V. cholerae, which is characterized by enhanced biofilm formation. Based on the idea of the insurance hypothesis (42), we propose that the functional diversity produced by phase variation between the smooth and rugose phenotype provides a selective advantage to V. cholerae for increased persistence in a heterogeneous and variable environment.
The protective nature of V. cholerae biofilms against grazing was not based simply on the physical inaccessibility of biofilm consortia. The decline of flagellate numbers observed on V. cholerae biofilms (Fig. 2) and the uncompromised persistence of VPS-deficient mutants (Fig. 4) indicated the existence of an additional chemical defense mechanism. The spatial concentration of bacteria on a surface allows synergistic interactions of cells in a semidiffusible environment. These high-density bacterial consortia are the prerequisite for cell density-dependent regulation, so-called quorum sensing, which synchronizes, for example, the secretion of extracellular effector molecules (43). According to the current model, information from the three quorum-sensing circuits in V. cholerae is channeled to the two component response regulator protein LuxO, which in turn regulates HapR and thereby controls gene expression (27). Although HapR has been demonstrated to control a range of phenotypes, including biofilm formation, our study suggests a distinctive environmental role for the HapR regulon, namely the involvement in the production of an antiprotozoal factor inhibiting flagellate activity and thus protecting the biofilm from grazing.
The antiprotozoal factor(s) of V. cholerae was determined to be an exoproduct because cell-free biofilm supernatants significantly reduced the feeding activity of the flagellates (Table 2). Besides being instrumental for communication by means of quorum sensing, biofilms enable cells to synergistically act to secrete defensive factors and thus are functionally critical for employing extracellular inhibitors against predators or competitors (13). As opposed to intracellular storage of predator-active compounds (44), diffusion of extracellular effector molecules must be considered a significant energy sink that can be minimized by the exopolymer matrix and high cell densities found in biofilms. Although the production of VPS facilitates the formation of biofilms and thus the avoidance from planktonic grazers, the density-dependent production of antiprotozoal factors seems to implement the effective protection of V. cholerae against biofilm grazers (compare refs. 45 and 46). Both high-density cell consortia mediated by VPS production and inhibitor secretion synchronized by quorum sensing indicate some degree of cellular cooperation to enhance antipredator fitness and therefore highlight the essential role of biofilm-specific cellular mechanisms in concentrating V. cholerae cells in the environment.
Although the identity of the antiprotozoal compound remains unknown, the fact that both toxigenic and nontoxigenic strains exhibit a significant level of inhibitory activity (Fig. 5) and that the cholera toxin CT has no impact on protozoan grazers (data not shown) points to the contribution of accessory and hitherto unknown cytotoxins to the environmental fitness of V. cholerae. Recent studies report that toxin-related genes are present in a wide range of nontoxigenic environmental strains (47), suggesting that environmental strains may have additional undiscovered virulence genes. Further studies are required to specify the benefits imparted to V. cholerae for possessing and maintaining virulence-associated genes in the environment and their putative role in the antagonistic interaction with protozoan predators.
The current model for the seasonal occurrence of cholera epidemics involves a number of abiotic factors favoring V. cholerae growth (e.g., temperature, pH, salinity, and dissolved organic carbon) and the members of the planktonic food web, such as algae and zooplankton (6). Recent findings also suggest the involvement of bacteriophages in the autoregulation of V. cholerae populations (48, 49). Our results introduce protozoan grazing as a powerful ecological agent in the control of environmental V. cholerae populations and identify biofilm-associated mechanisms that ensure the persistence and accumulation of V. cholerae in aquatic environments despite the imminent predatory pressure. We propose that protozoan grazing, as a major mortality factor of planktonic bacterial communities, contributes to the suppression and selective enrichment of toxigenic V. cholerae strains, which may have important implications for the evolution of pathogenic clones and the temporal dynamics of cholera epidemics.
Acknowledgments
We thank Ruiting Lan (University of New South Wales) and Douglas Bartlett (The Scripps Institution of Oceanography, La Jolla, CA) for providing isolates of V. cholerae, and Hartmut Arndt (University of Cologne, Cologne, Germany) for providing original cultures of R. nasuta and C. roenbergensis. We also thank one anonymous reviewer for his constructive comments on the manuscript. This work was supported by German Science Foundation (DFG) Grant Ma 2491/1-1 (to C.M.), the Otto Hahn Fellowship of the Max Planck Society (to C.M.), National Health and Medical Research Council Grant 222847 (to D.M. and S.K.), and the Centre for Marine Biofouling and Bio-Innovation at the University of New South Wales.
Author contributions: C.M., D.M., and S.K. designed research; C.M., A.M.M., and P.Y.Y. performed research; C.M. analyzed data; F.H.Y. contributed new reagents/analytic tools; and C.M. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviation: VPS, Vibrio polysaccharide.
References
- 1.Colwell, R. R. & Huq, A. (1994) Ann. N.Y. Acad. Sci. 740, 44-54. [DOI] [PubMed] [Google Scholar]
- 2.Chakraborty, S., Mukhopadhyay, A. K., Bhadra, R. K., Ghosh, A. N., Mitra, R., Shimada, T., Yamasaki, S., Faruque, S. M., Takeda, Y., Colwell, R. R. & Nair, G. B. (2000) Appl. Environ. Microbiol. 66, 4022-4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mukhopadhyay, A. K., Chakraborty, S., Takeda, Y., Nair, G. B. & Berg, D. E. (2001) J. Bacteriol. 183, 4737-4746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Khan, M. U., Shahidullah, M. D., Haque, M. S. & Ahmed, W. U. (1984) Trop. Geogr. Med. 36, 335-340. [PubMed] [Google Scholar]
- 5.Zo, Y. G., Rivera, I. N. G., Russek-Cohen, E., Islam, M. S., Siddique, A. K., Yunus, M., Sack, R. B., Huq, A. & Colwell, R. R. (2002) Proc. Natl. Acad. Sci. USA 99, 12409-12414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lipp, E. K., Huq, A. & Colwell, R. R. (2002) Clin. Microbiol. Rev. 15, 757-770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jürgens, K. & Matz, C. (2002) Antonie Leeuwenhoek 81, 413-434. [DOI] [PubMed] [Google Scholar]
- 8.Sherr, E. B. & Sherr, B. F. (2002) Antonie Leeuwenhoek 81, 293-308. [DOI] [PubMed] [Google Scholar]
- 9.Beardsley, C., Pernthaler, J., Wosniok, W. & Amann, R. (2003) Appl. Environ. Microbiol. 69, 2624-2630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chao, W. L., Ding, R. J. & Chen, R. S. (1988) Can. J. Microbiol. 34, 753-756. [DOI] [PubMed] [Google Scholar]
- 11.Davies, M. D., Long, J. A. H., Donald, M. & Ashbolt, N. J. (1995) Appl. Environ. Microbiol. 61, 1888-1896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sibille, I., Sime-Ngando, T., Mathieu, L. & Block, J. C. (1998) Appl. Environ. Microbiol. 64, 197-202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matz, C. & Kjelleberg, S. (2005) Trends Microbiol. 13, 302-307. [DOI] [PubMed] [Google Scholar]
- 14.Huq, A., Small, E. B., West, P. A., Huq, M. I., Rahman, R. & Colwell, R. R. (1983) Appl. Environ. Microbiol. 45, 275-283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Islam, M. S., Drasar, B. S. & Sack, R. B. (1994) J. Diarrhoeal Dis. Res. 12, 87-96. [PubMed] [Google Scholar]
- 16.Tamplin, M. L., Gauzens, A. L., Huq, A., Sack, D. A. & Colwell, R. R. (1990) Appl. Environ. Microbiol. 56, 1977-1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hall-Stoodley, L. & Stoodley, P. (2005) Trends Microbiol. 13, 7-10. [DOI] [PubMed] [Google Scholar]
- 18.Parsek, M. R. & Singh, P. K. (2003) Annu. Rev. Microbiol. 57, 677-701. [DOI] [PubMed] [Google Scholar]
- 19.Colwell, R. R., Huq, A., Islam, M. S., Aziz, K. M. A., Yunus, M., Khan, N. H., Mahmud, A., Sack, R. B., Nair, G. B., Chakraborty, J., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 1051-1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Artolozaga, I., Ayo, B., Latatu, A., Azúa, I., Unanue, M. & Iriberri, J. (2000) FEMS Microbiol. Ecol. 33, 191-196. [DOI] [PubMed] [Google Scholar]
- 21.Patterson, D. J. & Lee, W. Y. (2000) in The Flagellates: Unity, Diversity and Evolution, eds. Leadbeater, B. S. C. & Green, J. C. (Taylor & Francis, London), pp. 269-287.
- 22.Boenigk, J. & Arndt, H. (2000) Aquat. Microb. Ecol. 22, 243-249. [Google Scholar]
- 23.Paludan-Müller, C., Weichart, D., McDougald, D. & Kjelleberg, S. (1996) Microbiology 142, 1675-1684. [DOI] [PubMed] [Google Scholar]
- 24.Vaatanen, P. (1976) Walter Andre Nottback Found. Sci. Rep. 1, 1-58. [Google Scholar]
- 25.O'Toole, G. A., Pratt, L. A., Watnick, P. I., Newman, D. K., Weaver, V. B. & Kolter, R. (1999) Methods Enzymol. 310, 91-109. [DOI] [PubMed] [Google Scholar]
- 26.Casper-Lindley, C. & Yildiz, F. H. (2004) J. Bacteriol. 186, 1574-1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hammer, B. K. & Bassler, B. L. (2003) Mol. Microbiol. 50, 101-114. [DOI] [PubMed] [Google Scholar]
- 28.Yildiz, F. H., Liu, X. S., Heydorn, A. & Schoolnik, G. K. (2004) Mol. Microbiol. 53, 497-515. [DOI] [PubMed] [Google Scholar]
- 29.Macek, M., Carlos, G., Memije, P. & Ramirez, P. (1997) Aquat. Microb. Ecol. 13, 257-266. [Google Scholar]
- 30.Perez, M. E. M., Macek, M. & Galvan, M. T. C. (2004) Trop. Med. Int. Health 9, 133-140. [DOI] [PubMed] [Google Scholar]
- 31.Mourino-Perez, R. R., Worden, A. Z. & Azam, F. (2003) Appl. Environ. Microbiol. 69, 6923-6931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Worden, A. Z., Seidel, M., Smriga, S., Wick, A., Malfatti, F., Bartlett, D. & Azam, F. (July, 2005) Environ. Microbiol., 10.1111/j. 1462-2920.2005.00863.x. [DOI] [PubMed]
- 33.Fux, C. A., Costerton, J. W., Stewart, P. S. & Stoodley, P. (2005) Trends Microbiol. 13, 34-40. [DOI] [PubMed] [Google Scholar]
- 34.Yildiz, F. H. & Schoolnik, G. K. (1999) Proc. Natl. Acad. Sci. USA 96, 4028-4033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hall-Stoodley, L., Costerton, J. W. & Stoodley, P. (2004) Nat. Rev. Microbiol. 2, 95-108. [DOI] [PubMed] [Google Scholar]
- 36.Watnick, P. I. & Kolter, R. (1999) Mol. Microbiol. 34, 586-595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mitra, R. K., Nandy, R. K., Ramamurthy, T., Bhattacharya, S. K., Yamasaki, S., Shimada, T., Takeda, Y. & Nair, G. B. (2001) J. Med. Microbiol. 50, 268-276. [DOI] [PubMed] [Google Scholar]
- 38.Islam, M. S., Ahsan, S., Khan, S. I., Ahmed, Q. S., Rashid, M. H., Islam, K. M. N. & Sack, R. B. (2004) Microbiol. Immunol. 48, 229-235. [DOI] [PubMed] [Google Scholar]
- 39.Morris, J. G., Jr., Sztein, M. B., Rice, E. W., Nataro, J. P., Losonsky, G. A., Panigrahi, P., Tacket, C. O. & Johnson, J. A. (1996) J. Infect. Dis. 174, 1364-1368. [DOI] [PubMed] [Google Scholar]
- 40.Ali, A., Rashid, M. H. & Karaolis, D. K. R. (2002) Appl. Environ. Microbiol. 68, 5773-5778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matz, C., Deines, P. & Jürgens, K. (2002) FEMS Microbiol. Ecol. 39, 57-65. [DOI] [PubMed] [Google Scholar]
- 42.Yachi, S. & Loreau, M. (1999) Proc. Natl. Acad. Sci. USA 96, 1463-1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhu, J., Miller, M. B., Vance, R. E., Dziejman, M., Bassler, B. L. & Mekalanos, J. J. (2002) Proc. Natl. Acad. Sci. USA 99, 3129-3134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matz, C., Deines, P., Boenigk, J., Arndt, H., Eberl, L., Kjelleberg, S. & Jürgens, K. (2004) Appl. Environ. Microbiol. 70, 1593-1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Matz, C., Bergfeld, T., Rice, S. A. & Kjelleberg, S. (2004) Environ. Microbiol. 6, 218-226. [DOI] [PubMed] [Google Scholar]
- 46.Weitere, M., Bergfeld, T., Rice, S. A., Matz, C. & Kjelleberg, S. (2005) Environ. Microbiol. 7, 1593-1601. [DOI] [PubMed] [Google Scholar]
- 47.Purdy, A., Rohwer, F., Edwards, R., Azam, F. & Bartlett, D. H. (2005) J. Bacteriol. 187, 2992-3001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Faruque, S. M., Bin Naser, I., Islam, M. J., Faruque, A. S. G., Ghosh, A. N., Nair, G. B., Sack, D. A. & Mekalanos, J. J. (2005) Proc. Natl. Acad. Sci. USA 102, 1702-1707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Faruque, S. M., Islam, M. J., Ahmad, Q. S., Faruque, A. S. G., Sack, D. A., Nair, G. B. & Mekalanos, J. J. (2005) Proc. Natl. Acad. Sci. USA 102, 6119-6124. [DOI] [PMC free article] [PubMed] [Google Scholar]