Abstract
Improving the digestibility of forages provides a means to enhance animal performance and protect the environment against excessive animal waste. Increased lignin content during maturity, and corresponding changes in lignin composition, correlate with decreased digestibility of forages. These relationships have yet to be investigated in isogenic systems. By targeting three specific cytochrome P450 enzymes of the lignin pathway for antisense down-regulation, we generated transgenic alfalfa lines with a range of differences in lignin content and composition. There was a strong negative relationship between lignin content and rumen digestibility, but no relationship between lignin composition and digestibility, in these transgenic lines. Models for genetic manipulation of forage digestibility based on the changes in lignin composition that increase paper-pulping efficiency in trees are therefore invalid. Down-regulation of 4-coumarate 3-hydroxylase provided the largest improvements in digestibility yet seen in a forage crop.
Keywords: forage digestibility, lignin modification, metabolic engineering
Feeding and grazing studies have shown that small changes in forage digestibility significantly impact animal performance (1), and improving digestibility is an important goal of forage breeding programs. Lignin, a polymer of hydroxylated and methoxylated phenylpropane units (monolignols) linked by means of oxidative coupling (2), exerts a negative influence on digestibility.
Lignin is found in secondarily thickened plant cell walls and is critical for structural integrity of the wall and the strength of stems (3, 4). Angiosperm lignin contains two major monolignols, monomethoxylated guaiacyl (G) and dimethoxylated syringyl (S) units, polymerized through at least five different linkage types (5). Gymnosperm lignin is low in S units and highly condensed because the high G unit composition facilitates additional interunit bonding. Gymnosperm wood is therefore less amenable than angiosperm wood to the harsh chemical and physical treatments (pulping) necessary to remove lignin from cellulose during paper production (6), and pulping properties are improved by genetic modification of trees to increase the S/G ratio (5). In many forage crops, lignin content and S/G ratio increase with stem maturity (7, 8), and both parameters correlate negatively with forage digestibility in ruminant animals (8–12). The amount of G lignin also has been linked with reduced cell wall degradability in forages (13), although studies with synthetic lignins (14) have questioned effects of lignin composition on forage digestibility.
Most of the above studies used materials with different lignin contents and/or compositions as a result of divergent selection (15), natural genetic variation (16), delignification (17), or maturity (18). Such studies are complicated by the many uncontrolled genetic and developmental variables potentially affecting digestibility (19). Alfalfa (Medicago sativa L.) is an autotetraploid, autogamous forage legume, and inbred lines are therefore not available. Generating isogenic transgenic lines in which lignin content or composition is modified by altering expression of a single gene provides a strategy for better elucidating the lignin/digestibility relationship in alfalfa.
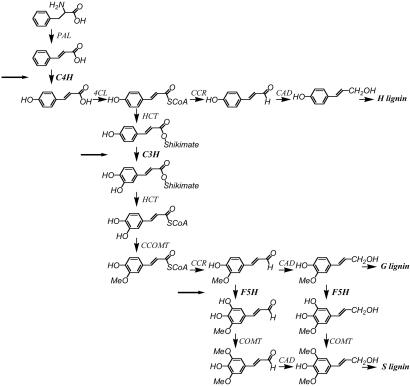
Lignin biosynthesis is now believed to proceed according to the pathway in Fig. 1 (20–24). Genes encoding all of the enzymes in Fig. 1 have been identified, and many of the enzymatic steps have been down-regulated in transgenic plants. The results of these studies (24–30) have generally confirmed the pathway in Fig. 1 and supported increasing S/G ratio as a means of improving paper pulping in trees. They have not, however, provided definitive information on the major factors impacting digestibility of forage crops.
Fig. 1.
Current scheme (52) for the biosynthesis of monolignols. PAL, L-phenylalanine ammonia-lyase; 4CL, 4-coumarate: CoA ligase; CCR, cinnamoyl CoA reductase; CAD, cinnamyl alcohol dehydrogenase; HCT, hydroxycinnamoyl CoA: hydroxycinnamoyl transferase. Targets for antisense down-regulation are marked with arrows.
To generate transgenic alfalfa lines with large changes in lignin content and composition, we targeted the three cytochrome P450 enzymes, cinnamate 4-hydroxylase (C4H), coumaroyl shikimate 3-hydroxylase [also known as coumarate 3-hydroxylase (C3H)] and coniferaldehyde 5-hydroxylase [also known as ferulate 5-hydroxylase (F5H)], that occupy strategically placed reactions in the formation of all monolignols, G-units, and S-units, respectively (Fig. 1). Down-regulation of C4H, C3H, or F5H produced plants with greatly reduced lignin without significant impact on composition, lignin-rich in p-hydroxyphenyl (H) units, or lignin rich in G-units with reduced S content, respectively. By using these plants, as well as previously generated alfalfa lines down-regulated in caffeic acid 3-O-methyltransferase (COMT) and caffeoyl CoA 3-O-methyltransferase (CCoAOMT), we have determined the relationships between lignin content, composition, and forage quality in alfalfa.
Materials and Methods
Plasmids and Alfalfa Transformation. Standard methods (31) were used to make all gene constructs. The shuttle vector pPTN1 (32) containing the bean PAL2 promoter was digested with BamHI and NdeI and blunt ended by using Turbo Pfu Polymerase (Stratagene). The Gateway cassette (33) was ligated to the blunt-ended pPTN1, which then was digested with EcoRI and HindIII to release the bean PAL2 promoter-Gateway cassettenos terminator. This cassette was ligated into pCAMBIA2200 digested with EcoRI and HindIII, resulting in the binary destination vector pCAMBIA2200-GW (see Fig. 4, which is published as supporting information on the PNAS web site).
The entire coding sequence of M. truncatula TC94739, an ortholog of Arabidopsis C3H (22), was amplified by PCR using forward (ATGGCTCTGTTTCTCACAATACCCCTTTCA) and reverse (CACCTTAGATATCAGCTGTCACACGTTTGTACAA) primers. Similarly, M. truncatula C4H represented by TC106704 was amplified by using forward (ATATGGATCTTCTCCTCCTTGAAAAGACC) and reverse (CACCTTAAAATGATCTTGGCTTAGCAACAATGGT) primers. Two F5H cDNAs, K1 and K10, were selected by screening an alfalfa stem cDNA library. The coding sequence of alfalfa F5H-K10 was amplified by PCR using forward (GCCACCATGGATTCGCTTCTAAAATTTCCAATCATG) and reverse (CACCTTAATCCAAAGGACACAAGACACGCTTAGT) primers. PCR-amplified P450 coding sequences were directionally cloned in the antisense orientation into pENTR/D-TOPO (Invitrogen) according to the manufacturer's protocol. They then were introduced into pCAMBIA2200-GW by using the LR clonase mix from Invitrogen, resulting in pCAMBIA2200-MtC3Has, pCAMBIA2200MtC4Has, and pCAMBIA2200-MsF5Has (Fig. 4). The three binary plasmids were introduced into Agrobacterium tumefaciens strain GV3101 (34) by electroporation. Leaf disk transformation of clonally propagated, tetraploid alfalfa cv Regen SY was performed as described in ref. 35, by using kanamycin (25 mg/liter) selection.
RNA Gel Blot Analysis. Total RNA was isolated from 500 mg of ground stem tissue (internodes 2–5) by using TRIREAGENT solution (Molecular Research Center, Cincinnati) according to the manufacturer's instructions. Ten micrograms of total RNA was separated on 1.2% formaldehyde-containing agarose gels and transferred onto a nylon membrane (Hybond-N+, Amersham Pharmacia Biosciences) by following standard procedures (31) and UV cross-linked by using a Stratalinker (Stratagene). The ≈1.5 kb C3H, C4H, and F5H coding sequences were labeled with a α-P32-dCTP labeling kit (Amersham Pharmacia), purified on Probe Quant G50 microcolumns, and used for hybridization as described in ref. 36.
Enzyme Extraction and Assay. Stem tissue from internodes 3–6 was harvested from greenhouse-grown alfalfa plants and ground in liquid nitrogen by using a mortar and pestle. Microsomes were isolated from 5 g of ground tissue as described in ref. 37 and assayed for C4H (37), C3H (22), and F5H (21) by using the described procedures. Various modifications to time of incubation, buffer pH, and nature of reductant (2-mercaptoethanol or glutathione) were made in attempts to measure F5H activity.
Histochemical Staining of Lignin. Alfalfa stem sections were made by using a vibratome (series 1000, Ted Pella, Inc., Redding, CA) and subjected to Maule staining as described in ref. 32. Photographs were taken by using an Olympus SZX stereomicroscope system with a SPOT RT color camera.
Determination of Lignin Content and Composition. Lignin content of stem material (internodes 1–5) was determined by the acetyl bromide method (38). Thioacidolysis (39, 40) was used to determine lignin composition. Thioacidolysis was performed by using ≈20 mg of extractive-free samples reacted with 15 ml of 0.2 M BF3 etherate in an 8.75:1 dioxane/ethanethiol mixture. Lignin-derived monomers were identified by gas chromatography mass spectrometry (GC/MS) and quantified by GC, as their trimethylsilyl derivatives. The GC/MS was performed on a Hewlett–Packard 5890 series II gas chromatograph with a 5971 series mass selective detector (column:HP-1; 60 m × 0.25 mm; 0.25-μm film thickness), and mass spectra were recorded in electron impact mode (70 eV) with 60–650 m/z scanning range.
Fractionation of Cell Wall Polysaccharides. Ground alfalfa tissue samples were boiled for 15 min in methanol at 80°C, then washed and stored in methanol. Cell wall polysaccharides were fractionated as described in ref. 41. Total sugar content in each fraction was determined by the phenol-sulfuric acid method (42) using glucose as standard.
Determination of Forage Quality. Vegetatively propagated alfalfa cuttings were grown in parallel in 1-gallon pots in the greenhouse. Aerial portions were harvested at the early bud stage to ensure material was matched developmentally, and dried in a 50°C oven for at least 72 h. The samples were then ground in a Thomas-Wiley model 4 Laboratory Mill (Lehman Scientific, Wrightsville, PA) with 1-mm sieves. Acid detergent fiber and neutral detergent fiber (NDF) were estimated by standard protocols (43). For NDF analysis, 0.35 g of ground samples were transferred to a F57 ANKOM filter bag (ANKOM Technology, Fairport, NY) and heated at 100°C for 1 h in an ANKOM Fiber Analyzer, according to the manufacturer's instructions. The samples were washed in near-boiling water, dried at 105°C for 6 h, and weighed to determine fiber loss. Acid detergent fiber was estimated sequentially on the material remaining after NDF analysis. The residue was then used for determination of acid detergent lignin (ADL) by incubation in 72% (vol/vol) sulfuric acid for 3 h, washing thoroughly, and drying at 105°C for 6 h, before weighing.
For determination of in vitro digestibility, ground tissue samples were dried at 105°C for 6 h before determining preextraction dry weights. The same procedure was used to obtain postextraction dry weights. Digestibility analysis (0.5 g samples) was performed by using F57 filter bags and the DAISY II incubator (ANKOM Technology) (44), following the manufacturer's instructions. For determination of in situ digestibility in fistulated steers, ground, dried tissue (5 g) was put into each preweighed ANKOM rumen in situ filter bag (10 × 20 cm; pore size = 50 μm). These bags were placed in a Mainstays mesh utility bag (60.96 × 91.44 cm; Pro-Mart Industries, Rancho Cucumanoga, CA) and then placed into the rumens of fistulated steers for 12, 24, 36, and 72 h of digestion. The five steers were placed on ad lib alfalfa hay while pastured in small pens with a low-volume forage base of volunteer winter annuals and dormant bermudagrass for 2 weeks before the trials. During the trials, they were fed only alfalfa. Each sample was in duplicate for each time point in each of the steers. The bags were removed from the rumen, thoroughly washed in a commercial washing machine, and freeze dried. Digestibility was calculated based on the sample dry weight difference before and after digestion.
Results
Transgenic Alfalfa Down-Regulated in Cytochrome P450 Genes. The model legume Medicago truncatula (45) is closely related to alfalfa, with very high sequence identity among orthologous genes (46). M. truncatula C4H and C3H sequences were mined from the TIGR Medicago gene index (www.tigr.org/tigr-scripts/tgi/T_index.cgi?species=medicago). Tentative consensus (TC) sequences 94739 and 106704, orthologs of C3H and C4H, respectively, existed as full-length clones in the Noble Foundation's M. truncatula EST library collection. Alfalfa F5H-K10, selected by screening a stem cDNA library, was 97% identical in its ORF to alfalfa F5H-K1 and 96% identical to M. truncatula TC110196. Full-length ORFs of M. truncatula C4H and C3H, and alfalfa F5H-K10, were cloned into a pCAMBIA binary vector in the antisense orientation, under control of the vascular tissue-specific bean PAL2 promoter (32, 47) (see Fig. 4), and introduced into alfalfa by Agrobacterium-mediated transformation and regeneration by means of somatic embryogenesis.
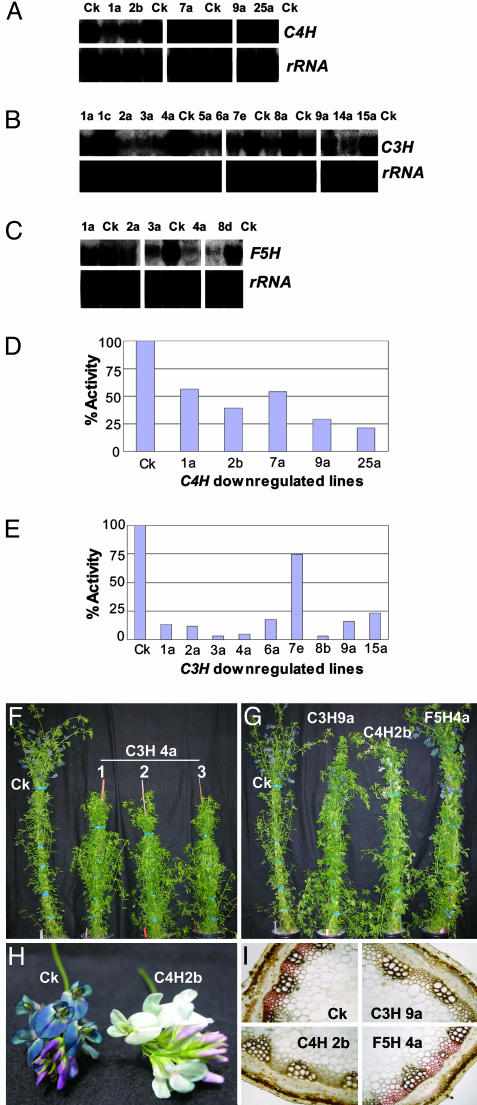
Transgene insertion was determined by PCR analysis for each of 76 C4H, 57 C3H, and 110 F5H lines that survived selection and transfer to the greenhouse. Of these, 41 C4H, 30 C3H, and 32 F5H lines represented independent transformation events because each was derived from an individual callus. These lines were analyzed for target P450 transcript levels by RNA gel blot analysis or RT-PCR. Five C4H, 11 C3H, and 5 F5H lines exhibited reduced target transcript levels in stem tissue (Fig. 2 A–C).
Fig. 2.
Molecular and visible phenotypes resulting from down-regulation of cytochrome P450 enzymes in alfalfa. (A–C) RNA gel blot analysis of stem tissue (internodes 1–5) from selected control lines and independent antisense transformants down-regulated in C4H (A), C3H (B), and F5H (C). Blots were probed with full-length cDNAs corresponding to the targeted transgene and 18S rRNA as loading control. (D and E) Corresponding enzyme activities for the target C4H and C3H P450s in alfalfa stem microsomes. Ck, independent control lines. Numbers refer to independent down-regulated transgenics. (F) Overall appearance and size comparisons of early bud stage empty vector-transformant and three clonally propagated C3H 4a lines of the same age. (G) Overall appearance and size comparisons of early bud stage empty vector-transformant and phenotypically normal C3H, C4H, and F5H down-regulated lines of the same age. (H) Flowers of an empty vector control (Ck) and C4H down-regulated plant. (I) Maule staining of cross sections through the 5th internodes of empty vector control line Ck48 and plants down-regulated in C3H (line 9a), C4H (line 2b), and F5H (line 4a).
Microsomal preparations were made from 5-g batches of stems from the individual down-regulated lines, and C4H, C3H, and F5H enzymatic activities were measured by using cinnamic acid, 4-coumaroyl shikimate, or coniferyl alcohol, respectively, as substrates. Products from assays performed in the presence or absence of NADPH were monitored by HPLC with diode array detection (see Fig. 5, which is published as supporting information on the PNAS web site). The results (Fig. 2 D and E) demonstrated strong down-regulation of the targeted C4H and C3H activities in several independent lines. The strongest reduction in C4H activity was to 21% of wild-type (WT) activity in line 25a, whereas C3H activity was reduced to 3% of WT activity in lines 3a and 8b and to 5% in line 4a. Surprisingly, we could not demonstrate F5H activity in microsomes from WT or transgenic plants using ferulic acid, coniferaldehyde, or coniferyl alcohol as potential substrates, using previously published protocols and variants thereof.
Phenotypes of Lignin-Modified Alfalfas. All plants with <30% of WT C4H activity, or <15% of WT C3H activity, appeared somewhat smaller than corresponding empty vector control lines or lines down-regulated in F5H (Fig. 2 F and G). However, C3H line 9a, with ≈20% of WT C3H activity, and C4H line 2b, with ≈40% of WT C4H activity, were of normal size (Fig. 2G). A striking loss of purple-blue flower pigmentation was seen in all C4H down-regulated lines (Fig. 2H). C3H lines 4a and 5a showed delayed flowering, by as many as 4 weeks in the case of 5a (see Table 4, which is published as supporting information on the PNAS web site).
Stem cross-sections of the most down-regulated lines for each transgene were treated with Maule reagent (48) (Fig. 2I). The bright red coloration throughout the vascular tissue was reduced in the C4H line to a dark brown coloration with more limited distribution between the major xylem cells, indicating an overall reduction in S lignin content. A similar coloration was observed in the C3H down-regulated line. The F5H down-regulated line maintained a similar staining pattern, but with reduced intensity, to that of the empty vector control.
The Regen SY genotype of alfalfa used for transformation is not lodging resistant. However, lodging scores were not significantly increased in P450 down-regulated plants grown in the greenhouse (data not shown).
Lignin and Forage Quality Parameters. Total forage samples (leaf plus stem) from internodes 1 (uppermost) to 5 were harvested from independent P450 down-regulated lines at the first bud stage. Lignin content was estimated by the acetyl bromide procedure and by total thioacidolysis yield. Acetyl bromide lignin levels of forage samples were significantly reduced in C4H and C3H down-regulated lines, but not in F5H down-regulated lines (Table 1). These results were even more apparent in the corresponding total thioacidolysis yields, and striking differences were observed in the thioacidolysis yields of the individual H, G, and S monomers. Thus, down-regulation of C4H resulted in a relative increase in the ratio of G to total units at the expense of S units, with a resultant drop in S/G ratio. A very similar pattern was seen for the F5H down-regulated lines, although total thioacidolysis yield was considerably higher. C4H and F5H down-regulation therefore have different effects on lignin content but cause similar changes in overall lignin composition in alfalfa. In contrast, down-regulation of C3H resulted in a massive increase in the proportion of H units in the lignin and a significant decrease in the ratio of G to total units.
Table 1. Lignin content and composition of control and transgenic alfalfa (leaf plus stem) down-regulated in C4H, C3H, or F5H.
| Acetyl bromide lignin, g/g dry wt
|
Thioacidolysis yield, μmol/g dry wt
|
Thioacidolysis monomer ratios
|
||||
|---|---|---|---|---|---|---|
| Plant line* | H/total | G/total | S/total | S/total | ||
| C4H | 0.06 ± 0.01 | 20.32 ± 21.96 | 0.04 ± 0.01 | 0.80 ± 0.02 | 0.16 ± 0.02 | 0.20 ± 0.03 |
| C3H | 0.07 ± 0.01 | 54.05 ± 35.63 | 0.48 ± 0.06 | 0.32 ± 0.04 | 0.20 ± 0.02 | 0.62 ± 0.05 |
| F5H | 0.10 ± 0.02 | 169.81 ± 24.20 | 0.03 ± 0.00 | 0.80 ± 0.04 | 0.17 ± 0.04 | 0.21 ± 0.05 |
| Control | 0.10 ± 0.01 | 149.32 ± 16.74 | 0.03 ± 0.01 | 0.63 ± 0.02 | 0.30 ± 0.05 | 0.47 ± 0.07 |
Lines analyzed were 1a and 2b (C4H; n = 2); 1a, 2a, 3a, 4a, 5a, and 9a (C3H; n = 6); and 1a and 4a (F5H; n = 2). Controls (n = 6) included nontranformed and empty vector lines. All lines were in the same clonally propagated genotype.
The same samples were analyzed for cell wall polysaccharide composition (Table 2). The levels of hemicellulose were reduced in the C3H lines, but α-cellulose (cellulose plus lignin) was relatively constant in control and transgenic lines. The constant α-cellulose level in plants with reduced lignin levels suggests compensatory cellulose accumulation, as reported previously in poplar plants down-regulated in 4CL (49).
Table 2. Cell wall polysaccharide composition of control and transgenic alfalfa (leaf plus stem) down-regulated in C4H, C3H, or F5H.
| Plant line* | Pectin (% dry wt) | Hemicellulose (% dry wt) | α-Cellulose (% dry wt) |
|---|---|---|---|
| C4H | 19.84 ± 3.79 | 39.08 ± 5.28 | 55.68 ± 4.02 |
| C3H | 19.21 ± 4.99 | 31.36 ± 1.93 | 56.17 ± 3.14 |
| F5H | 20.71 ± 5.77 | 36.38 ± 2.20 | 60.09 ± 7.18 |
| Control | 25.47 ± 4.32 | 35.28 ± 3.60 | 55.87 ± 2.39 |
Plant lines are as in Table 1.
Forage quality analysis was performed on stem material from highly down-regulated lines (Table 3), and on previously generated lines, in the same genetic background, down-regulated in COMT, CCoAOMT, or both enzymes (32). Down-regulation of COMT results in a strikingly reduced S/G ratio, whereas down-regulation of CCoAOMT reduces G lignin, but not S lignin, in alfalfa (32). Acid detergent fiber and NDF were slightly reduced in all transgenic as compared with control lines (Table 3). More striking differences were observed for ADL levels, which mirrored the lignin values obtained by the acetyl bromide and thioacidolysis approaches. ADL was not reduced in the F5H down-regulated line 4a. The reduced values for COMT and CCoAOMT lines relative to controls (Table 3) were as observed in ref. 28.
Table 3. Forage quality analysis of stem material from individual plant lines.
| Plant line | IVDMD | ADF | NDF | ADL |
|---|---|---|---|---|
| C3H 4a | 84.10 ± 0.32 | 49.62 ± 0.26 | 60.52 ± 0.31 | 6.52 ± 0.11 |
| C4H 2b | 78.01 ± 0.19 | 52.99 ± 0.34 | 65.42 ± 0.08 | 7.28 ± 0.12 |
| F5H 4a | 54.80 ± 0.32 | 57.77 ± 0.11 | 68.70 ± 0.39 | 13.34 ± 0.04 |
| C1-4 | 66.94 ± 0.61 | 49.42 ± 0.28 | 59.61 ± 0.62 | 9.84 ± 0.11 |
| CC2-305 | 66.62 ± 0.83 | 52.48 ± 0.17 | 64.71 ± 0.37 | 8.67 ± 0.15 |
| CK 48 | 56.26 ± 0.04 | 59.07 ± 0.17 | 69.60 ± 0.28 | 11.92 ± 0.19 |
| C3H 4a | 82.13 ± 0.41 | 52.90 ± 0.07 | 64.46 ± 0.41 | 7.49 ± 0.19 |
| C3H 5a | 77.98 ± 1.31 | 55.39 ± 0.04 | 67.82 ± 0.08 | 8.70 ± 0.25 |
| C3H 9a | 75.93 ± 0.29 | 56.72 ± 0.61 | 68.93 ± 0.24 | 8.70 ± 0.29 |
| CK 48 | 49.18 ± 0.16 | 61.31 ± 0.07 | 72.58 ± 0.15 | 13.42 ± .04 |
All values are expressed per g of dry weight and represent a single biological sample comprising 15-30 pooled, vegetatively propagated plants of each line. Analytical variation for triplicate samples is indicated. In addition to lines down-regulated in C3H, C4H, and F5H, lines down-regulated in COMT (C1-4) or CCoAOMT (CC2-305) were also included. CK 48 is an empty vector control line. The lower set of values for independent C3H lines and empty vector control were obtained from material regrown after cut back at a different time from the set of six lines above. ADF, acid detergent fiber; IVDMD, in vitro dry matter digestibility determined using rumen fluid.
Determination of forage quality parameters was repeated several times on regrown material after cut-back. Analysis was performed on stems (Table 3; see also Table 5, which is published as supporting information on the PNAS web site) or on total forage stems plus leaves (see Tables 6 and 7, which are published as supporting information on the PNAS web site). The relative differences in the various transgenic lines were confirmed.
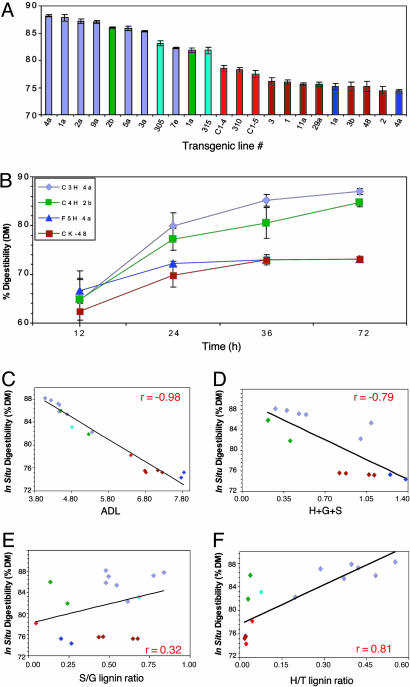
Relationship Between Forage Digestibility and Lignin Content/Composition. In vitro dry matter digestibility of stem or total forage was strikingly increased in the C4H and C3H lines and increased intermediately in the COMT and CCoAOMT lines, but was not much affected in the F5H lines (Tables 3, 5, and 7). To better pursue the lignin-digestibility relationship, multiple vegetative cuttings of individual C4H, C3H, COMT, CCoAOMT, and F5H lines, plus corresponding controls, were grown in the greenhouse to maturity (first bud stage) to generate sufficient forage material for in situ digestibility measurements in fistulated steers. For each of the 23 different lines used in the study, duplicate forage samples (stem material) were analyzed in five separate steers. The results (Fig. 3A) confirm a striking increase in in situ digestibility end points of stem material from the C3H and C4H lines, greater than the values for COMT or CCoAOMT down-regulated material observed previously (28) or in the present trial. The various empty vector control lines exhibited similar end point digestibility, indistinguishable from that of the F5H lines. This experiment was repeated on regrown material after cut-back of the plants, with essentially the same relative results (Table 6).
Fig. 3.
Digestibility relationships for P450 and OMT down-regulated alfalfa lines. (A) In situ digestibility of individual lines. (B) Digestibility kinetics for the most digestible line for each targeted transgene. (C) Relation between ADL content and in situ digestibility for the lines shown in A. (D) Relation between thioacidolysis yield and in situ digestibility. (E) Relation between S/G ratio and in situ digestibility. (F) Relation between H/T ratio and in situ digestibility. Purple, C3H; green, C4H; turquoise, CCoAOMT; red, COMT; brown, controls; blue, F5H. Results show average and SD (in A and B) for analysis of duplicate forage samples in five different steers.
Differences in digestibility for the most digestible line representative of each targeted transcript were apparent within 24 h of incubation in the rumen (Fig. 3B). F5H line 4a was more digestible than the control line at early time points but attained the same end-point digestibility value after 36 h.
There was a strong, negative, linear relationship (r = –0.98) between in situ digestibility and ADL level across all transgenic lines, irrespective of lignin composition (Fig. 3C). The less significant negative relationship between in situ digestibility and total thioacidolysis yield (T) (Fig. 3D) is consistent with thioacidolysis yield being a function of both lignin content and composition (39). There was no clear relationship between S/G ratio and in situ digestibility (Fig. 3E), and the positive relationship (r = 0.81) between H/T ratio and digestibility (Fig. 3F) can be explained by the contribution of the seven C3H lines, in which H/T ratio was related to reduced lignin and therefore increased digestibility; digestibility in the other lines varied greatly at a relatively constant, low H/T ratio. Taken together, the results demonstrate, for the isogenic material analyzed, that among the parameters measured, only lignin content significantly impacts forage digestibility in alfalfa.
The strong positive correlation between in vitro and in situ digestibility of the various transgenic lines (see Fig. 6, which is published as supporting information on the PNAS web site) suggests that future studies on digestibility of transgenic alfalfa forage will not require the expensive and time-consuming use of fistulated animals.
Discussion
We have determined the lignin:digestibility relationship in alfalfa using isogenic material and, at the same time, investigated optimal strategies for forage quality improvement. Antisense down-regulation of the three cytochrome P450 enzymes C4H, C3H, and F5H leads to striking but different changes in lignin content and composition in alfalfa. These changes do not reflect altered leaf:stem ratios, because the effects on total forage or stem material alone were the same. Reducing activity of the early pathway enzymes had a greater effect on lignin content than down-regulating S-lignin-specific F5H. Our inability to detect F5H activity in microsomes from whole stem extracts might result from the limited distribution of F5H to vascular tissue, whereas C4H and C3H, which are not specific for lignin biosynthesis, may be expressed in a larger number of cell types. Nevertheless, the two lines in which F5H transcripts were most strongly down-regulated exhibited a decrease in S lignin consistent with reduced F5H enzymatic activity (20).
The Arabidopsis ref8 mutant, in which C3H activity is totally abolished, exhibits an extreme dwarf phenotype (25). The present alfalfa C3H antisense lines, several of which had lost 95% or more of WT enzyme activity, exhibited H/T ratios ≈25-fold higher than those of control plants. Importantly, only those lines with the highest C3H down-regulation showed delayed growth. Thus, it is possible to produce alfalfa plants with strongly down-regulated C3H activity that appear phenotypically normal but still show striking changes in lignification, a conclusion not apparent from the phenotype of Arabidopsis ref8.
Arabidopsis ref8 exhibits increased cell wall digestibility with a crude mixture of endo- and exo-glucanases (50). However, it was not clear whether this increase was the result of reduced lignin content or altered lignin composition (50). The highly improved digestibility in alfalfa lines with equally strongly reduced lignin levels but closer to WT lignin composition after down-regulation of C4H suggests that lignin content is the major factor impacting digestibility of C3H-reduced alfalfa.
Improvements in alfalfa forage digestibility by down-regulation of the “late” pathway enzymes COMT and CAD (28–30) are considerably less than achieved here with C3H or C4H down-regulation. For development of agronomically viable lines, it may be necessary to further fine-tune the level of C3H or C4H expression to eliminate the possibility of reduced growth. Delayed flowering, as seen in some of the C3H lines, is advantageous for forage use (if it results in an ultimately increased biomass) but not for seed production. However, plants with less strong C3H down-regulation (e.g., line 9a) have excellent improvements in digestibility but maintain normal flowering. C4H down-regulated alfalfa lines not only showed a loss of flower coloration, possibly as a result of movement of the silencing signal from the vascular region of the stem to other parts of the plant (51), but also possessed a different floral scent, the chemical basis of which remains to be determined; together, these changes may have significant effects on pollination by honey bees.
The paper-pulping paradigm suggests that decreasing potential interunit linkages in lignin by increasing the S/G ratio is important for improving the chemical removal of lignin from cellulose. The present results indicate that the paper-pulping model does not apply to digestion of cell wall material by rumen microorganisms. In experiments that eliminated the potentially confounding variables encountered in previous studies, there was no relation between S/G ratio and digestibility, whereas total lignin content was very highly correlated and is thus the parameter most affecting digestibility.
The U.S. Dairy and Forage Research Center has estimated that a 10% increase in fiber digestibility would result in an annual $350 million increase in U.S. milk/beef production and a 2.8 million tons decrease in manure solids produced each year. The present work defines enzymatic targets that allow this type of improvement to be realized in alfalfa, the United States' major forage legume.
Supplementary Material
Acknowledgments
We thank Drs. Stephen Temple and Kiran Mysore for critical reading of the manuscript, Dennis Walker for assistance with forage quality analysis, and Dr. John Ralph (U.S. Dairy and Forage Research Center, Madison, WI) for providing coumaroyl and caffeoyl shikimate. This work was supported by Forage Genetics International and the Samuel Roberts Noble Foundation.
Author contributions: M.S.S.R. and R.A.D. designed research; M.S.S.R., F.C., G.S., L.J., and H.A. performed research; F.C. and H.A. contributed new reagents/analytic tools; M.S.S.R., F.C., and R.A.D. analyzed data; and R.A.D. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: ADL, acid detergent lignin; C3H, coumarate 3-hydroxylase; C4H, cinnamate 4-hydroxylase; COMT, caffeic acid 3-O-methyltransferase; CCoAOMT, caffeoyl CoA 3-O-methyltransferase; F5H, ferulate 5-hydroxylase; G, guaiacyl; H, p-hydroxyphenyl; NDF, neutral detergent fiber; S, syringyl.
Data deposition: The alfalfa F5H cDNA sequences reported in this paper have been deposited in the GenBank database [accession nos. DQ222911 (F5H-K1) and DQ222912 (F5H-K10)].
References
- 1.Casler, M. D. & Vogel, K. P. (1999) Crop Sci. 39, 12–20. [Google Scholar]
- 2.Boudet, A. M., Lapierre, C. & Grima-Pettenati, J. (1995) New Phytol. 129, 203–236. [DOI] [PubMed] [Google Scholar]
- 3.Chabannes, M., Ruel, K., Yoshinaga, A., Chabbert, B., Jauneau, A., Joseleau, J. P. & Boudet, A. M. (2001) Plant J. 28, 271–282. [DOI] [PubMed] [Google Scholar]
- 4.Jones, L., Ennos, A. R. & Turner, S. R. (2001) Plant J. 26, 205–216. [DOI] [PubMed] [Google Scholar]
- 5.Boerjan, W., Ralph, J. & Baucher, M. (2003) Annu. Rev. Plant Biol. 54, 519–546. [DOI] [PubMed] [Google Scholar]
- 6.Whetten, R. & Sederoff, R. (1991) Forest Ecol. Manage. 43, 301–316. [Google Scholar]
- 7.Jung, H. G. & Vogel, K. P. (1986) J. Anim. Sci. 62, 1703–1712. [DOI] [PubMed] [Google Scholar]
- 8.Buxton, D. R. & Russell, J. R. (1988) Crop Sci. 28, 553–558. [Google Scholar]
- 9.Sewalt, V. J. H., Glasser, W. G., Fontenot, J. P. & Allen, V. G. (1996) J. Sci. Food Agric. 71, 195–203. [Google Scholar]
- 10.Albrecht, K. A., Wedin, W. F. & Buxton, D. R. (1987) Crop Sci. 27, 735–741. [Google Scholar]
- 11.Jung, H. G., Mertens, D. R. & Payne, A. J. (1997) J. Dairy Sci. 80, 1622–1628. [DOI] [PubMed] [Google Scholar]
- 12.Grabber, J. H., Jung, G. A., Abrams, S. M. & Howard, D. B. (1992) Crop Sci. 32, 806–810. [Google Scholar]
- 13.Jung, H. G. & Deetz, D. A. (1993) in Forage Cell Wall Structure and Digestibility, eds. Jung, H. G., Buxton, D. R., Hatfield, R. D. & Ralph, J. (Am. Soc. of Agronomy, Crop Sci. Soc. of Am., and Soil Sci. Soc. of Am., Madison, WI), pp. 315–346.
- 14.Grabber, J. H., Ralph, J., Hatfield, R. D. & Quideau, S. (1997) J. Agric. Food Chem. 45, 2530–2532. [Google Scholar]
- 15.Méchin, V., Argillier, O., Menanteau, V., Barriére, Y., Mila, I., Pollet, B. & Lapierre, C. (2000) J. Sci. Food Agric. 80, 574–580. [DOI] [PubMed] [Google Scholar]
- 16.Casler, M. D. (1987) Crop Sci. 27, 931–934. [Google Scholar]
- 17.Jung, H. G., Valdez, F. R., Hartfield, R. D. & Blanchette, R. A. (1992) J. Sci. Food. Agric. 58, 347–355. [Google Scholar]
- 18.Reeves, J. B., III (1987) J. Dairy Sci. 70, 1583–1594. [Google Scholar]
- 19.Titgemayer, E. C., Cochran, R. C., Towne, E. G., Armendariz, C. K. & Olson, K. C. (1996) J. Anim. Sci. 74, 648–657. [DOI] [PubMed] [Google Scholar]
- 20.Humphreys, J. M. & Chapple, C. (2002) Curr. Opin. Plant Biol. 5, 224–229. [DOI] [PubMed] [Google Scholar]
- 21.Humphreys, J. M., Hemm, M. R. & Chapple, C. (1999) Proc. Natl. Acad. Sci. USA 96, 10045–10050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schoch, G., Goepfert, S., Morant, M., Hehn, A., Meyer, D., Ullmann, P. & Werck-Reichart, D. (2001) J. Biol. Chem. 276, 36566–36574. [DOI] [PubMed] [Google Scholar]
- 23.Hoffmann, L., Maury, S., Martz, F., Geoffroy, P. & Legrand, M. (2003) J. Biol. Chem. 278, 95–103. [DOI] [PubMed] [Google Scholar]
- 24.Nair, R. B., Bastress, K. L., Ruegger, M. O., Denault, J. W. & Chapple, C. (2004) Plant Cell 16, 544–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Franke, R., Humphreys, J. M., Hemm, M. R., Denault, J. W., Ruegger, M. O. & Chapple, C. (2002) Plant J. 30, 33–45. [DOI] [PubMed] [Google Scholar]
- 26.Patzlaff, A., McInnis, S., Courtenay, A., Surman, C., Newman, L. J., Smith, C., Bevan, M. W., Mansfield, S., Whetten, R. W., Sederoff, R. R. & Campbell, M. M. (2003) Plant J. 36, 743–754. [DOI] [PubMed] [Google Scholar]
- 27.Halpin, C. & Boerjan, W. (2003) Trends Plant Sci. 8, 363–365. [DOI] [PubMed] [Google Scholar]
- 28.Guo, D., Chen, F., Wheeler, J., Winder, J., Selman, S., Peterson, M. & Dixon, R. A. (2001) Transgenic Res. 10, 457–464. [DOI] [PubMed] [Google Scholar]
- 29.Baucher, M., BernardVailhe, M. A., Chabbert, B., Besle, J. M., Opsomer, C., VanMontagu, M. & Botterman, J. (1999) Plant Mol. Biol. 39, 437–447. [DOI] [PubMed] [Google Scholar]
- 30.Chen, L., Auh, C.-K., Dowling, P., Bell, J., Chen, F., Hopkins, A., Dixon, R. A. & Wang, Z.-Y. (2003) Plant Biotech. J. 1, 437–449. [DOI] [PubMed] [Google Scholar]
- 31.Sambrook, J., Fritsch, E. F. & Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 32.Guo, D., Chen, F., Inoue, K., Blount, J. W. & Dixon, R. A. (2000) Plant Cell 13, 73–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karimi, M., Inze, D. & Depicker, A. (2002) Trends Plant Sci. 7, 193–195. [DOI] [PubMed] [Google Scholar]
- 34.Koncz, C. & Schell, J. (1986) Mol. Gen. Genet. 204, 383–396. [Google Scholar]
- 35.Thomas, M. R., Johnson, L. B. & White, F. F. (1990) Plant Sci. 69, 189–198. [Google Scholar]
- 36.Reddy, M. S. S., Ghabrial, S. A., Redmond, C. T., Dinkins, R. D. & Collins, G. B. (2001) Phytopathology 91, 831–838. [DOI] [PubMed] [Google Scholar]
- 37.Blount, J. W., Korth, K. L., Masoud, S. A., Rasmussen, S., Lamb, C. & Dixon, R. A. (2000) Plant Physiol. 122, 107–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hatfield, R. D., Ralph, J. & Grabber, J. H. (1999) Crop Sci. 39, 27–37. [Google Scholar]
- 39.Lapierre, C., Monties, B. & Rolando, C. (1985) J. Wood Chem. Technol. 5, 277–292. [Google Scholar]
- 40.Lapierre, C., Pollet, B. & Rolando, C. (1995) Res. Chem. Intermed. 21, 397–412. [Google Scholar]
- 41.Kamisaka, S., Takeda, D., Takahashi, K. & Shibata, K. (1990) Physiol. Plant. 78, 1–7. [Google Scholar]
- 42.Dubois, M., Gilles, K. A., Hamilton, J. K., Rebers, P. A. & Smith, F. (1956) Anal. Chem. 28, 350–356. [Google Scholar]
- 43.Goering, H. K. & Van Soest, P. J. (1970) Forage Fiber Analysis (U.S. Government Printing Office, Washington, DC).
- 44.Vogel, K. P., Pederson, J. F., Masterson, S. D. & Toy, J. J. (1999) Crop Sci. 39, 276–279. [Google Scholar]
- 45.May, G. D. & Dixon, R. A. (2004) Curr. Biol. 14, R180–R181. [DOI] [PubMed] [Google Scholar]
- 46.Aziz, N., Paiva, N. L., May, G. D. & Dixon, R. A. (2005) Planta 221, 28–38. [DOI] [PubMed] [Google Scholar]
- 47.Liang, X., Dron, M., Schmid, J., Dixon, R. A. & Lamb, C. J. (1989) Proc. Natl. Acad. Sci. USA 86, 9284–9288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lewis, N. G. & Yamamoto, E. (1990) Annu. Rev. Plant Physiol. Plant Mol. Biol. 41, 455–496. [DOI] [PubMed] [Google Scholar]
- 49.Hu, W.-J., Harding, S. A., Lung, J., Popko, J. L., Ralph, J., Stokke, D. D., Tsai, C.-J. & Chiang, V. L. (1999) Nat. Biotech. 17, 808–812. [DOI] [PubMed] [Google Scholar]
- 50.Franke, R., Hemm, M. R., Denault, J. W., Ruegger, M. O., Humphreys, J. M. & Chapple, C. (2002) Plant J. 30, 47–59. [DOI] [PubMed] [Google Scholar]
- 51.García-Pérez, R. D., Houdt, H. V. & Depicker, A. (2003) Plant J. 38, 594–602. [DOI] [PubMed] [Google Scholar]
- 52.Hoffmann, L., Besseau, S., Geoffroy, P., Ritzenthaler, C., Meyer, D., Lapierre, C., Pollet, B. & Legrand, M. (2004) Plant Cell 16, 1446–1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.