Abstract
Within the mammalian testis, specialized tight junctions between somatic Sertoli cells create basal and apical polarity within the cells, restrict movement of molecules between cells, and separate the seminiferous epithelium into basal and adluminal compartments. These tight junctions form the basis of the blood–testis barrier, a structure whose function and dynamic regulation is poorly understood. In this study, we used microarray gene expression profiling to identify genes with altered transcript levels in a mouse model for conditional androgen insensitivity. We show that testosterone, acting through its receptor expressed in Sertoli cells, regulates the expression of claudin 3, which encodes a transient component of newly formed tight junctions. Sertoli cell-specific ablation of androgen receptor results in increased permeability of the blood–testis barrier to biotin, suggesting claudin 3 regulates the movement of small molecules across the Sertoli cell tight junctions. These results suggest that androgen action in Sertoli cells regulates germ cell differentiation, in part by controlling the microenvironment of the seminiferous epithelium. Our studies also indicate that hormonal strategies for male contraception may interfere with the blood–testis barrier.
Keywords: androgen receptor, testosterone, tight junctions
Spermatogenesis in mammals requires the actions of a complex assortment of peptide and steroid hormones. These hormonal messengers are critical not only for regulation of germ cell differentiation, but also for the proliferation and function of the somatic cell types required for proper development of the testis (1, 2). These cells include the interstitial steroidogenic Leydig cells, whose primary function is to produce testosterone (3), the myoid cells that surround the seminiferous tubules and secrete basal lamina components (4), and the Sertoli cells, whose direct contact with proliferating and differentiating germ cells within the seminiferous tubules makes them essential for providing both physical and nutritional support for spermatogenesis (5). Each of these cell types is essential for unimpaired male fertility and is a direct target of one or more of the reproductive hormones.
Genetic and endocrine ablation studies suggest that androgens, testosterone and its derivatives, regulate several steps in mammalian spermatogenesis (1, 6, 7). In sexually mature mice, the androgen receptor (Ar) is expressed in Sertoli cells in a stage-dependent pattern (8), and androgen withdrawal experiments support the notion that androgens exert their influence during the stages of highest androgen receptor expression (9–11). Sertoli cell-specific ablation of the gene encoding the androgen receptor indicates that androgens are required for progression of germ cells through meiosis I, again during the transition from round to elongating spermatids, and finally during the terminal stages of spermiogenesis (12–14).
Mammalian spermatogenesis is a continuous process that occurs within a tubular seminiferous epithelium formed by a radial clustering of somatic Sertoli cells. Diploid germ cells, which include spermatogonial stem cells, transit-amplifying spermatogonia, and preleptotene spermatocytes, reside in the basal compartment in direct contact with the basal lamina and the somatic Sertoli cells. Further progression of germ cell differentiation occurs after the movement of the premeiotic cells off the basal lamina and into the adluminal compartment of the epithelium. As the germ cells move into the adluminal compartment, tight junctions (TJs) between adjacent Sertoli cells form behind the germ cells creating the blood–testis barrier (BTB) (Fig. 1D) (15, 16). The BTB segregates the meiotic and postmeiotic cells into the immunologically privileged adluminal compartment. The androgen receptor is expressed at highest levels during stages when new TJs form and premeiotic cells move from the basal to the adluminal compartment of the seminiferous epithelium (8, 16, 17).
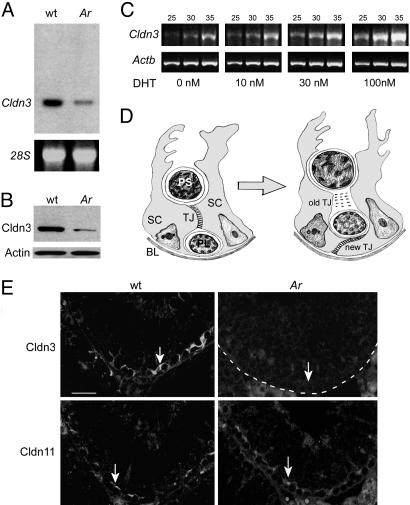
Fig. 1.
Cldn3 expression is regulated by androgens. (A Upper) Northern blot analysis on total RNA from testes of 2-month-old wild-type and Arinvflox(ex1-neo)/Y;Tg (Amh-Cre) mice and probed with a 32P-labeled Cldn3 cDNA fragment. (Lower) 28S rRNA was used as a loading control. (B) Western blot analysis of total testis protein extracts prepared from wild-type and Ar mutant mice probed with anti-Cldn3 antibody (Upper) and an anti-actin antibody as a loading control (Lower). (C) Semiquantitative RT-PCR analysis of Cldn3 expression in immortalized Sertoli-like TM4 cells transiently transfected with a cDNA encoding the androgen receptor. RNA was harvested from cells 48 h after treatment with varying doses of dihydrotestosterone (DHT), reverse transcribed, and amplified by PCR with primers specific for Cldn3 (Upper) or Actb (Lower). Reactions were terminated after 25, 30, and 35 cycles. (D) Schematic drawing of a transverse section through the seminiferous epithelium. The epithelium is segregated into basal and adluminal compartments by the formation of TJs between neighboring Sertoli cells (15). As germ cells move from the basal to the adluminal compartment, new TJs form and old TJs are disassembled. Expression of the androgen receptor in Sertoli cell nuclei is maximal during the stages of new TJ formation (8, 16). SC, Sertoli cell; PL, preleptotene spermatocyte; PS, pachytene spermatocyte; BL, basal lamina. (E) Immunofluorescence detection of Cldn3 and Cldn11 on serial sections from wild-type and Ar mutant testes. Both proteins are localized to TJs in the basal compartment of the seminiferous epithelium, although the staining of Cldn3 (arrow in Upper Left) appears to extend beyond that of the staining of Cldn11 (arrow in Lower Left). Expression of Cldn3 is absent in the region of Sertoli cell TJs in Ar mutants (Upper Right). The basal lamina of the tubule is outlined with a dotted line. Expression of Cldn11 is retained in Ar mutant testis (Lower Right). (Scale bar: 50 μm.)
TJs are specialized anchoring junctions composed of several integral and peripheral membrane proteins (18, 19). The membrane-spanning proteins include the occludins, claudins, and junction adhesion molecules (20–22). Occludin, present in mouse Sertoli TJs but not human or guinea pig, appears to be essential for TJ function in the testis of mice, because a targeted mutation in the occludin gene causes male sterility (23). More than 20 different claudins have been described since the initial discovery of Cldn1 and Cldn2 in the late 1990s (21). Among these proteins, claudins 1, 3, 4, 5, 7, 8, and 11 have been shown to be expressed in the testis (reviewed in ref. 24). Targeted deletion of Cldn11 has revealed that it is essential for normal Sertoli cell TJs, and the myelin sheaths of oligodendrocytes, because the mice have both reproductive and CNS defects (25). Two junction adhesion molecules, Jam1 and Jam2, are expressed in the testis (22, 26). Associated with the cytoplasmic tails of the membrane spanning proteins are several peripheral membrane proteins, including zonula occludens, cingulin, and symplekin (24). Almost nothing is known about the regulation of the Sertoli cell TJ components and their assembly and disassembly during the cycle of the seminiferous epithelium, although TGFβ3 has been shown to affect the expression of Cldn11 in cell culture (27).
We have previously investigated the function of the androgen receptor in Sertoli cells by creating mice with a hypomorphic inverted floxed allele of Ar (Arinvflox(ex1-neo)) and crossing them with mice expressing Cre recombinase driven by the Sertoli cell-specific promoter of the anti-Müllerian hormone gene (Amh). In this model of incomplete and conditional androgen insensitivity, Ar levels are reduced in all cells where it is normally expressed because of an aberrant and incomplete splicing reaction and are completely absent in Sertoli cells because of loxP-mediated inversion of the first exon. The mice are azoospermic and show a block in the transition from round to elongating spermatids (14). Accompanying the arrest are increased germ cell apoptosis and sloughing of spermatids into the lumen of the seminiferous epithelium.
In hopes of discovering the mechanisms by which androgens control spermatogenesis, it is necessary first to identify transcriptional targets of the androgen receptor. In this study we used microarray gene expression profiling to investigate the transcriptional consequences after the loss of androgen signaling in Sertoli cells. We found that claudin 3 (Cldn3) is down-regulated in Arinvflox(ex1-neo)/Y;Tg (Amh-Cre) mice, suggesting that androgens may regulate the assembly or function of Sertoli cell TJs. We tested this hypothesis and found that the permeability of the BTB is altered in mice with conditional ablation of Ar expression in Sertoli cells.
Materials and Methods
Mice. The creation of Arinvflox(ex1-neo)/Y and Tg (Amh-Cre) mice has been described in ref. 14. C57BL6/J mice were generated in our colony. All animal studies were approved by the University of Washington's Institutional Animal Care and Use Committee. All experimental data were collected from a minimum of 3 animals, and in some cases as many as 12 animals, of each genotype. Two or more replicates of all experiments were performed.
Immunofluorescence Microscopy. Mouse testes from 8-week-old wild-type and Arinvflox(ex1-neo)/Y;Tg(Amh-Cre) males were incubated overnight in Bouins fixative at 4°C and prepared for immunofluorescence microscopy as described in ref. 28. Testis paraffin sections were incubated overnight with rabbit anti-Cldn3, anti-Cldn11, or anti-occludin antibodies (Zymed Laboratories). A goat anti-rabbit secondary antibody conjugated to Alexa Fluor 568 (Molecular Probes) was used at a dilution of 1/2,000. The slides were stained with DAPI and mounted in prolonged anti-fade mounting medium (Molecular Probes).
Molecular Biology and Biochemistry. RNA extraction, the preparation of testis protein extracts, and Western and Northern blotting were done as described in ref. 29. Protein extracts were prepared from seminiferous tubule fragments identified by transillumination as described in ref. 30. Rabbit anti-Cldn3 primary antibody (Zymed Laboratories) was detected by using a horseradish peroxidase-conjugated goat anti-rabbit secondary antibody (Bio-Rad) and the ECL Western blotting detection system (Amersham Pharmacia). Rabbit anti-actin antibody (Sigma) was used as a control. A 32P-radiolabeled 700 bp SalI-EcoRI fragment containing the Cldn3 cDNA (generously provided by Mikio Furuse, Kyoto University, Kyoto) was used for Northern blotting. Testis total RNA was reverse-transcribed and amplified by PCR (RT-PCR) by using the Access RT-PCR system (Promega) as described in ref. 28. Cldn3 primers: 5′-TTTTCCTGTTGGCGGCTCTG-3′ and 5′-CGAGGTTTCTTTGTCCATTCGG-3′.
Cell Culture. TM4 cells, originally isolated from freshly dispersed mouse testicular cells and selected for their morphology and hormone responsiveness (31), were purchased from the American Type Culture Collection and cultured in a 1:1 mixture of DMEM and F12 media containing 7% charcoal-treated FBS (HyClone) at 37°C. The cells were transiently transfected with an expression plasmid containing the human cytomegalovirus promoter fused to a full-length murine Ar cDNA by using Lipofectamine 2000 (Invitrogen). Approximately 6 h after transfection, dihydrotestosterone was added at a final concentration of 0, 10, 30, or 100 nM and incubated for 48 h at 37°C. RNA was extracted from the cells with TRIzol (Invitrogen).
Microarray Analysis. Total testis RNA from 2-month-old wild-type and Arinvflox(ex1-neo)/Y;Tg (Amh-Cre) mice was used for microarray analysis with Affymetrix (Santa Clara, CA) MOE430A arrays. Microarray hybridization was performed by following the manufacturer's standard protocol. Separate testis samples were collected and used for duplicate microarray hybridization. Data acquisition was achieved by using Affymetrix mas 5.0, and RNA levels were quantified by using the default analysis parameters. All probe sets from each microarray were scaled to a target signal of 125. Comparison between wild-type and Arinvflox(ex1-neo)/Y;Tg (Amh-Cre) testes was performed by using genespring 6.1 (Silicon Genetics, Redwood City, CA) with the default normalization scheme. Transcripts were considered differentially expressed if the signal level was >100 in either the wild-type or Arinvflox(ex1-neo)/Y;Tg (Amh-Cre) samples and demonstrated a 2-fold or greater difference in expression.
Biotin Tracer Studies. The permeability of the BTB was assessed by using a biotin tracer (32). We anesthetized wild-type 15- and 25-day-old pups, and wild-type and Arinvflox(ex1-neo)/Y;Tg (Amh-Cre) adults, exposed their testes, gently created a small opening in the tunica albuginea with fine forceps, and using a small pipet, injected into the interstitium of one testis 50 μl of 10 mg/ml EZ-Link Sulfo-NHS-LC-Biotin (Pierce Chemical Co.) freshly diluted in PBS containing 1 mM CaCl2. The other testis was injected similarly with 50 μl of 1 mM CaCl2 in PBS as a control. After 30 min the animals were euthanized, and their testes were immediately removed and frozen on dry ice. Cryosections of 5-μm thickness were prepared as described in ref. 32, incubated with rabbit anti-occludin antibody (Zymed) at 1:200 dilution in PBS for 30 min, washed four times for 30 min with PBS, then treated with a mixture of Alexa Fluor 488 goat anti-rabbit antibody and Alexa Fluor 568 streptavidin for 30 min at 25°C. The sections were rinsed twice with PBS for 10 min, stained with DAPI, rinsed four times with PBS for 20 min, and mounted in prolonged antifade mounting medium.
Results
We performed gene expression profiling by using oligonucleotide microarrays on testicular RNA isolated from the testes of 2-month-old Arinvflox(ex1-neo)/Y;Tg (Amh-Cre) and wild-type littermate control mice. More than 60 genes exhibiting a 2-fold or greater change in expression level relative to controls were identified in Arinvflox(ex1-neo)/Y;Tg (Amh-Cre) mice (S. M. Eacker, R.W.H., J.E.S., M.D.G., and R.E.B., unpublished data). Among these genes, Cldn3 RNA levels were reduced 10-fold. We confirmed the decrease by Northern blot analysis (Fig. 1 A) and used an anti-Cldn3 antibody and Western blotting to demonstrate that the level of the Cldn3 protein was similarly reduced (Fig. 1B).
It is believed that androgens control spermatogenesis by inducing a transcriptional response in Sertoli cells upon binding the androgen receptor. However, recent findings suggest that a nongenomic pathway may also be triggered when Ar is bound by its ligand (33). To determine whether Cldn3 is transcriptionally regulated by testosterone, we transfected the TM4 Sertoli-like cell line with an Ar expression construct, treated the cells with dihydrotestosterone (DHT), and assayed Cldn3 mRNA by semiquantitative RT-PCR. Cldn3 mRNA was up-regulated by DHT in cell culture, suggesting that androgens directly regulate the expression of Cldn3 in Sertoli cells (Fig. 1C).
The Cldn3 gene encodes a four-pass integral membrane protein of 219 amino acids and is a known component of TJs in several tissues (18, 34). To verify that Cldn3 is localized to Sertoli cell TJs, we assayed Cldn3 protein localization in testis sections by immunofluorescence microscopy. As expected, Cldn3 was detected in a belt-like pattern near the basal lamina of the epithelium in the region of the TJs (Fig. 1E). Expression of Cldn11, a known component of the Sertoli TJs (25), partially overlapped with Cldn3, consistent with Cldn3 being a component of the Sertoli cell TJs. Interestingly, the staining pattern of Cldn3 extended more toward the basal lamina than the staining pattern of Cldn11, suggesting that the two proteins might have different functions. We did not detect Cldn3 by immunofluorescence in the seminiferous epithelium of Arinvflox(ex1-neo)/Y;Tg (Amh-Cre) mice (Fig. 1E), confirming our Northern and Western blot analysis. However, Cldn11 staining was still detected, suggesting that the TJ structures remain intact in the mutant.
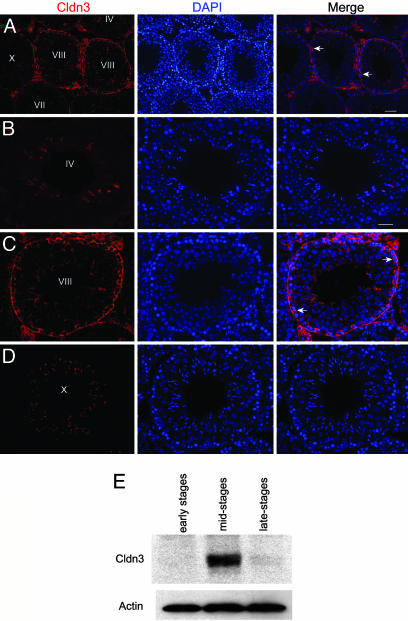
Unlike the blood–brain barrier, the BTB is a dynamic structure that must regularly break down and reform as premeiotic germ cells move from the basal to the adluminal compartment of the epithelium during stages VIII–IX of the spermatogenic cycle (16). Once formed, the TJs remain intact during an entire cycle of the seminiferous epithelium. We were therefore surprised to find that although Cldn3 is associated with newly formed TJs (Fig. 2 A and C), it was undetectable in old TJs at either early (Fig. 2B, Stage IV) or late (Fig. 2D, Stage X) stages. Other TJ components, such as Cldn11 and occludin, were detected at all stages of the cycle (data not shown). To confirm that stage-specific detection of Cldn3 by immunofluorescence was not an artifact resulting from masking of the Cldn3 epitope during early and late stages, we used transillumination (30) to dissect out whole tubule segments representative of early, middle, and late stages, prepared protein extracts from the segments, and performed Western blotting with an anti-Cldn3 antibody. Convincingly, Cldn3 protein was present in extracts prepared from mid-stage tubules but absent from early and late stage tubules (Fig. 2E). This example of a claudin shows a transient association with TJs at the time of their formation.
Fig. 2.
Expression of Cldn3 is restricted to newly formed TJs. (A–D) Immunofluorescence localization of Cldn3 in adult testis. Nuclei were stained with DAPI and the tubules staged based on the particular association of germ cells within a tubule (44). The arrows indicate regions of Cldn3 detection. High magnification images show the absence of Cldn3 in Stages IV and X and its presence in Stage VIII. (Scale bars: A, 50 μm; B, 20 μm for B–D). (E) Western analysis of Cldn3 in extracts prepared from seminiferous tubule fragments identified by transillumination (30). Actin was used as a loading control.
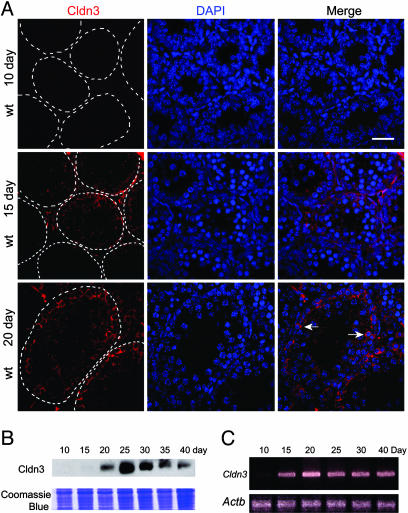
TJs form during puberty coincident with a rise in testosterone and expression of the androgen receptor in Sertoli cells (35). To determine whether Cldn3 is expressed during initial TJ formation in pubertal animals, we killed mice at various ages and examined Cldn3 expression. We first detected a weak and diffuse pattern of Cldn3 expression by immunofluorescence at 15 days postpartum (Fig. 3A). By day 20 postpartum, Cldn3 was concentrated in the TJs near the basal lamina. Consistent with this finding, we first detected Cldn3 RNA by RT-PCR analysis, and Cldn3 protein by Western analysis, at day 15 postpartum (Fig. 3 B and C).
Fig. 3.
Cldn3 expression is developmentally regulated. (A) Immunofluorescence detection of Cldn3 in testis sections during puberty. Cldn3 is first detected at day 15 postpartum and is diffusely distributed. By day 20 postpartum, Cldn3 is localized to Sertoli cell TJ. The sections were counterstained with DAPI. The basal lamina of the tubules is outlined with a dashed line. (Scale bar: 50 μm.) (B) Western blot showing that Cldn3 protein is first detected at day 15 postpartum. A portion of the gel was stained with Coomassie blue as a control for equal loading of sample. (C) RT-PCR (35 cycles) showing that Cldn3 RNA is first detected at day 15 postpartum. Actb was used as a control.
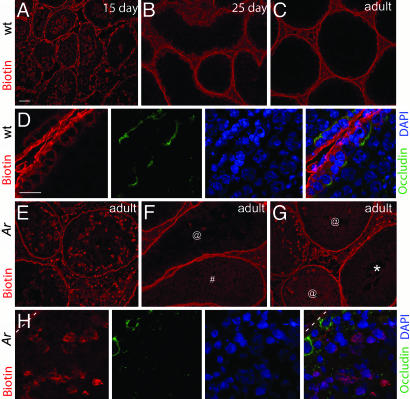
The detection of Cldn11 indicates that TJ structures are present in the seminiferous epithelium of Arinvflox(ex1-neo)/Y;Tg (Amh-Cre) mice. However, the absence of Cldn3 suggests that the TJs may have altered or compromised function. To test whether the BTB in Arinvflox(ex1-neo)/Y;Tg (Amh-Cre) mutants has increased permeability to small molecules (<600 Da), we injected a biotin tracer into the interstitial space of the testis of live anesthetized mice. Thirty minutes after injection we euthanized the mice, prepared cryosections from their testes, and determined how far the biotin tracer had permeated into the seminiferous tubule by using streptavidin conjugated to Alexa Fluor 568. The BTB was detected with an anti-occludin antibody (20, 36). In 15-day-old wild-type mice, the biotin tracer penetrated the seminiferous tubule, as expected, because the BTB has yet to fully form (Fig. 4A). However, in testes from 25-day-old and adult wild-type mice, the biotin tracer was present in the interstitial spaces and basal compartment but excluded from the adluminal compartment (Fig. 4 B–D). In contrast, in adult Arinvflox(ex1-neo)/Y;Tg (Amh-Cre) mice, the biotin tracer passed through the BTB and entered the adluminal compartment (Fig. 4 E–H), even though occludin staining showed that TJ structures were present (Fig. 4H). Together, these data suggest that androgens regulate the permeability of the BTB by controlling the stage-specific expression of Cldn3.
Fig. 4.
Loss of androgen signaling in Sertoli cells increases the permeability of the BTB. (A) Penetration of the seminiferous tubules of a 15-day-old wild-type mouse by a biotin tracer 30 min after injection into the interstitial space. (B and C) Restriction of the biotin tracer to the interstitial space and basal compartment of 25-day-old and adult mice. (D) High magnification image of TJ and exclusion of the biotin tracer from the adluminal compartment of a sexually mature mouse. The TJs were stained with anti-occludin antibody, and the nuclei were counterstained with DAPI. (E) Penetration of biotin into the adluminal compartment of several seminiferous tubules 30 min after it was injected into the interstitial space of an adult Ar mutant. In some tubules, penetration was considerable and was detected as aggregates throughout the adluminal compartment. (F and G) Examples of seminiferous tubules from adult Ar mutants showing lesser degrees of biotin diffusion into the adluminal compartment. @, tubules with moderate biotin accumulation; #, light diffuse staining throughout the tubule; *, tubule with no biotin detected in the adluminal region. (H) High magnification image of TJ and penetration of biotin into the adluminal compartment of a seminiferous tubule from an adult Ar mutant. The basal lamina is outlined with a dashed line. (Scale bars: A, 50 μm for A–C and E–G; D, 20 μm for D and H.)
Discussion
Identification of Ar target genes is essential to understanding how androgen action in Sertoli cells supports spermatogenesis. Our studies demonstrate that Cldn3 is a transcriptional target of Ar in Sertoli cells. Cldn3 mRNA levels are significantly decreased in mice containing Sertoli cell-specific ablation of Ar, Cldn3 mRNA is induced in the TM4 Sertoli-like cell line after androgen treatment, and Cldn3 protein is detected by immunostaining during the highest levels of Ar expression in Sertoli cells. The presence of a putative androgen response element in the promoter region of the Cldn3 gene (J.M. and R.E.B., unpublished data) suggests that it is a direct transcriptional target of the androgen receptor.
We used a biotin tracer to show that the permeability of the BTB is altered in Arinvflox(ex1-neo)/Y;Tg (Amh-Cre) mice. We propose that the increased permeability of the BTB is due to the loss of Cldn3 protein. However, it is possible that the altered expression of other Ar-regulated genes is responsible for the increase in permeability. Selective ablation of Cldn3 in Sertoli cells will be necessary to directly demonstrate the importance of Cldn3 in regulating the permeability of the BTB. Interestingly, neonatal exposure of rats to diethylstilbestrol (DES) causes a reduction in the expression of Ar (37) and delays the formation of the BTB during puberty (38). Our data suggest the possibility that the mechanism of DES action may involve altered developmental expression of Cldn3.
The BTB has classically been considered an immunological barrier. However, our studies clearly demonstrate that the BTB also functions to regulate the movement of small molecules between the testicular lymph and the adluminal compartment of the seminiferous epithelium. Similar findings have been demonstrated for the blood–brain barrier, where mutation of Cldn5 increases the permeability of the blood–brain barrier to molecules <800 Da (39), and for the epidermal barrier, where mutation of Cldn1 results in transepidermal water loss leading to severe dehydration and wrinkling of the skin (32). In both Cldn1 and Cldn5 null animals, the remaining claudin proteins maintain the anastomosing arrays of the TJs. Our observations are similar, despite the loss of Cldn3, TJ structures containing Cldn11 and occludin are retained around the Sertoli cells. Interestingly, in Cldn11 mutants, which are sterile, the membrane-spanning occludin protein is present, although the linear arrays of TJs observed by freeze-fracture electron microscopy are absent (25).
An unexpected and intriguing finding in our studies was the transient association of Cldn3 with the newly formed TJs at the time that germ cells move from the basal to the adluminal compartment. This transient expression is in contrast to Cldn11 and occludin, which remain associated with the TJs during the entire cycle of the seminiferous epithelium (25, 36). It is possible that Cldn3 is required at the site of newly formed TJs to restrict passage of molecules into the adluminal compartment specifically at that time. Alternatively, although Cldn3 expression is restricted to newly formed TJs, the TJs formed in its absence may be structurally abnormal. Further investigation is required to determine what leads to the loss of Cldn3 in old TJs in wild-type animals.
Androgen receptor expression in Sertoli cells is required for several steps in germ cell differentiation, including progression through meiosis and haploid spermatid differentiation (12–14). However, despite the critical importance of androgens in supporting spermatogenesis, little is known about how androgen action in somatic Sertoli cells mediates progression of germ cell differentiation. Androgen action could trigger a transcriptional response in Sertoli cells leading to instructive signaling to the germ cells. Alternatively, androgens could regulate the microenvironment within the seminiferous epithelium, creating conditions that are permissive, rather than instructive, for germ cell differentiation. Our studies suggest that androgens may regulate the microenvironment of epithelium during the time of new TJ formation. TJs have been shown in other tissues to regulate the selective transport of ions and nutrients from the bloodstream and interstitial fluid (40). The provocative demonstration that rat germ cells transplanted into a mouse testis differentiate with rat cell cycle kinetics (41), and the recent demonstration that embryonic stem cells can differentiate into germ cells in vitro in the absence of the seminiferous epithelium (42, 43) support the hypothesis that Sertoli cells provide a permissive environment that facilitates germ cell differentiation.
Acknowledgments
We thank M. Sharma, A. Dearth, and G. Martin of the University of Washington Keck Imaging Center for invaluable technical advice; D. Pouchnik for microarray hybridization and scanning; and S. Eacker, M. Olson, and K. Swisshelm for critical review of the manuscript. This research was supported by National Institute of Child Health and Human Development/National Institutes of Health Grant HD12629 as part of the Specialized Cooperative Centers Program in Reproduction Research.
Author contributions: J.M., R.W.H., and R.E.B. designed research; J.M., R.W.H., and J.E.S. performed research; J.M., R.W.H., J.E.S., M.D.G., and R.E.B. analyzed data; and J.M., M.D.G., and R.E.B. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: Ar, androgen receptor; BTB, blood–testis barrier; Cldn3, claudin 3; TJ, tight junctions.
References
- 1.Sharpe, R. M. (1994) in The Physiology of Reproduction, eds. Knobil, E. & Neill, J. D. (Raven, New York).
- 2.McLachlan, R. I., O'Donnell, L., Meachem, S. J., Stanton, P. G., de, K., Pratis, K. & Robertson, D. M. (2002) J. Androl. 23, 149–162. [PubMed] [Google Scholar]
- 3.Mendis-Handagama, S. M. (1997) Histol. Histopathol. 12, 869–882. [PubMed] [Google Scholar]
- 4.Maekawa, M., Kamimura, K. & Nagano, T. (1996) Arch Histol. Cytol. 59, 1–13. [DOI] [PubMed] [Google Scholar]
- 5.Griswold, M. D. (1998) Semin. Cell Dev. Biol. 9, 411–416. [DOI] [PubMed] [Google Scholar]
- 6.McLachlan, R. I., O'Donnell, L., Meachem, S. J., Stanton, P. G., de Kretser, D. M., Pratis, K. & Robertson, D. M. (2002) Recent Prog. Horm. Res. 57, 149–179. [DOI] [PubMed] [Google Scholar]
- 7.Holdcraft, R. W. & Braun, R. E. (2004) Int. J. Androl. 27, 335–342. [DOI] [PubMed] [Google Scholar]
- 8.Zhou, Q., Nie, R., Prins, G. S., Saunders, P. T., Katzenellenbogen, B. S. & Hess, R. A. (2002) J. Androl. 23, 870–881. [PubMed] [Google Scholar]
- 9.Russell, L. D. & Clermont, Y. (1977) Anat. Rec. 187, 347–366. [DOI] [PubMed] [Google Scholar]
- 10.Ghosh, S., Sinha-Hikim, A. P. & Russell, L. D. (1991) Tissue Cell 23, 613–630. [DOI] [PubMed] [Google Scholar]
- 11.Kerr, J. B., Maddocks, S. & Sharpe, R. M. (1992) Cell Tissue Res. 268, 179–189. [DOI] [PubMed] [Google Scholar]
- 12.Chang, C., Chen, Y.-T., Yeh, S.-D., Xu, Q., Wang, R.-S., Guillou, F., Lardy, H. & Yeh, S. (2004) Proc. Natl. Acad. Sci. USA 101, 6876–6881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.De Gendt, K., Swinnen, J. V., Saunders, P. T., Schoonjans, L., Dewerchin, M., Devos, A., Tan, K., Atanassova, N., Claessens, F., Lecureuil, C., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 1327–1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Holdcraft, R. W. & Braun, R. E. (2004) Development (Cambridge, U.K.) 131, 459–467. [DOI] [PubMed] [Google Scholar]
- 15.Dym, M. & Fawcett, D. W. (1970) Biol. Reprod. 3, 308–326. [DOI] [PubMed] [Google Scholar]
- 16.Russell, L. (1977) Am. J. Anat. 148, 313–328. [DOI] [PubMed] [Google Scholar]
- 17.Bremner, W. J., Millar, M. R., Sharpe, R. M. & Saunders, P. T. (1994) Endocrinology 135, 1227–1234. [DOI] [PubMed] [Google Scholar]
- 18.Tsukita, S., Furuse, M. & Itoh, M. (2001) Nat. Rev. Mol. Cell Biol. 2, 285–293. [DOI] [PubMed] [Google Scholar]
- 19.Schneeberger, E. E. & Lynch, R. D. (2004) Am. J. Physiol. 286, C1213–C1228. [DOI] [PubMed] [Google Scholar]
- 20.Furuse, M., Hirase, T., Itoh, M., Nagafuchi, A., Yonemura, S. & Tsukita, S. (1993) J. Cell Biol. 123, 1777–1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Furuse, M., Sasaki, H., Fujimoto, K. & Tsukita, S. (1998) J. Cell Biol. 143, 391–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin-Padura, I., Lostaglio, S., Schneemann, M., Williams, L., Romano, M., Fruscella, P., Panzeri, C., Stoppacciaro, A., Ruco, L., Villa, A., et al. (1998) J. Cell Biol. 142, 117–127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Saitou, M., Furuse, M., Sasaki, H., Schulzke, J. D., Fromm, M., Takano, H., Noda, T. & Tsukita, S. (2000) Mol. Biol. Cell 11, 4131–4142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lui, W. Y., Mruk, D., Lee, W. M. & Cheng, C. Y. (2003) Biol. Reprod. 68, 1087–1097. [DOI] [PubMed] [Google Scholar]
- 25.Gow, A., Southwood, C. M., Li, J. S., Pariali, M., Riordan, G. P., Brodie, S. E., Danias, J., Bronstein, J. M., Kachar, B. & Lazzarini, R. A. (1999) Cell 99, 649–659. [DOI] [PubMed] [Google Scholar]
- 26.Aurrand-Lions, M., Duncan, L., Ballestrem, C. & Imhof, B. A. (2001) J. Biol. Chem. 276, 2733–2741. [DOI] [PubMed] [Google Scholar]
- 27.Lui, W. Y., Lee, W. M. & Cheng, C. Y. (2003) Biol. Reprod. 68, 1597–1612. [DOI] [PubMed] [Google Scholar]
- 28.Buaas, F. W., Kirsh, A. L., Sharma, M., McLean, D. J., Morris, J. L., Griswold, M. D., de Rooij, D. G. & Braun, R. E. (2004) Nat. Genet. 36, 647–652. [DOI] [PubMed] [Google Scholar]
- 29.Braun, R. E., Peschon, J. J., Behringer, R. R., Brinster, R. L. & Palmiter, R. D. (1989) Genes Dev. 3, 793–802. [DOI] [PubMed] [Google Scholar]
- 30.Kotaja, N., Kimmins, S., Brancorsini, S., Hentsch, D., Vonesch, J. L., Davidson, I., Parvinen, M. & Sassone-Corsi, P. (2004) Nat. Methods 1, 249–254. [DOI] [PubMed] [Google Scholar]
- 31.Mather, J. P. (1980) Biol. Reprod. 23, 243–252. [DOI] [PubMed] [Google Scholar]
- 32.Furuse, M., Hata, M., Furuse, K., Yoshida, Y., Haratake, A., Sugitani, Y., Noda, T., Kubo, A. & Tsukita, S. (2002) J. Cell Biol. 156, 1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker, W. H. (2003) Curr. Top. Dev. Biol. 56, 25–53. [DOI] [PubMed] [Google Scholar]
- 34.Mitic, L. L., Van Itallie, C. M. & Anderson, J. M. (2000) Am. J. Physiol. 279, G250–G254. [DOI] [PubMed] [Google Scholar]
- 35.Nagano, T. & Suzuki, F. (1976) Anat. Rec. 185, 403–417. [DOI] [PubMed] [Google Scholar]
- 36.Cyr, D. G., Hermo, L., Egenberger, N., Mertineit, C., Trasler, J. M. & Laird, D. W. (1999) Endocrinology 140, 3815–3825. [DOI] [PubMed] [Google Scholar]
- 37.Sharpe, R. M., Atanassova, N., McKinnell, C., Parte, P., Turner, K. J., Fisher, J. S., Kerr, J. B., Groome, N. P., Macpherson, S., Millar, M. R. & Saunders, P. T. (1998) Biol. Reprod. 59, 1084–1094. [DOI] [PubMed] [Google Scholar]
- 38.Toyama, Y., Ohkawa, M., Oku, R., Maekawa, M. & Yuasa, S. (2001) J. Androl. 22, 413–423. [PubMed] [Google Scholar]
- 39.Nitta, T., Hata, M., Gotoh, S., Seo, Y., Sasaki, H., Hashimoto, N., Furuse, M. & Tsukita, S. (2003) J. Cell Biol. 161, 653–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Goodenough, D. A. (1999) Proc. Natl. Acad. Sci. USA 96, 319–321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Franca, L. R., Ogawa, T., Avarbock, M. R., Brinster, R. L. & Russell, L. D. (1998) Biol. Reprod. 59, 1371–1377. [DOI] [PubMed] [Google Scholar]
- 42.Geijsen, N., Horoschak, M., Kim, K., Gribnau, J., Eggan, K. & Daley, G. Q. (2004) Nature 427, 148–154. [DOI] [PubMed] [Google Scholar]
- 43.Toyooka, Y., Tsunekawa, N., Akasu, R. & Noce, T. (2003) Proc. Natl. Acad. Sci. USA 100, 11457–11462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Russell, L. D., Ettlin, R. A., SinhaHikim, A. P. & Clegg, E. D. (1990) Histological and Histopathological Evaluation of the Testis (Cache River, Clearwater, FL).