Abstract
Mutations in lamins cause premature aging syndromes in humans, including the Hutchinson–Gilford Progeria Syndrome (HGPS) and Atypical Werner Syndrome. It has been shown that HGPS cells in culture undergo age-dependent progressive changes in nuclear architecture. However, it is unknown whether similar changes in nuclear architecture occur during the normal aging process. We have observed that major changes of nuclear architecture accompany Caenorhabditis elegans aging. We found that the nuclear architecture in most nonneuronal cell types undergoes progressive and stochastic age-dependent alterations, such as changes of nuclear shape and loss of peripheral heterochromatin. Furthermore, we show that the rate of these alterations is influenced by the insulin/IGF-1 like signaling pathway and that reducing the level of lamin and lamin-associated LEM domain proteins leads to shortening of lifespan. Our work not only provides evidence for changes of nuclear architecture during the normal aging process of a multicellular organism, but also suggests that HGPS is likely a result of acceleration of the normal aging process. Because the nucleus is vital for many cellular functions, our studies raise the possibility that the nucleus is a prominent focal point for regulating aging.
Keywords: lamin, LEM domain, nuclear envelope, nuclear lamina
In eukaryotic cells, the nucleus and the cytoplasm are separated by the nuclear envelope, which consists of inner and outer nuclear membranes, nuclear pore complexes, and the nuclear lamina. The nuclear lamina underlies the inner nuclear membrane and is composed of lamins, which are type V intermediate filament proteins, and lamin-associated proteins (1). In metazoan cells, the nuclear lamina is required for maintaining nuclear shape, organization of chromatin, DNA replication, RNA transcription, cell cycle progression, nuclear migration, and cellular development and differentiation (see reviews in refs. 2–4).
Mutations in lamins and lamin-associated proteins cause a variety of heritable human diseases that are collectively called laminopathies, ranging from muscular dystrophy to accelerated aging (see reviews in refs. 5–8). Fibroblast cells from laminopathic patients frequently display irregular nuclear shapes, abnormal composition of nuclear lamina proteins, and changes in chromatin organization (1). Among the laminopathies, Hutchinson–Gilford Progeria Syndrome (HGPS) is a premature aging disorder that is most commonly caused by a silent point mutation in the human lamin A gene (LMNA), which creates a new splice isoform that lacks 50 residues in its C-terminal domain (9, 10). Goldman et al. (11) have recently shown that HGPS cells exhibit age-dependent progressive alterations of nuclear architecture, which they hypothesize would ultimately lead to premature aging in HGPS patients. These studies raise the question whether there is any age-dependent alteration of nuclear architecture in individuals undergoing the normal aging process.
The nematode Caenorhabditis elegans offers an excellent model system to investigate possible changes of nuclear architecture during the normal aging process. C. elegans has a relatively short lifespan. Like humans, aging C. elegans animals experience stochastic deterioration of tissue integrity, especially in muscles (12, 13). The evolutionarily conserved insulin/IGF-1-like signaling pathway is involved in regulating the lifespan of C. elegans (see a recent review in ref. 14). In particular, mutations in the daf-2 insulin receptor gene and the age-1 phosphatidylinositol 3-kinase gene lead to an extension of lifespan, and this lifespan extension phenotype requires the function of the FOXO transcription factor DAF-16 and the heat-shock transcription factor HSF-1 (14). The various mutants in the insulin/IGF-1-like signaling pathway provide us with powerful genetic tools to study age-related processes.
In this study, we characterized the major changes of nuclear architecture that accompany C. elegans aging. We show that the nuclear architecture in most nonneuronal cell types undergoes progressive and stochastic alteration as the animal ages and that the rate of this alteration is affected by mutations in the insulin/IGF-1 like signaling pathway. Furthermore, we show that reducing the levels of lamin and lamin-associated LEM domain proteins can lead to shortening of the lifespan.
Materials and Methods
C. elegans Strains. Strains were maintained and manipulated under standard conditions as described by Brenner (15). The following strains were used: N2, daf-16(mu86) I, emr-1(gk119) I, lmn-1(tm1502) I/hT2(qIs48) (I; III) (tm1502 was obtained from Shohei Mitani, Tokyo Women's Medical University School of Medicine, Tokyo), age-1(hx546) II, and daf-2(e1370) III.
The following strains carrying integrated reporter transgenes were used to visualize the nuclear envelope. All integrated strains were generated by γ-irradiation except for YG003 and 720519, which were generated by bombardment. They were all subsequently out-crossed at least three times with the wild-type N2 strain.
lmn-1::gfp: LW0697(ccIs4810) X, LW0698(ccIs4811) X, and LW0700(ccIs4812) X, containing [pJKL380.4(lmn-1p::-lmn-1::gfp::lmn-1 3′UTR)+pMH86(dpy-20(+)] (16).
gfp::lmn-1: LW0709(jjIs0709) I and LW0696(jjIs0696) (not on X), containing [pDRNL1(lmn-1p::gfp::lmn-1::unc-54 3′UTR) +pMH86(dpy-20(+)] (ref. 16 and this work).
emr-1::gfp: LW0699(YG003 out-crossed three times) and LW0275(720519 out-crossed three times), containing [p720–4(lmn-1p::emr-1::gfp::unc-54 3′UTR)+unc-119(+)] (this work).
npp-1::gfp: LW0271(jjIs0271), containing [pnpp-1 (npp-1p::-npp-1::gfp::npp-1 3′UTR)+pMH86(dpy-20(+)] (this work, plasmid pnpp-1 is a gift from Aaron Schetter and Ken Kemphues, Cornell University).
Lifespan Assays. Lifespan assays were carried out by using a slightly modified protocol of Lee et al. (17). Animals from single-day egglays (day 0) were grown on NGM plates at 16°C until they reached young adult stage. Fifty to 100 animals (≈30 animals per plate) were then transferred to NGM plates containing 0.05 mg/ml 5-fluoro 2′-deoxyuridine (FUDR) to prevent growth of progeny and were allowed to grow on these plates at 25°C unless noted. Animals were scored every day until they no longer responded to a gentle prodding with a platinum wire, which is considered dead. Animals that exploded were censored from the experiments. Control experiments lacking FUDR showed no effect of FUDR on lifespan.
For lifespan assays of animals undergoing RNA interference (RNAi), gravid adults were allowed to egglay overnight on dsRNA-expressing bacteria on RNAi plates (17). When they reached young adulthood, 50–100 of them were transferred to fresh RNAi plates with 5-fluoro 2′-deoxyuridine (≈30 animals per plate). Freshly induced RNAi bacteria were added to the plates every 5 days. Lifespan was scored as described above. All lifespan assays of animals undergoing RNAi were carried out at 20°C.
For all lifespan experiments, assays were repeated at least twice. Data were analyzed by using the Kaplan–Meier Survival Analysis of spss software (version 11, SPSS, Chicago) for statistical analysis.
RNAi Constructs. The following plasmids were transformed to HT115(DE3) (18) and used in the RNAi feeding experiments: pJKL483.1 for lmn-1 (19) and pAJZ1.2 for lem-2 (this work, cloning information available upon request). The hsf-1 RNAi clone is from the Ahringer library (20). L4440 (gift from Andy Fire, Stanford University) was used as a negative control in all experiments.
Fluorescence and Electron Microscopy. Age-synchronized N2 animals were fixed and prepared for indirect immunofluorescence staining with anti-lamin antibodies as described (16). Direct observation of GFP epifluorescence in transgenic animals carrying various integrated GFP markers was carried out by mounting 5–10 animals per day per strain on 2% agarose pads with ENL buffer (50 mM NaCl/5 mM EDTA/0.33 mM Levamisole) and visualized either on a Leica DMRA2 compound microscope, where the images were captured by a Hamamatsu Orca-ER camera using openlab software (version 3.0.9, Improvision), or on a Leica TCS SP2 confocal microscope. Subsequent image analysis was performed by using photoshop 7.0 (Adobe Systems, San Jose).
Transmission electron microscopy (TEM) of C. elegans was carried out essentially as described (21). Adult C. elegans were collected in PBS and washed twice in PBS. The animals were cut with sharp forceps behind either the pharynx or the tail to allow rapid penetration of solutions and immediately subjected to fixation in PBS (pH 7.5) containing 2.5% paraformaldehyde and 2.5% glutaraldehyde overnight at 4°C. The animals were washed three times with PBS and mounted in 3.4% low melting point agarose (Sigma) to control the horizontal orientation of the worms in the final Epon blocks. The agarose blocks were incubated at 22°C in 0.1 M sodium cacodylate (pH 7.5) for 10 min, and for 1 h in 0.1 M sodium cacodylate containing 1% OsO4 and 1.5% reduced K4Fe(CN)6. The blocks were then washed four times, 10 min each, with 0.1 M sodium cacodylate. The sample was then washed twice, 10 min each, with double distilled H2O and then incubated in 30%, 50%, 70%, 80%, 90%, and 95% ethanol for 10 min each, followed by three washes, 20 min each, in 100% ethanol. Samples were then washed twice, 10 min each, in propylene oxide. The samples were then incubated in a series of graded Epon in propylene oxide solutions overnight at 4°C (1:4, 1:1, 1:3), followed by overnight incubation at 4°C in fresh 100% Epon. The Epon block was polymerized for 3 days at 60°C. They were then sectioned horizontally by using a Diatome diamond knife, to give fine slices of 80–90 nm of longitudinal view of each worm from head to tail. The sections were picked up on 200 mesh thin bar copper grids and stained with uranyl acetate and lead citrate. Samples were viewed with an electron microscope (Philips Technai 12) equipped with a MegaView II charge-coupled device camera.
Western Blot Analysis. Age-synchronized animals (40–50 animals per time point) were collected and subjected to SDS/PAGE analysis. Proteins were transferred to Immobilon P membranes (Millipore) and probed with rabbit polyclonal anti-lamin antibodies (16) and the mouse monoclonal anti-actin antibody JLA20 (22). Both antibodies were used in 1:2,000 dilutions. JLA20 was obtained from the Developmental Studies Hybridoma Bank, developed under the auspices of the National Institute of Child Health and Human Development and maintained by The University of Iowa Department of Biological Sciences (Iowa City).
Results
Aging Wild-Type Animals Show Specific Alterations of the Nuclear Lamina. To observe any age-related changes of the nuclear lamina in wild-type C. elegans animals, we generated integrated transgenic lines that carry GFP fused to the single C. elegans lamin encoded by lmn-1 (16); this allowed direct observation of the GFP signal in live animals. We obtained multiple integrated transgenic lines carrying either the C-terminal lmn-1::gfp fusion construct (three lines) or the N-terminal gfp::lmn-1 fusion construct (two lines) (see Materials and Methods). We out-crossed all transgenic lines with wild-type N2 background multiple times to eliminate possible mutations generated during integration. As shown in Table 1, which is published as supporting information on the PNAS web site, all transgenic lines had a similar lifespan to that of wild-type N2 animals. We conclude that the transgenic fusion proteins have no effect on lifespan.
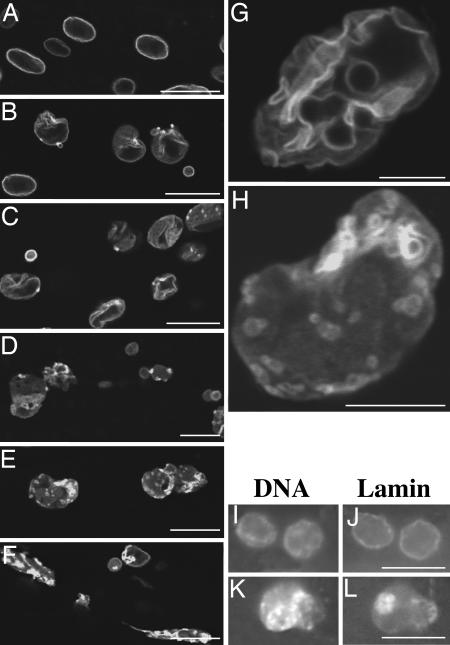
We then monitored for changes in nuclear morphology by LMN-1::GFP in stage-synchronized transgenic animals every other day starting from the L2 stage until the animals died by using both compound and confocal microscopy (a total of 20–30 animals on average were examined at each time point, see Materials and Methods). We found very few changes of nuclear lamina morphology in neuronal nuclei (data not shown), consistent with previous findings by Herndon et al. (13) that the nervous system remains rather intact in old animals. Instead, we found that body wall muscle, hypodermal, and intestinal nuclei exhibited progressive and stochastic changes of nuclear lamina morphology as animals aged. As shown in Fig. 1A, most nuclei in young animals (day 4) had the LMN-1::GFP signal smoothly distributed around the periphery (class I nuclei, Fig. 2). Over time, an increasing number of nuclei started to show a rather convoluted nuclear periphery, with multiple folds and, sometimes, bright dots of GFP signals (Fig. 1 B, C, and G; class II nuclei in Fig. 2). As the animals became older and less mobile, they started to accumulate nuclei with significantly increased intranuclear and decreased nuclear peripheral LMN-1::GFP, as well as nuclei that showed increasingly abnormal shape and/or extensive stretching and fragmentation (Fig. 1 D–F and H; class III nuclei in Fig. 2). The specific nuclear morphology adopted by different nuclei (even of the same cell type) was variable both among different animals and within individual animals (data not shown). However, the total number of class I nuclei decreased, whereas the number of class II and III nuclei increased as wild-type animals aged (Fig. 2). Previously, Herndon et al. (13) have grouped wild-type animals into three classes based on their locomotory activities, from the highly mobile class A animals, to the increasingly less mobile class B and C animals. We observed that the class C animals tend to accumulate more class III nuclei as compared to class A and B animals (data not shown).
Fig. 1.
The nuclear lamina undergoes progressive changes in aging nematodes. The nuclear lamina morphology as revealed in live C. elegans animals by LMN-1::GFP in days 4 (A),8(B), 12 (C), 15 (D) and 18 (E and F) animals under a confocal microscope. (G and H) Higher magnification confocal images of typical nuclei in days 12 (G) and 18 (H). Notice the folding of the nuclear lamina in G and the less distinct nuclear periphery and increased intranuclear localization of LMN-1::GFP in H. DAPI (I and K) and anti-LMN-1 antibody (J and L) staining of days 4 (I and J) and 14 (K and L) animals. Animals used in this experiment and the experiments in Figs. 2 and 5 were grown at 16°C from eggs until day 4. They were then shifted to 25°C and allowed to grow at 25°C until death. (Scale bars, 5 μmin G and H and 10 μmin A–F and I–L.)
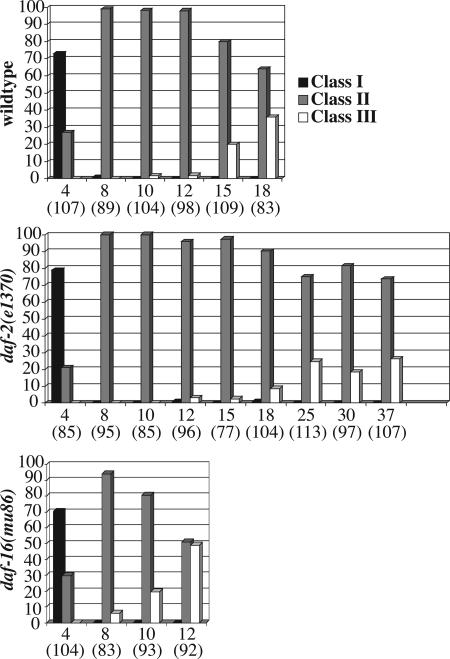
Fig. 2.
Summary of nuclear morphology changes in aging animals of different genetic backgrounds. Nuclear morphology of wild-type, daf-2(e1370), and daf-16(mu86) animals was monitored by using ccIs4810(lmn-1::gfp). Multiple images were taken of the middle part of the worm (excluding the head and the tail) by a confocal or a compound microscope. Nuclei from the collected images were counted and grouped into three different classes (class I–III) based on their morphology and GFP signal distribution (see Results). Neuronal nuclei were excluded from the counting. The nuclei counted are primarily hypodermal. Data were derived from a total of 20–30 animals on each of the days indicated (pooled from two to three experiments). y axis, percent of nuclei in a given category (black column, class I; gray column, class II; white column, class III). x axis, days (total number of nuclei scored).
To rule out the possibility that the observed age-related changes in nuclear lamina morphology were an artifact of the LMN-1::GFP used, we fixed age-synchronized populations of wild-type N2 animals and stained them with anti-Ce-lamin antibodies. We observed similar age-related changes of Ce-lamin distribution to those observed by using LMN-1::GFP. As shown in Fig. 1 I–K, although most nuclei of young animals at day 4 had distinct nuclear peripheral Ce-lamin staining (Fig. 1 I and J), nuclei in old animals at day 14 showed less distinct nuclear peripheral Ce-lamin staining and relatively more intranuclear staining with abnormal accumulation of the LMN-1 signal in bright foci or blobs (Fig. 1 K and L). In addition to staining fixed animals, we also observed live animals under differential interference contrast optics and found that the nuclear periphery in older animals became progressively less well defined (data not shown), similar to observations previously reported by Garigan et al. (12). Therefore, we conclude that wild-type animals undergo stochastic and progressive alterations of their nuclear lamina as they aged.
Aging Wild-Type Animals Show Alterations of Other Aspects of Nuclear Architecture. To examine whether the nuclear envelope in general underwent progressive alteration in aging wild-type animals, we followed other components of the nuclear envelope by generating integrated transgenic lines carrying GFP fused to Ce-emerin, an integral inner nuclear membrane protein encoded by emr-1 (19) and NPP-1, a component of the nuclear pore complex encoded by npp-1 (A. Schetter and K. Kemphues, personal communication). As shown in Fig. 6, which is published as supporting information on the PNAS web site, both proteins showed age-dependent changes in distribution, similar to those observed by using LMN-1::GFP. Thus, different nuclear envelope components undergo progressive alterations in aging C. elegans animals.
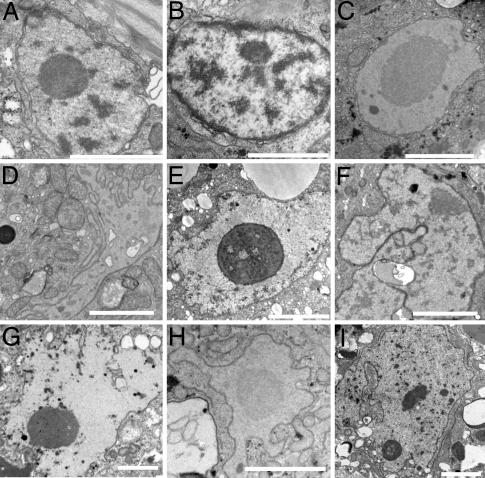
To further analyze the nuclear and chromatin phenotypes in aging animals, wild-type worms that were grown at 25°C to different ages were fixed, embedded, sectioned, stained, and viewed by TEM. At day 4 of development the nuclear membranes, nuclear pore complexes and heterochromatin of muscle (Fig. 3 A–C), hypodermal, and pharyngeal nuclei (data not shown) looked normal and similar to that of nuclei from larval stages (data not shown). At day 8, ≈30% of the nuclei started to have a convoluted appearance (n = 47), which was accompanied by heterochromatin loss from the nuclear periphery (Fig. 3 D–F). By day 12, most nuclei were highly lobulated and peripheral heterochromatin could not be detected (Fig. 3 G–I and Fig. 7, which is published as supporting information on the PNAS web site). However, condensed chromatin could be observed away from the nuclear periphery (Figs. 3 G and I and 7). In addition, 21% of (57 of 273) of the muscle nuclei looked highly lobulated or fragmented (Figs. 3 G and I and 7). Again, these changes in nuclear morphology were accompanied with changes in heterochromatin loss from the nuclear periphery. Higher magnification of day 12 or older nuclei showed additional phenotypes including the presence of mitochondria in grooves of the nuclear envelope, abnormal looking chromatin, dark inclusions of unknown material, and additional layers of nuclear membranes that indicated membrane proliferation (Fig. 7).
Fig. 3.
Thin section electron microscope pictures demonstrating the gradual deterioration of nuclear and nuclear envelope structures with age. Muscle nuclei from anterior (B, F, and H), middle (A, D, and I), and posterior (C, E, and G) of the worm. The nuclei were from days 4 (A–C),8(D–F), and 12 (G–I) animals grown at 25°C. (Scale bars, 2 μm.)
All together, these results demonstrate specific changes in nuclear architecture in aging animals.
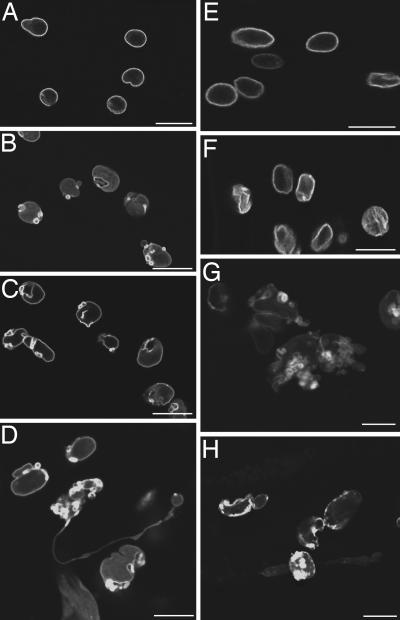
The Insulin/IGF-1-Like Signaling Pathway Influences the Rate of Age-Related Nuclear Architecture Changes. The insulin/IGF-1-like signaling pathway plays a major role in regulating the lifespan of C. elegans (14). Previous studies have shown that mutations in this pathway change the rate at which tissues age (12, 13). To examine whether this pathway also affects the rate of age-related changes in nuclear architecture, we introduced the integrated lmn-1::gfp transgene (ccls4810) into the long-lived mutant daf-2(e1370) and the short-lived mutant daf-16(mu86) (23, 24). We then examined the nuclear envelope morphology in aging animals of different genotypes by following LMN-1::GFP distribution. We found that the nuclear envelope in both daf-2(e1370) and daf-16(mu86) mutant animals underwent age-related morphological changes and that the characteristics of these changes in the mutants were similar to those of wild-type animals (Fig. 4). However, the rate of changes in these mutant animals was significantly different from that of wild-type animals. As shown in Figs. 2 and 4, although both daf-2(e1370) and daf-16(mu86) mutant animals started to accumulate class II nuclei at about the same age, the age onset of significant accumulation of class III nuclei in these mutant animals differed from that of wild-type animals. In wild-type animals, significant accumulation of class III nuclei (20.2%) was detected on day 15 (Fig. 2). A similar portion of class III nuclei (24.8%) was detected in long-lived daf-2(e1370) mutant animals only at day 25, whereas 10-day-old short-lived daf-16(mu86) mutant animals accumulated 19.4% class III nuclei (Fig. 2). Similar observations were obtained in the long-lived age-1(hx546) (25) and the short-lived hsf-1(RNAi) (26) mutant animals (data not shown). Therefore, we conclude that the activity of the insulin/IGF-1-like signaling pathway influences the rate of age-related changes of the nuclear envelope.
Fig. 4.
Mutants in the insulin/IGF-1-like signaling pathway exhibit altered rate of nuclear envelope deterioration. (A–D) LMN-1::GFP in daf-2(e1370) mutant animals at days 4 (A), 8 (B), 20 (C), and 37 (D). (E–H) LMN-1::GFP in daf-16(mu86) mutant animals at days 4 (E),8(F), 10 (G), and 12 (H). (Scale bars, 10 μm in all panels.)
Reducing the Level of Nuclear Lamina Components Results in Shortened Lifespan. Given that mutations in the human LMNA gene cause HGPS and that the accumulation of the mutant LMNA protein is correlated with progressive changes of nuclear architecture in HGPS cells (9–11), we next examined by Western blot analysis whether there were age-correlated changes of Ce-lamin level or modification state. We saw no dramatic changes in either the mobility or the level of Ce-lamin from aging animals (Fig. 8, which is published as supporting information on the PNAS web site).
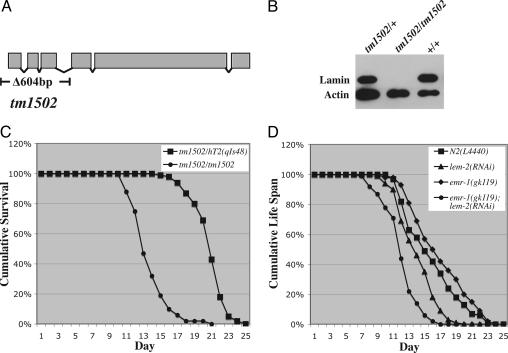
To further investigate whether Ce-lamin plays any role in the aging process, we examined the consequences of reducing the level of Ce-lamin on lifespan. We obtained an lmn-1 deletion allele, tm1502, from the National Bioresource Project for the nematode. tm1502 deletes 604 base pairs, including the translation start site and the first three exons of the lmn-1 gene (Fig. 5A). We out-crossed and balanced the tm1502 mutation. tm1502/tm1502 homozygous animals are viable but sterile. Their survival to adulthood is presumably due to the maternal load of the Ce-lamin protein from the heterozygous tm1502/+ mother, as we have shown that Ce-lamin proteins are very stable (16). Adult tm1502/tm1502 animals had no detectable Ce-lamin on Western blots (Fig. 5B). However, they showed no morphological or movement defects. They appeared to have a normal-looking vulva, and they contained all of the differentiated cells derived from the single postembryonic mesodermal lineage, the M lineage (data not shown). The normal vulva and the normal M lineage suggest that postembryonic cell divisions in somatic tissues are normal in tm1502/tm1502 mutant animals. Despite the apparently normal soma, tm1502/tm1502 animals lived significantly shorter compared to tm1502/+ animals (Fig. 5C and Table 1).
Fig. 5.
Animals with reduced level of Ce-lamin or the two LEM domain proteins Ce-emerin and Ce-MAN1 have a shorter lifespan. (A) Diagram of the lmn-1 genomic structure depicting the location of the tm1502 mutation. (B) Western blots of extracts from adult N2, tm1502/+ and tm1502/tm1502 animals probed with antibodies against Ce-lamin and actin. (C) Lifespan of tm1502/+ and tm1502/tm1502 animals at 20°C. (D) Lifespan of emr-1(gk119); lem-2(RNAi) animals at 20°C (see Materials and Methods). N2(L4440) and emr-1(gk119) refer to wild-type and emr-1(gk119) single mutant animals fed on control bacteria with the empty RNAi feeding vector L4440, respectively. lem-2(RNAi) and emr-1(gk119); lem-2(RNAi) refer to wild-type and emr-1(gk119) single mutant animals fed on bacteria expressing ds-RNA against lem-2, respectively.
We also specifically reduced Ce-lamin levels during postembryonic development to bypass its essential requirement for normal embryonic development. We did this by feeding wild-type animals from hatching through postembryonic development with bacteria expressing dsRNA against lmn-1. More than 80% of the lmn-1(RNAi) animals generated this way were fertile and they produced close to 100% dead embryos, as expected by the loss of maternal Ce-lamin function (16). These fertile animals showed no detectable morphological defects (n > 200). However, they exhibited a shortened lifespan compared with wild-type animals fed with control bacteria (Table 1). Thus, Ce-lamin is critical for the normal lifespan of C. elegans.
We have previously shown that Ce-lamin interacts with two LEM domain proteins Ce-emerin and Ce-MAN1, encoded by emr-1 and lem-2, respectively (19, 27). Because reducing the level of Ce-lamin led to a decrease in lifespan (Fig. 5C and Table 1), we also examined the consequences of reducing both Ce-emerin and Ce-MAN1 on the lifespan of the animals. We have obtained a deletion allele of emr-1 from the C. elegans Gene Knockout Consortium, gk119, which has the entire coding region of emr-1 deleted. emr-1(gk119) mutant animals are fertile with a normal lifespan (Fig. 5D and Table 1). However, when we fed emr-1(gk119) mutant animals from hatching throughout postembryonic development with bacteria expressing dsRNA against lem-2, the resulting emr-1(gk119); lem-2(RNAi) animals were sterile and had a significantly shorter lifespan than that of emr-1(gk119) animals fed on control bacteria (Fig. 5D and Table 1). Feeding wild-type animals with bacteria expressing dsRNA against lem-2 also resulted in a reduction of lifespan, but to a lesser degree (Fig. 5D and Table 1). Therefore, we conclude that nuclear envelopes with normal Ce-lamin and the two LEM domain proteins Ce-emerin and Ce-MAN1 are critical for the normal lifespan of C. elegans animals.
Discussion
Our studies demonstrate that wild-type C. elegans animals undergo age-related changes of the nuclear architecture. We have observed changes of nuclear shape, abnormal distribution of nuclear envelope proteins and loss of peripheral heterochromatin in increasingly older animals. These age-related changes of nuclear architecture are not restricted to specific areas of the animal, and they primarily occur in nonneuronal cells, including muscle, hypodermal, and intestinal cells. Furthermore, the specific changes we observed varied among individual cells within an animal and among different individual animals. These findings are consistent with previous findings that C. elegans animals undergo age-dependent deterioration of nonneuronal tissues, especially in muscle cells, and that the age-related tissue deterioration in C. elegans has a strong stochastic component (12, 13). We have shown that the insulin/IGF-1-like signaling pathway influences the rate of age-related nuclear architecture changes (Figs. 2 and 4). This same signaling pathway has also been shown to influence rates of tissue degeneration in aging animals (12, 13). Thus, the insulin/IGF-1-like signaling pathway affects lifespan by influencing multiple aspects of aging.
The age-related changes of nuclear architecture described here are very similar to the progressive changes of nuclear architecture observed by Goldman et al. (11) using fibroblast cells of HGPS patients, including abnormal nuclear shape and loss of peripheral heterochromatin. Thus, we propose that HGPS patients are very likely undergoing an acceleration of the normal aging process. We have found no or few changes of nuclear architecture in the nervous system of aging C. elegans animals. This finding could potentially explain why HGPS patients show no defects in their mental and intellectual abilities despite the presence of multiple premature aging symptoms.
HGPS is caused by mutations in the human LMNA gene (9, 10). It has been shown that the progressive changes of nuclear architecture in HGPS cells are caused by the accumulation of a mutant lamin A protein that has an internal 50 aa-truncation near the C terminus (11, 28). Indeed, elimination of this mutant protein in HGPS cells can lead to reversal of the cellular phenotypes, including changes of nuclear architecture (28). These findings raise the question of what causes the changes of nuclear architecture in wild-type C. elegans animals. Using Western blot analysis, we observed no apparent changes in the level or mobility of the Ce-lamin protein in old animals (Fig. 8). However, we cannot rule out the possibility that a fraction of the Ce-lamin protein pool undergoes biochemical changes, such as changes due to oxidative damage, posttranslational modifications, or aberrant splicing in aging animals.
Despite the lack of detectable changes in the level of Ce-lamin in aging animals, we found that reducing the level of Ce-lamin during postembryonic development resulted in a shortening of lifespan (Fig. 5C and Table 1). In addition, reducing the level of two redundant Ce-lamin interacting LEM-domain proteins, Ce-emerin and Ce-MAN1, also resulted in a shortening of lifespan (Fig. 5D). We have previously shown that Ce-lamin, the two LEM domain proteins and Ce-BAF form a lamina-dependent network required to regulate nuclear organization and assembly (27, 29, 30). Our results presented in this study suggest that these proteins are also required for normal longevity. The future challenge will be to understand how lamin and its interacting LEM-domain proteins are involved in the normal aging process.
The nucleus is vital for many cellular functions, such as DNA replication, transcription, and RNA processing. The nuclear envelope plays critical roles in regulating many of these events as well as maintaining genomic stability and regulating protein/RNA transport in and out of the nucleus (1, 31). We have observed changes of nuclear shape, abnormal distribution of nuclear envelope proteins, and loss of peripheral heterochromatin in aging C. elegans animals. These changes could potentially result in alterations in transcription, RNA processing, nuclear import and export, and increased genomic instability. Because similar nuclear architecture changes are also observed in cells of HGPS patients and HGPS is caused by mutations in the LMNA gene, which encodes a critical component of the nuclear envelope, our results raise the possibility that the nucleus is a prominent focal point for regulating aging and that lamin plays a critical role in this process. We would also like to propose that compromised nuclear architecture and nuclear function could be a central cause of aging.
Supplementary Material
Acknowledgments
We thank Sylvia Lee, Shohei Mitani, Aaron Schetter, Jeff Stambough, and the C. elegans Gene Knockout Consortium for strains and clones; Carol Bayles for help with the confocal microscope; Ken Kemphues, Sylvia Lee, and members of the Liu laboratory for helpful discussions and suggestions; and Ken Kemphues and Sylvia Lee for critical comments on the manuscript. Some strains used in this study were obtained from the C. elegans Genetics Center (CGC), which is supported by a grant from the National Institutes of Health National Center for Research Resources. This work was supported in part by National Institutes of Health Grant R01 GM066953 and by a grant from the Muscular Dystrophy Association (to J.L.), the USA–Israel Binational Science Foundation, the Cooperation Program in Cancer Research of the Deutsches Krebsforschungszentrum, Israel's Ministry of Science and Technology, and the Israel Ministry of Health (to Y.G.). A.J.Z. was partly supported by National Institutes of Health Grant T32 GM07617.
Author contributions: Y.G. and J.L. designed research; E.H., Y.D., E.N., A.J.Z., N.F., A.M., and J.L. performed research; Y.G. and J.L. analyzed data; and Y.G. and J.L. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: HGPS, Hutchinson–Gilford Progeria Syndrome; RNAi, RNA interference; TEM, transmission electron microscopy.
References
- 1.Gruenbaum, Y., Margalit, A., Goldman, R. D., Shumaker, D. K. & Wilson, K. L. (2005) Nat. Rev. Mol. Cell. Biol. 6, 21–31. [DOI] [PubMed] [Google Scholar]
- 2.Goldman, R. D., Gruenbaum, Y., Moir, R. D., Shumaker, D. K. & Spann, T. P. (2002) Genes Dev. 16, 533–547. [DOI] [PubMed] [Google Scholar]
- 3.Hutchison, C. J. (2002) Nat. Rev. Mol. Cell. Biol. 3, 848–858. [DOI] [PubMed] [Google Scholar]
- 4.Gruenbaum, Y., Goldman, R. D., Meyuhas, R., Mills, E., Margalit, A., Fridkin, A., Dayani, Y., Prokocimer, M. & Enosh, A. (2003) Int. Rev. Cytol. 226, 1–62. [DOI] [PubMed] [Google Scholar]
- 5.Wilson, K. L., Zastrow, M. S. & Lee, K. K. (2001) Cell 104, 647–650. [PubMed] [Google Scholar]
- 6.Worman, H. J. & Courvalin, J. C. (2002) Trends Cell Biol. 12, 591–598. [DOI] [PubMed] [Google Scholar]
- 7.Herrmann, H. & Foisner, R. (2003) Cell Mol. Life Sci. 60, 1607–1612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mounkes, L. C. & Stewart, C. L. (2004) Curr. Opin. Cell Biol. 16, 322–327. [DOI] [PubMed] [Google Scholar]
- 9.Eriksson, M., Brown, W. T., Gordon, L. B., Glynn, M. W., Singer, J., Scott, L., Erdos, M. R., Robbins, C. M., Moses, T. Y., Berglund, P., et al. (2003) Nature 423, 293–298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Sandre-Giovannoli, A., Bernard, R., Cau, P., Navarro, C., Amiel, J., Boccaccio, I., Lyonnet, S., Stewart, C. L., Munnich, A., Le Merrer, M., et al. (2003) Science 300, 2055. [DOI] [PubMed] [Google Scholar]
- 11.Goldman, R. D., Shumaker, D. K., Erdos, M. R., Eriksson, M., Goldman, A. E., Gordon, L. B., Gruenbaum, Y., Khuon, S., Mendez, M., Varga, R., et al. (2004) Proc. Natl. Acad. Sci. USA 101, 8963–8968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Garigan, D., Hsu, A. L., Fraser, A. G., Kamath, R. S., Ahringer, J. & Kenyon, C. (2002) Genetics 161, 1101–1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Herndon, L. A., Schmeissner, P. J., Dudaronek, J. M., Brown, P. A., Listner, K. M., Sakano, Y., Paupard, M. C., Hall, D. H. & Driscoll, M. (2002) Nature 419, 808–814. [DOI] [PubMed] [Google Scholar]
- 14.Kenyon, C. (2005) Cell 120, 449–460. [DOI] [PubMed] [Google Scholar]
- 15.Brenner, S. (1974) Genetics 77, 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu, J., Rolef Ben-Shahar, T., Riemer, D., Treinin, M., Spann, P., Weber, K., Fire, A. & Gruenbaum, Y. (2000) Mol. Biol. Cell 11, 3937–3947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee, S. S., Lee, R. Y., Fraser, A. G., Kamath, R. S., Ahringer, J. & Ruvkun, G. (2003) Nat. Genet. 33, 40–48. [DOI] [PubMed] [Google Scholar]
- 18.Timmons, L., Court, D. L. & Fire, A. (2001) Gene 263, 103–112. [DOI] [PubMed] [Google Scholar]
- 19.Gruenbaum, Y., Lee, K. K., Liu, J., Cohen, M. & Wilson, K. L. (2002) J. Cell Sci. 115, 923–929. [DOI] [PubMed] [Google Scholar]
- 20.Kamath, R. S., Fraser, A. G., Dong, Y., Poulin, G., Durbin, R., Gotta, M., Kanapin, A., Le Bot, N., Moreno, S., Sohrmann, M., et al. (2003) Nature 421, 231–237. [DOI] [PubMed] [Google Scholar]
- 21.Cohen, M., Tzur, Y. B., Neufeld, E., Feinstein, N., Delannoy, M. R., Wilson, K. L. & Gruenbaum, Y. (2002) J. Struct. Biol. 140, 232–240. [DOI] [PubMed] [Google Scholar]
- 22.Lin, J. J. (1981) Proc. Natl. Acad. Sci. USA 78, 2335–2339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kenyon, C., Chang, J., Gensch, E., Rudner, A. & Tabtiang, R. (1993) Nature 366, 461–464. [DOI] [PubMed] [Google Scholar]
- 24.Lin, K., Dorman, J. B., Rodan, A. & Kenyon, C. (1997) Science 278, 1319–1322. [DOI] [PubMed] [Google Scholar]
- 25.Friedman, D. B. & Johnson, T. E. (1988) Genetics 118, 75–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hsu, A. L., Murphy, C. T. & Kenyon, C. (2003) Science 300, 1142–1145. [DOI] [PubMed] [Google Scholar]
- 27.Liu, J., Lee, K. K., Segura-Totten, M., Neufeld, E., Wilson, K. L. & Gruenbaum, Y. (2003) Proc. Natl. Acad. Sci. USA 100, 4598–4603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scaffidi, P. & Misteli, T. (2005) Nat. Med. 11, 440–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Margalit, A., Segura-Totten, M., Gruenbaum, Y. & Wilson, K. L. (2005) Proc. Natl. Acad. Sci. USA 102, 3290–3295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Margalit, A., Liu, J., Fridkin, A., Wilson, K. L. & Gruenbaum, Y. (2005) Novartis Found. Symp. 264, 231–240. [PubMed] [Google Scholar]
- 31.Liu, B., Wang, J., Chan, K. M., Tjia, W. M., Deng, W., Guan, X., Huang, J., Li, K. M., Chau, P. Y., Chen, D. J., et al. (2005) Nat. Med. 11, 780–785. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.