Abstract
Wiskott–Aldrich syndrome protein (WASP)-homology domain 2 (WH2) is a small and widespread actin-binding motif. In the WASP family, WH2 plays a role in filament nucleation by Arp2/3 complex. Here we describe the crystal structures of complexes of actin with the WH2 domains of WASP, WASP-family verprolin homologous protein, and WASP-interacting protein. Despite low sequence identity, WH2 shares structural similarity with the N-terminal portion of the actin monomer-sequestering thymosin β domain (Tβ). We show that both domains inhibit nucleotide exchange by targeting the cleft between actin subdomains 1 and 3, a common binding site for many unrelated actin-binding proteins. Importantly, WH2 is significantly shorter than Tβ but binds actin with ≈10-fold higher affinity. WH2 lacks a C-terminal extension that in Tβ4 becomes involved in monomer sequestration by interfering with intersubunit contacts in F-actin. Owing to their shorter length, WH2 domains connected in tandem by short linkers can coexist with intersubunit contacts in F-actin and are proposed to function in filament nucleation by lining up actin subunits along a filament strand. The WH2-central region of WASP-family proteins is proposed to function in an analogous way by forming a special class of tandem repeats whose function is to line up actin and Arp2 during Arp2/3 nucleation. The structures also suggest a mechanism for how profilin-binding Pro-rich sequences positioned N-terminal to WH2 could feed actin monomers directly to WH2, thereby playing a role in filament elongation.
Keywords: x-ray crystallography, isothermal titration calorimetry, nucleotide exchange
The actin cytoskeleton plays an essential role in many cellular functions, including intracellular transport and the control of cell shape and polarity (1). In the cell, a vast number of actin-binding proteins (ABPs) direct the location, rate, and timing for actin assembly into different structures, such as filopodia, lamellipodia, stress fibers, and focal adhesions. ABPs are commonly multidomain proteins, containing signaling domains and structurally conserved actin-binding motifs. One of the most abundant actin-binding motifs is Wiskott–Aldrich syndrome protein (WASP)-homology domain 2 (WH2) (2). The hematopoietic-specific protein, WASP, and its ubiquitously expressed ortholog N-WASP form part of a family that also includes the three WASP-family verprolin homologous protein (WAVE/SCAR) isoforms: WAVE1, WAVE2, and WAVE3 (1, 3). Members of this family activate Arp2/3-dependent actin nucleation and branching in response to signals mediated by Rho-family GTPases. Although the domain structure of these proteins varies, reflecting different modes of regulation, they all share a common C-terminal WH2 central-acidic region (CA region) (Fig. 1A), which constitutes the smallest fragment necessary for Arp2/3 activation (4). WH2 is also present in members of the WASP-interacting protein (WIP) family, which form complexes with WASP/N-WASP and modulate their functions in vivo (5, 6). Members of this family include WIP, CR16, and WIRE (or WICH) in mammals and verprolin in yeast.
Fig. 1.
Sequences and actin-binding affinities of WH2 and Tβ.(A) Domain structure of WASP (B, basic region; GBD, GTPase-binding domain; C, C region; A, acidic region). (B) Sequence alignment of Tβ and WH2. The sequence accession numbers are Q546P5 (Tβ4), O97428 (ciboulot), P18281 (actobindin), O43312 (MIM), Q53TA9 (WIP), P50552 (VASP), P42768 (WASP), O00401 (N-WASP), Q9Y6W5 (WAVE2), and Q9U4F1 (Spire). Conserved amino acids are colored according to their chemical characteristics (green, hydrophobic; blue, basic; red, acidic; yellow, small amino acid). The insert in the linker region of Tβ is shown in pink. Consensus profilin-binding sequences are shown in purple. Alignment of the CA region underlines the similarity between the C region and WH2. (C) Actin-binding affinities determined by ITC (see also Table 2). Highlighted in green are the WH2 domains whose structures were determined, whereas the amino acids that are seen in the structures are shown in bold in B.
It has been proposed, based on sequence analysis, that WH2 forms part of an extended family with the thymosin β domain (Tβ) (7). However, this view is controversial, in part because of the different biological functions and low sequence similarity of WH2 and Tβ (8). The actin-bound structures of the N-terminal half of ciboulot domain 1 (9) and that of a hybrid protein consisting of gelsolin domain 1 and the C-terminal half of Tβ4 (10) have been reported. These structures have been combined into a model of Tβ4–actin (10), and, although both Tβ4 and ciboulot belong in the Tβ family, their structures have been described as representatives of WH2 (9, 10).
Here we report the crystal structures of complexes of actin with the WH2 domains of WASP, WAVE2, and WIP, in parallel with a biochemical characterization of WH2 and Tβ. Important structural and biochemical differences set apart WH2 and Tβ in accordance with their specialized cellular functions. The structures of WH2–actin shed light on the molecular principles supporting the role of WH2 in filament nucleation and elongation.
Materials and Methods
Proteins and Peptides. Actin was prepared and labeled with acrylodan as described in ref. 11. Ultrapure-grade bovine pancreatic DNase I was purchased from BioWorld (Dublin, OH). Peptides corresponding to WASP430–458, WAVE2433–464, WAVE2450–464, WIP29–60, WIP29–46, WIP46–63, MIM724–755, Tβ42–44, Tβ42–33, Tβ418–44, ciboulot10–43, ciboulot49–81, and ciboulot87–119 were synthesized on an ABI431 peptide synthesizer and then purified by HPLC. The concentrations of the peptides were determined by amino acid analysis (Dana–Farber Cancer Institute, Boston).
Crystallization, Data Collection, and Structure Determination. WH2–actin–DNase I complexes were prepared at a 1:1:1.5 molar ratio, dialyzed against G-buffer (2 mM Tris, pH 7.5/0.2 mM CaCl2/0.2mM ATP/1 mM NaN3), and concentrated to ≈10 mg/ml using a Centricon device (Millipore). The complexes were crystallized at 20°C, using the hanging drop vapor diffusion method. The 2-μl hanging drops consisted of a 1:1 (vol/vol) mixture of protein solution and a well solution containing 13–14% polyethylene glycol 3350, 50 mM Na cacodylate (pH 6.8–7.2), and 100 mM Na formate. The crystals were flash-frozen in propane, with 25% glycerol as a cryoprotectant. X-ray data sets were collected at Industrial Macromolecular Crystallography Association Collaborative Access Team Beamline 17-ID at the Advance Photon Source (Argonne, IL). The data sets were indexed and scaled with the program hkl-2000 (HKL Research, Charlottesville, VA). The structures were determined by molecular replacement using ccp4 program amore and the structure of actin–DNase I (12) as a search model. Model building and refinement were done with the program coot (13), and the ccp4 program refmac (Table 1, which is published as supporting information on the PNAS web site).
Isothermal Titration Calorimetry (ITC). ITC measurements were done with a VP-ITC system (MicroCal, Northampton, MA). To determine the ΔH and Ka of WH2–actin association, WH2 peptides at a 100 μM were titrated in 10-μl injections into 1.44 ml of 10 μM actin in G-buffer at 25°C. The duration of the injections was 10 s, with an interval of 3 min between injections. The heat of binding was corrected for the small exothermic heat of injection, determined by injecting WH2 peptides into buffer. Data were analyzed with MicroCal's origin program (Table 2, which is published as supporting information on the PNAS web site).
Nucleotide Exchange Assay. The rate of fluorescence decay upon release of 1,N6-etheno-ATP, (ε-ATP) from G-actin was measured at 20°C. G-actin was dialyzed against G-buffer containing ε-ATP instead of ATP. Aliquots (20 μl) of ε-ATP–G-actin were mixed with WH2 peptides, then 180 μl of G-buffer containing 1.11 mM ATP was added, and the reaction was monitored on a Carry Eclipse spectrofluorometer at 412 nm with excitation at 360 nm. The final concentrations were 1 μM (actin) and 0, 0.5, 1, 2, 4, and 8 μM (WH2 peptides). The nucleotide exchange rate was estimated from the initial slope of the fluorescence decay and expressed as a fraction of the exchange rate in the absence of peptide.
Results and Discussion
Structures of WH2–Actin. According to a sequence analysis of a large number of WH2-containing proteins (8), WH2 can be defined as an ≈20-aa sequence, including a predicted amphiphilic α-helix at the N terminus and a LKKT-related sequence (Fig. 1B). In most proteins, the limits of WH2 are determined by the presence of low-order sequences rich in Pro and Gly residues that flank both ends of the domain. In some proteins, including WIP and MIM (missing in metastasis), WH2 occurs at the N or C terminus, further confining the boundaries of the domain. Based on this analysis, peptides corresponding to the WH2 domains of various proteins, including WASP, WAVE2, WIP, and MIM, were synthesized. To ensure that all of the structural–functional determinants of WH2 were included, the size of the peptides was conservatively set to ≈32 aa, starting before the predicted α-helix and ending according to the C terminus of MIM (Materials and Methods).
The complexes of actin with WH2 failed to crystallize, forming thick bundles under most crystallization conditions. DNase I was then used as a cofactor in the crystallization. DNase I binds with high affinity to actin subdomains 2 and 4 (12), whereas most ABPs bind on the opposite side, in the cleft between subdomains 1 and 3 (14), allowing certain ABPs to form ternary complexes with actin-DNase I. The formation of the ternary complexes of WH2–actin–DNase I was investigated by monitoring the fluorescence of acrylodan–actin (Table 3, which is published as supporting information on the PNAS web site). According to these measurements, DNase I does not affect the actin-binding affinities of the WH2 domains of WASP and WAVE2 and has just a minor effect on the binding of those of WIP and MIM. In agreement with studies on WASP and MIM (15, 16), we also find that WH2 binds ATP–actin with higher affinity than ADP–actin.
Crystals of the ternary complexes of actin–DNase I with the WH2 domains of WASP, WAVE2 and WIP were obtained under conditions somewhat related to those of actin–DNase I (12). However, the crystals belong to different space groups and diffract the x-rays to higher resolution than that of the original actin–DNase I structure (Table 1). A higher-resolution structure (1.85 Å) of actin–DNase I was also determined in a different space group than the original structure and was later used for comparative purposes. The conformations of actin and DNase I are very similar among the structures, which all contain ATP bound in the catalytic site of actin. In none of the structures does WH2 interact directly with DNase (Fig. 5, which is published as supporting information on the PNAS web site). Interestingly, whereas all of the amino acids in the WH2 domain of WIP are visible in the structure, those of WASP and WAVE become disordered after the LKKT sequence (Fig. 2). As shown below, these differences in the structures correlate with differences in the sequences, leading to the identification of two types of WH2: short and long (Fig. 1B).
Fig. 2.
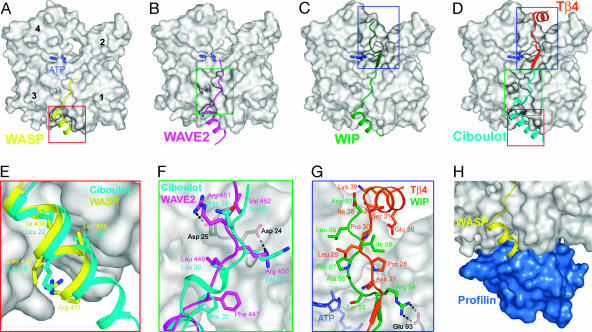
WH2–actin structures. (A–C) Structures of the WH2 domains of WASP, WAVE2, and WIP determined as ternary complexes with actin (gray) and DNase I (see Fig. 5). (D) Superimposition of the structures of ciboulot (9) and Tβ4 (10), which together represent Tβ–actin. (E–G) Close-view comparisons of different parts of the WH2–actin and Tβ–actin structures shown in A–D. (H) Partial overlap between the actin-binding sites of profilin (blue) and WH2.
The N-terminal portion of all three WH2 domains consists of a three-turn amphiphilic α-helix, with its hydrophobic side embedded within the cleft formed between actin subdomains 1 and 3. A similar α-helix in the first domain of Drosophila ciboulot is one helical turn longer (9) (Fig. 2E). In contrast, NMR studies show that in actin-bound Tβ4 the α-helix is only two turns long (17), which can be explained by a breakage in the periodicity of hydrophobic amino acids in the sequence (Fig. 1B). The length of the α-helix is critical because it may determine whether WH2 can coexist with intersubunit contacts in F-actin (see below) and whether it can bind actin simultaneously with profilin. Indeed, profilin, which feeds ATP–actin monomers to growing actin filaments, appears to coexist, at least transiently, with the shorter α-helix of Tβ4 (18), but its binding site on actin (19) partially overlaps with the longer α-helix of WH2 (Fig. 2H).
The N-terminal α-helices of the WH2 domains studied here contain a conserved Arg at the N terminus and a conserved Gly at the C terminus, both absent in Tβ proteins (Fig. 1B). The Arg is the first amino acid of WH2 that interacts with actin directly, whereas the presence of the Gly shifts the position of the linker that connects to the LKKT sequence forward, relative to Tβ proteins. In Tβ proteins, the linker is also 3 aa longer than in WH2 and forms a helical turn (Figs. 1B and 2F). The conserved Leu in the LKKT motif and a second hydrophobic amino acid in the linker, two amino acids N-terminal to this Leu, bind together in a hydrophobic pocket in actin (Fig. 2F). Because of their longer linker, the corresponding two amino acids are five residues apart in Tβ proteins (Figs. 1B and 2F), which, according to our measurements of binding affinities (see below), seems to constrain this important interaction into a less favorable conformation.
The Lys residues in the LKKT sequence do not interact with actin directly, except in WAVE where these two positions are occupied by Arg, which form salt bridges with actin residues Asp-24 and Asp-25 (Fig. 2F). Surrounding the LKKT binding site, actin presents a number of negatively charged amino acids, including Asp-24, Asp-25, Glu-99, Glu-100, Glu-334, and a series of amino acids at the N terminus of actin, which are not seen in the structures. Positively charged amino acids in the LKKT motif may engage in long-range electrostatic interactions with these amino acids in actin. The last amino acid in the LKKT motif is frequently Thr or Val, although Ala is also common (Fig. 1B). This amino acid binds in a small pocket in actin, which would be incompatible with a larger side chain.
As previously noticed (8), the amino acid sequence of Tβ is well conserved after the LKKT motif, but that of WH2 diverges widely (Fig. 1B). Depending on the specific sequence, WH2 can either end immediately after the LKKT motif, as in WASP and WAVE, or follow a path along the nucleotide cleft in actin, as in WIP and MIM (structure not shown). Therefore, from the viewpoint of its interactions with actin, WH2 has a variable length, ranging from ≈17 aa (short) to ≈27 aa (long) (Fig. 2). The five amino acids that follow the LKKT sequence play a key role in determining whether the chain interacts with actin. In WIP and MIM, these amino acids are similar to those in Tβ4 and accordingly follow a path similar to that of Tβ4 (10), forming a β-strand that runs parallel to a β-sheet in actin subdomain 1. The side chains of these amino acids make a number of interactions with actin, including a salt bridge between actin residue Glu-93 and the amino acid corresponding to Arg-54 in WIP (Fig. 2G). After this position, WIP (and all of the long WH2 domains) present a Ser residue, which is missing in Tβ, allowing these proteins to penetrate deeper into the nucleotide cleft of actin than Tβ4 (Fig. 2G). From this point on, the backbone of WIP takes a different path from that of Tβ4, but the two proteins converge again at the top of actin subdomains 2 and 4 so that the position of Asp-60 of WIP roughly coincides with Ser-31 of Tβ4 (Fig. 2D). Asp-60 of WIP corresponds to the C-terminal Ser-755 of MIM and marks the end of WH2, which in WIP is followed by a low-order sequence. In Tβ4, however, Ser-31 marks the beginning of a C-terminal α-helix that binds on top of actin subdomains 2 and 4 (10). Strong sequence identity suggests that this α-helix is conserved in all three domains of ciboulot (Fig. 1B) and tetraTβ. However, the α-helix is not seen in the structure of ciboulot–actin (9) (Fig. 2D), possibly because of the use of latrunculin A in the crystallization, which sterically hinders some of the interactions observed with Tβ4 (10). Therefore, the C-terminal α-helix, which constitutes a characteristic feature of Tβ, does not exist in WH2 (Figs. 1B and 2).
Actobindin, which ref. 20 describes as a member of the Tβ family, shares most of the features of a long WH2 domain (Fig. 1B), including the lack of a C-terminal α-helix, a short linker region, and the presence of an extra Ser residue in the sequence that binds along the nucleotide cleft in actin. Actobindin could therefore be the only known protein consisting uniquely of tandem WH2 domains.
Actin-Binding Function of WH2 and Tβ. The actin-binding function of WH2 and Tβ was investigated by using ITC (Fig. 1C; see also Table 2 and Fig. 6, which are published as supporting information on the PNAS web site), a method that avoids reported effects on binding affinity measurements resulting from the use of fluorescence probes (15). The results are somewhat in agreement with reported values for WASP (15), WIP (6), MIM (16), and Tβ4 (21). However, the measurements here aim to establish a direct correlation between different parts of the structures of WH2 and Tβ and their role in actin binding under identical conditions.
There is significant variation in the actin-binding affinities of the WH2 domains analyzed here. The strongest binder, WAVE, binds actin with 5-fold higher affinity than the weakest binder, WASP. This difference could be explained by the presence of the two Arg residues in the LKKT sequence of WAVE, which form salt bridges with actin (Fig. 2F). Supporting this view, WAVE450–464, starting from the LKKT sequence to the end, shows higher binding affinity than the equivalent WIP peptide (WIP46–63), which, according to the structures, has a larger binding interface.
Our measurements (Fig. 1C), together with published affinity values for various WH2 (6, 15, 16) and Tβ (21, 22) domains, consistently show that WH2 binds actin with ≈10-fold higher affinity than Tβ. Owing to the presence of the C-terminal α-helix, Tβ has a larger actin-binding interface than WH2 (Fig. 2D). How can a larger binding interface be reconciled with the lower binding affinity of Tβ? To assess the contribution of the C-terminal α-helix and to obtain a direct comparison between WH2 and Tβ, ciboulot and Tβ4 peptides were synthesized that lack the C-terminal α-helix and match the length of long-type WH2 domains. Thus, Tβ42–33 binds actin with 4-fold lower affinity than full-length Tβ4, indicating that the C-terminal α-helix has only a moderate contribution to the binding affinity of Tβ. This conclusion is further supported by the fact that Tβ418–44, starting from the LKKT sequence to the end, fails to bind actin. Similarly, the N-terminal portion of WIP (WIP29–46) accounts for most of its binding affinity, whereas WIP46–63 binds very weakly. Therefore, it can be concluded that hydrophobic contacts involving the N-terminal α-helices of WH2 and Tβ play a predominant role in the interaction with actin over electrostatic interactions within their C-terminal portions. It is also possible that, by stabilizing the actin structure, the binding of the N-terminal α-helices allosterically potentiates the binding of the C-terminal portions of WH2 and Tβ.
Despite missing their C-terminal α-helices, the peptides corresponding to the three domains of ciboulot all bind actin, although with somewhat lower affinity than the equivalent Tβ42–33 peptide. This result is not surprising given the high degree of sequence similarity with Tβ4 (Fig. 1B). Therefore, a previous report (9) that the second and third domains of ciboulot do not bind actin may reflect an incorrect definition of domain boundaries. In agreement with this view, all four domains of tetraTβ, a ciboulot-related protein in Caenorhabditis elegans, bind actin with affinities similar to those found here for ciboulot (22).
Control of Actin Nucleotide Exchange by WH2 and Tβ. As with the study of binding affinities, we aim to establish a relationship between nucleotide exchange inhibition and different parts of the structures of WH2 and Tβ (Fig. 3). Tβ4 (21) and MIM (16) are known to inhibit nucleotide exchange on actin. In Tβ4, the finding that the C-terminal α-helix binds on top of actin subdomains 2 and 4 led to the suggestion that, by locking the nucleotide cleft, this interaction could account for nucleotide exchange inhibition (10). However, we find that the WH2 domains of WIP and MIM, which lack C-terminal α-helices, inhibit nucleotide exchange to the same extent as Tβ4. Moreover, Tβ42–33, which lacks the C-terminal α-helix, inhibits nucleotide exchange similarly to full-length Tβ4 after accounting for the 4-fold-lower actin-binding affinity of this peptide (Fig. 1C).
Fig. 3.
Inhibition of nucleotide exchange on actin by WH2 and Tβ. ε-ATP–actin (1 μM) was mixed with 0, 0.5, 1, 2, 4, or 8 μM WASP430–458 (▪), WAVE433–464 (♦), WIP29–60 (▴), WIP29–46 (▵), MIM724–755 (×), Tβ42–44 (□), or Tβ42–33 (♦), and the fluorescence decay was monitored after addition of 1 mM ATP (see Materials and Methods).
As described above, the WH2 domains of WASP and WAVE do not follow a path along the nucleotide cleft in actin, as do those of WIP and MIM, yet they also efficiently inhibit nucleotide exchange. Therefore, a path along the actin nucleotide cleft is not necessary for nucleotide exchange inhibition, suggesting that this function resides within the N-terminal part of WH2 and Tβ. In agreement with this notion, WIP29–46, corresponding to the N-terminal α-helix, inhibits nucleotide exchange as potently as the full-length WH2 motif after taking into consideration the lower actin-binding affinity of this peptide. Therefore, we conclude that nucleotide exchange inhibition is a function of the N-terminal α-helix of WH2 and Tβ, which binds between actin subdomains 1 and 3, away from the nucleotide site. Because nucleotide exchange requires the dynamic opening/closing of the nucleotide cleft, inhibition of nucleotide exchange most likely results from a stabilization of the actin structure. Binding of an α-helix in the cleft between actin subdomains 1 and 3 is an effective way to produce such a rigid state, because it falls near the hinge for domain opening in actin (consisting of α-helix Ile-136 to Gly-146 in actin) (23).
Structure–Function Relationship of WH2 and Tβ. The proposed classification of Tβ and WH2 into a unified family with a common ancestor (7) is controversial (8), as the two domains are too short and their sequences too divergent to warrant a relationship. Sequence identity within the separated families is ≈50% and ≈30% for Tβ and WH2, respectively, but drops <15% when comparing sequences across domains. In metazoa, Tβ proteins consist of a single copy of Tβ, with Tβ4 as the prototypical member of this family. However, in arthropoda and nematoda, Tβ occurs only in the form of tandem repeats, such as in Drosophila ciboulot and C. elegans tetraTβ (24). In contrast, WH2 is found from yeast to human in the form of single or multiple copies within large multidomain proteins.
We have established here that WH2 is significantly shorter, in particular in WASP/WAVE (≈17 aa), than Tβ (≈44 aa). The extra amino acids of Tβ form a C-terminal α-helix that adapts this domain for its actin-monomer-trapping function by capping both ends of actin (10), a function that would be at odds with the role of WH2 in filament nucleation (see below). Yet there are similarities between the N-terminal portions of Tβ (9) and WH2 (Fig. 2). At the N terminus, both domains present an amphiphilic α-helix whose hydrophobic side interacts in the cleft between actin subdomains 1 and 3. Although there is no significant sequence conservation within this α-helix, the periodicity of hydrophobic amino acids involved in the interaction with actin is well conserved. This region also accounts for most of the actin-binding affinity (Fig. 1C) and nucleotide exchange inhibition (Fig. 3) of both domains. However, the WH2 domains analyzed here bind actin with ≈10-fold higher affinity than the Tβ domains. Within the N-terminal region, the most important differences that could account for such a gap in binding affinity occur in the linker (Figs. 1B and 2F). The differences in the linker are also important when considering a potential relationship between the two domains, because the binding of an α-helix in the cleft between actin subdomains 1 and 3 is a common feature among multiple unrelated ABPs (14), and, in at least one more protein, gelsolin, the α-helix occurs in addition to a LKKT-related sequence (10) (Fig. 7, which is published as supporting information on the PNAS web site). This example raises the question as to whether the relationship between WH2 and Tβ resulted from convergent or divergent evolution. In this regard, it is important to note that the extra amino acids that characterize long WH2 domains (compared with short WH2 domains) are well conserved in Tβ and interact similarly with actin. Therefore, long WH2 domains may represent an evolutionary intermediate between WH2 and Tβ, lending support to the proposed relationship between these two families (7).
Implications for Actin Filament Nucleation and Elongation. WH2 occurs in a large number of cytoskeletal proteins, including WASP/WAVE, WIP, vasodilator-stimulated phosphoprotein (VASP), and spire, which generally function in actin filament nucleation and/or elongation. In these proteins, WH2 is present as one or multiple copies, most commonly C-terminal to Pro-rich sequences. Although the distinction between filament nucleation and elongation is not always clear because of intrinsic limitations of the so-called pyrene–actin assay (25), published data strongly suggest that WH2 plays a role in both processes. Spire, for instance, a protein involved in filament nucleation (26), contains four tandem WH2 domains (Fig. 1B), which appear to function as a ruler by determining the size of filament nuclei according to the number of WH2 repeats. This finding led to the suggestion that the WH2 domains of spire line up actin subunits along a filament strand of the actin double helix, thereby generating nuclei for actin assembly (26). The Pro-rich sequences that precede WH2 domains comprise consensus profilin-binding sites (27). Profilin is the major actin monomer carrier in the cell (1). By delivering actin monomers to growing actin filaments, profilin stimulates actin filament elongation by VASP (28) and WASP/WAVE (29–31). The role of profilin in elongation appears to be common among cytoskeletal proteins. Formins, for instance, function in a similar way, promoting elongation in a profilin-dependent manner by combining an actin binding FH2 domain C-terminal to a Pro-rich FH1 domain, which mediates profilin binding (25).
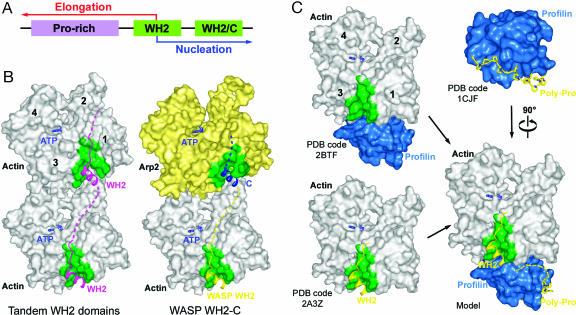
The WH2–actin structures show how WH2 could play a role in both nucleation and elongation (Fig. 4A). The WH2 domains of spire, similar to those in WASP/WAVE, are of the short kind (≈17 aa), and are connected by short linkers, which has two important implications. First, the actin-binding interface of these WH2 domains does not interfere with intersubunit contacts in F-actin and can thereby coexist with the actin filament, as illustrated by a superimposition of a WH2–actin structure onto two consecutive subunits of Holmes' filament model (32) (Fig. 4B). Second, consecutive WH2 domains must bind actin subunits along the same filament strand because the size of the linkers cannot support connections between actin subunits on opposite filaments strands. The actin subunits brought together by this type of interaction can form the nucleus for a new filament, as observed in spire (26).
Fig. 4.
Structural basis for the role of WH2 in filament nucleation and elongation. (A) In most proteins WH2 occurs in the form of tandem repeats C-terminal to Pro-rich sequences that support the binding of profilin–actin. The structures of WH2–actin suggest that this basic arrangement may allow WH2 to function in filament nucleation and elongation, two processes that are tightly connected but that can be conceptually separated. (B) Tandem repeats of short WH2 domains connected by short linkers can function to line up actin monomers along a filament strand, thereby playing a role in filament nucleation. Given the existing similarity between the C region of WASP/WAVE and WH2 (see Fig. 1B), the WH2 C region (C) may represent a specialized form of tandem repeat whose role is to add an actin subunit at the barbed end of Arp2 during Arp2/3 nucleation. (C) A superimposition of the structures of profilin–actin (19) and profilin–polyPro (36) with the structures of WH2–actin illustrates how profilin bound to the last consensus profilin-binding site (27) could deliver its actin directly to WH2, playing a role in filament elongation. A partial overlap between the actin-binding sites of profilin and WH2 may then help release profilin from the barbed end of the growing filament. Such a mechanism may constitute the basis for the so-called actoclampin model of actin polymerization (35).
As noticed before (9, 10), the C region of WASP/WAVE proteins bears similarity with WH2 (Fig. 1B). Therefore, WH2 C region could represent a specialized form of tandem repeat involved in Arp2/3 nucleation. Like WH2, the N-terminal portion of the C region forms an amphiphilic α-helix (33). Mutation of Leu-471 on the hydrophobic side of this α-helix impairs Arp2/3-mediated nucleation. As demonstrated here, the α-helix of WH2 binds in the cleft between actin subdomains 1 and 3, accounting for most of the actin-binding affinity and nucleotide exchange inhibition of WH2. This cleft in actin constitutes a “hot spot” for numerous ABPs (14). Although Arp2 is partially disordered in the structure of Arp2/3 complex (34), a model based on the structure of actin suggests that this cleft is conserved in Arp2, whereas a C-terminal extension of the polypeptide chain partially occupies the cleft in Arp3. Therefore, by analogy with tandem WH2 domains, WH2 C region is proposed to line up an actin subunit and Arp2 along a strand of the daughter filament branch during Arp2/3 nucleation (Fig. 4B).
The Pro-rich regions preceding WH2 domains may facilitate filament elongation in two possible ways: by increasing the local concentration of profilin–actin, consistent with the stimulation of assembly by profilin-WASP/WAVE (29–31) and/or by mediating actin monomer transfer directly to WH2 via profilin. The latter corresponds to the “loading” step in the so-called actoclampin motor model (35), in which an actin-binding domain (or clamp) translocates processively at the growing filament end in response to ATP hydrolysis by actin. We propose that in proteins such as VASP (Fig. 1B) WH2 forms such a clamp. Because WH2 has higher affinity for ATP–actin than ADP–actin (Table 3), ATP hydrolysis by actin could provide the energy necessary for the processive stepping of WH2-based motors. The structures of WH2–actin, together with those of profilin–actin (19) and profilin-polyPro (36), suggest that profilin bound to the consensus Pro-rich sequence immediately N-terminal to WH2 could deliver its actin directly to WH2, contributing to barbed-end filament elongation (Figs. 1B and 4C). A slight overlap between the actin-binding sites of profilin and WH2 (Fig. 2H) and/or allosteric effects may then release profilin from the growing filament.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grant GM073791. Use of the Advanced Photon Source was supported by the U.S. Department of Energy, Basic Energy Sciences, Office of Science, under Contract W-31-109-Eng-38. Use of Industrial Macromolecular Crystallography Association Collaborative Access Team Beamline 17-ID was supported through a contract with the University of Chicago.
Author contributions: D.C. and R.D. designed research; D.C., F.K., and R.D. performed research; P.G. contributed new reagents/analytic tools; D.C., F.K., Z.G., K.L., and R.D. analyzed data; and R.D. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: WASP, Wiskott–Aldrich syndrome protein; WH2, WASP homology domain 2; WAVE, WASP-family verprolin homologous protein; WIP, WASP-interacting protein; ABP, actin-binding protein; Tβ, thymosin β domain; ITC, isothermal titration calorimetry; VASP, vasodilator-stimulated phosphoprotein; C region, central region; CA region, central-acidic region.
Data deposition: The atomic coordinates reported in this paper were deposited in the Protein Data Bank, www.pdb.org (PDB ID codes 2A40, 2A41, 2A42, and 2A3Z).
References
- 1.Pollard, T. D. & Borisy, G. G. (2003) Cell 112, 453–465. [DOI] [PubMed] [Google Scholar]
- 2.Symons, M., Derry, J. M., Karlak, B., Jiang, S., Lemahieu, V., McCormick, F., Francke, U. & Abo, A. (1996) Cell 84, 723–734. [DOI] [PubMed] [Google Scholar]
- 3.Bompard, G. & Caron, E. (2004) J. Cell Biol. 166, 957–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Machesky, L. M. & Insall, R. H. (1998) Curr. Biol. 8, 1347–1356. [DOI] [PubMed] [Google Scholar]
- 5.Moreau, V., Frischknecht, F., Reckmann, I., Vincentelli, R., Rabut, G., Stewart, D. & Way, M. (2000) Nat. Cell. Biol. 2, 441–448. [DOI] [PubMed] [Google Scholar]
- 6.Martinez-Quiles, N., Rohatgi, R., Anton, I. M., Medina, M., Saville, S. P., Miki, H., Yamaguchi, H., Takenawa, T., Hartwig, J. H., Geha, R. S. & Ramesh, N. (2001) Nat. Cell. Biol. 3, 484–491. [DOI] [PubMed] [Google Scholar]
- 7.Paunola, E., Mattila, P. K. & Lappalainen, P. (2002) FEBS Lett. 513, 92–97. [DOI] [PubMed] [Google Scholar]
- 8.Edwards, J. (2004) FEBS Lett. 573, 231–233. [DOI] [PubMed] [Google Scholar]
- 9.Hertzog, M., van Heijenoort, C., Didry, D., Gaudier, M., Coutant, J., Gigant, B., Didelot, G., Preat, T., Knossow, M., Guittet, E. & Carlier, M. F. (2004) Cell 117, 611–623. [DOI] [PubMed] [Google Scholar]
- 10.Irobi, E., Aguda, A. H., Larsson, M., Guerin, C., Yin, H. L., Burtnick, L. D., Blanchoin, L. & Robinson, R. C. (2004) EMBO J. 23, 3599–3608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Marriott, G., Zechel, K. & Jovin, T. M. (1988) Biochemistry 27, 6214–6220. [DOI] [PubMed] [Google Scholar]
- 12.Kabsch, W., Mannherz, H. G., Suck, D., Pai, E. F. & Holmes, K. C. (1990) Nature 347, 37–44. [DOI] [PubMed] [Google Scholar]
- 13.Emsley, P. & Cowtan, K. (2004) Acta Crystallogr. D 60, 2126–2132. [DOI] [PubMed] [Google Scholar]
- 14.Dominguez, R. (2004) Trends Biochem. Sci. 29, 572–578. [DOI] [PubMed] [Google Scholar]
- 15.Marchand, J. B., Kaiser, D. A., Pollard, T. D. & Higgs, H. N. (2001) Nat. Cell. Biol. 3, 76–82. [DOI] [PubMed] [Google Scholar]
- 16.Mattila, P. K., Salminen, M., Yamashiro, T. & Lappalainen, P. (2003) J. Biol. Chem. 278, 8452–8459. [DOI] [PubMed] [Google Scholar]
- 17.Domanski, M., Hertzog, M., Coutant, J., Gutsche-Perelroizen, I., Bontems, F., Carlier, M. F., Guittet, E. & van Heijenoort, C. (2004) J. Biol. Chem. 279, 23637–23645. [DOI] [PubMed] [Google Scholar]
- 18.Yarmola, E. G., Parikh, S. & Bubb, M. R. (2001) J. Biol. Chem. 276, 45555–45563. [DOI] [PubMed] [Google Scholar]
- 19.Schutt, C. E., Myslik, J. C., Rozycki, M. D., Goonesekere, N. C. & Lindberg, U. (1993) Nature 365, 810–816. [DOI] [PubMed] [Google Scholar]
- 20.Hertzog, M., Yarmola, E. G., Didry, D., Bubb, M. R. & Carlier, M. F. (2002) J. Biol. Chem. 277, 14786–14792. [DOI] [PubMed] [Google Scholar]
- 21.De La Cruz, E. M., Ostap, E. M., Brundage, R. A., Reddy, K. S., Sweeney, H. L. & Safer, D. (2000) Biophys. J. 78, 2516–2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van Troys, M., Ono, K., Dewitte, D., Jonckheere, V., De Ruyck, N., Vandekerckhove, J., Ono, S. & Ampe, C. (2004) Mol. Biol. Cell 15, 4735–4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Graceffa, P. & Dominguez, R. (2003) J. Biol. Chem. 278, 34172–34180. [DOI] [PubMed] [Google Scholar]
- 24.Manuel, M., Kruse, M., Muller, W. E. & Le Parco, Y. (2000) J. Mol. Evol. 51, 378–381. [DOI] [PubMed] [Google Scholar]
- 25.Higgs, H. N. (2005) Trends Biochem. Sci. 30, 342–353. [DOI] [PubMed] [Google Scholar]
- 26.Quinlan, M. E., Heuser, J. E., Kerkhoff, E. & Mullins, R. D. (2005) Nature 433, 382–388. [DOI] [PubMed] [Google Scholar]
- 27.Purich, D. L. & Southwick, F. S. (1997) Biochem. Biophys. Res. Commun. 231, 686–691. [DOI] [PubMed] [Google Scholar]
- 28.Kang, F., Laine, R. O., Bubb, M. R., Southwick, F. S. & Purich, D. L. (1997) Biochemistry 36, 8384–8392. [DOI] [PubMed] [Google Scholar]
- 29.Suetsugu, S., Miki, H. & Takenawa, T. (1998) EMBO J. 17, 6516–6526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yang, C., Huang, M., DeBiasio, J., Pring, M., Joyce, M., Miki, H., Takenawa, T. & Zigmond, S. H. (2000) J. Cell Biol. 150, 1001–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yarar, D., D'Alessio, J. A., Jeng, R. L. & Welch, M. D. (2002) Mol. Biol. Cell 13, 4045–4059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holmes, K. C., Popp, D., Gebhard, W. & Kabsch, W. (1990) Nature 347, 44–49. [DOI] [PubMed] [Google Scholar]
- 33.Panchal, S. C., Kaiser, D. A., Torres, E., Pollard, T. D. & Rosen, M. K. (2003) Nat. Struct. Biol. 10, 591–598. [DOI] [PubMed] [Google Scholar]
- 34.Robinson, R. C., Turbedsky, K., Kaiser, D. A., Marchand, J. B., Higgs, H. N., Choe, S. & Pollard, T. D. (2001) Science 294, 1679–1684. [DOI] [PubMed] [Google Scholar]
- 35.Dickinson, R. B., Southwick, F. S. & Purich, D. L. (2002) Arch. Biochem. Biophys. 406, 296–301. [DOI] [PubMed] [Google Scholar]
- 36.Mahoney, N. M., Rozwarski, D. A., Fedorov, E., Fedorov, A. A. & Almo, S. C. (1999) Nat. Struct. Biol. 6, 666–671. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.