Abstract
Whereas linear electron flow (LEF) in photosynthesis produces both ATP and NADPH, the cyclic electron flow (CEF) around photosystem I has been shown to produce only ATP. Two alternative routes have been shown for CEF; NAD(P)H dehydrogenase (NDH)- and ferredoxin:plastoquinone oxidoreductase (FQR)-dependent flows, but their physiological relevance has not been elucidated in detail. Meanwhile, because C4 photosynthesis requires more ATP than does C3 photosynthesis to concentrate CO2, it has not been clear how the extra ATP is produced. In this study, to elucidate whether CEF contributes to the additional ATP needed in C4 photosynthesis, we estimated the amounts of PGR5, which participates in FQR-dependent flow, and NDH-H, a subunit of NDH, in four C4 species. Although the expression profiles of PGR5 did not correlate well with the additional ATP requirement, NDH was greatly expressed in mesophyll cells in the NAD-malic enzyme (ME) species, and in bundle-sheath cells in NADP-ME species, where there is a strong need for ATP in the respective cells. Our results indicate that CEF via NDH plays a central role in driving the CO2-concentrating mechanism in C4 photosynthesis.
Keywords: ATP synthesis, ferredoxin:plastoquinone oxidoreductase, NAD(P)H dehydrogenase, PGR5, plant
During plant photosynthesis, both ATP and NADPH are produced by the linear electron flow (LEF) using both photosystem (PS)I and PSII. Alternatively, ATP can also be produced by cyclic electron flow (CEF) without production of NADPH. Because the ratio of ATP/NADPH required for CO2 fixation changes, depending on activities of photorespiration or nitrate assimilation to glutamate, balancing between CEF and LEF is important to perform C3 photosynthesis effectively (1-4).
There are two known pathways of CEF around PSI; chloroplastic NAD(P)H dehydrogenase (NDH)-dependent flow and ferredoxin:plastoquinone oxidoreductase (FQR)-dependent flow. NDH complex was identified on the basis of high homology with respiratory complex I in bacteria and mitochondria, when plastid genomes of tobacco (5) and liverwort (6) were completely sequenced. To elucidate the function of NDH complex, several laboratories inactivated the tobacco ndh genes by the plastid transformation technique. Although NDH-inactivated tobacco plants showed normal growth under standard conditions (7-10), environmental stress, such as heat, high light, or decreased air humidity, causes retarded growth, suggesting that the absence of the ATP produced by NDH-dependent CEF makes it more difficult to adapt to changing environmental conditions (9, 11, 12).
On the contrary, the molecular identity of FQR had not been revealed. However, a mutant analysis of Arabidopsis identified the protein PGR5, which is essential for FQR-mediated CEF, although PGR5 itself has no known redox motif (13).
Involvement of PGR5 in FQR-mediated CEF is supported by the report that ssr2016 in Synechocystis sp. PCC 6803, homologue of Arabidopsis pgr5, is also required for FQR activity (14). Double mutants defective in both NDH- and FQR-dependent flows showed very retarded growth, suggesting that CEF is always essential for efficient C3 photosynthesis (15). However, molecular function of PGR5 and the regulation of both flows remain uncertain, and most studies on NDH complex and PGR5 were performed by using C3 plants, thus there is little knowledge about molecular function of NDH and PGR5 in C4 plants.
Although C4 plants have evolved the CO2-concentrating mechanism, which can achieve a high CO2 concentration that minimizes the energy loss resulting from the oxygenase reaction of ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco), it costs two additional ATPs to fix one CO2 compared with C3 photosynthesis. Although the extra ATP cannot be provided by LEF because it generates both ATP and NADPH at a fixed ratio, the enhancement of CEF is a possible solution for supplying the extra ATP. Interestingly, two subtypes of C4 photosynthesis, NAD-ME and NADP-ME, have been shown to have different cell-specific ATP requirements. The ATP/NADPH ratio required in NAD-ME species is higher in mesophyll cells than in bundle-sheath cells, but the opposite is true for NADP-ME species (16, 17). This cell-type-specific ATP requirement suggests that the activity of CEF should be higher in mesophyll cells of NAD-ME species and in bundle-sheath cells of NADP-ME species, if CEF plays a critical role in supplying the additional ATP needed in C4 photosynthesis.
Indirect evidence linking CEF to the extra ATP requirement in C4 photosynthesis is higher PSI/PSII ratios in the mesophyll cells of NAD-ME species and in the bundle-sheath cells of NADP-ME species (17). Because CEF uses only PSI, whereas LEF uses both PSI and PSII, a higher PSI/PSII ratio indicates higher CEF activity compared with LEF activity. Therefore, higher PSI/PSII ratios in the mesophyll cells of NAD-ME species and in the bundle-sheath cells of NADP-ME species are considered reasonable, in view of higher relative requirements for ATP (17).
However, little evidence is available on what CEF participates in mesophyll and bundle-sheath cells, especially in NAD-ME-type species. In this study, we determined the amounts of NDH and PGR5 in isolated mesophyll and bundle-sheath cells from NAD-ME and NADP-ME C4 plants to evaluate the correlation between the ATP requirement and the CEF activity in the respective cells.
Materials and Methods
Plants. Seeds of Amaranthus hybridus, Portulaca oleracea, Zea mays, and Portulaca grandiflora were purchased from a local nursery. NDH-B-disrupted tobacco has been reported in ref. 8. All of the plants were grown in fertilized soil in a growth chamber at 28°C, with a 14-h photoperiod at a photon flux density of 100 μmol quanta m-2·s-1 up to 3 weeks.
Preparations of Mesophyll Cells. Leaf slices (1 g fresh weight) were incubated in a digestion medium containing 0.5 M sorbitol, 2% (wt/vol) cellulase (Onozuka R-10, Yakult Pharmaceutical, Tokyo), 0.5% (wt/vol) macerozyme (Onozuka R-10, Yakult Pharmaceutical), 5 mM 2-(N-morpholino) ethanesulfonic acid (Mes)-KOH (pH 5.5), and 1 mM CaCl2. After 4 h of digestion at 25°C under dim light, the medium was discarded and the tissue was washed with gentle shaking in sorbitol medium containing 0.5 M sorbitol, 5 mM Mes-KOH (pH 6.0), and 1 mM CaCl2. The suspension was passed through an 80-μm nylon mesh, and the filtrate was centrifuged at 300 × g for 5 min. The pellet was suspended in 5 ml of a sucrose medium containing 0.5 M sucrose, 5 mM Mes-KOH (pH 6.0), and 1 mM CaCl2, and the sorbitol medium was slowly added to the suspension. After centrifugation at 300 × g for 5 min, the mesophyll protoplasts were collected from the interface between the sorbitol and sucrose layers.
Preparations of Bundle-Sheath Strands. Bundle-sheath strands were prepared according to the protocol of Höfer et al. (18) with some modifications. The leaf tissues (10 g) were cut into small pieces (2-3 mm) with a sharp razor blade, and homogenized in a Waring blender (Nissei AM-8, Nihonseiki Kaisha, Tokyo) at high speed for 20 s in 100 ml of homogenizing medium (0.3 M sorbitol/50 mM Mes-KOH, pH 6.1/1 mM MgCl2/1 mM MnCl2/2 mM EDTA/30 mM KCl/0.25 mM KH2PO4). The suspension was filtered through an 80-μm nylon mesh, and the retained tissues were suspended in 100 ml of homogenizing medium. This procedure was repeated until most of the adhering mesophyll cells were released from the bundle-sheath strands, as assessed by light microscopic observation. These bundle-sheath strands were used to prepare bundle-sheath proteins and for the assay of total RNA and chlorophyll.
Chlorophyll Measurement. The chlorophyll a/b ratio was determined by using the method of Arnon (19).
Preparation of Anti-NDH-H Antiserum. The coding regions of tobacco ndhH were cloned into a pET21-d vector (Novagen). Recombinant protein containing a His tag was expressed in Escherichia coli strain Rosetta-gami B (DE3) (Novagen) as inclusion bodies, purified on a nickel-column, and used for the immunization of rabbits. The resulting antiserum recognized NDH-H in the thylakoid fraction from wild-type tobacco, but no corresponding band was detected in that from the NDH-B-disrupted tobacco (see Fig. 5).
Fig. 5.
Estimation of NDH and PGR5 in C3 and C4 plants. Ten micrograms of total cell (TC) or thylakoid membrane (TM) fraction proteins were subjected to SDS/PAGE and immunodetected by antibodies against NDH-H or PGR5. Proteins were extracted from wild-type tobacco (Nt), ndhB-disrupted tobacco (ΔB), Amaranthus hybridus (Ah), P. oleracea (Po), Z. mays (Zm) and P. grandiflora (Pg).
Protein Electrophoresis and Immunoblotting. Total cell proteins were extracted in SDS sample buffer (62.5 mM Tris·HCl, pH 6.8/2.5% SDS/10% (wt/vol) glycerol/2.5% 2-mercaptoethanol) from whole leaves of 3-week-old plants. Thylakoid membrane fraction of tobacco was prepared from osmotically ruptured chloroplasts (8) by centrifugation and also lysed in SDS sample buffer. SDS/PAGE was carried out according to the method of Laemmli (20). For the immunoblot analysis, 10 μg of proteins were subjected to SDS/PAGE, blotted onto PROTORAN nitrocellulose membrane (Schleicher and Schüll, Dassel, Germany) and detected by using Western Lightning Chemiluminescence Reagent Plus (PerkinElmer Life Sciences). For quantitative analyses, exposed films were scanned and analyzed with the public-domain program nih imagej (available at http://rsb.info.nih.gov/ij). These experiments were repeated at least three times.
PCR Cloning of pgr5s. For the RT-PCR experiment, total RNA was isolated from young leaves of four C4 species by using an RNeasy Plant mini kit (Qiagen, Valencia, CA), and first-strand cDNAs were synthesized by using SuperScript III reverse transcriptase (Invitrogen) according to the manufacturer's instructions. For cloning of the pgr5 fragments, degenerate PCR primers (see Table 1, which is published as supporting information on the PNAS web site) were designed from conserved amino acid residues (or sequences) identified in an alignment of three higher-plant PGR5 sequences (accession nos: AAD24646, BAD10341, and AW133320). The first PCR was performed with the forward primer and reverse primer. For nested PCR, the first PCR products were used as templates with the forward primer and reverse-nested primer. The PCR conditions were as follows: 94°C for 4 min and then 35 cycles of 94°C for 15 s, 37°C for 1 min, 72°C for 30 s, and, finally, 72°C for 5 min. RACE (5′- and 3′-) were performed with a GeneRacer kit (Invitrogen) according to the manufacturer's instructions. The gene-specific primers used for 5′- and 3′-RACE are shown in Table 2, which is published as supporting information on the PNAS web site. The pgr5 homologues (Amaranthus hybridus, P. oleracea, Z. mays, and P. grandiflora) were registered in the nucleotide sequence databases (accession nos. AB218879-AB218882).
Quantitative Real-Time RT-PCR (qPCR). By using gene-specific primers (see Table 3, which is published as supporting information on the PNAS web site) and first-strand cDNAs from mesophyll and bundle-sheath cells, qPCR was performed by using a DNA Engine Opticon Continuous Fluorescence Detection system (MJ Research, Cambridge, MA) and the DyNAmo SYBR green qPCR kit (Finnzymes, Helsinki) with the following cycling parameters: 95°C for 10 s, 55°C for 20 s, and 72°C for 20 s. The amplification of actin cDNA as the internal control was performed with a primer pair based on conserved sequences among various higher plants (Table 3). To ensure the absence of nonspecific PCR products, a melting-curve analysis and agarosegel electrophoresis were performed after each run. The data obtained were analyzed by using the program opticon (MJ Research).
Measurement of in Vivo NDH Activity. Chlorophyll fluorescence was measured with a PAM-2000 portable fluorometer (Walz Effeltrich, Germany). A transient increase in fluorescence after actinic illumination was measured as NDH-dependent CEF activity, as reported in ref. 8.
Results and Discussion
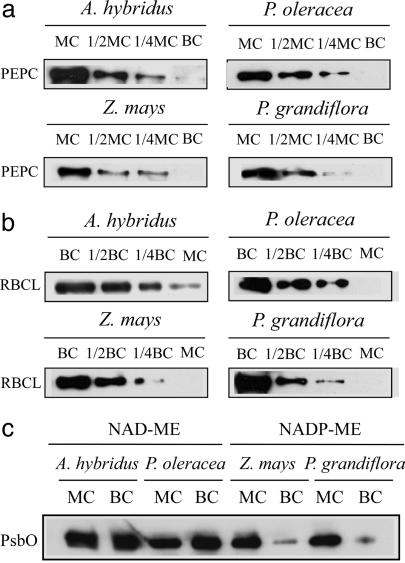
Mesophyll and bundle-sheath preparations were obtained from the NAD-ME species Amaranthus hybridus (21) and P. oleracea (22) and from the NADP-ME species Z. mays (22) and P. grandiflora (22). Their purity was estimated by immunoblot analysis of the mesophyll-specific phosphoenolpyruvate carboxylase and the bundle-sheath-specific Rubisco (Fig. 1 a and b), and we confirmed that cross-contamination was <20% in all preparations. This result was supported by a marked decrease in the extrinsic 33-kDa proteins in PSII in the bundle-sheath preparations of NADP-ME species (Fig. 1c) and by a chlorophyll a/b ratio that represented the PSI/PSII ratio (see Table 4, which is published as supporting information on the PNAS web site), as shown in previous reports (16, 17, 22).
Fig. 1.
Estimation of cross-contamination. (a) Immunoblot analysis of the mesophyll-specific phosphoenolpyruvate carboxylase (PEPC) and (b) bundle-sheath-specific Rubisco (RBCL). Soluble proteins were extracted from mesophyll preparations (MC) and bundle-sheath preparations (BC). For the 1/1 samples, 4 μg of proteins were loaded per lane. As judged from a dilution series of immunoblot analysis (1/1, 1/2, and 1/4), the amounts of PEPC in all MC were ≈4-fold more than those in the corresponding BC, suggesting that all MC contained <20% contamination. Likewise, all BC contained <20% contamination based on the results of immunoblot analysis of Rubisco. (c) Immunoblot analysis of extrinsic 33-kDa proteins (PsbO) in the PSII. Equal amounts of proteins (10 μg) extracted from MC and BC were loaded per lane. The PsbO proteins in the PS II were greatly decreased in the BC of NADP-ME species, as in previous reports (16, 17, 22).
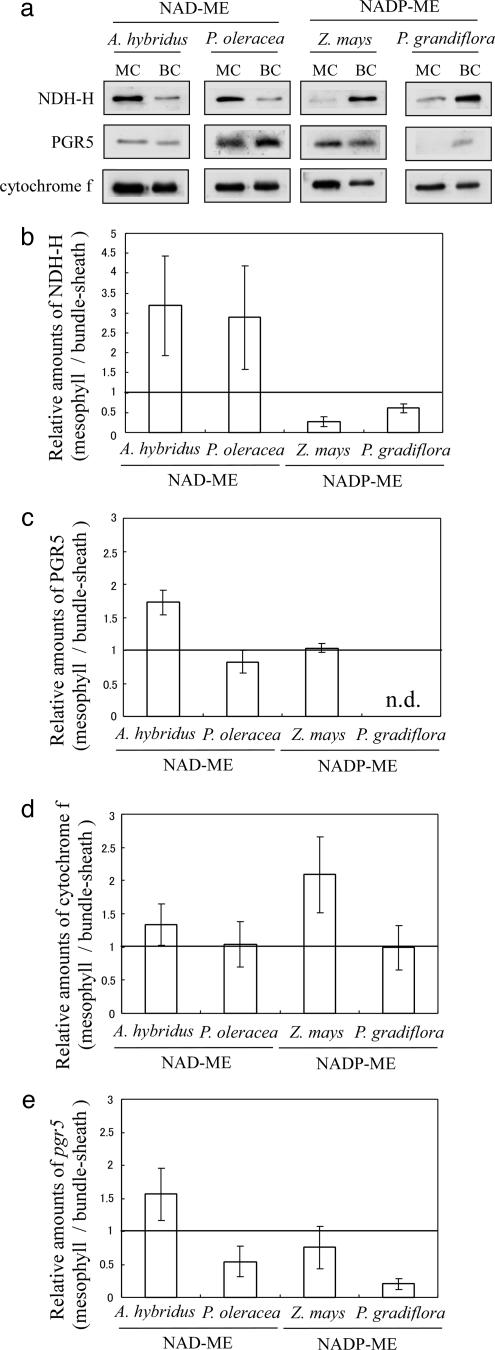
No significant difference was found in the contents of cytochrome b6/f between mesophyll and bundle-sheath preparations (Fig. 2 a and d), consistent with previous results (23). Because the intersystem electron carrier cytochrome b6/f complex participates in both LEF and CEF, its contents should not be affected by PSI/PSII and show good correlation with PSI (24).
Fig. 2.
Relative amounts of CEF-related enzymes and transcripts of mesophyll cells to bundle-sheath cells. (a) Immunoblot analysis of NDH-H, PGR5, and cytochrome f. (b-d) Densitometric analysis of immunoblots using polyclonal antibodies against NDH-H (b), PGR5 (c), and cytochrome f (d) proteins. Each value represents the relative intensity of mesophyll preparations to bundle-sheath preparations. Data are means ± SD (n = 3). (e) Relative pgr5 gene transcripts of mesophyll preparations to bundle-sheath preparations. The amounts of pgr5 gene transcripts were estimated by qPCR as described in Materials and Methods, normalized to the corresponding actin mRNA levels. Data are the means ± SD (n = 5).
Immunoblot analysis of NDH showed that the content of NDH-H in mesophyll preparations of NAD-ME species was ≈3-fold higher than that in bundle-sheath preparations (Fig. 2 a and b). In contrast, in NADP-ME species, the ratio of NDH-H contents in mesophyll to bundle-sheath preparations was only 0.27 (Z. mays) and 0.61 (P. grandiflora), which is consistent with previous reports that NDH complex is greatly accumulated in bundle-sheath cells of the NADP-ME-type Sorgum bicolor and Z. mays (25, 26). These results indicate that NDH-H is highly accumulated in cells that require a high level of ATP synthesis; i.e., mesophyll-sheath cells of NAD-ME species and bundle-sheath cells of NADP-ME species. This result suggests that the CEF via NDH plays an essential role in supplying additional ATP in C4 photosynthesis.
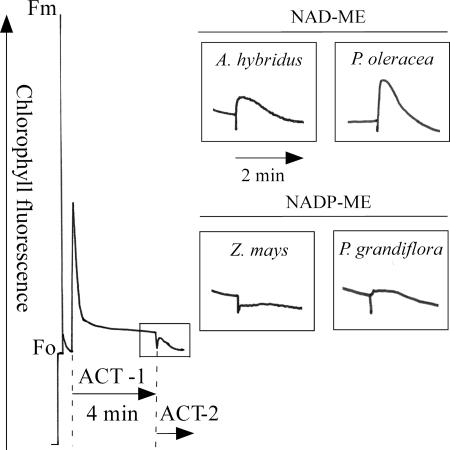
To estimate the active participation of NDH, we measured the transient increase in chlorophyll fluorescence of PSII after turning off actinic illumination, which represents electron transport from stromal electron donors to plastoquinone via NDH (7-9, 27, 28). The NDH-dependent increase in chlorophyll fluorescence was much greater in NAD-ME-type plants than in NADP-ME-type plants (Fig. 3), even though NADP-ME-type plants contain a large amount of NDH-H in bundle-sheath cells. This is because a low content of PSII in bundle-sheath cells of NADP-ME species (16, 17, 22, 29) results in a low yield of chlorophyll fluorescence of PSII. In fact, when we observed chlorophyll fluorescence in cross sections of P. grandiflora leaves by using a MicroFluorCam (Photon Systems Instruments, Bruno, Czech Republic), the level of fluorescence emitted from the mesophyll cells was much higher than that from the bundle-sheath cells (data not shown). This observation has also been reported in S. bicolor in confocal microscopy (30).
Fig. 3.
In vivo measurement of NDH activity. After the actinic light illumination (ACT1; 85 μmol photons m-2·s-1), the transient increases in chlorophyll fluorescence were measured under dim light (Act2; 3 μmol m-2·s-1), recorded, and are shown in the enlarged boxes. This transient increase represents the reduction of the plastoquinone pool by the reducing equivalents accumulated during actinic illumination, confirmed by the experimental result that the transient increase disappeared when illuminated by far-red light after actinic illumination.
Whereas the amount of NDH depended on the cell type, immunoblot analysis showed that PGR5 was accumulated rather uniformly (Fig. 2 a and c). To confirm this observation at the transcription level, we cloned full-length pgr5 genes from the four C4 species by PCR and compared their deduced amino acid sequences by using the clustalw algorithm (31) (Fig. 4). Because their C-terminal of 64 deduced amino acid sequences showed high homology (ranging from 84% to 93% identity) with Arabidopsis PGR5 protein, we regarded those as pgr5 homologues. In addition, the N-terminal sequences varied considerably among plant species, and the corresponding sequence was not found in prokaryotic Synechocystis, suggesting that those sequences are the putative chloroplast transit peptides. Furthermore, they were predicted to be localized in chloroplast by the program targetp (32) (www.cbs.dtu.dk/services/TargetP), the same as Arabidopsis PGR5. These results suggested that PGR5 homologues were correctly isolated from the four C4 species.
Fig. 4.
Sequence comparison of higher plant pgr5 genes. The deduced amino acid sequences from Arabidopsis thaliana (AAK53038), Amaranthus hybridus (AB218879), P. oleracea (AB218880), Z. mays (AB218881), P. grandiflora (AB218882), and Synechocystis sp. PCC6803 (S77542) were compared by using the CLUSTALW analysis algorithm and visually refined by using the BioEdit sequence-alignment editor (34). Identical residues are shaded in black, and similar residues are in gray. Dashes are gaps introduced to maximize the alignment.
Analysis of pgr5 transcripts by qPCR showed similar expression profiles (Fig. 2e) with corresponding protein expression (Fig. 2 a and c). The amounts of PGR5 in mesophyll preparations of P. grandiflora were below the detection limit of immunoblot analysis (Fig. 2 a and c), as supported by a low ratio of mesophyll/bundle-sheath preparations in qPCR analysis (Fig. 2e) and weak immunosignal in bundle-sheath preparations of P. grandiflora (Fig. 2a). Because neither the protein nor mRNA levels of PGR5 showed a clear correlation with the ratio of ATP/NADPH required for CO2 fixation in the respective cells, we concluded that the FQR-dependent CEF may contribute slightly to extra ATP production for C4 photosynthesis.
On the other hand, immunoblot analysis showed that NDH in the whole leaves of the four C4 species was accumulated more than that in the C3 plant tobacco (Fig. 5). Although the ratios of chlorophyll/protein in the total cell (TC) were not significantly changed among tobacco and C4 species, tobacco NDH was only detectable in the thylakoid membrane (TM) fraction, where the ratio of chlorophyll/protein was concentrated >10-fold compared with TC (data not shown). In addition, an anti-NDH-H antiserum was supposed to have the highest cross-reactivity with tobacco, because it was raised against tobacco NDH-H protein. Thus, the NDH in the C4 leaves should be accumulated at least 10-fold higher than that in tobacco. However, the amount of tobacco PGR5 was comparable with that in C4 species (Fig. 5). Similar results were also seen for the C3 plant Arabidopsis and the C4 plant Setaria viridis (data not shown). These experimental data also support the notion that the NDH-dependent pathway is the primary route to produce extra ATP in C4 photosynthesis.
It should be noted that the water-water cycle is also the alternative electron pathway with capability to provide the extra ATP. Overall reaction of the water-water cycle is the reduction of dioxygen to water in PSI by the electrons generated in PSII from water and can produce ATP but not NADPH (33). It has been proposed that operation of the water-water cycle is indispensable to activate the CO2-fixation cycle in C4 photosynthesis, because oxygen at low concentration (half-saturation; 0.15 kPa) is required to initiate the CO2 fixation in maize.§ However, after the leaves attain the steady state of photosynthesis, no oxygen is required to maintain the CO2 fixation. In addition, the water-water cycle can operate only in the cells where the electrons from PSII are available. Thus, the generation of ATP through the water-water cycle appears not to contribute so much to the extra ATP required for steady-state C4 photosynthesis.
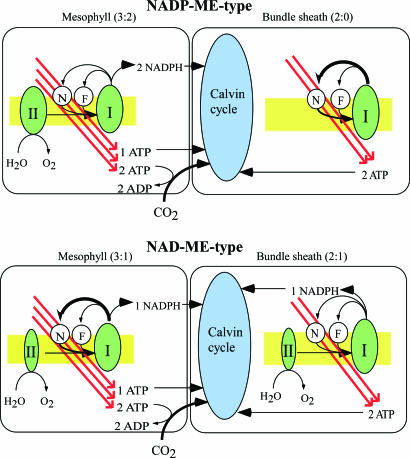
The relationship between C4-cell types and CEF is summarized in Fig. 6. The evolution of C4 photosynthesis from C3 photosynthesis in different phylogenetic plants would require the alteration of several key enzymes and regulatory factors. From an energy viewpoint, the key enzyme for evolution of C4 plants must be the activation of NDH.
Fig. 6.
Schematic model of ATP:NADPH production in the electron-transport chain and consumption in CO2 concentration and fixation. The energy requirements in each cell type are based on calculations by Edwards and Walker (16). II, I, N, and F represent PS II, PS I, NDH, and FQR, respectively. Values in parentheses represent the production ratio of ATP:NADPH in each cell type. This model implies that the NDH-dependent pathway meets the need for the extra ATP in C4 photosynthesis. In reality, CO2 is transported from mesophyll to bundle sheath in the form of C4-carbonic acids, whereas ATP and NADPH are transported in the form of triosephosphate and malate.
FQR-dependent flow is a major route of CEF around PSI in the C3 plant Arabidopsis (13, 15), also supported by the finding that NDH-dependent flow is indispensable only under photooxidative conditions in which the demand for ATP increases (9-12). In contrast, a comparison of the NDH and PGR5 in both NAD- and NADP-ME-type C4 plants clearly showed that NDH is the main contributor of the extra ATP required for C4 photosynthesis to concentrate CO2 in bundle-sheath cells. Why is NDH, rather than FQR, greatly expressed in ATP-requiring cells in C4 plants? One possible explanation is that the NDH pathway is more independent of the LEF than the FQR pathway, in which the plastoquinone reduction site of the cytochrome b6/f complex may be shared with the Q cycle, perhaps resulting in severe interference with linear flow when the rate of CEF increases. An alternative advantage of the NDH pathway is that the electron flow through NDH likely produces an extra pH gradient via the putative proton pump. In addition, the pool size of NADPH and triosephosphate, which can generate NADPH in the Calvin cycle, is larger than that of reduced ferredoxin in chloroplasts.
Supplementary Material
Acknowledgments
We thank Dr. Shikanai (Kyushu University, Fukuoka, Japan) for kindly providing the antibodies against PGR5 and cytochrome f, Dr. T. Furumoto for suggestions on the isolation of mesophyll cells, and Dr. E. Yamamoto for help with the analysis of MicroFluorCam. The antiserum against spinach PsbO was kindly provided by the late A. Watanabe (University of Tokyo). This study was supported, in part, by Grant-in-Aid for Basic Research C 136440646 (to T.E.), a Center of Excellence Scientific Research Grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan, Research for the Future Program Grant JSPS-RFTF 00L01606 (to F.S.), and a fellowship from the Japan Society for the Promotion of Science (to A.T.).
Author contributions: A.T., K.A., T.E., and F.S. designed research; A.T. and M.K. performed research; A.T. analyzed data; and A.T., K.A., T.E., and F.S. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: CEF, cyclic electron flow; FQR, ferredoxin:plastoquinone oxidoreductase; LEF, linear electron flow; NDH, NAD(P)H dehydrogenase; ME, malic enzyme; PEPC, phosphoenolpyruvate carboxylase; PS, photosystem; qPCR, quantitative real-time RT-PCR.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database (accession nos. AB218879-AB218882).
Footnotes
Ohwaki, T. & Asada, K. (2003) Plant Biol. 329 (abstr.).
References
- 1.Bukhov, N. & Carpentier, R. (2004) Photosynth. Res. 82 17-33. [DOI] [PubMed] [Google Scholar]
- 2.Cruz, J. A., Avenson, T. J., Kanazawa, A., Takizawa, K., Edwards, G. E. & Kramer D. M. (2004) J. Exp. Bot. 56 395-406. [DOI] [PubMed] [Google Scholar]
- 3.Johnson, G. N. (2005) J. Exp. Bot. 56 407-416. [DOI] [PubMed] [Google Scholar]
- 4.Kramer, D. M., Avenson, T. J. & Edwards, G. E. (2004) Trends Plant Sci. 9 349-357. [DOI] [PubMed] [Google Scholar]
- 5.Shinozaki, K., Ohme, M., Tanaka, M., Wakasugi, T., Hayashida, N., Matsubayashi, T., Zaita, N., Chunwongse, J., Obokata, J., Yamaguchi-Shinozaki, K., et al. (1986) EMBO J. 5, 2043-2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ohyama, K., Fukuzawa, H., Kohchi, T., Shirai, H., Sano, T., Sano, S., Umesono, K., Shiki, Y., Takeuchi, M., Chang, Z., et al. (1986) Nature 322 572-574. [Google Scholar]
- 7.Burrows, P. A., Sazanov, L. A., Svab, Z., Maliga, P. & Nixon, P. J. (1998) EMBO J. 17 868-876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shikanai, T., Endo, T., Hashimoto, T., Yamada, Y., Asada, K. & Yokota, A. (1998) Proc. Natl. Acad. Sci. USA 95 9705-9709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Horváth, E. M., Peter, S. O., Joët, T., Rumeau, D., Cournac, L., Horváth, G. V., Kavanagh, T. A., Schäfer, C., Peltier, G. & Medgyesy, P. (2000) Plant Physiol. 123 1337-1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takabayashi, A., Endo, T., Shikanai, T. & Sato, F. (2002) Biosci. Biotech. Biochem. 66 2107-2111. [DOI] [PubMed] [Google Scholar]
- 11.Endo, T., Shikanai, T., Takabayashi, A., Asada, K. & Sato, F. (1999) FEBS Lett. 457 5-8. [DOI] [PubMed] [Google Scholar]
- 12.Li, X.-G., Duan, W., Meng, Q.-W., Zou, Q. & Zhao, S.-J. (2004) Plant Cell Physiol. 45 103-108. [DOI] [PubMed] [Google Scholar]
- 13.Munekage, Y., Hojo, M., Meurer, J., Endo, T., Tasaka, M. & Shikanai, T. (2002) Cell 110 361-371. [DOI] [PubMed] [Google Scholar]
- 14.Yeremenko, N., Jeanjean, R., Prommeenate, P., Krasikov, V., Nixon, P. J., Vermaas, W. F., Havaux, M. & Matthijs, H. C. (2005) Plant Cell Physiol., 46 1433-1436. [DOI] [PubMed] [Google Scholar]
- 15.Munekage, Y., Hashimoto, M., Miyake, C., Tomizawa, K., Endo, T., Tasaka, M. & Shikanai, T. (2004) Nature 429 579-582. [DOI] [PubMed] [Google Scholar]
- 16.Edwards, G. E. & Walker, D. A. (1983) C3, C4: Mechanisms, and Cellular and Environmental Regulation, of Photosynthesis (Blackwell Scientific, Oxford).
- 17.Hatch, M. D. (1987) Biochim. Biophys. Acta 895 81-106. [Google Scholar]
- 18.Höfer, M. U., Santore, U. J. & Westhoff, P. (1992) Planta 186 304-312. [DOI] [PubMed] [Google Scholar]
- 19.Arnon, D. (1949) Plant Physiol. 24 1-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laemmli, U. K. (1970) Nature 227 680-685. [DOI] [PubMed] [Google Scholar]
- 21.Sage, R. F. & Monson, R. K. (1999) C4 Plant Biology (Academic, San Diego), p. 480.
- 22.Pfündel, E., Nagel, E. & Meister., A. (1996) Plant Physiol. 112 1055-1070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kubicki, A., Steinmüller, K. & Westhoff, P. (1994) Plant Mol. Biol. 25 669-679. [DOI] [PubMed] [Google Scholar]
- 24.Oswald, A., Streubel, M., Ljungberg, U., Hermans, J., Eskins, K. & Westhoff, P. (1990) Eur. J. Biochem. 190 185-194. [DOI] [PubMed] [Google Scholar]
- 25.Kubichi, A., Funk, E., Westhoff, P. & Steinmüller, K. (1996) Planta 199 276-281. [Google Scholar]
- 26.Darie, C. C., Biniossek, M. L., Winter, V., Mutschler, B. & Haehnel, W. (2005) FEBS Lett. 272 2705-2716. [DOI] [PubMed] [Google Scholar]
- 27.Kofer, W., Koop, H. U., Wanner, G. & Steinmüller, K. (1998) Mol. Gen. Genet. 258 166-173. [DOI] [PubMed] [Google Scholar]
- 28.Asada, K., Heber, U. & Schreiber, U. (1993) Plant Cell Physiol. 34 39-50. [Google Scholar]
- 29.Ivanov, B., Asada, K., Kramer, D. M. & Edwards, G. (2005) Planta 220 572-581. [DOI] [PubMed] [Google Scholar]
- 30.Edwards, G. E., Furbank, R.T., Hatch, M. D. & Osmond, C. B. (2001) Plant Physiol. 125 46-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thompson, J. D., Higgins, D. G. & Gibson, T. J. (1994) Nucleic Acids Res. 22 4673-4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emanuelsson, O., Nielsen, H., Brunak, S. & von Heijne, G. (2000) J. Mol. Biol. 300 1005-1016. [DOI] [PubMed] [Google Scholar]
- 33.Asada, K. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 601-639. [DOI] [PubMed] [Google Scholar]
- 34.Hall, T. A. (1999) Nucleic Acids Symp. Ser. 41 95-98. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.