Abstract
Phosphatidylinositol 3 (PI3)-kinase enhancer (PIKE) is a nuclear GTPase that enhances PI3-kinase activity in a GTP-dependent manner. Both PIKE-L and -A isoforms contain GTPase, pleckstrin homology (PH), ADP ribosylation factor-GTPase-activating protein, and two ankyrin repeats domains, and C-terminal ADP ribosylation factor-GTPase-activating protein activates its internal GTPase activity. However, whether PH domain modulates the intramolecular action and subsequently influences its downstream signalings remains elusive. Here we show that PH domain from PIKE-L robustly binds PI(3,4,5)P3 and exclusively resides in the nucleus. By contrast, the mutant (K679,687N), unable to bind phosphoinositol lipids, translocates to the cytoplasm. This mutation substantially compromises the stimulatory effects on PI3-kinase by PIKE-L. Surprisingly, PH domain from PIKE-A distributes in the cytoplasm. Similar mutation in PH domain of PIKE-A abolishes its binding to PI(3,4,5)P3 and significantly decreases its activation of Akt. Moreover, amplified PIKE-A from human cancers contains mutations and highly stimulates Akt kinase activity, correlating with its GTPase activity. Thus, phosphatidylinositols regulate PIKE GTPase activity, mediating its downstream PI3-kinase/Akt signaling through a feedback mechanism by binding to its PH domain.
Keywords: pleckstrin homology domain, nuclear translocation, negative feedback, GTPase activation
Phosphatidylinositol 3 (PI3)-kinase enhancer (PIKE) is a recently identified brain-specific nuclear GTPase that binds PI3-kinase and stimulates its lipid kinase activity. PIKE activates nuclear PI3-kinase in a GTP-dependent manner (1). Nerve growth factor elicits membrane-associated 4.1N nuclear translocation and interaction with PIKE, which was initially identified as a 4.1N-binding partner in a yeast two-hybrid screening. The binding of 4.1N to PIKE prevents its interactions with nuclear PI3-kinase, leading to inactivation of nuclear PI3-kinase. To date, three forms of PIKE have been characterized, PIKE-S, PIKE-L, and PIKE-A. PIKE-S is the initially identified shorter isoform (1). PIKE-L, a longer isoform of PIKE gene, differs from PIKE-S by C-terminal extension containing ADP ribosylation factor-GTPase activating protein (Arf-GAP) and two ankyrin repeats domains. In contrast to the exclusive nuclear localization of PIKE-S, PIKE-L occurs in both the nucleus and the cytoplasm (2). In addition, we show that WT merlin, binds PIKE-L and inhibits PI3-kinase activity. This suppression of PI3-kinase activity results from merlin disrupting the binding of PIKE-L to PI3-kinase (3). Recently, a third PIKE isoform, PIKE-A, was identified in human glioblastoma multiforme brain cancers. We have reported that PIKE-A is coamplified with cyclin-dependent kinase 4 in a variety of human cancers. Unlike the brain-specific PIKE-L and -S isoforms, PIKE-A distributes in various tissues. PIKE-A contains the same domains present in PIKE-L but lacks N-terminal proline-rich domain, which binds PI3-kinase and PLC-γ1. Instead, PIKE-A specifically binds to active Akt and up-regulates its activity in a GTP-dependent manner, mediating human cancer cell invasion and preventing apoptosis (4, 5).
The pleckstrin homology (PH) domain is a common piece in the structural patchwork of signaling proteins. The PH domain predominantly binds different phosphatidylinositol lipids. Accumulating evidence demonstrates that phosphoinositide lipids regulate the enzymatic activity of the binding targets through association with PH domain. For instance, phosphatidylinositols bind the PH domain of dynamin and activate its GTPase activity, which is important in dynamin function during vesicle budding (6). Phosphoinositides can convert Arf-GTP into Arf-GDP by binding to PH domain of centaurin β family of Arf-GAPs, including ASAP1 and 2 (7-9). PIKE-A contains a PH domain, which separates the N-terminal GTPase and C-terminal Arf-GAP domains. It has been shown that PIKE-A (also called GGAP2) has high GTPase activity through direct intramolecular interaction of the N-terminal GTPase domain and the C-terminal GAP domain. By contrast, in the absence of Arf-GAP domain, the N-terminal GTPase domain alone has very low activity, suggesting that Arf-GAP domain is necessary to activate it (10). However, whether PH domain in PIKE plays any role in mediating its internal GTPase activity and subsequently affects its downstream signalings including PI3-kinase/Akt remains obscure.
In the present work, we report that phosphoinositides manipulate the subcellular distribution of PIKE through its PH domain. In addition, they also affect the GTPase activity of PIKE through its C-terminal Arf-GAP domain and regulate PI3-kinase/Akt signaling cascade. Our findings indicate that PIKE mediates the stimulatory activity on its downstream effectors through a feedback loop by phosphoinositides binding to its PH domain.
Materials and Methods
Cells and Reagents. HEK293 and human glioblastoma SF188, MO67 cells were maintained in DMEM, and neuroblastoma NGP-127 and bone sarcoma CRL-2098 cells were cultured in RPMI medium 1640, supplemented with 10% FBS, 2 mg/ml glutamine, and 100 units of penicillin-streptomycin at 37°C with 5% CO2 atmosphere in a humidified incubator. Mouse monoclonal anti-hemagglutinin (anti-HA), anti-Myc, and anti-GST-horseradish peroxidase Abs were from Sigma. Mouse monoclonal anti-Ser-473, anti-Akt, and anti-phospho-glycogen synthase kinase (GSK)3α/β (Ser-21/9) Abs were from Cell Signaling Technology (Beverly, MA). Rabbit polyclonal anti-p85 and -p110 Abs were from Santa Cruz Biotechnology. Anti-PIKE Ab was raised against GST-PIKE-L (amino acids 1,095-1,186) protein. Protein A/G-conjugated agarose beads were from Calbiochem. Glutathione Sepharose 4B was supplied by Amersham Pharmacia. GST-GSK3 fusion protein and Crosstide were from Cell Signaling Technology. Chemicals not listed above were from Sigma. Phosphoinositide-4-phosphate beads were from Echelon (Salt Lake City).
Assay of GTPase Activity. In vitro GTPase assays were performed as described in ref. 11. In brief, purified GST-GTPase of PIKE (0.5 μg of each) were washed thoroughly with loading buffer (20 mM Tris·HCl, pH 8/2 mM EDTA/10 mM DTT) and were incubated with [α-32P]GTP (0.1 μM) for 30 min at 30°C in 50 μl of the loading buffer. The resin was rinsed twice with ice-cold loading buffer followed by resuspension in the reaction buffer (20 mM Tris·HCl, pH 8/10 mM MgCl2/10 mM DTT) with or without the C-terminal GST-GAP domain (1 μg each) of PIKE-L. The GTP hydrolysis was conducted at room temperature. Samples were taken at the indicated times and immediately solubilized in the elution buffer (0.2% SDS/5 mM EDTA/5mMGTP/5 mM GDP) by heating at 65°C for 2 min. The eluted GTP and GDP were separated by TLC on polyethyleneimine-cellulose plates as described in ref. 12.
In Vitro Phosphoinositol Lipids-Binding Assay. A variety of agarose-conjugated phosphoinositol lipids (25 μl) were incubated with 3 μg of purified recombinant GST-PH protein in 500 μl of binding buffer (10 mM Hepes/150 mM NaCl/0.25% Triton X-100) at 4°C for 2.5 h. The beads were washed three times with the binding buffer, and the associated proteins were subjected to SDS/PAGE and analyzed by immunoblotting assay with anti-GST-horseradish peroxidase Ab. For lipids competition assay, gradually increased water-soluble phosphatidylinositol-3,4,5,-trisphosphate [PI(3,4,5)P3] or phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] was used under the same binding condition.
GTP-Loading Assays. The assays were performed essentially as described in ref. 13 with minor modification. HEK293 cells were transfected with GFP-PIKE constructs and metabolically labeled in phosphate-free DMEM with 0.5 mCi/ml (1 Ci = 37 GBq) [32P]H3PO4 for 3-4 h at 37°C. Cells were lysed in 0.5 ml of lysis buffer (20 mM Tris·HCl, pH7.4/150 mM NaCl/1 mM MgCl2/1% Triton X-100). Extracts were rocked at 4°C for 30 min and centrifuged for 5 min at 12,000 × g. Supernatant fluid (0.5 ml) was added to 20 μl of protein A/G Sepharose, rocked with 5 μg of anti-GFP Ab for 30 min at 4°C, and then washed three times with lysis buffer and once with PBS. Succeeding steps were the same as described.
Results
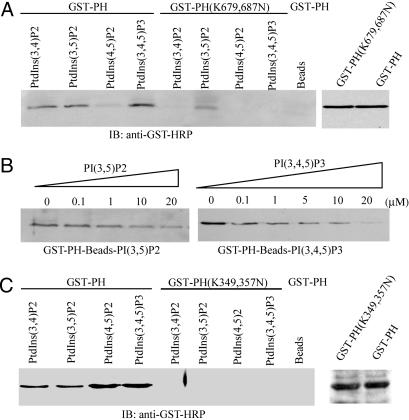
PH Domain of PIKE Possesses Distinct Phosphoinositol Lipids-Binding Activities. PH domain in different proteins displays distinct phosphoinositol lipids-binding specificities (14). In vitro-binding assay shows that PIKE PH domain robustly binds to agarose beads-conjugated PI(3,4,5)P3, followed by PI(3,5)P2 and PI(3,4)P2. By contrast, it does not bind PI(4,5)P2 at all. Mutation of two Lys-679 and -687 residues, which are well conserved among the PH domains from different proteins (9), into asparagines in PH domain compromises its binding activity to most of phosphoinositides, although it still weakly associates with PI(3,5)P2, demonstrating that PIKE PH domain specifically binds 3′-phosphate-phosphoinositol lipids (Fig. 1A). Competition assay reveals that the gradually increased concentrations of free PI(3,4,5)P3 abolish beads-bound GST-PH recombinant protein, underscoring that the interaction is specific (Fig. 1B Right). However, the same amount of free PI(3,5)P2 is unable to completely disrupt PI(3,5)P2-bound protein, indicating that the interaction between PI(3,5)P2 and PH domain might be stronger than that of PI(3,4,5)P3 (Fig. 1B Left). This finding is consistent with the result that K679,687N mutation fails to totally block PI(3,5)P2 binding to PH domain.
Fig. 1.
PH domain of PIKE possesses distinct phosphoinositol lipids-binding activities. (A) PIKE-L PH domain WT, but not mutant, binds to 3′-phosphate phosphoinositol lipids. Purified GST-PH WT and (K679,687N) mutant recombinant proteins were incubated with agarose-conjugated phosphoinositol lipid beads. After washing, the associated proteins of the beads were analyzed by immunoblotting with anti-GST-horseradish peroxidase Ab. WT PH domain strongly binds 3′-phosphate phosphoinositol lipids, but not PI(4,5)P2, and (K679,687N) mutation completely eliminates the binding. However, the mutant still weakly associates with PI(3,5)P2 (Left). (B) PI(3,4,5)P3 specifically binds to PH domain. GST-PH fusion proteins were coupled to PI(3,4,5)P3-conjugated beads. Different [PI(3,4,5)P3] were incubated with the complex at 4°C for 3 h. PI(3,4,5)P3 (20 μM) completely abolishes PH domain binds to PI(3,4,5)P3-conjugated beads (Right). However, the same concentration of PI(3,5)P2 fails (Left). (C) PIKE-A PH domain W,T but not (K349,357N) mutant, binds to phosphoinositol lipids. As a control, agarose beads do not bind to GST-PH fusion protein (Left). The expression of WT and mutant GST-PH domain from PIKE-A is confirmed (Right). The above experiments were repeated three times.
PIKE-L is brain specific, whereas PIKE-A is ubiquitously expressed and amplified in a variety of human cancers (4, 5). PH domains of these two isoforms are slightly different. PH domain from PIKE-A omits 20 aa in the C terminus, and a small exon is skipped during RNA splicing in PIKE-A (data not shown). Surprisingly, it strongly binds to all phosphoinositides used, and K349,357N mutation completely disrupts its binding (Fig. 1C Left), indicating that PH domains from PIKE-L and -A isoforms possess distinct phosphoinositol lipid-binding behaviors.
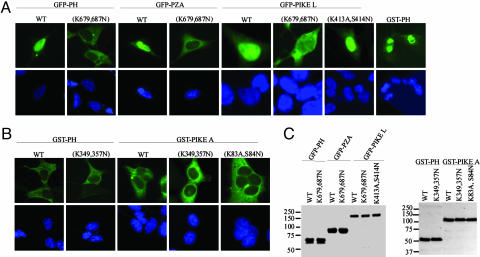
PH Domain of PIKE Dictates Its Nuclear Localization. To explore whether PH domain plays any role in mediating PIKE subcellular localization, we prepared a variety of GFP-tagged constructs and transfected into HEK293 cells. Strikingly, the PH domain of PIKE-L exclusively distributes in the nucleus, whereas K679,687N mutant selectively resides in the cytoplasm. Similar subcellular distribution occurs to the PH-Arf-GAP (Zinc finger)-Ank (PZA) fragment of PIKE-L. GST-tagged PH domain also displays the same results as GFP-PH does (Fig. 2A, rightmost image). HA-tagged proteins and different cell lines also exhibit identical subcellular distribution (data not shown). These observations demonstrate that the PH domain of PIKE-L specifically localizes in the nucleus regardless of the polypeptide tags. Interestingly, GFP-PH or PZA aggregates and forms a bright spot outside the nucleus, resembling the centrosome. Immunofluorescent staining with γ-tubulin Ab, a centrosome-specific marker, suggests the spot is not the centrosome (data not shown). Moreover, both W T and dominant-negative (K413A,S415N) PIKE-L predominantly localize in the nucleus of transfected HEK293 cells. Strikingly, PIKE-L (K679,687N) mutant evidently translocates into the cytoplasm (Fig. 2 A). Sequence analysis shows that both K679 and K687 residues are in the putative nuclear localization signal motif. Mutating them into asparagine residues coincidently abolishes phosphoinositol lipid-binding activity and nuclear localization. PIKE-S consists of a partial PH domain, and it does not bind to phosphoinositol lipids (data not shown). Nevertheless, PIKE-S selectively resides in the nucleus, indicating that the N terminus also contributes to its nuclear localization. In addition to comprising the whole PIKE-S sequence, PIKE-L contains a full module of PH domain and C-terminal Arf-GAP-Ank repeats extension. The cytoplasmic translocation by PIKE-L (K679,687N) mutant demonstrates that PH domain dictates PIKE subcellular localization. In contrast to the nuclear localization of PIKE-L PH domain, PH domain from PIKE-A selectively localizes in the cytoplasm, as does its mutant (K349,357N). We observed the similar cytoplasmic distribution for full-length PIKE-A regardless of GTPase dead (K83A,S84N) or phosphoinositol lipids binding crippled mutant (K349,357N). The nucleus was stained with DAPI (Fig. 2B, leftmost image). The expression of the transfected proteins is verified (Fig. 2C).
Fig. 2.
PH domain of PIKE dictates its nuclear localization. (A) PIKE-L PH domain resides in the nucleus. GFP-tagged PH, PZA, and full-length PIKE-L were transfected into HEK293 cells. WT proteins and dominant-negative PIKE-L (K413A,S414N) predominantly distribute in the nucleus. The nuclei were stained with DAPI (Lower, leftmost image). By contrast, PH domain and PZA fragment mutants, unable to bind phosphatidylinositols, exclusively localize in the cytoplasm, whereas PIKE-L (K679,687N) distributes in both the cytoplasm and the nucleus. GST-tagged PH domain also exclusively resides in the nucleus (Upper, rightmost image). (B) PIKE-A PH domain and its full-length proteins localize in the cytoplasm, no matter whether it binds phosphatidylinositol lipids or not (leftmost images). (C) The expression of GST- and GFP-tagged constructs is verified.
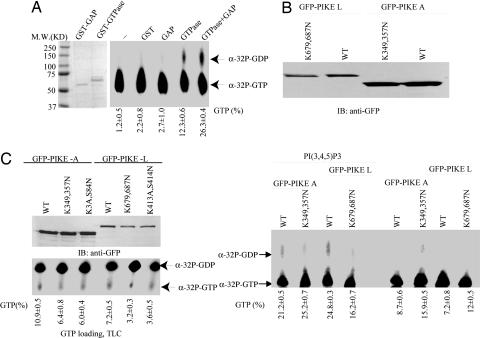
Phosphatidylinositol Lipids Mediate PIKE GTPase Activity In Vivo. The Arf-GAP domain in GGAPs, homologs of PIKE, displays potent catalytic activity on its internal GTPase domain by stimulating bound GTP to hydrolyze into GDP (10). PIKE GTPase assay demonstrates that its GTPase domain alone slowly hydrolyzes bound GTP into GDP. Addition of Arf-GAP domain markedly enhances its hydrolytic activity, confirming the previous finding (10). As a control, GST or GST-GAP itself is unable to hydrolyze GTP (Fig. 3A Right). The in vitro GTPase assay with full-length PIKE-L and -A proteins shows that both WT and GTPase mutant of PIKE protein reveal negligible GTPase activity in the absence of phosphatidylinositol lipid. Introduction of 25 μM water-soluble PI(3,4,5)P3 significantly increases the GTPase activity of WT PIKE-L but not that of K412A,S413N mutant. PI(3,4,5)P3 also exhibits the similar stimulatory effects on PIKE-A (Fig. 3B). The remnant weak GTPase activity in both mutant samples with PI(3,4,5)P3 might result from nonspecific GTPases, which were coimmunoprecipitated with GFP-PIKE mutants and activated by PI(3,4,5)P3. The observation that PI(3,4,5)P3 triggers PIKE GTPase to hydrolyze the bound GTP into GDP suggests that phosphatidylinositol lipids may regulate PIKE conformation through PH domain, leading to the C-terminal Arf-GAP domain accessible to its N-terminal GTPase domain and accelerating its intrinsic GTPase activity. To explore whether phosphatidylinositol lipids binding may influence PIKE GTPase activity in vivo, we performed metabolic labeling of HEK293 cells with [32P]H3PO4, which were transfected with a variety of PIKE-L and -A constructs. Guanine nucleotides bound to immunoprecipitated PIKE-L and -A were analyzed on TLC. Quantitative analysis with phosphoimager demonstrates that both WT PIKE-L and -A bind more GTP than GTPase-dead and lipids nonbinding mutants (Fig. 3C). Thus, phosphatidylinositol lipids regulate PIKE GTPase activity in vivo.
Fig. 3.
PIKE-L and -A have different GTPase activities. (A) In vitro GTPase assay with purified GST-fusion proteins. The purified recombinant proteins were incubated with [α-32P]GTP at 30°C for 30 min. The guanine nucleotides were separated on TLC. GAP robustly stimulates PIKE GTPase domain to hydrolyze GTP into GDP. (B) In vitro GTPase assay of full-length PIKE-L and -A. PIKE-L and -A constructs were transfected into HEK293 cells. Transfected proteins were immunoprecipitated and washed three times with binding buffer. The GTP hydrolysis assay was performed in the presence or absence of PI(3,4,5)P3. The reaction mixture was analyzed on TLC. Both PIKE-L and -A proteins exhibit negligible GTPase activity without PI(3,4,5)P3. Adding PI(3,4,5)P3 substantially increases the GTPase activity in both WT PIKE-L and -A. Experiments were repeated three times. (C) In vivo GTP-loading assay. Various PIKE-L and -A constructs were transfected into HEK293 cells and metabolically labeled with [32P]H3PO4 for 4 h in phosphate-free medium. Transfected proteins were immunoprecipiated with anti-GFP Ab. After extensive washing, precipitated protein-bound guanine nucleotides were analyzed on TLC. (Lower) WT PIKE binds more GTP than mutant PIKE does. (Upper) Expression of transfected GFP-tagged proteins is verified.
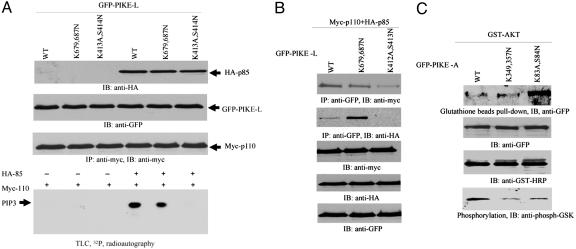
PH Domain of PIKE Mediates Its Activation of PI3-Kinase and Akt. Phosphatidylinositol lipids bind to PH domain of PIKE and regulate its GTPase activity, which might mediate the stimulatory effect of PIKE on its downstream targets. Accordingly, we examined the effect of both WT PIKE-L and (K679,687N) mutant on PI3-kinase. We cotransfected a variety of GFP-PIKE-L constructs into HEK293 cells with HA-p85 and Myc-p110, respectively. In the absence of the regulatory subunit of PI3-kinase, p85, no PI3-kinase activity was observed, despite which PIKE-L construct was transfected, consistent with the previous reported results (1, 15). The strongest PI3-kinase activity occurs when WT PIKE-L and both subunits are cotransfected. However, when dominant-negative PIKE-L (K413A,S414N) was used, PI3-kinase is completely abolished, confirming our previous findings (1). Interestingly, transfection of (K679,687N) mutant significantly decreases PI3-kinase activity (Fig. 4A, bottom image). Coimmunoprecipitation assay demonstrates that WT PIKE-L and K679,687N mutant robustly bind to both subunits of PI3-kinase. By contrast, dominant-negative PIKE-L (K412A,S413N) selectively interacts with PI3-kinase catalytic subunit p110 but not the regulatory subunit p85 (Fig. 4B, first and second images from top). This observation suggests that the GTPase activity is indispensable for PIKE-L to bind p85, which might provide molecular mechanism accounting for its dominant-negative effect to PI3-kinase.
Fig. 4.
PH domain of PIKE mediates its activation of PI3-kinase and Akt. (A) PI3-kinase assay in PIKE-L-transfected HEK293 cells. GFP-PIKE-L WT, (K679,687N), and (K413A,S414N) constructs were transfected into HEK293 cells with myc-p110 and HA-p85, respectively. PI3-kinase was pulled down with anti-myc Ab. PI3-kinase assay was performed in the presence of [γ-32P]ATP. In the absence of HA-p85, no PI3-kinase activity is observed. WT PIKE-L strongly provokes PI3-kinase activity. In contrast, PIKE-L(K413A,S414N) completely inhibits it. Strikingly, K679,687N mutation decreases PI3-kinase activity (bottom image). The expression of all transfected constructs is verified (first, second, and third images from top). (B) PIKE-L binds PI3-kinase. GFP-PIKE-L WT, K679,687N, and K413A,S414N constructs were transfected into 293 cells with myc-p110 and HA-p85, respectively. PIKE-L was immunoprecipitated with anti-GFP Ab. The beads-associated proteins were analyzed with anti-HA or anti-Myc Ab. All PIKE-L constructs interact with the catalytic subunit p110. However, PIKE-L (K413A,S414N) mutant does not bind to the regulatory subunit p85. Interestingly, K679,687N mutant binds to more p85 than WT PIKE-L (first and second images from top). (C) PIKE-A binds to Akt. GST-Akt was transfected into HEK293 cells with PIKE-A WT, (K349,357N), and (K83A,S84N) mutants, respectively. Akt was pulled down with glutathione beads, and the associated proteins were analyzed by immunoblotting assay with anti-GFP Ab. In the absence of EGF, PIKE-A (K83A,S84N) more potently binds to Akt than WT PIKE-A does. PIKE-A (K349,357N) reveals the comparable binding affinity to Akt as WT (top image). The expression of transfected constructs is verified (second and third images from top). In vitro Akt kinase assay with GSK3 demonstrates that WT presents highest activity than other two mutants (bottom image).
PIKE-A potently associates with Akt and promotes its kinase activity in a guanine nucleotide-dependent manner (4). Compared with WT, dominant-negative PIKE-A (K83A,S84N) robustly interacts with Akt, fitting with previous finding that PIKE-A specifically binds active Akt, but dominant-negative PIKE-A binds both active and inactive Akt (4). The binding by PIKE-A (K349,357N) mutant to Akt is comparable with its WT counterpart (Fig. 4C, top image), indicating that phosphoinositol lipids might be dispensable for the interaction between PIKE-A and Akt. However, WT PIKE-A provokes much higher Akt kinase activity than dominant-negative PIKE-A (K83A,S84N) and PIKE-A (K349,357N) mutants, as manifested by GSK phosphorylation, a well-characterized Akt substrate (Fig. 4C, bottom image). PIKE-L and -A mutants, unable to bind phosphoinositol lipids, demonstrate compromised stimulatory effects on the downstream targets of PI3-kinase and Akt, which tightly correlate with their in vivo GTP-loading activities (Fig. 3C), supporting that GTP-bound status mediates the catalytic activity of PIKE on PI3-kinase and Akt signaling cascades in cells.
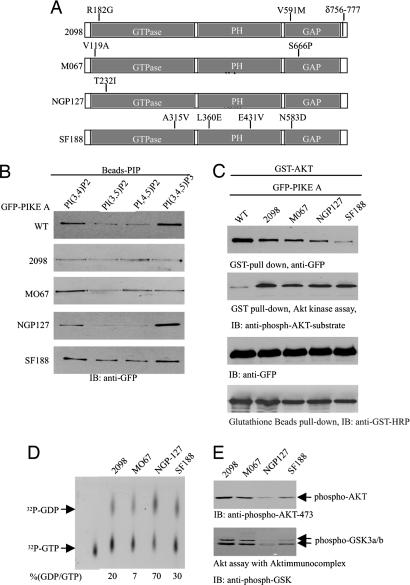
Various Patient-Derived PIKE-A Mutants Display Different GTPase Activities and Akt Activation Effects. RT-PCR of PIKE-A and DNA sequence analysis demonstrate that several amino acids in PIKE-A are mutated in bone sarcoma CRL-2098, NGP-127 neuroblastoma cell, and M067 and SF188 glioblstoma cell lines. Moreover, 21 residues (amino acids 756-777) are truncated in PIKE-A from 2,098 cells (Fig. 5A). In vitro lipid-binding assay reveals that PIKE-A from NGP-127 and SF188 cells displays the similar binding pattern as WT counterpart. However, PIKE-A from 2098 and MO67 exhibits substantially decreased binding affinity to PI(3,4,5)P3 compared to WT PIKE-A (Fig. 5B). Coimmunoprecipitation analysis demonstrates that WT exhibits much stronger binding affinity to active Akt than all PIKE-A mutants from cancer cell lines (Fig. 5C, top image). Nevertheless, Akt kinase activity assay demonstrates that PIKE-A mutants possess higher stimulatory activity than WT PIKE-A (Fig. 5C, second image from top).
Fig. 5.
Various patient-derived PIKE-A mutants display different phosphoinositol lipids-binding activities and Akt-activation effects. (A) PIKE-A from different human cancer cell lines contains various mutations. (B) PIKE-A WT and patient-derived mutants display different phosphatidylinositols-binding activities. Cell lysate from transfected HEK293 cells was incubated with phosphatidylinositols-conjugated beads. (C) PIKE-A mutants stimulate stronger Akt activity than WT PIKE-A. GST-Akt and various GFP-tagged PIKE-A constructs were cotransfected into 293 cells. Akt was pulled down with glutathione beads, and associated proteins were analyzed by immunoblotting with anti-GFP Ab. Compared with control, EGF stimulation elicits much stronger Akt binding by PIKE-A WT and mutants (top image). In vitro kinase assay shows that all mutants provoke much higher Akt activity than WT PIKE-A does regardless of EGF stimulation (second image from top). (D) In vitro GTPase assay. PIKE-A was immunoprecipitated from four human cancer cell lines and incubated with [α-32P]GTP at 30°C for 30 min. The guanine nucleotides were analyzed on TLC. PIKE-A from NGP-127 possesses the highest GTPase activity of the counterparts from the other three cell lines. (E) Akt activities in human cancer cells couple to PIKE-A GTPase activity. The cell lysates from cancer cells were analyzed by immunoblotting with antiphospho-Akt 473, and Akt kinase activity was evaluated with GSK3. PIKE-A from NGP-127 binds lowest amount of GTP, displaying weakest catalytic activity on Akt and its physiological substrate GSK3.
To assess PIKE-A GTPase activity in human cancer cells, we immunoprecipitated the overexpressed PIKE-A, which is due to PIKE gene amplification on chromosome 12q13.3, and conducted in vitro GTPase assay with the immunocomplex. PIKE-A from NGP-127 hydrolyzes GTP into GDP more potently than the other three cells, and PIKE-A from 2098, MO67, and SF188 cells binds more GTP than NGP-127 does (Fig. 5D Upper). Immunoblotting reveals that Akt phosphorylation in 2098, MO67, and SF188 cells is stronger than that in NGP-127 cells. The pattern tightly couples to the corresponding GTPase activity of PIKE-A, verifying the previous finding that PIKE-A up-regulates Akt kinase activity in a GTP-dependent manner. Moreover, GSK3 phosphorylation, a well-characterized physiological substrate of Akt, correlates with Akt activation status (Fig. 5E). The discrepancy on Akt activities by PIKE-A mutants in HEK293 and cancer cells might result from various cellular contexts in these different cells. For example, cancer cells may carry many gene mutations that are unavailable in HEK293 cells and contribute to up-regulate Akt activity.
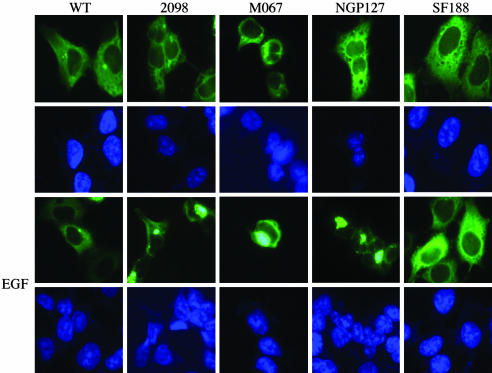
EGF Regulates PIKE-A Mutants Perinuclear Aggregation. PIKE-A mutants display phosphatidylinositol lipids-binding activities similar to WT counterpart, but they demonstrate much stronger stimulatory effect on Akt activity than WT PIKE-A does. Phosphatidylinositols might regulate PIKE subcellular distribution (Fig. 2). Accordingly, we monitored the subcellular localization of a variety of PIKE-A proteins. Both WT PIKE-A and SF188 reveal the similar cytoplasmic distribution, no matter whether the cells were treated with EGF or not; by contrast, EGF stimulates PIKE-A in 2098, MO67, and NGP-127 cells to aggregate in perinuclear zone (Fig. 6). Immunofluorescent staining of PIKE-A on human cancer cells with anti-PIKE-A Ab demonstrates that EGF elicits endogenous PIKE-A aberrant assembly in perinuclear region in 2098, MO67, and NGP-127 cells, whereas PIKE-A in SF188 cells remains the same regardless of EGF treatment (data not shown).
Fig. 6.
Patient-derived PIKE-A mutants demonstrate perinuclear congregation. PIKE-A mutants aggregate in the perinuclear zone. GFP-PIKE-A was transfected in to 293 cells and stimulated with or without 50 ng/ml EGF for 10 min. Both WT and SF188 PIKE-A reside in the cytoplasm regardless of EGF treatment. By contrast, EGF treatment triggers PIKE-A from 2098, MO67, and NGP-127 perinuclear zone aggregation.
Discussion
Our findings demonstrate that PH domain in PIKE potently binds phosphoinositols and plays a critical role in mediating its subcellular localization. Moreover, PH domain and phosphoinositols coupling might mediate the internal GTPase activity of PIKE through its C-terminal Arf-GAP domain. Thus, phosphoinositols regulate PIKE GTPase activity, manipulating its stimulatory effect on PI3-kinase and Akt. Previously, we showed that nerve growth factor elicits PLC-γ1 nuclear translocation and association with PIKE and acts as a guanine nucleotide exchange factor for PIKE through its SH3 domain (16). The GTP-bound PIKE subsequently mediates the activation of nuclear PI3-kinase. Our results indicate that activated PI3-kinase-generated D3-phosphoinositides feed back by binding to PH domain of PIKE-L and incur Arf-GAP domain of PIKE to stimulate the GTPase domain and hydrolyze the bound GTP into GDP, resulting in inactivation of PIKE-L and PI3-kinase. Phosphatidylinositols might exert a feedback self-regulatory mechanism on PIKE GTPase through intramolecular and intermolecular models. For intermolecular model, PI3-kinase-generated PI(3,4,5)P3 binds the PH domain of PIKE-L and triggers the translocation of PIKE-L to the cytoplasmic and nuclear membrane, where GTPase domain of PIKE-L is accessible to Arf-GAP domain from a neighbor PIKE-L molecule. For intramolecular model, PI3-kinase-generated PI(3,4,5)P3 binds PH domain of PIKE-L and induces its conformational change, which makes the N-terminal GTPase domain accessible to its own C-terminal Arf-GAP domain.
PH domain from PIKE-L exclusively occurs in the nucleus. Mutation of both K679 and K687 residues into N, abolishing its phosphoinositol lipids-binding activity, leads to cytoplasmic translocation. It is tempting to speculate that nuclear phosphoinositol lipids might tether PIKE-L in the nucleus through binding to PH domain. For example, insulin receptor substrate 3 is localized both at the plasma membrane and in the nucleus, and its PH domain localizes in the nucleus. Mutations within PH domain preventing insulin receptor substrate 3 intracellular localization result in an inhibition of insulin receptor substrate 3-induced glucose uptake (17). It has been shown before that nuclear PI(3,4,5)P3 facilitates translocation of proteins with PH domain. For example, both nuclear PI3-kinase and PI(3,4,5)P3 are necessary for the nuclear translocation of PKC-ζ (18). In addition to phosphoinositol lipids, PH domain also directly binds a number of proteins. For instance, TCL1 bind to Akt PH domain, and increases its kinase activity and as a consequence enhances substrate phosphorylation. Moreover, TCL1 promotes the transport of Akt1 to the nucleus (19, 20). Therefore, it is possible that PH domain of PIKE-L could directly bind some nuclear partners. Mutation of lysine residues in PH domain of PIKE-L might abolish the association, resulting its cytoplasmic distribution. The observation that PIKE-L PH domain fails to translocate to the cytoplasm in wortmannin-pretreated cells (data not shown), suggesting that phosphoinositol lipids are not implicated in dictating PH domain nuclear localization.
PH domain in PIKE-A is 20 residues shorter than the counterpart in PIKE-L but exhibits potent binding activity to phosphoinositol lipids similar to PIKE-L. Surprisingly, both PIKE-A and its PH domain mainly reside in the cytoplasm, suggesting that PI(3,4,5)P3 binding alone is not sufficient for its nuclear localization, for which these 20 residues are essential. Sequence analysis reveals that PH domain in PIKE-L contains two putative nuclear localization signal (NLS) motifs. The first is a bipartite motif KRSGNSLNKEWKKK, which also is conserved in PIKE-A. The second one should be within the 20 residues, AKRKMWKLKSFGSLRNIYKA, which is absent from PIKE-A isoform. Because PH domain from PIKE-A exclusively resides in the cytoplasm, it suggests that both NLS motifs are necessary for PIKE PH domain nuclear localization. K679,687N mutation in PH domain, abolishing its binding to phosphoinositol lipids and disrupting the first NLS, results in the cytoplasmic translocation of the PIKE-L PH domain.
Although PIKE-A possesses different mutations from different human cancer cells, they all potently enhance Akt kinase activity compared with WT counterpart, when cotransfected with GST-Akt (Fig. 5). Interestingly, PIKE-A from different human cancer cells displays distinct subcellular distributions upon EGF stimulation (Fig. 6). Presumably, different phosphoinositol lipids-binding affinities affect PIKE-A GTPase activities and subcellular localization. Conversely, both Akt and its substrate GSK phosphorylation are stronger in these three cells than those in NGP-127 cells, even though PIKE gene is amplified in all four cells. PIKE-A escalates Akt kinase activity, which is GTP-dependent. Stronger Akt signaling in these cells might be because PIKE-A binds more GTP than it does in NGP-127 cells. It is also possible that different cellular effectors in these cell lines contribute to regulate Akt activity in addition to PIKE-A. Further investigation to dissect the relationship between PIKE-A GTPase activity and tumorigenic effect will provide insight into the molecular mechanism of how PIKE-A mediates cell proliferation. Collectively, our data demonstrate that phosphoinositol lipids regulate PIKE-L nuclear localization through interacting with its PH domain. Further, phosphoinositol lipids bind to PIKE GTPase and mediate its stimulatory effect on PI3-kinase and Akt signalings.
Acknowledgments
We thank Dr. Randy Hall (Emory University) for critical reading of the manuscript. This work was supported by National Institutes of Health Grant R01 NS045627 (to K.Y.).
Author contributions: K.Y. designed research; Y.H. and Z.L. performed research; Y.H. and K.Y. analyzed data; and K.Y. wrote the paper.
Conflict of interest statement: No conflicts declared.
This paper was submitted directly (Track II) to the PNAS office.
Abbreviations: PI3, phosphatidylinositol 3; PIKE, PI3-kinase enhancer; PI(3,4,5)P3, phosphatidylinositol-3,4,5,-trisphosphate; PI(4,5)P2, phosphatidylinositol 4,5-bisphosphate; PH, pleckstrin homology; Arf-GAP, ADP ribosylation factor-GTPase-activating protein; PZA, PH-Arf-GAP (Zinc finger)-Ank; GSK, glycogen synthase kinase; HA, hemagglutinin.
References
- 1.Ye, K., Hurt, K. J., Wu, F. Y., Fang, M., Luo, H. R., Hong, J. J., Blackshaw, S., Ferris, C. D. & Snyder, S. H. (2000) Cell 103, 919-930. [DOI] [PubMed] [Google Scholar]
- 2.Rong, R., Ahn, J. Y., Huang, H., Nagata, E., Kalman, D., Kapp, J. A., Tu, J., Worley, P. F., Snyder, S. H. & Ye, K. (2003) Nat. Neurosci. 6, 1153-1161. [DOI] [PubMed] [Google Scholar]
- 3.Rong, R., Tang, X., Gutmann, D. H. & Ye, K. (2004) Proc. Natl. Acad. Sci. USA 101, 18200-18205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ahn, J. Y., Rong, R., Kroll, T. G., Van Meir, E. G., Snyder, S. H. & Ye, K. (2004) J. Biol. Chem. 279, 16441-16451. [DOI] [PubMed] [Google Scholar]
- 5.Ahn, J. Y., Hu, Y., Kroll, T. G., Allard, P. & Ye, K. (2004) Proc. Natl. Acad. Sci. USA 101, 6993-6998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scaife, R. M. & Margolis, R. L. (1997) Cell Signal. 9, 395-401. [DOI] [PubMed] [Google Scholar]
- 7.Jacques, K. M., Nie, Z., Stauffer, S., Hirsch, D. S., Chen, L. X., Stanley, K. T. & Randazzo, P. A. (2002) J. Biol. Chem. 277, 47235-47241. [DOI] [PubMed] [Google Scholar]
- 8.Nie, Z., Stanley, K. T., Stauffer, S., Jacques, K. M., Hirsch, D. S., Takei, J. & Randazzo, P. A. (2002) J. Biol. Chem. 277, 48965-48975. [DOI] [PubMed] [Google Scholar]
- 9.Kam, J. L., Miura, K., Jackson, T. R., Gruschus, J., Roller, P., Stauffer, S., Clark, J., Aneja, R. & Randazzo, P. A. (2000) J. Biol. Chem. 275, 9653-9663. [DOI] [PubMed] [Google Scholar]
- 10.Xia, C., Ma, W., Stafford, L. J., Liu, C., Gong, L., Martin, J. F. & Liu, M. (2003) Mol. Cell. Biol. 23, 2476-2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vitale, N., Moss, J. & Vaughan, M. (1996) Proc. Natl. Acad. Sci. USA 93, 1941-1944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bollag, G. & McCormick, F. (1995) Methods Enzymol. 255, 161-170. [DOI] [PubMed] [Google Scholar]
- 13.Rosen, L. B., Ginty, D. D., Weber, M. J. & Greenberg, M. E. (1994) Neuron 12, 1207-1221. [DOI] [PubMed] [Google Scholar]
- 14.Luttrell, L. M., Hawes, B. E., Touhara, K., van Biesen, T., Koch, W. J. & Lefkowitz, R. J. (1995) J. Biol. Chem. 270, 12984-12989. [DOI] [PubMed] [Google Scholar]
- 15.Piccione, E., Case, R. D., Domchek, S. M., Hu, P., Chaudhuri, M., Backer, J. M., Schlessinger, J. & Shoelson, S. E. (1993) Biochemistry 32, 3197-3202. [DOI] [PubMed] [Google Scholar]
- 16.Ye, K., Aghdasi, B., Luo, H. R., Moriarity, J. L., Wu, F. Y., Hong, J. J., Hurt, K. J., Bae, S. S., Suh, P. G. & Snyder, S. H. (2002) Nature 415, 541-544. [DOI] [PubMed] [Google Scholar]
- 17.Maffucci, T., Razzini, G., Ingrosso, A., Chen, H., Iacobelli, S., Sciacchitano, S., Quon, M. J. & Falasca, M. (2003) Mol. Endocrinol. 17, 1568-1579. [DOI] [PubMed] [Google Scholar]
- 18.Neri, L. M., Martelli, A. M., Borgatti, P., Colamussi, M. L., Marchisio, M. & Capitani, S. (1999) FASEB J. 13, 2299-2310. [PubMed] [Google Scholar]
- 19.Laine, J., Kunstle, G., Obata, T., Sha, M. & Noguchi, M. (2000) Mol. Cell 6, 395-407. [DOI] [PubMed] [Google Scholar]
- 20.Pekarsky, Y., Koval, A., Hallas, C., Bichi, R., Tresini, M., Malstrom, S., Russo, G., Tsichlis, P. & Croce, C. M. (2000) Proc. Natl. Acad. Sci. USA 97, 3028-3033. [DOI] [PMC free article] [PubMed] [Google Scholar]