Abstract
Resistance to inhibitors of cholinesterase (Ric) 8A is a guanine nucleotide exchange factor that activates certain G protein α-subunits. Genetic studies in Caenorhabditis elegans and Drosophila melanogaster have placed RIC-8 in a previously uncharacterized G protein signaling pathway that regulates centrosome movements during cell division. Components of this pathway include G protein subunits of the Gαi class, GPR or GoLoco domain-containing proteins, RGS (regulator of G protein signaling) proteins, and accessory factors. These proteins interact to regulate microtubule pulling forces during mitotic movement of chromosomes. It is unclear how the GTP-binding and hydrolysis cycle of Gαi functions in the context of this pathway. In mammals, the GoLoco domain-containing protein LGN (GPSM2), the LGN- and microtubule-binding nuclear mitotic apparatus protein (NuMA), and Gαi regulate a similar process. We find that mammalian Ric-8A dissociates Gαi-GDP/LGN/NuMA complexes catalytically, releasing activated Gαi-GTP in vitro. Ric-8A-stimulated activation of Gαi caused concomitant liberation of NuMA from LGN. We conclude that Ric-8A efficiently utilizes GoLoco/Gαi-GDP complexes as substrates in vitro and suggest that Ric-8A-stimulated release of Gαi-GTP and/or NuMA regulates the microtubule pulling forces on centrosomes during cell division.
Keywords: cell division, G protein, GoLoco, GPR, guanine nucleotide exchange
A nontraditional G protein signaling pathway is thought to direct centrosome/chromosome movements during cell division in multicellular organisms. In apparent lieu of regulation by G protein-coupled receptors, G protein subunits of the Gαi class work with a unique guanine nucleotide exchange factor, Ric-8 (resistance to inhibitors of cholinesterase 8), regulator of G protein signaling (RGS) GTPase-activating proteins, GoLoco domain-containing guanine nucleotide dissociation inhibitor (GDI) proteins, and other factors to regulate microtubule “pulling” forces on separating chromosomes during mitosis (for review, see refs. 1–4). Two microtubule-based structures control centrosome and chromosome movements. The mitotic spindle connects the two polar centrosomes and pulls sister chromatids toward the poles during anaphase. Aster microtubules link both centrosomes to the plasma membrane (cell cortex), the sites where G protein-regulated shortening/pulling of these microtubules is thought to occur during anaphase. Greater Gαi-mediated pulling activity at the posterior pole of the cell moves the entire mitotic spindle posteriorly to help define the characteristic asymmetric cleavage plane of the one-cell embryo (5).
Caenorhabditis elegans GPR1/2, Drosophila melanogaster Pins, and mammalian LGN (mammalian Pins or GPSM2) and AGS3 (GPSM1) each contain one or more conserved GoLoco domains at their carboxyl termini, and have amino-terminal regulatory domains that bind species-specific accessory factors known to regulate cell division. GPR or GoLoco domains have been described enzymatically as Gαi-class GDIs that bind Gα-GDP and inhibit release of the nucleotide (for review, see refs. 6 and 7). Their involvement in cell division pathways has been appreciated from genetic studies. C. elegans one-cell embryos with attenuated gpr1/2, gao/gpa16, ric-8, or the gene that encodes the GPR1/2-binding protein LIN5 all undergo a similar symmetric cell division that causes inappropriate distribution of cell lineage determinants and dislocation of cells within the developing embryo, ultimately leading to embryonic lethality. This shared mutant phenotype strongly indicates that these gene products interact in a common pathway to regulate cell division (for review, see refs. 3 and 4).
Alternative models describe the interplay of Ric-8, RGS, GoLoco, Gα, and their accessory proteins in regulating aster microtubule forces during cell division (3, 8–11). One model predicts that GoLoco protein-Gα-GDP is the active complex that regulates the microtubule pulling forces. A second model theorizes that GoLoco protein-Gα-GDP is a substrate for the guanine nucleotide exchange activity of Ric-8. Ric-8-stimulated nucleotide exchange leads to production of free Gα-GTP and signals force generation (1, 3, 4, 8, 9, 11). This hypothesis is attractive, given the opposed phenotypes of C. elegans rgs7 and ric-8 and that mammalian Ric-8A does not activate Gαβγ heterotrimers, but only free Gα-subunits, in vitro (12). In the context of G protein-mediated cell division, this model assigns the traditional Gβγ-subunit function to GoLoco proteins and the guanine nucleotide exchange activities of G protein-coupled receptors to Ric-8.
We have taken a biochemical approach using purified components to address the major unresolved question regarding these two models. Is GoLoco-bound Gαi a substrate for Ric-8A-mediated guanine nucleotide exchange? We demonstrate that mammalian Ric-8A recognizes LGN- and AGS3-Gαi-GDP complexes as substrates in vitro. Ric-8A dissociates GoLoco protein/Gαi-GDP complexes while activating Gαi catalytically in the presence of guanosine 5′-[γ-thio]triphosphate (GTP[γS]). Activation of the Gαi/LGN/nuclear mitotic apparatus protein (NuMA) complex by Ric-8A also stimulates release of NuMA from LGN. Dynamic release of NuMA from LGN has been proposed to be a mechanism of aster microtubule regulation during cell division (13).
Materials and Methods
Molecular Cloning. Rat LGN and AGS3 were cloned by yeast two-hybrid screening of a rat brain embryonic cDNA library with a Gαo bait (12). The full-length AGS3 and LGN cDNAs were subcloned into the baculovirus transfer vector pFastBac HTa, which was used in the Bac-to-Bac system to create recombinant baculoviruses per the manufacturer's instructions (Invitrogen). The 3′ 600 nucleotides of the AGS3 and LGN cDNAs were subcloned using PCR into pQE81L (Qiagen, Valencia, CA) to create His-6-tagged AGS3short and LGNshort Escherichia coli expression constructs. A pQE30 His-6-tagged expression construct encoding amino acids 1818–2001 of human NuMA [NuMA LGN-binding domain (BD)] was described in ref. 14.
Protein Purification. Ric-8A was purified from Sf9 cells as described in ref. 12. Myristoylated and unmodified Gαi-1 were purified from E. coli by the methods of Lee et al. (15). His-6-AGS3short and His-6-LGNshort were expressed in BL21 DE3 E. coli in T7/ampicillin medium by inducing log-phase cultures with 0.5 mM IPTG for 4 h at 25°C. Cells were pelleted and lysed in E. coli lysis buffer [20 mM Hepes, pH 8.0/150 mM NaCl/10 mM imidazole/5 mM 2-mercaptoethanol/protease inhibitor mixture (3.3 μg/ml leupeptin/3.3 μg/ml lima bean trypsin inhibitor/2.3 μg/ml PMSF/2.1 μg/ml Nα-tosyl-l-lysine-chloromethyl ketone/2.1 μg/ml N-tosyl-l-phenylalanine-chloromethyl ketone)] by treatment with lysozyme (40 mg/liter of culture) and freeze/thaw. The lysate was clarified by centrifugation and applied to a Ni-nitrilotriacetic acid (NTA) agarose (Qiagen) column (0.33-ml bed volume/liter of culture). The column was washed with lysis buffer containing 20 mM imidazole and eluted with lysis buffer containing 50 mM NaCl and 150 mM imidazole. The Ni-NTA column eluates were applied to a 1-ml Hi-trap Q column (Qiagen) equilibrated with buffer A (20 mM Hepes/50 mM NaCl/1 mM EDTA/1 mM DTT/protease inhibitor mixture) and eluted with a 0–400 mM linear NaCl gradient. LGNshort flowed through the column before application of the salt gradient, and AGS3short eluted at ≈150 mM NaCl. Pools of both proteins were concentrated by using 10,000 molecular weight cutoff Ultrafree concentration devices (Millipore) and exchanged into buffer A containing 100 mM NaCl.
The baculovirus encoding full-length His-6-LGN was used to infect Sf9 cells (2 × 106 cells per ml) in IPL41 medium containing 10% heat-inactivated FBS, 0.1% pluronic acid, and 10 μg/ml gentamicin. Cells were grown for 48 h at 27°C, harvested, and lysed in buffer A containing 150 mM NaCl (125 ml/liter of culture) by nitrogen cavitation using a Parr bomb. The lysates were clarified and supplemented with purified myristoylated or unmodified Gαi-1 (1 mg/liter of culture) and applied rapidly to a 4-ml Ni-NTA agarose column (Qiagen). The column was washed with buffer A containing 150 mM NaCl and 10 mM imidazole, and His-6-LGN/Gαi complexes were eluted with buffer A containing 150 mM NaCl and 150 mM imidazole. The amount of LGN present in the eluate was estimated by Bradford assay, and a 2-fold molar equivalent of Gαi-1 per GoLoco domain (4 total) was added. This mixture was applied to a Hi-trap Q column and eluted with a linear 100–500 mM NaCl gradient in buffer A. Fractions containing the protein complex were concentrated in a 100,000 molecular weight cutoff (MWCO) Ultrafree concentrator (Millipore) and supplemented with 5-fold and 1-fold molar equivalents of purified NuMA (LGN BD) and Gαi-1, respectively. This mixture was warmed to 30°C for 5 min, cooled to 15°C over 30 min, further concentrated, and gel-filtered at 0.4 ml/min over tandem Superdex 75/200 HR 10/30 columns (GE Healthcare) in buffer A containing 150 mM NaCl. Fractions containing the Gαi/LGN/NuMA (LGN BD) complex were pooled and concentrated in a 100,000 MWCO Ultrafree concentration device. LGNshort and AGS3short/Gαi complexes were prepared similarly from individually purified proteins by gel filtration.
Protein Complex Dissociation Assays. Purified Gαi-1/GoLoco complexes (5 or 10 μM) were incubated in 220-μl reactions containing gel filtration buffer supplemented with 50 or 100 μM GDP or GTP[γS] and the indicated concentrations of MgCl2. Ric-8A was added when indicated, and the incubations were continued for various times at 25°C or 30°C. The reaction mixtures were centrifuged at 15,000 × g for 5 min at 4°C, and the proteins in the supernatants were resolved over tandem Superdex 75/200 HR 10/30 columns at 0.4 ml/min. Eluted fractions were resolved by SDS/PAGE and visualized by staining.
GTP[γS]-Binding Assays. Intrinsic and Ric-8A-stimulated GTP[γS]-binding reactions were initiated by addition of 200 nM free Gαi-1 or 50 nM GoLoco protein/Gαi-1 complex to reaction buffer (20 mM NaHepes, pH 8.0/100 mM NaCl/2 mM MgCl2/1 mM EDTA/1 mM DTT/0.05% Genapol C-100) containing 10 μM [35S]GTP[γS] (10,000 cpm/pmol) and indicated concentrations of Ric-8A at 30°C. Duplicate aliquots were removed at the indicated times, and ice-cold buffer containing 20 mM Tris·HCl (pH 7.7), 100 mM NaCl, 2 mM MgCl2, 0.05% polyoxyethylene 10-lauryl ether (C12E10), and 1 mM GTP was added before filtration through BA-85 nitrocellulose. Filters were washed (20 mM Tris·HCl, pH 7.7/100 mM NaCl/2 mM MgSO4), dried, and subjected to scintillation counting. The Bio-Rad protein detection reagent (Bradford assay) was used to quantify protein concentrations.
GDP Release Assays. Myristoylated Gαi-1 and unmodified Gαi-1 (100 nM) were allowed to bind 10 μM [α32P]GDP (specific activity of 40,000 cpm/pmol) for 1 h at 30°C in 20 mM NaHepes, pH 8.0/0.5 mM DTT/0.05% C12E10/4% glycerol/5 mM EDTA/0.8 mM MgCl2. The reactions were cooled, and LGNshort (0–5 μM) was added to Gαi-1-[α32P]GDP for 5 min at 4°C. Release of GDP was initiated by the addition of 0 or 200 nM Ric-8A in reaction buffer (20 mM NaHepes/1 mM DTT/100 mM NaCl/2 mM MgCl2/100 μM GTP[γS]). Reactions containing myristoylated or unmodified Gαi-1 were conducted at 25°C and 30°C, respectively. Duplicate aliquots were removed at 2, 4, 6, 8, 12, 20, 30, and 60 min and quenched with AlF–4-containing quench buffer (20 mM Tris·HCl, pH 7.7/100 mM NaCl/30 mM MgCl2/30 μM AlCl3/5 mM NaF/50 μM GDP). Quenched reactions were filtered onto BA-85 nitrocellulose, washed with AlF–4 quench buffer, dried, and subjected to scintillation counting. Activities plotted as a function of time were fit as single exponentials by using origin 6.0 (Microcal Software, Northampton, MA). Calculated reaction rates were graphed logarithmically against LGNshort concentration to generate inhibition curves.
Results
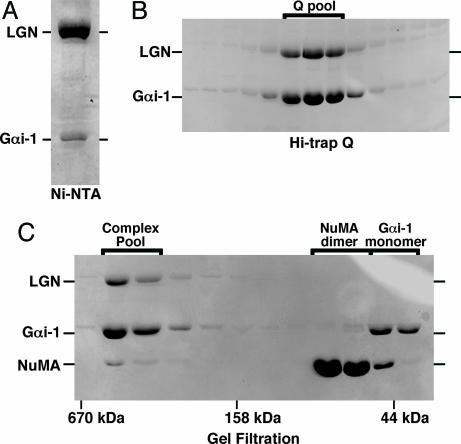
Purification of the Gαi-1/LGN/NuMA (LGN BD) Complexes. To assess whether GoLoco-bound Gαi is a substrate for Ric-8A, we first purified recombinant Gαi-1/LGN/(NuMA) complexes. Myristoylated or unmodified Gαi-1 purified from E. coli (15) was added directly to a soluble lysate created from His-6-LGN baculovirus-infected Sf9 cells. The lysate supplemented with Gαi-1 was adsorbed to a Ni-NTA agarose column, which was washed and eluted with imidazole (Fig. 1A). Attempts to purify active His-6-tagged LGN or His-6-tagged AGS3 in the absence of Gαi were marginally successful (the protein aggregated rapidly; not shown). The Ni-NTA column eluate was immediately supplemented with an additional quantity of purified Gαi-1, applied to a Hi-trap Q column (GE Healthcare), and eluted with a linear gradient of NaCl (Fig. 1B).
Fig. 1.
Purification of the LGN/Gαi-1/NuMA (LGN BD) complex. (A) A soluble Sf9 cell lysate containing rat His-6-LGN was supplemented with purified myristoylated Gαi-1 from E. coli and rapidly adsorbed to Ni-NTA agarose. The resin was eluted with imidazole, and a portion of the eluate was resolved by SDS/PAGE; the gel was stained with Coomassie brilliant blue. (B) The eluate containing LGN/Gαi-1 was immediately supplemented with a 5-fold molar excess (to LGN) of myristoylated Gαi-1, loaded onto a Hi-trap Q column (GE Healthcare), and resolved with a linear NaCl gradient. Fractions of the eluate were resolved by SDS/PAGE. The fractions that were pooled are noted (Q pool). (C) The Q pool of LGN/Gαi-1 complex was incubated with a 4-fold molar excess of NuMA (LGN BD) and a molar equivalent of myristoylated-Gαi-1, ultracentrifuged, and gel-filtered over tandem Superdex 75/200 columns. Fractions of the gel filtration eluate were resolved by SDS/PAGE and stained. Fractions containing the LGN/Gαi-1/NuMA complex were pooled (Complex Pool) and used for subsequent assays.
To prepare Gαi-1/LGN/NuMA complex and to estimate its mass, the Hi-trap Q Gαi/LGN pool was incubated with excess NuMA (LGN BD) purified from E. coli and a molar equivalent of Gαi-1. This mixture was concentrated and gel-filtered over tandem Superdex 75/200 HR 10/30 columns that had been calibrated with protein sizing standards (Bio-Rad). Fig. 1C shows a Coomassie blue-stained SDS gel of fractions eluted from the gel filtration columns. The mass of the Gαi-1/LGN/NuMA complex was estimated to be ≈265 kDa. There appeared to be more Gαi-1 than LGN in the complex. Both of these observations are consistent with the previously calculated stoichiometry of an AGS3short/Gαi-1 complex (i.e., 4 Gαi-1:1 AGS3short) (16).
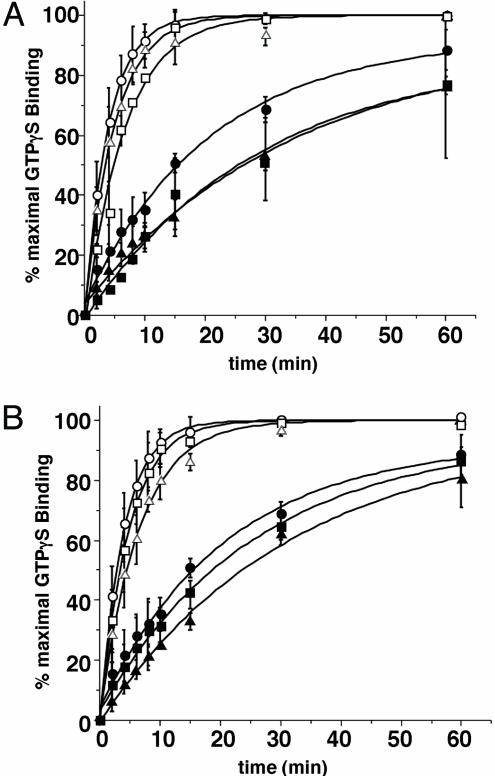
GTP-Binding Kinetics of GoLoco/Gαi-1 Complexes. Experiments were conducted to compare the effect of Ric-8A on the nucleotide exchange rate of free Gαi-1 with the Gαi-1/GoLoco complex. Unmodified Gαi-1, LGN/Gαi-1, and LGN/NuMA/Gαi-1 (Fig. 2A), as well as LGNshort/Gαi-1 and AGS3short/Gαi-1 (Fig. 2B) complexes, were incubated with or without Ric-8A in reactions containing 10 μM [35S]GTP[γS] (≈10,000 cpm/pmol) and 10 mM MgCl2 at 30°C; binding of GTP[γS] to Gαi-1 was measured as described in refs. 17 and 18. Because it was difficult to quantify precisely the amount of Gαi-1 present in each Gαi/GoLoco protein complex (estimated at one or fewer Gαi per GoLoco domain), the data are presented as a percentage of maximal observed Ric-8A-stimulated GTP[γS] binding; the calculated rates are shown in Table 1. Each GoLoco protein inhibited intrinsic and Ric-8A-stimulated GTP[γS] binding rather weakly. The presence of the amino-terminal tetratricopeptide repeat domains of LGN and/or the inclusion of NuMA in the complex had no noteworthy effect on GDI activities. Ric-8A accelerated GTP[γS] binding to Gαi-1 in each complex comparably to its capacity to stimulate GTP[γS] binding to free Gαi-1 (4.5- to 7-fold range vs. 5.3-fold stimulation). Thus, GoLoco proteins and NuMA have little ability to inhibit Ric-8A-stimulated nucleotide exchange on Gαi-1.
Fig. 2.
GTP[γS] binding to GoLoco protein/Gαi-1 complexes. (A) Full-length LGN/Gαi-1 (triangles), LGN/Gαi-1/NuMA (squares) complexes (50 nM), and free Gαi-1 (circles) (200 nM) were used to examine intrinsic (filled symbols) and 200 nM Ric-8A-stimulated (open symbols) rates of GTP[γS] binding. (B) LGNshort/Gαi-1 (triangles), AGS3short/Gαi-1 (squares) complexes (50 nM), and free Gαi-1 (circles) (200 nM) were used to examine intrinsic (filled symbols) and 200 nM Ric-8A-stimulated (open symbols) rates of GTP[γS] binding. Each reaction contained 10 mM MgCl2 and 10 μM[35S]GTP[γS] (10,000 cpm/pmol). Results are shown as the mean of triplicate experiments ± standard deviation.
Table 1. GTP[γS] binding to GoLoco protein/Gαi-1 complexes.
| Kobs, min-1 | Ric-8A-stimulated Kobs, min-1 | Percent GoLoco inhibition (intrinsic) | Percent GoLoco inhibition (Ric-8A-stimulated) | Fold Ric-8A stimulation | |
|---|---|---|---|---|---|
| Gαi-1 | 0.048 ± 0.006 | 0.253 ± 0.004 | — | — | 5.3 |
| Gαi/LGN | 0.029 ± 0.005 | 0.202 ± 0.011 | 39.6 | 20.2 | 7.0 |
| Gαi/LGN/NuMA | 0.034 ± 0.007 | 0.153 ± 0.012 | 29.2 | 39.5 | 4.5 |
| Gαi/LGNshort | 0.031 ± 0.003 | 0.155 ± 0.005 | 35.4 | 38.7 | 5.0 |
| Gαi/AGS3short | 0.039 ± 0.004 | 0.212 ± 0.006 | 18.8 | 16.2 | 5.4 |
Reactions rates (Kobs) were calculated from single exponential rate equations generated by using the origin 6.0 (Microcal Software) curve-fitting program.
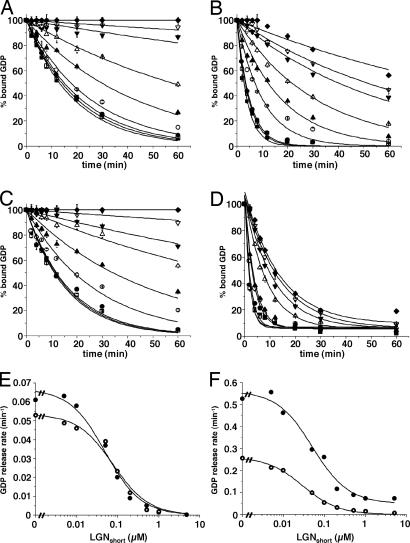
Ric-8A-Stimulated Release of GDP from Myristoylated and Unmodified Gαi/LGNshort Complexes. Lipidation of Gα-subunits dramatically affects many of their properties (19). The influence of myristoylation of Gαi-1 on the ability of Ric-8A to activate Gα-subunits or to overcome GoLoco-mediated inhibition of GDP release from Gα proteins is not known. We first noted that the rate of dissociation of GDP from myristoylated Gαi-1 is faster than the comparable rate observed with the unmodified protein (0.059 ± 0.006 min–1 vs. 0.029 ± 0.006 min–1) at 25°C in the presence of 1 mM free Mg+2 (data not shown and M. E. Linder, personal communication). These rates become nearly equivalent when the unmodified Gαi-1 reaction temperature is increased to 30°C (Fig. 3 A and C). LGNshort (0–5 μM) inhibited dissociation of GDP from myristoylated and unmodified Gαi-1 to similar extents (Fig. 3 A, C, and E). However, the rate of Ric-8A-stimulated dissociation of GDP from Gαi-1 was ≈2-fold greater with the myristoylated protein than with the unmodified subunit (0.56 ± 0.1 min–1 vs. 0.27 ± 0.02 min–1) (Fig. 3 B, D, and F). Whereas LGNshort completely inhibited Ric-8A-stimulated release of GDP from unmodified Gαi-1, it was strikingly less efficacious at inhibiting dissociation of nucleotide from the myristoylated protein (Fig. 3 E and F). There was also a modest loss of potency of LGN when interacting with myristoylated Gαi-1 (IC50 = 50 nM vs. 27 nM for the unmodified protein). Thus, myristoylation greatly enhances the ability of Ric-8A to use Gαi and GoLoco-bound Gαi as substrates, but it has little effect on the interaction between Gαi-1 and LGN in the absence of guanine nucleotide exchange factor.
Fig. 3.
Release of GDP from complexes containing myristoylated and unmodified Gαi-1 and LGNshort. Myristoylated Gαi-1 and unmodified Gαi-1 (100 nM) were bound to [α-32P]GDP and incubated with LGNshort (0, ▪; 5 nM, □;10 nM, •; 50 nM, ○; 100 nM, ▴; 200 nM, ▵; 500 nM, ▾ ;1 μM, ▿;5 μM, ♦) at 4°C for 5 min. Release of GDP from unmodified Gαi-1 was initiated at 30°C by adding 0 (A) or 200 nM (B) Ric-8A. Release of GDP from myristoylated Gαi-1 was initiated at 25°C by adding 0 (C) or 200 nM (D) Ric-8A. Duplicate aliquots of each reaction mixture were removed and quenched at the indicated times with AlF–4 quench buffer. Results are shown as the mean of duplicate experiments (± standard deviation). The intrinsic (E) and Ric-8A-stimulated (F) rates of GDP release from myristoylated Gαi(•) and unmodified Gαi (○) are plotted logarithmically against LGNshort concentration by using origin 6.0 (Microcal Software).
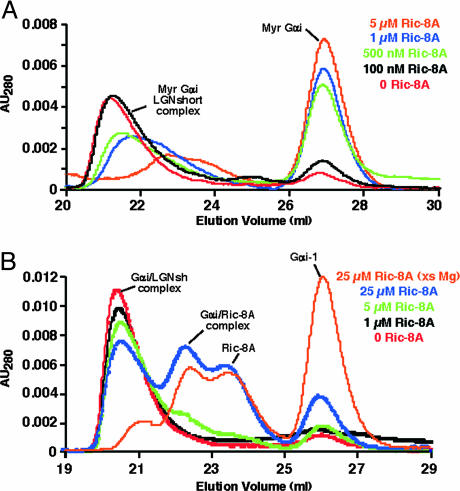
To explore further the influence of myristoylation of Gαi on activation of GoLoco-bound Gαi-GDP by Ric-8A, an assay was developed to monitor dissociation of the Gαi-GDP/GoLoco protein complex. Ric-8A was added to reaction mixtures containing 5 μM preformed Gαi-GDP/LGNshort complex and excess GDP or GTP[γS]. The reaction products were then separated at indicated times by gel filtration. Fig. 4A shows that as little as 500 nM Ric-8A was efficient at dissociating and releasing the majority of Gαi-GTP[γS] from 5 μM myristoylated Gαi-GDP/LGNshort complex (<20 μM Gαi-1) during a 15-min incubation at 25°C in the presence of 1 mM free Mg+2 (green, Fig. 4A). A lower concentration of Ric-8A (100 nM) had little effect (black, Fig. 4A). In contrast, 5 or even 25 μM Ric-8A was only partially effective at dissociating the Gαi-GDP/LGNshort complex containing unmodified Gαi-1 under similar conditions (green and blue, Fig. 4B). Only when the nucleotide exchange reaction was conducted at 30°C and in the presence of 10 mM MgCl2 did Ric-8A (25 μM) become efficient at dissociating and releasing Gαi-GTP[γS] from 5 μMGαi-GDP/LGNshort complex (orange, Fig. 4B). Thus, Ric-8A catalyzes nucleotide exchange on myristoylated Gαi-GDP/GoLoco complexes efficiently in vitro. Given the modest concentrations of protein (10-fold more complex than Ric-8A) and free Mg+2 (1 mM) that were used, we suggest that Gαi-GDP/GoLoco complexes are plausible substrates for Ric-8A-mediated activation in vivo.
Fig. 4.
Ric-8A activates the LGNshort/myristoylated Gαi-1 complex catalytically. (A) Myristoylated Gαi-1/LGNshort complex (≈5 μM) was incubated with 50 μM GTP[γS], ≈1 mM free Mg+2, and 0 (red), 100 nM (black), 500 nM (green), 1 μM (blue), or 5 μM (orange) Ric-8A for 15 min at 25°C. (B) Unmodified Gαi-1/LGNshort complex (≈5 μM) was incubated with 50 μM GTP[γS], ≈1 mM free Mg+2, and 0 (red), 1 μM (black), 5 μM (green), or 25 μM (blue) Ric-8A for 15 min at 25°C or ≈10 mM free Mg+2 and 25 μM Ric-8A (orange) for 15 min at 30°C. Reaction mixtures were loaded onto tandem Superdex 75/200 columns and resolved. The UV absorbance traces of the column eluates are shown.
Activation of the Gαi-1/LGN/NuMA Complex. Gαi and NuMA bind LGN cooperatively in vivo, and stabilization of microtubules by NuMA is precluded when NuMA is bound to LGN (13, 14, 20, 21). Thus, dynamic release of NuMA from LGN at the sites of aster microtubule anchorage (the plasma membrane and/or centrosomes) may be the mechanism to regulate aster microtubule forces during cell division. Given that Ric-8A dissociates activated Gαi from Myr-Gαi-1/LGNshort (Fig. 4), we tested whether Ric-8A would also stimulate dissociation of NuMA from LGN contained in complexes with Gαi-1. Purified Myr-Gαi-GDP/LGN/NuMA complex (10 μM; ≤40 μM Myr-Gαi-1) was incubated with 100 μM GDP or GTP[γS] and 1 mM free Mg+2 with or without 1 μM Ric-8A for 10 or 30 min at 30°C. GTP[γS], but not GDP, stimulated a small amount of dissociation of the complex (Fig. 5 A and C). In the presence of GDP, Ric-8A caused little complex dissociation, resulting in the formation of scant amounts of Ric-8A/Gαi-1, free Gαi-1 monomer, and NuMA (LGN BD) dimer (Fig. 5B). However, when Ric-8A and GTP[γS] were included in the reaction, most of the complex dissociated, and most of the myristoylated Gαi-1 and NuMA were released as GTP[γS]-bound monomers and dimers, respectively (Fig. 5D). These experiments demonstrate that Ric-8A-stimulated binding of GTP to Gαi-1 contained in Myr-Gαi-GDP/LGN/NuMA complexes releases both LGN and NuMA, providing a preliminary biochemical understanding of how the Gαi·GTP switch may dynamically regulate the interactions of NuMA with LGN and microtubules.
Fig. 5.
Ric-8A dissociates the LGN/Gαi-1/NuMA complex, activating myristoylated Gαi-1 and liberating NuMA. LGN/Gαi-1/NuMA complex (10 μM, ≈40 μM Gαi-1) was incubated for 10 (red) or 30 (blue) min at 30°C with 100 μM GDP (A), 100 μM GTP[γS] (B), 100 μM GDP and 1 μM Ric-8A (C), or 100 μM GTP[γS] and 1 μM Ric-8A (D). The reaction mixtures were loaded onto tandem Superdex 75/200 columns and resolved. Fractions from each experiment were analyzed by SDS/PAGE. Each silver-stained gel is shown below the UV absorbance of the gel filtration eluate. The positions of gel filtration mass standards and the fractions where NuMA dimer appeared are noted.
Discussion
G protein regulation of the mitotic spindle and aster microtubule pulling forces during asymmetric cell division is essential for establishing and maintaining differentiated tissues and cell types in multicellular organisms (for review, see refs. 1–3 and references therein). Alternate mechanisms for regulation of the conventional G protein switch are involved because G protein-coupled receptors (GPCRs) and Gβγ-subunits do not appear to participate directly. Instead, GPR or GoLoco GDI proteins may serve a βγ-like function, and Ric-8 may act like a GPCR to catalyze nucleotide exchange on GoLoco-bound Gαi-GDP. It is not known how nucleotide hydrolysis by the Gα protein intersects with microtubule-dependent chromosome/centrosome movements.
To test directly whether Ric-8A can activate GoLoco-bound Gαi-GDP, we prepared stable complexes of Gαi-1-GDP bound to LGNshort, AGS3short, full-length LGN, or LGN/NuMA and saw that Ric-8A stimulated binding of GTP[γS] to each Gαi-GDP/GoLoco complex, markedly overcoming the GDI activity of each GoLoco protein (Fig. 2). This observation contrasts with our previous finding that Ric-8A does not activate Gαi-GDP-βγ heterotrimers in vitro, and, to our knowledge, it is the first direct demonstration that Gαi-1 bound to a GoLoco domain-containing GDI protein can be activated by Ric-8A.
It was proposed previously that GoLoco-bound Gα-GDP is a much poorer substrate for Ric-8 than is free Gα-GDP, because high concentrations of the C. elegans GPR1/2 GoLoco-domain inhibited Ric-8-stimulated binding of GTP[γS] to GAO in vitro (10). We also note that high concentrations of LGNshort inhibit both intrinsic and Ric-8A-stimulated release of GDP from Gαi-1 (Fig. 3). However, LGNshort inhibits Ric-8A-stimulated GDP release from unmodified Gαi-1 completely but is less potent and strikingly less efficacious when myristoylated Gαi-1 is used. Thus, although myristoylation of Gαi-1 does not affect GoLoco GDI activity (Fig. 3E), which is consistent with a recent report (22), lipid modification greatly improves the ability of Gαi-1 to serve as a substrate for Ric-8A-stimulated nucleotide exchange regardless of whether Gαi is bound to GoLoco or is free.
These findings led us to test whether myristoylation of Gαi-1 enhanced the ability of Ric-8A to cause dissociation of Gαi-GDP/GoLoco complexes. Substoichiometric amounts of Ric-8A dissociated myristoylated Gαi-GDP/LGNshort complexes catalytically in the presence of GTP[γS], whereas a superstoichiometric amount of Ric-8A dissociated unmodified Gαi-GDP/LGNshort complexes only partially under the same conditions (Fig. 4). Collectively, these studies show that the net effect of myristoylation of Gαi-1 in the presence of a nucleotide dissociation inhibitor (GoLoco) and activator (Ric-8A) is to accelerate the observed rate of production of Gαi-GTP.
The amino termini of LGN, AGS3, and C. elegans GPR1/2 mediate interactions with accessory proteins that are important for regulating cell division (11, 14, 23). LGN recruits NuMA to the plasma membrane and pericentriolar regions during mitosis (20, 21). Binding of NuMA and Gαi-GDP to LGN is cooperative, and NuMA is unable to interact with microtubules when bound to LGN (20, 21). Thus, the potential of NuMA to regulate aster microtubule pulling forces may be realized by dynamic binding and release of NuMA from LGN (13, 21). This regulation could be achieved by Ric-8A-stimulated activation of Gα and concomitant dissociation of GTP-Gα and NuMA from LGN. The experiments described in this report demonstrate this reaction (Fig. 5). Further study will be required to provide evidence of a direct interaction between Ric-8A and the Gαi/LGN/NuMA protein complex in mammalian cells. Interestingly, a small amount of a ternary complex containing C. elegans RIC-8, GPR1/2, and GAO was isolated by immunoprecipitation (10). This complex may be a reaction intermediate representing the initial transitory interaction of Ric-8 with Gα-GDP/GoLoco.
We envision models in which one cellular function of Ric-8A is to dissociate Gαi-GDP/GoLoco complexes by stimulation of nucleotide exchange. G protein control of asymmetric cell division involves cycling of Gαi between its GDP- and GTP-bound forms, as evidenced by the fact that (in C. elegans) both RIC-8 and RGS7 influence the pathway in opposed fashion (8). It remains speculative whether Gαi-GDP/GoLoco or the production of Gαi-GTP from a GoLoco scaffold activates signaling. It stands to reason that Gαi-GTP must dissociate from GoLoco at some point during signaling. If multiple rounds of cycling between Gαi-GDP/GoLoco and liberated Gαi-GTP are required to complete cell division, then Ric-8A-stimulated dissociation of a Gαi/GoLoco complex could be responsible for either terminating or activating the signal. In either context, RGS-facilitated hydrolysis of GTP by Gα ensues. The resultant Gαi-GDP could rebind to GoLoco (and not βγ) to complete one round of the cycle. Rapid cycling of this process may be necessary to regulate the pulling forces on microtubules appropriately during a round of chromosome segregation (see Fig. 6, which is published as supporting information on the PNAS web site, for these proposed models). Regulation of other Gα or Gα/GoLoco-mediated signaling pathways by Ric-8A is also worth considering, given the number of distinct Gα binding partners of mammalian Ric-8A and Ric-8B (12, 24, 25) and the many processes that appear to be regulated by RIC-8 in C. elegans (26–28).
Supplementary Material
Acknowledgments
We thank Andrejs Krumins and Tom Wilkie for reading the manuscript critically and Jonathan Hopkins, Lindsey Gardner, Kevin Vale, and Philip Ramirez for providing excellent technical support. This work was supported by National Institutes of Health Grant GM34497 and the Raymond and Ellen Willie Distinguished Chair in Molecular Neuropharmacology (to A.G.G.) and American Heart Association (Texas Affiliate) Postdoctoral Fellowship 0325033Y (to G.G.T.).
Author contributions: G.G.T. designed research; G.G.T. performed research; G.G.T. contributed new reagents/analytic tools; G.G.T. and A.G.G. analyzed data; and G.G.T. and A.G.G. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: NuMA, nuclear mitotic apparatus protein; Ric, resistance to inhibitors of cholinesterase; BD, binding domain; GTP[γS], guanosine 5′-[γ-thio]triphosphate; RGS protein, regulator of G protein signaling; GDI, guanine nucleotide dissociation inhibitor; NTA, nitrilotriacetic acid.
References
- 1.Manning, D. R. (2003) Sci. STKE 2003, pe35. [DOI] [PubMed] [Google Scholar]
- 2.Goldstein, B. (2003) Curr. Biol. 13, R879–R880. [DOI] [PubMed] [Google Scholar]
- 3.Hampoelz, B. & Knoblich, J. A. (2004) Cell 119, 453–456. [DOI] [PubMed] [Google Scholar]
- 4.Siderovski, D. P. & Willard, F. S. (2005) Int. J. Biol. Sci. 1, 51–66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gotta, M. & Ahringer, J. (2001) Nat. Cell Biol. 3, 297–300. [DOI] [PubMed] [Google Scholar]
- 6.Willard, F. S., Kimple, R. J. & Siderovski, D. P. (2004) Ann. Rev. Biochem. 73, 925–951. [DOI] [PubMed] [Google Scholar]
- 7.Blumer, J. B., Cismowski, M. J., Sato, M. & Lanier, S. M. (2005) Trends Pharmacol. Sci. 26, 470–476. [DOI] [PubMed] [Google Scholar]
- 8.Hess, H. A., Roper, J.-C., Grill, S. W. & Koelle, M. R. (2004) Cell 119, 209–218. [DOI] [PubMed] [Google Scholar]
- 9.Couwenbergs, C., Spilker, A. C. & Gotta, M. (2004) Curr. Biol. 14, 1871–1876. [DOI] [PubMed] [Google Scholar]
- 10.Afshar, K., Willard, F. S., Colombo, K., Johnston, C. A., McCudden, C. R., Siderovski, D. P. & Gonczy, P. (2004) Cell 119, 219–230. [DOI] [PubMed] [Google Scholar]
- 11.Srinivasan, D. G., Fisk, R. M., Xu, H. & van den Heuvel, S. (2003) Genes Dev. 17, 1225–1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tall, G. G., Krumins, A. M. & Gilman, A. G. (2003) J. Biol. Chem. 278, 8356–8362. [DOI] [PubMed] [Google Scholar]
- 13.Du, Q. & Macara, I. G. (2004) Cell 119, 503–516. [DOI] [PubMed] [Google Scholar]
- 14.Du, Q., Stukenberg, P. T. & Macara, I. G. (2001) 3, 1069–1075. [DOI] [PubMed]
- 15.Lee, E., Linder, M. E. & Gilman, A. G. (1994) Methods Enzymol. 237, 146–163. [DOI] [PubMed] [Google Scholar]
- 16.Adhikari, A. & Sprang, S. R. (2003) J. Biol. Chem. 278, 51825–51832. [DOI] [PubMed] [Google Scholar]
- 17.Krumins, A. M. & Gilman, A. G. (2002) Methods Enzymol. 344, 673–685. [DOI] [PubMed] [Google Scholar]
- 18.Sternweis, P. C. & Robishaw, J. D. (1984) J. Biol. Chem. 259, 13806–13813. [PubMed] [Google Scholar]
- 19.Linder, M., Pang, I., Duronio, R., Gordon, J., Sternweis, P. & Gilman, A. (1991) J. Biol. Chem. 266, 4654–4659. [PubMed] [Google Scholar]
- 20.Du, Q., Taylor, L., Compton, D. A. & Macara, I. G. (2002) Curr. Biol. 12, 1928–1933. [DOI] [PubMed] [Google Scholar]
- 21.Kisurina-Evgenieva, O., Mack, G., Du, Q., Macara, I., Khodjakov, A. & Compton, D. A. (2004) J. Cell Sci. 117, 6391–6400. [DOI] [PubMed] [Google Scholar]
- 22.McCudden, C. R., Willard, F. S., Kimple, R. J., Johnston, C. A., Hains, M. D., Jones, M. B. & Siderovski, D. P. (2005) Biochim. Biophys. Acta 1745, 254–264. [DOI] [PubMed] [Google Scholar]
- 23.Blumer, J. B., Bernard, M. L., Peterson, Y. K., Nezu, J.-i., Chung, P., Dunican, D. J., Knoblich, J. A. & Lanier, S. M. (2003) J. Biol. Chem. 278, 23217–23220. [DOI] [PubMed] [Google Scholar]
- 24.Klattenhoff, C., Montecino, M., Soto, X., Guzman, L., Romo, X., Garcia, M. D., Mellstrom, B., Naranjo, J. R., Hinrichs, M. V. & Olate, J. (2003) J. Cell. Physiol. 195, 151–157. [DOI] [PubMed] [Google Scholar]
- 25.Von Dannecker, L. E. C., Mercandante, A. F. & Malnic, B. (2005) J. Neurosci. 25, 3793–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Schade, M. A., Reynolds, N. K., Dollins, C. M. & Miller, K. G. (2005) Genetics 169, 631–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Reynolds, N. K., Schade, M. A. & Miller, K. G. (2005) Genetics 169, 651–670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller, K. G., Emerson, M. D., McManus, J. R. & Rand, J. B. (2000) Neuron 27, 289–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.