Abstract
Many clinically useful antibiotics interfere with protein synthesis in bacterial pathogens by inhibiting ribosome function. The sites of action of known drugs are limited in number, are composed primarily of ribosomal RNA (rRNA), and coincide with functionally critical centers of the ribosome. Nucleotide alterations within such sites are often deleterious. To identify functional sites and potential sites of antibiotic action in the ribosome, we prepared a random mutant library of rRNA genes and selected dominant mutations in 16S rRNA that interfere with cell growth. Fifty-three 16S rRNA positions were identified whose mutation inhibits protein synthesis. Mutations were ranked according to the severity of the phenotype, and the detrimental effect of several mutations on translation was verified in a specialized ribosome system. Analysis of the polysome profiles of mutants suggests that the majority of the mutations directly interfered with ribosome function, whereas a smaller fraction of mutations affected assembly of the small ribosomal subunit. Twelve of the identified mutations mapped to sites targeted by known antibiotics, confirming that deleterious mutations can be used to identify antibiotic targets. About half of the mutations coincided with known functional sites in the ribosome, whereas the rest of the mutations affected ribosomal sites with less clear functional significance. Four clusters of deleterious mutations in otherwise unremarkable ribosomal sites were identified, suggesting their functional importance and potential as antibiotic targets.
Keywords: 16S rRNA, 30S subunit, resistance, ribosome, translation
With a molecular mass of ≈2.5 million Da and >50 different RNA and protein building blocks, the ribosome represents one of the largest and most complex enzymes in the cell. Ribosomal RNA (rRNA) accounts for two-thirds of the ribosome and is responsible for its main functions in protein synthesis: interpretation of genetic information and polymerization of amino acids into a polypeptide. rRNA is also intimately involved in known auxiliary activities of the ribosome, such as nascent peptide release, binding of translation factors, GTP hydrolysis, etc. (reviewed in ref. 1).
The ribosome is the predominant antibiotic target in the bacterial cell. A large variety of natural and synthetic antibiotics interfere with translation by binding to rRNA and preventing the correct placement of ribosomal ligands, corrupting rRNA structure, or affecting conformational flexibility of rRNA (2). Advantages of the ribosome as an antibiotic target may stem from its RNA-based design. The multiplicity of rRNA genes in microbial genomes makes it difficult for a microorganism to develop resistance by mutating the drug-binding site (3). Furthermore, RNA offers fewer mutational options than protein enzymes (3 versus 19, respectively), which makes it more difficult for a microbial pathogen to “find” a mutation that would reduce antibiotic binding without compromising functional integrity of the enzyme. In the clinical setting, resistance to protein synthesis inhibitors is usually associated with the acquisition of resistance genes (often originating in the antibiotic producers) rather than mutation of target sites (4, 5), illustrating the high cost of fitness and the difficulty of acquiring and fixing rRNA mutations.
Known ribosomal inhibitors act on a fairly limited number of sites usually located within the ribosomal functional centers. Mapping the sites of the drug action has played an important role in the identification and characterization of functionally critical regions of the ribosome (6, 7). However, given the enormous size and functional complexity of the ribosome, the number of possible antibiotic targets and sites of functional importance is likely to exceed those currently known. The occasional serendipitous discovery of antibiotics acting on novel ribosomal sites and subsequent recognition of the functional significance of such sites support this notion (8, 9). Nonetheless, only a few attempts have been made to identify new sites of antibiotic action and to understand the activity of the associated centers in the ribosome (10).
The performance of the rRNA sites critical for ribosome functions, structure, or assembly critically depends on their chemical makeup. Therefore, nucleotide alterations at such sites are expected to be deleterious for the cell. The distribution of the recognized ribosomal functional sites and the sites of antibiotic action clearly correlate with the location of the known deleterious mutations in rRNA (11). Thus, deleterious mutations in rRNA serve as hallmarks of both functionally important ribosomal centers and antibiotic sites. In a search for putative functional sites in the ribosome that can be targeted by new antibiotics, we used a combination of random mutagenesis and negative selection to identify a variety of deleterious mutations in the rRNA of small and large ribosomal subunits of bacterial ribosomes. Here we describe the isolation of a collection of mutations in 16S rRNA that highlight structure-sensitive rRNA centers that can be targeted by new antibiotics, including rRNA regions with previously unrecognized functional significance.
Materials and Methods
Enzymes and Chemicals. All of the antibiotics were from Sigma, enzymes were from Fermentas (Hanover, MD) or New England Biolabs, and chemicals were from Fisher Scientific.
Generation of Segment–Mutant rRNA Libraries. The Escherichia coli mutator strain XL-1 red (Stratagene) was cotransformed with the kanamycin resistance (Kanr) plasmid pLG857 (12), carrying a temperature-sensitive λ repressor gene (cI857) and ampicillin resistance (Ampr) plasmid pLK45 that carries the E. coli rrnB operon under the control of the λ PL promoter (13). Transformants were selected at 30°C on LB agar plates containing 100 μg/ml ampicillin and 50 μg/ml kanamycin. Several hundred colonies were washed from the plates. Cells were propagated for 24 h at 30°C in 100 ml of LB broth supplemented with antibiotics, and randomly mutated plasmid was prepared.
The unique restriction sites of pLK45, KpnI, ApaI, and XbaI were used for a fragment exchange to generate segment–mutant pLK45 libraries where only specific segments of the plasmid would carry random mutations. The rrnB segment flanked by KpnI and ApaI restriction sites, encompassing the 5′ transcribed spacer and the 930-nucleotide-long 5′ region of the 16S rRNA gene, was PCR amplified by using a low-mutation-frequency Triple Master PCR system (Eppendorf) and a pair of primers, ATAACCATCTGCGGTGATACTGAG and CGAATTAAACCACATGCTCCACCGC. The amplified PCR fragment was treated with DpnI to remove the template, cut with KpnI and ApaI, and cloned into WT pLK45 cut with the same restriction enzymes and treated with calf intestine phosphatase. The resulting segment–mutant library A was transformed into highly competent POP2136 cells, which carried a chromosomal copy of cI857 repressor (14). Transformed cells were grown overnight in ampicillin-LB at 30°C without prior plating. The analogous procedure was used to produce segment–mutant library B, which carried a randomly mutagenized ApaI–XbaI segment that encompassed a 611-nucleotide-long 3′ segment of the 16S rRNA gene and 182 nucleotides of the 16S/23S spacer. Primers used for PCR amplification of this segment were GGGAGTACGGCCGCAAGGTTAAAAC and CGTGAAAGGGCGGTGTCCTGGGCC. Segment–mutant libraries were enriched in clones carrying deleterious mutations by using negative selection, essentially as described in ref. 15, and total plasmid was prepared (for details, see Supporting Materials and Methods, which is published as supporting information on the PNAS web site).
Screening Segment–Mutant Libraries for Clones with Deleterious rRNA Mutations. The segment–mutant libraries A or B, enriched in clones with deleterious rRNA mutations, were transformed into fresh POP2136 cells and plated on LB/ampicillin/agar plates. Plates were incubated overnight at 30°C. By using a robotic colony picker, 8,000–12,000 colonies were picked from the plates and inoculated individually into 90 μl of LB/ampicillin medium in 384-well plates. After growth at 30°C for 48 h, each plate was replicated by using a 384-pin replicator (Boekel, Feasterville, PA) into two new plates, one with LB/ampicillin medium and the other with LB/ampicillin medium supplemented with 15 μg/ml erythromycin. Plates were incubated overnight at 30°C (LB/ampicillin plate) or at 42°C (LB/ampicillin/erythromycin plate). The A600 of the cultures in the plate wells was read by using a SpectraMax Plus384 plate reader (Molecular Devices).
Plasmids were prepared from clones that exhibited poor growth at 42°C, and mutant rDNA segments were sequenced from the same pairs of primers that were used for the construction of the corresponding libraries.
The severity of phenotypes conferred by rRNA mutations was tested by transforming plasmids into JM109 cells (16) plated at 37°C or into POP2136 cells plated at 30°C or 42°C.
Testing Mutants in the Specialized Ribosome System. Individual 16S rRNA mutations were introduced by fragment exchange by using restriction enzymes KpnI and CpoI into the pKF207 plasmid, which contains, under the control of an arabinose-inducible promoter, the 16S rRNA gene with a mutated anti-Shine–Dalgarno sequence 5′-GGGGU-3′ (17, 18). KLF10 cells [F – ara Δ(gpt – lac)5 λ(Φ Pant–SDAUCCC–lacZ) KanR srlR301::Tn10 Δ(recA–srl)306] that carry a chromosomally encoded lacZ reporter gene with an altered Shine–Dalgarno sequence (5′-AUCCC-3′) were transformed with the resulting plasmids and were plated onto LB/agar plates supplemented with 100 μg of ampicillin. The β-galactosidase activity was determined by using the conventional procedure (19), with some modifications (see Supporting Materials and Methods).
Polysome Analysis. Polysomes were analyzed following published protocols (20), with minor modifications described in Supporting Materials and Methods. The ratio of mutant to WT 16S rRNA in the gradient fractions was determined by primer extension as described in ref. 3, using a primer, AAGGGCCATGATGACTTGA, specific for the pLK45 resident mutation C1192U. Primer-extension products were separated on 12% denaturing polyacrylamide gel and quantified by using a PhosphorImager (Molecular Dynamics).
Results
Selection of Deleterious Mutations in rRNA. Our strategy for identification of functionally and structurally critical sites in the rRNA of the small ribosomal subunit that could be used as antibiotic targets was based on mapping deleterious mutations in 16S rRNA. Conditionally expressed rRNA genes were randomly mutagenized, mutant libraries were enriched in clones with deleterious rRNA mutations, such clones were identified by replica plating, and mutations were mapped by sequencing.
Mutations were generated in the E. coli rrnB operon in the pLK45 plasmid, where it is expressed under the control of the λ PL promoter (13). The plasmid was propagated in E. coli strain POP2136, which carries a chromosomal copy of the temperature-sensitive λ repressor gene. At 30°C, the expression of mutant rRNA genes is abolished; at 42°C, the repressor is inactivated, and expression of the plasmid-borne rrnB is induced. The rrnB operon in pLK45 carries a spectinomycin resistance mutation, C1192T, in the 16S rRNA gene and an erythromycin resistance mutation, A2058G, in the 23S rRNA gene that permitted monitoring of the amount of plasmid-encoded rRNA in the cell. After 3–4 h of induction, plasmid-encoded 16S rRNA accounted for 40–60% of the total cellular 16S rRNA (ref. 13 and data not shown).
Random mutations were introduced into the pLK45 plasmid by propagating it in the E. coli mutator strain XL-1 red. To avoid counterselection of deleterious rRNA mutations, pLK45 was cotransformed into the mutator cells together with plasmid pLG857 that encodes temperature-sensitive λ repressor (12), and cells were grown at 30°C to prevent expression of mutant rRNA. Under the exploited mutagenesis conditions, the expected frequency of mutations is 1 per 2,000 base pairs (21). The initial plasmid library prepared from XL-1 red mutant cells contained ≈1012 mutant plasmid molecules. Sequencing of the rRNA operon in plasmids prepared from several random clones confirmed the expected frequent presence of multiple mutations. Therefore, to reduce the number of mutations per clone and to facilitate subsequent mutation mapping, secondary (“segment–mutant”) libraries were generated where only a specific portion of the rRNA operon would carry mutations. A 1,103-bp-long KpnI–ApaI segment of mutant pLK45 containing the 5′ external transcribed spacer of rrnB and 930 nucleotides of the 5′ portion of the 16S rRNA gene (library A) or a 793-bp-long ApaI–XbaI fragment containing the remaining 3′ segment of the 16S rRNA gene and a part of the 16S-23S intergenic spacer (library B) was introduced into otherwise WT pLK45 by restriction fragment exchange. Each of the segment–mutant libraries contained ≈5 × 104 clones. To increase representation of deleterious mutations, the libraries were subjected to one round of negative selection, which raised the frequency of clones with deleterious mutations to ≈8%.
Next, 12,000 individual clones of segment–mutant library A and 8,000 clones of library B were individually tested in 384-well plates for their ability to grow at 30°C (noninduced conditions) or 42°C (induced). About 400 clones that grew poorly at 42°C were selected from each library, and, after retesting the phenotypes, the mutated segments of the plasmid-borne rrnB were sequenced in a total of 200 clones (Fig. 1; see also Table 1, which is published as supporting information on the PNAS web site). The majority of the sequenced clones contained individual point mutations. Some of the mutations were repeatedly found in several independent clones, whereas others were represented in only one sequenced clone. Clones that contained more than one mutation were excluded from further analysis.
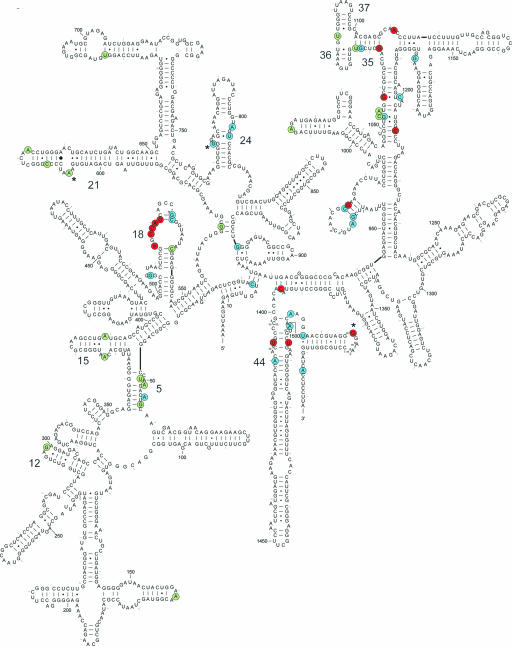
Fig. 1.
Dominant deleterious mutations in E. coli 16S rRNA selected from a random mutant library. Mutant positions are colored according to the severity of the phenotype: red, strongly deleterious; blue, moderately deleterious; green, mildly deleterious. Single nucleotide deletions within a stretch of identical nucleotides are marked by asterisks. The relevant helices of 16S rRNA are marked by their numbers according to ref. 31. The 16S rRNA secondary structure (38) was retrieved (26) and simplified for clarity of presentation.
Deleterious Mutations Identified in 16S rRNA. A total of 53 individual point mutations were identified after screening the segment–mutant libraries that span the entire length of the 16S rRNA gene. Even though the adjacent transcribed spacers of rrnB accounted for ≈20% of the combined mutagenized segments represented in two segment–mutant libraries, deleterious mutations were confined exclusively to the mature 16S rRNA sequence (Fig. 1). Of the 53 point mutations, 50 were base substitutions and 3 were single-base deletions.
To rank the mutations, the severity of deleterious phenotypes was assessed by using the transformation assay. In POP2136 cells, 16S rRNA transcribed from the pLK45 plasmid at 42°C accounts for 40–60% of the cellular rRNA. Dominant mutations that severely impair protein synthesis prevent colony formation upon transforming the corresponding plasmids into POP2136 cells and incubating the plates at 42°C. E. coli JM109 cells lack the λ repressor altogether; as a result, pLK45-encoded 16S rRNA accumulates to up to 85% of the total cellular 16S rRNA. Consequently, even moderately deleterious dominant rRNA mutations are expected to impair formation of JM109 colonies. Thus, according to their ability to transform POP2136 cells (at 42°C) or JM109 cells (at 37°C), all of the mutations were grouped into three major classes. The first class (red in Fig. 1 and in Movie 1, which is published as supporting information on the PNAS web site) included 14 strongly deleterious mutations that failed to transform either POP2136 or JM109 cells. Plasmids carrying any of the 21 moderately deleterious mutations (blue in Fig. 1) could transform the POP2136 strain but would prevent colony formation in JM109 cells. Finally, 18 mutations of the third class, mildly deleterious (green in Fig. 1), slowed the growth of POP2136 cells in liquid culture at 42°C but did not prevent colony formation in POP2136 or JM109 cells.
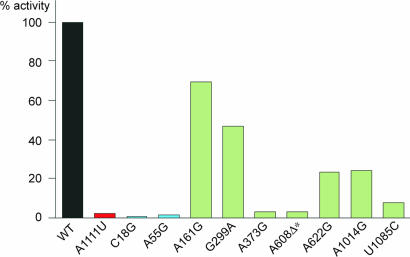
Effect of Selected rRNA Mutations on Protein Synthesis. To verify that selected rRNA mutations prevent small ribosomal subunits from participating in protein synthesis, 10 individual mutations were tested in a specialized ribosome system (17, 22). These mutations were engineered in plasmid pKF207, which codes for the 16S rRNA gene with an altered anti-Shine–Dalgarno sequence (GGGGU), and mutant 16S rRNAs were expressed in E. coli strain KLF10, which carries a chromosomal copy of the β-galactosidase gene (lacZ) with a ribosome binding site (AUCCC) recognized by pKF207-encoded 16S rRNA (18). The 30S subunits assembled with the plasmid-encoded 16S rRNA translate only lacZ but not other cellular genes. Therefore, mutations in the 16S rRNA gene in pKF207 do not affect cell growth, whereas the level of β-galactosidase activity reflects the capacity of mutant 16S rRNA to support protein synthesis. In comparison with the control 16S rRNA, which carried only alterations in the Shine–Dalgarno region, all of the mutations engineered in the pKF207 16S rRNA gene reduced reporter expression from ≈2- to 100-fold, confirming that the selected mutations interfered with protein synthesis (Fig. 2). In good correlation with the severity of the phenotype, highly and moderately deleterious mutations (A1111U, C18G, and A55G) dramatically reduced expression of the reporter, whereas mildly deleterious mutations had a more variable effect that ranged from intermediate (A161G) to strong (A373G) inhibition of translation.
Fig. 2.
Protein synthesis activity of individual 16S rRNA mutants in the specialized ribosome system. Activity of the LacZ reporter (Miller units) in cells expressing 16S rRNA that carried altered anti-Shine–Dalgarno region but no other mutations (WT) was taken as 100%. Bars representing protein synthesis activity of individual mutants are colored according to the scheme used in Fig. 1: red, strongly deleterious; blue, moderately deleterious; green, mildly deleterious.
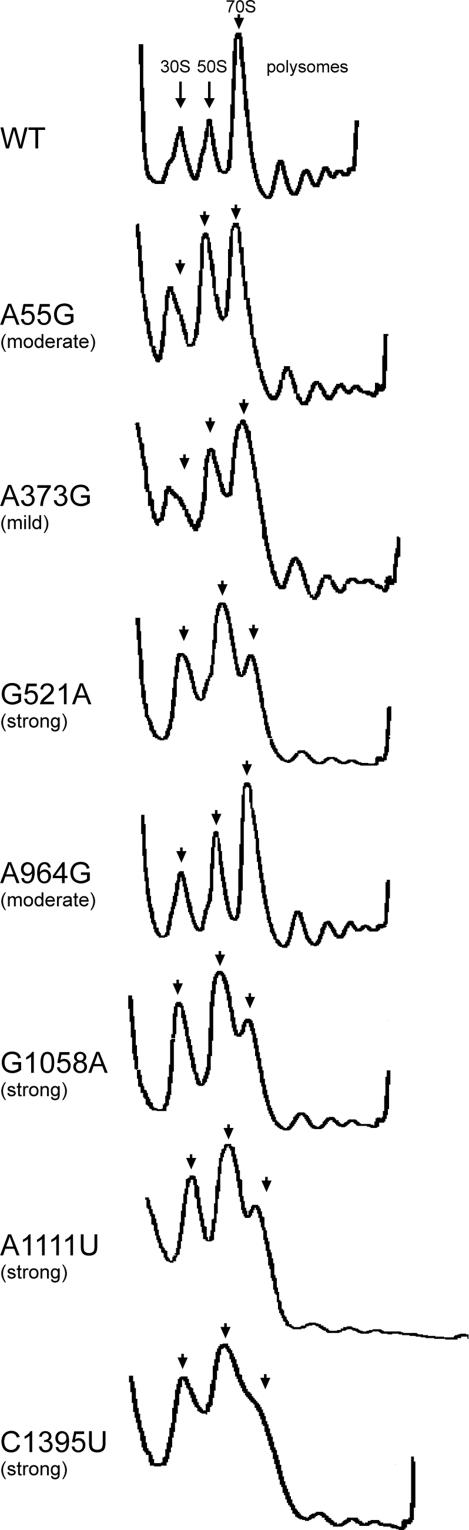
The 16S rRNA mutations can interfere with protein synthesis either directly (by affecting the structure and function of the small ribosomal subunit) or indirectly (by disrupting the rRNA sites critical for the subunit assembly). Both possibilities open up interesting opportunities for development of protein synthesis inhibitors. To understand the general trend of the mode of action of deleterious mutations, we analyzed polysome profiles in several selected POP2136 clones expressing strongly, moderately, or mildly deleterious mutations (Fig. 3). In the only analyzed mildly deleterious mutant, A373G, and in one moderately deleterious mutant, A55G, the accumulation of material that sedimented at around 25S and likely represented the aberrant or stalled assembly complexes was clearly seen. Appearance of a slowly sedimenting material was accompanied by a decreased abundance of 70S ribosomes as compared with free subunits. Primer extension analysis of the A373G mutant showed that mutant 16S rRNA was prevalent in the material sedimenting at 25S (93%), was reduced in the 30S peak (61%), and was notably underrepresented in peaks of 70S ribosomes and polysomes (27% and 22%, respectively), indicating that perturbed ribosome assembly is the primary cause of inhibition of translation and cell growth in these mutants. None of the other five strongly or moderately deleterious mutants that were tested (G521A, A964G, G1058A, A1111U, and C1395U) showed any indication of assembly defects. However, all of these mutants showed a marked decrease in the amounts of 70S ribosomes and polysomes relative to free subunits, which is compatible with the idea that the mutations affect functions of the 30S subunits in translation. Thus, it appears that the majority of the identified strongly deleterious and probably moderately deleterious 16S rRNA mutations are associated with functional defects, whereas a smaller number of the mutations may interfere with ribosome assembly.
Fig. 3.
Sucrose gradient profiles of polysomes prepared from cells transformed with WT or mutant pLK45 plasmids. Positions of 30S subunits, 50S subunits, and 70S ribosomes are indicated by arrowheads. The severity of deleterious phenotype conferred by the mutation (strong, moderate, or mild) is indicated.
Discussion
The main goal of this work was to map a variety of functionally important sites in the rRNA of the small ribosomal subunit that represent potential antibiotic targets. By mapping deleterious mutations in E. coli 16S rRNA, we have identified rRNA sites that are critical for efficient translation and as such could be targeted by antibiotics.
If our main concept, that deleterious rRNA mutations coincide with the potential sites of antibiotic action, is correct, then at least some of the mutations selected from a random library should fall close to the sites of action of known drugs. Indeed, 12 of 53 of the deleterious mutations identified in 16S rRNA clustered around the sites targeted by well characterized antibiotics such as aminoglycosides, tetracycline, or streptomycin (Fig. 4, which is published as supporting information on the PNAS web site) (23–25), thus validating our approach. The rest of the mapped mutations were in the rRNA regions not targeted by the known drugs and, thus, could be used to identify new antibiotic targets.
Our experiments were designed to isolate rRNA mutations whose effects would resemble those of an antibiotic bound to the corresponding rRNA sites. When cells are treated with antibiotics, only a certain fraction of ribosomes carries the drug. Potent antibiotics should effectively inhibit protein synthesis, even when a relatively large fraction of ribosomes remains drug-free. The experimental system we used mimicked this situation because, in POP2136 cells transformed with the pLK45 plasmid, mutant ribosomes accounted for approximately one-half of the ribosomal population. Therefore, the isolated dominant mutations identify ribosomal sites whose distortion diminished cellular translation despite the presence of WT ribosomes. Accordingly, we expect that cells will remain sensitive to the drugs targeted against identified sites even if some of multiple rrn alleles in the cell acquire resistance mutations.
Deleterious mutations that were identified in our screening highlight functionally important nucleotides in rRNA. Direct involvement in the function should lead to evolutionary conservation of an rRNA residue. Indeed, the majority of deleterious mutations (48 of 53) are at the nucleotides that show >98% conservation in bacterial 16S rRNA. Therefore, identified nucleotide residues critical in the E. coli ribosome may be functionally important in other bacteria as well and could potentially represent targets for broad-spectrum antibiotics. It should be emphasized, however, that, in the absence of experimental data, a mere conservation of a nucleotide is a relatively weak predictor of the extent of its functional importance. More than 600 positions in 16S rRNA exhibit 98% or more conservation (26). A number of mutations engineered at conserved rRNA sites, including those that show conservation across the evolutionary domains, had only weak or even no growth defects (27). In addition, 42 of the positions that we identified are conserved in the human cytoplasmic ribosomes, which raises the question of whether drugs targeted against such sites could be selective. However, as the structures of the ribosome–drug complexes show, antibiotics form multiple contacts with a number of rRNA residues, and conservation of one or even several nucleotides is not sufficient to eliminate drug selectivity. As an example, peptidyl transferase-targeting antibiotics, many of which show excellent selectivity, act on the ribosomal site, which includes many universally conserved nucleotides.
The deleterious mutations were unevenly distributed in the structure of the 30S subunit. Although extensive areas of the subunit were virtually mutation-free, several rRNA sites were characterized by clustering of the mutations. Many of the moderate and, even more so, highly deleterious mutations clustered at the functionally charged interface side of the subunit, generally following the path of mRNA and coinciding with the sites of action of several known antibiotics (28–30) (see Movie 1). A number of these mutations were localized within known functional sites involved in interactions with tRNA and mRNA, accuracy control, or other ribosome activities (Fig. 5, which is published as supporting information on the PNAS web site). Importantly, however, a number of mutations clustered in several regions of less obvious functional significance, regions that can be viewed as putative new antibiotic targets. We identified four such regions. One region, which included mutations in helices 5 (U49C, A51G, A55G, and G57A) and 15 (A373G and A389G), was located at the lower part of the interface side of the subunit (Fig. 6, which is published as supporting information on the PNAS web site). Although the majority of mutations in this cluster produced only a mild deleterious effect in POP2136 cells (Fig. 1), two tested mutations, A55G and A373G, almost completely precluded translation of the reporter protein in the specialized ribosome system (Fig. 2). This seeming controversy is easily reconciled by the observation that A55G and A373G mutations interfere with assembly of 30S subunits. A reduced rate of assembly of the plasmid-encoded 30S subunit may only marginally decrease the total amount of functional ribosomes in the cell when chromosome-encoded WT ribosomes are present, but it may entirely eliminate translation of the reporter, which entirely relies on the plasmid-encoded 16S rRNA. Our data implicate this otherwise unremarkable region of the ribosome as an important player in ribosome biogenesis and underscores it as one of the putative antibiotic targets in the ribosome.
Three mutations, transitions A802G and U804C and a deletion of one of four Gs (773–776), are located in close vicinity of each other in the middle portion of helix 24, which constitutes a part of the intersubunit bridge B7b (31) (Fig. 7, which is published as supporting information on the PNAS web site). This region, which upon subunit association makes a contact with protein L2, was proposed to be part of a signal pathway linking the decoding center of the small ribosomal subunit with the catalytic center of the large subunit (31). Clustering of moderately deleterious mutations in this part of helix 24 strongly supports its functional significance. The rRNA mutations, as well as the likely binding of small organic molecules at the internal loop of helix 24, may also affect the overall structure of the hairpin, including its apex loop, which may contribute to the binding of tRNA in P and E sites, subunit association, and translation initiation (32, 33).
Mutations in helix 21 (C614A, A622G, and deletion of one A in a triple-A cluster 607–609) and the G299A mutation in helix 12 converge at the back of the 30S subunit (Fig. 8, which is published as supporting information on the PNAS web site). These deleterious mutations are located in close proximity to the ribosomal proteins S4 and S16, whereas the bottom part of helix 21 interacts with ribosomal protein S8. Proteins S4 and S8 are among the primary assembly proteins (34–36). Thus, similar to the earlier discussed site comprising helices 5 and 15, which might be involved in subunit assembly, the drugs targeted against this site are expected to interfere with formation of functional 30S subunits.
The fourth rRNA site comprises elements of helices 35–37 of the 3′ major domain of 16S rRNA (mutations G1068A, G1072A, U1073C, U1085C, and A1111U) (Fig. 9, which is published as supporting information on the PNAS web site). This site is located on the back (solvent) side of the neck of the subunit substantially far from the known functional centers that occupy the interface side. Finding mutations with a strong deleterious effect (G1068A and A1111U) here was unexpected. One of the mutations that was studied in more detail, A1111U, did not interfere with the subunit assembly but dramatically reduced the fraction of plasmid-encoded 16S rRNA in 70S ribosomes and polysomes (Fig. 3), which indicates severe functional defects associated with this mutation. This conclusion is supported by the inability of mutant 30S subunits to translate the reporter mRNA in the specialized ribosome system (Fig. 2). Accordingly, targeting antibiotics toward this rRNA region is expected to strongly inhibit translation.
How complete is the set of deleterious mutations in 16S rRNA that we have identified? The complexity of the segment–mutant libraries (≈50,000 clones) suggests good coverage of the mutation sequence space. However, a notable bias in favor of transitions versus transversions during in vivo mutagenesis as well as the use of negative selection, which was essential for the success of the project, should inevitably reduce the library complexity. Our collection of point mutations includes 6 of 22 deleterious mutations that were previously engineered or selected in E. coli 16S rRNA by using plasmid systems similar to the one used in our experiments (Fig. 6) (see ref. 11 and K. L. Triman, personal communication). Thus, we estimate that ≈30% of all deleterious mutations in 16S rRNA were obtained in this study. Because deleterious mutations tend to cluster in rRNA sites of high functional significance, we believe that we have found most of the functional sites, despite the fact that our screen was not saturated. Consistent with this assertion, we obtained at least one mutation in all of the sites specified by the 22 deleterious mutations isolated previously.
The resident mutations present in plasmid pLK45 (C1192U in 16S rRNA and A2058G in 23S rRNA) were commonly considered to be silent (13). However, a recent report indicated that they may exhibit synthetic lethality when combined with specific other mutations (37). Although we have not tested all of the selected mutations in segregation from the resident pLK45 mutations, 10 of the individual deleterious mutations tested in the specialized ribosome system showed severe defects in translation, and several other mutations from our collection were previously individually engineered in 16S rRNA and shown to inhibit cell growth (Table 1). Therefore, we believe that the majority of the mutations we isolated are genuinely deleterious.
In conclusion, we have isolated an extended collection of deleterious mutations in bacterial 16S rRNA. Clusters of mutations uncovered putative functional regions of the ribosome unrecognized previously. Investigation of these regions may reveal unexpected ribosomal activities and aid in the development of new antibiotics.
Supplementary Material
Acknowledgments
We thank Dr. Robyn Hickerson for preparing the movie and providing valuable advice and help, Sonia Larsen for help in the early stages of the project, and Shannon Foley for help in preparing the manuscript. A.S.M. thanks Drs. Harry Noller and Albion Baucom for advice and help during his sabbatical stay at the University of California, Santa Cruz. This work was supported by National Institutes of Health Grant U19 AI56575 (to A.S.M.).
Author contributions: A.Y. performed research; A.S.M. designed research; K.F. contributed new reagents/analytic tools; and A.Y. and A.S.M. analyzed data and wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviation: rRNA, ribosomal RNA.
References
- 1.Ramakrishnan, V. (2002) Cell 108, 557–572. [DOI] [PubMed] [Google Scholar]
- 2.Cundliffe, E. (1990) in The Ribosome: Structure, Function, and Evolution, eds. Hill, W. E., Dahlberg, A., Garrett, R. A., Moore, P. B., Schlessinger, D. & Warner, J. R. (Am. Soc. for Microbiology, Washington, DC), pp. 479–490.
- 3.Sigmund, C. D., Ettayebi, M., Borden, A. & Morgan, E. A. (1988) Methods Enzymol. 164, 673–690. [DOI] [PubMed] [Google Scholar]
- 4.Farrell, D. J., Douthwaite, S., Morrissey, I., Bakker, S., Poehlsgaard, J., Jakobsen, L. & Felmingham, D. (2003) Antimicrob. Agents Chemother. 47, 1777–1783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wright, G. D. (2003) Curr. Opin. Chem. Biol. 7, 563–569. [DOI] [PubMed] [Google Scholar]
- 6.Cundliffe, E. (1987) Biochimie 69, 863–869. [DOI] [PubMed] [Google Scholar]
- 7.Garrett, R. A. & Rodriguez-Fonseca, C. (1996) in Ribosomal RNA: Structure, Evolution, Processing, and Function in Protein Biosynthesis, eds. Zimmermann, R. A. & Dahlberg, A. E. (CRC, Boca Raton, FL), pp. 327–355.
- 8.Adrian, P. V., Mendrick, C., Loebenberg, D., McNicholas, P., Shaw, K. J., Klugman, K. P., Hare, R. S. & Black, T. A. (2000) Antimicrob. Agents Chemother. 44, 3101–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Belova, L., Tenson, T., Xiong, L., McNicholas, P. M. & Mankin, A. S. (2001) Proc. Natl. Acad. Sci. USA 98, 3726–3731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Laios, E., Waddington, M., Saraiya, A. A., Baker, K. A., O'Connor, E., Pamarathy, D. & Cunningham, P. R. (2004) Arch. Pathol. Lab. Med. 128, 1351–1359. [DOI] [PubMed] [Google Scholar]
- 11.Triman, K. L. & Adams, B. J. (1997) Nucleic Acids Res. 25, 188–191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Remaut, E., Stanssens, P. & Fiers, W. (1981) Gene 15, 81–93. [DOI] [PubMed] [Google Scholar]
- 13.Powers, T. & Noller, H. F. (1990) Proc. Natl. Acad. Sci. USA 87, 1042–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rottmann, N., Kleuvers, B., Atmadja, J. & Wagner, R. (1988) Eur. J. Biochem. 177, 81–90. [DOI] [PubMed] [Google Scholar]
- 15.Tenson, T., Herrera, J. V., Kloss, P., Guarneros, G. & Mankin, A. S. (1999) J. Bacteriol. 181, 1617–1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yanisch-Perron, C., Vieira, J. & Messing, J. (1985) Gene 33, 103–119. [DOI] [PubMed] [Google Scholar]
- 17.Lee, K., Holland-Staley, C. A. & Cunningham, P. R. (1996) RNA 2, 1270–1285. [PMC free article] [PubMed] [Google Scholar]
- 18.Abdi, N. N. & Fredrick, K. (2005) RNA 11, 1624–1632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller, J. H. (1992) A Short Course in Bacterial Genetics: Laboratory Manual (Cold Spring Harbor Lab. Press, Woodbury, NY).
- 20.Ron, E. Z., Kohler, R. E. & Davis, B. D. (1966) Science 153, 1119–1120. [DOI] [PubMed] [Google Scholar]
- 21.Greener, A., Callahan, M. & Jerpseth, B. (1997) Mol. Biotechnol. 7, 189–195. [DOI] [PubMed] [Google Scholar]
- 22.Hui, A. & de Boer, H. A. (1987) Proc. Natl. Acad. Sci. USA 84, 4762–4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Carter, A. P., Clemons, W. M., Brodersen, D. E., Morgan-Warren, R. J., Wimberly, B. T. & Ramakrishnan, V. (2000) Nature 407, 340–348. [DOI] [PubMed] [Google Scholar]
- 24.Pioletti, M., Schlunzen, F., Harms, J., Zarivach, R., Gluhmann, M., Avila, H., Bashan, A., Bartels, H., Auerbach, T., Jacobi, C., et al. (2001) EMBO J. 20, 1829–1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brodersen, D. E., Carter, A. P., Clemons, W. M., Jr., Morgan-Warren, R. J., Murphy, F. V., Ogle, J. M., Tarry, M. J., Wimberly, B. T. & Ramakrishnan, V. (2001) Cold Spring Harbor Symp. Quant. Biol. 66, 17–32. [DOI] [PubMed] [Google Scholar]
- 26.Cannone, J. J., Subramanian, S., Schnare, M. N., Collett, J. R., D'Souza, L. M., Du, Y., Feng, B., Lin, N., Madabusi, L. V., Muller, K. M., et al. (2002) BMC Bioinformatics 3, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Triman, K. L. (1995) Adv. Genet. 33, 1–39. [DOI] [PubMed] [Google Scholar]
- 28.Wimberly, B. T., Brodersen, D. E., Clemons, W. M., Jr., Morgan-Warren, R. J., Carter, A. P., Vonrhein, C., Hartsch, T. & Ramakrishnan, V. (2000) Nature 407, 327–339. [DOI] [PubMed] [Google Scholar]
- 29.Schluenzen, F., Tocilj, A., Zarivach, R., Harms, J., Gluehmann, M., Janell, D., Bashan, A., Bartels, H., Agmon, I., Franceschi, F. & Yonath, A. (2000) Cell 102, 615–623. [DOI] [PubMed] [Google Scholar]
- 30.Yusupova, G. Z., Yusupov, M. M., Cate, J. H. & Noller, H. F. (2001) Cell 106, 233–241. [DOI] [PubMed] [Google Scholar]
- 31.Yusupov, M. M., Yusupova, G. Z., Baucom, A., Lieberman, K., Earnest, T. N., Cate, J. H. & Noller, H. F. (2001) Science 292, 883–896. [DOI] [PubMed] [Google Scholar]
- 32.Tapprich, W. E., Goss, D. J. & Dahlberg, A. E. (1989) Proc. Natl. Acad. Sci. USA 86, 4927–4931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee, K. S., Varma, S., SantaLucia, J., Jr., & Cunningham, P. R. (1997) J. Mol. Biol. 269, 732–743. [DOI] [PubMed] [Google Scholar]
- 34.Mizushima, S. & Nomura, M. (1970) Nature 226, 1214–1218. [DOI] [PubMed] [Google Scholar]
- 35.Jagannathan, I. & Culver, G. M. (2003) J. Mol. Biol. 330, 373–383. [DOI] [PubMed] [Google Scholar]
- 36.Gregory, R. J. & Zimmermann, R. A. (1986) Nucleic Acids Res. 14, 5761–5776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rodriguez-Correa, D. & Dahlberg, A. E. (2004) RNA 10, 28–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Woese, C. R., Magrum, L. J., Gupta, R., Siegel, R. B., Stahl, D. A., Kop, J., Crawford, N., Brosius, J., Gutell, R., Hogan, J. J. & Noller, H. F. (1980) Nucleic Acids Res. 8, 2275–2293. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.