Abstract
The regulation of glycogen metabolism is critical for the maintenance of glucose and energy homeostasis in mammals. Glycogen synthase, the enzyme responsible for glycogen production, is regulated by multisite phosphorylation in yeast and mammals. We have previously identified PAS kinase as a physiological regulator of glycogen synthase in Saccharomyces cerevisiae. We provide evidence here that PAS kinase is an important regulator of mammalian glycogen synthase. Glycogen synthase is efficiently phosphorylated by PAS kinase in vitro at Ser-640, a known regulatory phosphosite. Efficient phosphorylation requires a region of PAS kinase outside the catalytic domain. This region appears to mediate a direct interaction between glycogen synthase and PAS kinase, thereby targeting kinase activity to this substrate specifically. This interaction is regulated by the PAS kinase PAS domain, raising the possibility that this interaction (and phosphorylation event) is modulated by the cellular metabolic state. This mode of regulation provides a mechanism for metabolic status to impinge directly on the cellular decision of whether to store or use available energy.
Keywords: phosphorylation
Glycogen, a large branched polymer of glucose, acts as a reserve of carbon and energy in a variety of organisms. In mammals, the most important stores are found in the liver and skeletal muscle (1). Liver glycogen is required to efficiently buffer blood glucose levels during fasting, whereas muscle glycogen is primarily used locally as a fuel for muscle contraction (2). Dysregulation of glycogen metabolism has been implicated in the development of many diseases, including type 2 diabetes mellitus (3, 4).
The synthesis of glycogen is primarily controlled through regulation of the enzyme glycogen synthase, which catalyzes bulk glycogen synthesis (5–7). The muscle isoform of glycogen synthase is inactivated by reversible phosphorylation that occurs at nine distinct sites within the enzyme, but full activity can be restored in the presence of the allosteric activator, glucose-6-phosphate (G6P) (8–10). In rabbit muscle glycogen synthase, the best characterized form of the enzyme, the phosphorylation sites are clustered at the N and C termini (Fig. 6, which is published as supporting information on the PNAS web site). The N-terminal sites Ser-7 (site 2) and Ser-10 (site 2a) and the C-terminal sites Ser-640 (site 3a) and Ser-644 (site 3b) appear to be the most important regulatory sites (11–13). Phosphorylase kinase and casein kinase I are likely physiological kinases for Ser-7 and Ser-10, respectively (12, 13). Glycogen synthase kinase-3 (GSK-3) phosphorylates the C-terminal regulatory sites and, in vitro, catalyzes sequential phosphorylation of sites 4, 3c, 3b, and 3a (14, 15). GSK-3, however, is not the sole kinase that phosphorylates C-terminal regulatory sites, and, as described below, GSK-3-independent mechanisms must also exist. Glycogen synthase expressed in COS cells or Rat1 fibroblasts can be efficiently phosphorylated and inactivated even when Ser-7 and Ser-10 are mutated to alanine, and the GSK-3 recognition motif is disrupted by serine-to-alanine substitutions at sites 3c, 4, and/or 5 (16). These serine-to-alanine substitutions block GSK-3-mediated phosphorylation of the important regulatory sites Ser-640 and Ser-644. The fact that the mutant enzyme could still be phosphorylated at Ser-640 and/or Ser-644 suggests that Ser-640 and/or Ser-644 kinases other than GSK-3 must exist. We have since purified a specific Ser-640 kinase and shown that this preparation contained a member of the dual-specificity tyrosine-phosphorylated and -regulated kinase (DYRK) family, DYRK1A, and a WD-40 repeat domain protein, Han11 (17).
Glycogen synthase from the budding yeast Saccharomyces cerevisiae is regulated by phosphorylation in a manner analogous to the mammalian enzyme (18, 19). We recently identified a pair of parologous protein kinases that are genetically required for maintenance of normal glycogen stores in S. cerevisiae (20). These kinases, named PSK1 and PSK2, are members of a family of PAS domain-containing serine/threonine kinases. This family includes orthologous fly, mouse, and human PAS kinase (PASK) genes (21, 22). The PAS domain of PASK appears to have a regulatory function, because deletion or inactivation of this domain increases kinase activity toward all substrates studied (21, 23). PAS domains frequently bind small-molecule ligands or cofactors [such as heme, flavin mononucleotide, or dioxin (24)], which serve regulatory roles for PAS domain function. NMR analysis of the PASK PAS domain shows that it is also capable of binding specific small ligands that elicit a conformational change in the PAS domain (23). Similar conformational changes are believed to modulate histidine kinase activity in the well studied FixL/FixJ oxygen sensor of Rhizobia sp. (25). A model has been proposed whereby the PAS domain of PASK would bind some ligand, perhaps a small metabolite, thereby regulating kinase activity (26).
Genetic and proteomic screens using yeast PASK identified a number of substrates and implicated this kinase in the regulation of carbohydrate metabolism and translation (20). We found that a yeast strain lacking the PSK1 and PSK2 genes accumulates ≈5- to 10-fold excess glycogen relative to a wild-type strain. We also showed that yeast PASK phosphorylates glycogen synthase in vitro and that strains lacking the PASK genes had elevated glycogen synthase activity, consistent with impaired ability to phosphorylate glycogen synthase in vivo (20). Because glycogen synthesis and translation are two processes tightly regulated in response to nutrient availability and because PAS domains are frequently involved in metabolic sensing, a role for PASK in the cellular response to metabolic status was proposed. Indeed, it was recently demonstrated that mammalian PASK plays a role in the cellular response to nutrients. The catalytic activity of PASK in pancreatic islet β-cells is rapidly increased in response to glucose addition, and PASK is required for the glucose-responsive expression of some β-cell genes, including preproinsulin (27).
In the present work, we provide further evidence of a role for mammalian PASK in metabolic control by implicating this enzyme in the control of glycogen synthase activity. Recombinant human PASK (hPASK) phosphorylates purified muscle glycogen synthase, causing robust inactivation. Furthermore, hPASK interacts directly with glycogen synthase when expressed in cultured cells and this interaction and the phosphorylation of glycogen synthase by human PASK (hPASK) are inhibited by glycogen. We propose that PASK is a regulator of mammalian glycogen synthesis.
Materials and Methods
Isolation of Enzymes. Purification of the G6P-independent form (I-form) and the G6P-dependent form (D-form) of rabbit muscle glycogen synthase. The largely dephosphorylated (I-form) and phosphorylated (D-form) of glycogen synthase were prepared from rabbit skeletal muscle as described by de Paoli-Roach et al. (28).
Expression and purification of full-length and truncated hPASK. The expression and purification of Δ955 and full-length hPASK has been described in ref. 21. The Δ444 variant of hPASK was expressed and purified identically. In short, His6-tagged hPASK was expressed in Sf-9 cells by using recombinant adenovirus and purified by using Ni-NTA and MonoQ chromatography.
Expression and purification of DYRK2. Human DYRK2 was expressed in Escherichia coli as a glutatione S-transferase fusion protein from the vector pGEX-4T1 (Amersham Biosciences, which is now GE Healthcare) and purified over glutathione Sepharose as described in ref. 17.
Phosphorylation and Inactivation of Glycogen Synthase. Recombinant hPASK (≈0.15 μg) was incubated for 30 min at 30°C with 200 μM ATP/5 mM MgCl2/1 mM DTT/50 mM Tris·HCl, pH 7.4, to allow autophosphorylation and activation of hPASK. Duplicate reactions were set up, one reaction receiving [γ-32P]ATP (3,000–4,000 dpm/pmol) and the other receiving unlabeled ATP. Purified rabbit muscle glycogen synthase (2 μg, I-form) was then added, bringing the final incubation volume to 50 μl. The final glycogen synthase concentration was ≈0.5 μM, and the hPASK concentration was ≈20 pM. At intervals, 5-μl aliquots were mixed with either 160 μl of stop mix [50 mM Tris·HCl/25 mM KF/20 mM EDTA/1 mM DTT/1% (wt/vol) glycogen, pH 7.8], in the case of the reactions with unlabeled ATP, or with 20 μl of SDS-PAGE sample buffer containing 20 mM EDTA, for reactions with [γ-32P]ATP. Aliquots (30 μl) from the reactions terminated in stop mix were assayed for glycogen synthase activity as described (11), both in the presence and in the absence of G6P. The phosphorylation stoichiometry was determined by SDS/PAGE analysis of the reactions terminated with SDS/PAGE sample buffer. The band corresponding to glycogen synthase was excised from the gel, and the radioactivity incorporated was determined with a liquid scintillation counter.
Phosphorylation of Glycogen Synthase and Uridine-Cytidine Kinase 2 (UCK2) by hPASK and the Δ955-hPASK and Δ444-hPASK Truncation Mutants in the Presence and Absence of Glycogen and Phosphorylation of Histone and Phosphorylation Site Mapping. Phosphorylation of glycogen synthase and UCK2 was carried out essentially as described above, except that the reactions all contained [γ-32P]ATP and were terminated by addition of SDS/PAGE sample buffer. UCK2 and glycogen synthase were used at 0.5 μM in the phosphorylation reaction, and hPASK and the Δ444 and Δ955 truncation mutants were present at 20 pM. Phosphorylation with DYRK2 was carried out as described in ref. 17, except that the glycogen synthase concentration was 0.5 μM. Rabbit liver glycogen, when present, was added from an aqueous stock at the concentrations indicated (see Fig. 5). The extent of phosphorylation was determined by analysis of dried gels using a Bio-Rad GS-250 Molecular Imager.
Fig. 5.
Phosphorylation of glycogen synthase by hPASK is specifically inhibited by glycogen. (A) I-form glycogen synthase from rabbit muscle was subjected to phosphorylation in the presence of [γ-32P]ATP by wild-type hPASK, hPASK ΔN955, or hPASK ΔN444, or DYRK2 in the presence of the indicated glycogen concentrations. After incubation for 30 min at 30°C, samples were subjected to SDS/PAGE and autoradiography was used to determine phosphorylation stoichiometry. (B) As in A but with UCK2 as substrate.
For histone phosphorylation, histone type III (Sigma) was used at a concentration of 0.3 mg/ml, and reactions were terminated by spotting aliquots to P-81 phosphocellulose paper (Whatman) and washing with 1% (vol/vol) phosphoric acid. Blank reactions containing no histone were included to control for autophosphorylation of hPASK. The papers were dried under a heating lamp, and 32P incorporation was determined by liquid scintillation counting. The phosphorylated residue of glycogen synthase was identified as described in ref. 20. The identification of peptides by mass spectrometry was confirmed by automated Edman sequencing.
Results
PASK Phosphorylates and Inactivates the Muscle Isoform of Glycogen Synthase. We recently identified glycogen synthase as a phosphorylation substrate for PASK in the budding yeast Saccharomyces cerevisiae (20). This finding prompted us to consider whether mammalian glycogen synthase was likewise a physiological substrate of hPASK. Muscle glycogen synthase is inhibited by phosphorylation but is activated by binding of the allosteric activator, G6P. G6P has little effect on the catalytic rate of dephosphorylated glycogen synthase, but addition of sufficient G6P can overcome the inactivating effects of phosphorylation and restore nearly full activity to the phosphorylated enzyme. The activity of glycogen synthase measured in the absence of G6P relative to the activity measured in the presence of this compound is referred to as the activity ratio and serves as an index of the phosphorylation state of the enzyme. The phosphorylated and dephosphorylated forms of glycogen synthase have been referred to as the D- and I-forms, respectively (6).
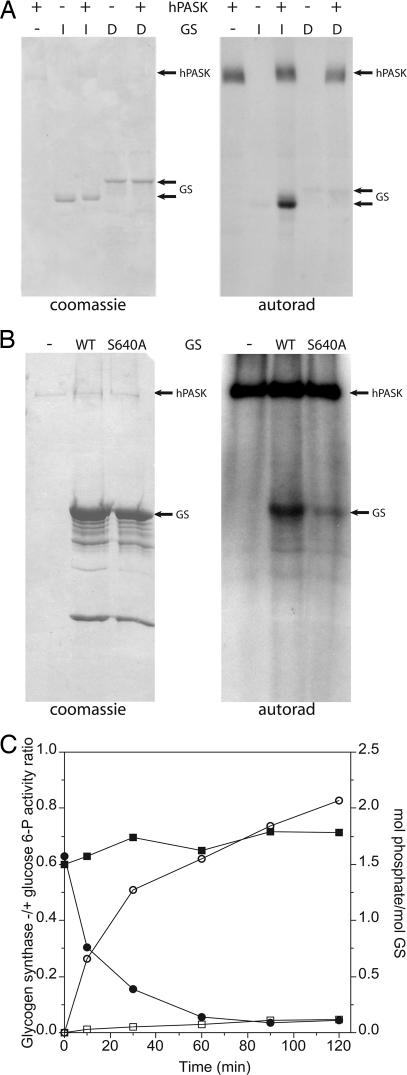
Glycogen synthase was purified from rabbit skeletal muscle as the D-form or I-form. This protein was incubated with [γ-32P]ATP with or without the addition of recombinant hPASK. Phosphorylation of the I-form but not the D-form was detected (Fig. 1A). The D-form is heavily phosphorylated at known regulatory sites (activity ratio of ≤0.1), indicating that hPASK was likely phosphorylating a physiologically relevant site in glycogen synthase available in the I-form but already occupied in the D-form.
Fig. 1.
hPASK phosphorylates rabbit muscle glycogen synthase (GS) at Ser-640. (A) I-form (dephosphorylated) or D-form (phosphorylated) glycogen synthase purified from rabbit muscle was incubated with [γ-32P]ATP and hPASK or buffer for 30 min at 30°C. The reactions were subjected to SDS/PAGE followed by staining with Coomassie brilliant blue (Left), or the gel was dried and subjected to autoradiography (Right). (B) hPASK was incubated with [γ-32P]ATP and buffer, wild-type glycogen synthase, or S640A glycogen synthase for 30 min at 30°C. Glycogen synthase was expressed and purified from recombinant E. coli. The reactions were subjected to SDS/PAGE followed by staining with Coomassie brilliant blue (Left), or the gel was dried and subjected to autoradiography (Right). (C) I-form glycogen synthase was subjected to phosphorylation by hPASK (open and filled circles) or buffer (open and filled squares) in the presence of ATP. At the times indicated, samples were subjected to SDS/PAGE for determining phosphorylation stoichiometry (open symbols) and glycogen synthase activity assay (filled symbols).
To determine which site(s) in glycogen synthase was being phosphorylated, we phosphorylated I-form glycogen synthase with hPASK. We subjected the phosphorylated protein to in-gel trypsinolysis followed by MALDI mass spectrometry. We detected a mass corresponding to the phosphorylated form of a peptide comprising residues 638–649 of glycogen synthase. This peptide contains two potential phosphorylation sites, Ser-640 and Ser-644. To determine which of these sites is phosphorylated by hPASK, we generated serine-to-alanine mutations at both sites by using PCR-based mutagenesis. These mutants and the wild-type protein were expressed and purified from E. coli. The S644A mutation had no effect on hPASK-dependent phosphorylation (data not shown). The S640A mutation, however, greatly reduced phosphorylation of glycogen synthase by hPASK, although some residual phosphorylation remained (Fig. 1B). To further confirm Ser-640 as a principle site of phosphorylation, we used a phospho-specific antibody that specifically recognizes glycogen synthase phosphorylated at this residue. Purified glycogen synthase (I-form) was incubated with or without hPASK. The reaction products were resolved by SDS/PAGE and analyzed by immunoblotting by using the phospho-specific antibody or by using antibody to total glycogen synthase protein (Fig. 7, which is published as supporting information on the PNAS web site). As expected, hPASK was shown to phosphorylate Ser-640. These data are further supported by analysis of hPASK-phosphorylated glycogen synthase by cyanogen bromide cleavage (Fig. 8, which is published as supporting information on the PNAS web site).
To better quantify the phosphorylation of glycogen synthase by hPASK and to determine the effects of this phosphorylation on enzyme activity, we performed the following experiment. Purified I-form glycogen synthase was incubated with ATP alone or with ATP plus hPASK. Parallel incubations were carried out by using [γ-32P]ATP to determine phosphorylation stoichiometry or unlabeled ATP to allow determination of the ratio of glycogen synthase activity –G6P to +G6P after phosphorylation. At intervals, aliquots were removed and resolved on an SDS/PAGE gel to determine phosphorylation stoichiometry or used in a glycogen synthase assay. After 120 min of incubation, hPASK had catalyzed robust inactivation of glycogen synthase while incorporating ≈2 mol of phosphate per mol of glycogen synthase (Fig. 1C). The low level of glycogen synthase phosphorylation seen in the absence of added hPASK did not affect activity and is the result of trace contamination of the enzyme preparation with phosphorylase kinase (29).
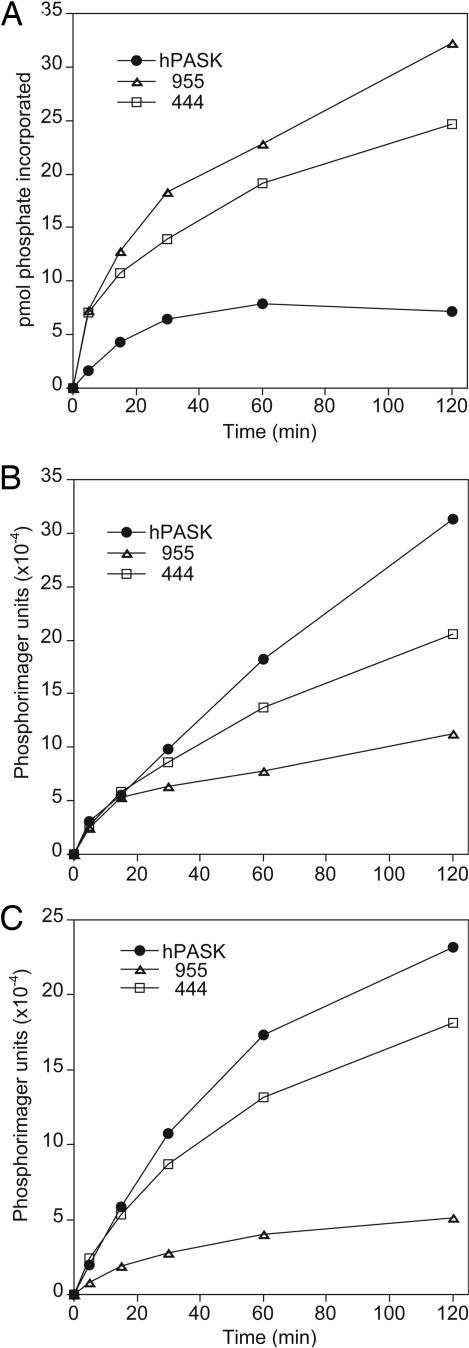
Efficient Phosphorylation of Substrates Requires the hPASK N Terminus. Deletion of the N-terminal noncatalytic region, including the PAS domain, has been shown to greatly increase the activity of hPASK toward nonphysiological peptide substrates (21, 23). Accordingly, we observed that such a deletion (Δ955) caused substantially increased hPASK activity toward histone, a nonphysiological hPASK substrate (Fig. 2A). Surprisingly, with glycogen synthase as the substrate, the Δ955 mutant exhibited substantially reduced kinase activity (Fig. 2B). Similarly, the phosphorylation of the human UCK2 protein, a recently identified substrate of hPASK (B.P. and S. L. McKnight, personal communication), was substantially more efficient when the full-length protein was used relative to the Δ955 mutant (Fig. 2C). We hypothesized that, although the PAS domain clearly inhibits catalytic activity, there might be some region of the hPASK N terminus that is required for the efficient phosphorylation of certain substrates. To test this hypothesis, we constructed a truncation mutant, Δ444, that lacks the PAS domain but retains the rest of the N-terminal sequence. The Δ444 mutant was more active toward glycogen synthase and UCK2 than was the Δ955 construct, consistent with the presence of a positive regulatory sequence. The Δ444 mutant was also more active toward histone than was the full-length hPASK, as expected by loss of the PAS domain. We interpret this result as implying that the hPASK midregion, the region between residues 444 and 955, is required for the efficient phosphorylation of some substrates, such as glycogen synthase and UCK2, but not important for the phosphorylation of others. This region contains no previously recognized structural or functional domains or sequences. It is interesting to note that the phosphorylation of all nonphysiological substrates we have tested to date is independent of this regulatory region.
Fig. 2.
Phosphorylation of glycogen synthase requires the hPASK midregion. Histone (type III) (A), I-form rabbit muscle glycogen synthase (B), or UCK2 (C) was subjected to phosphorylation in the presence of [γ-32P]ATP by wild-type hPASK, hPASK ΔN955, or hPASK ΔN444. After incubation for the time indicated, samples were subjected to SDS/PAGE and autoradiography was used to determine phosphorylation stoichiometry.
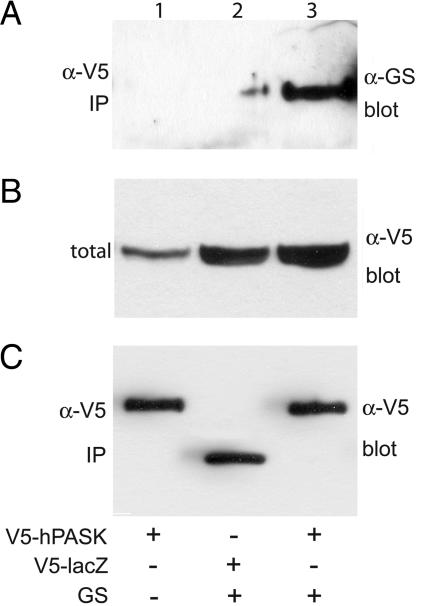
The hPASK/Glycogen Synthase Interaction Occurs in Cells and Is Negatively Regulated by the PAS Domain. The observation that efficient phosphorylation of glycogen synthase and UCK2 required the hPASK midregion led us to propose a role for this region in substrate recognition and binding. To address this issue, we introduced V5-tagged hPASK, wild-type muscle glycogen synthase, or both into HEK293 cells by cotransfection. The cells were cultured for an additional 24 h, followed by lysis and immunoprecipitation with anti-V5 antibody. As shown in Fig. 3, hPASK efficiently coimmunoprecipitates glycogen synthase.
Fig. 3.
hPASK and glycogen synthase (GS) interact in cultured cells. HEK293 cells were transfected as indicated with CMV-based vectors encoding V5-tagged, wild-type hPASK (lanes 1 and 3) or LacZ (lane 2) and either empty vector (lane 1) or glycogen synthase (lanes 2 and 3). After 24 h, the cells were harvested and subjected to immunoprecipitation (IP) with α-V5 antibody. Immunoprecipitates (A and C) and crude lysate (B) were analyzed by Western blotting with α-glycogen synthase antibody (A and B) or α-V5 antibody (C).
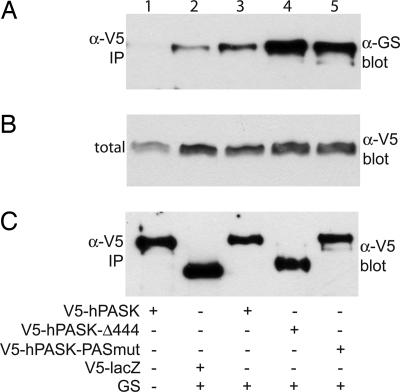
The one described domain in the N-terminal noncatalytic region of hPASK is the PAS domain, which has been shown to inhibit kinase activity (21, 23). We sought to determine the effect of this domain on the interaction between hPASK and glycogen synthase in cells. We functionally eliminated this domain in two ways. First, we used the Δ444 deletion mutant as described. Second, we generated a triple-point mutant of hPASK (LGY155–157EDI) that destroys the PAS domain fold (C. Amezcua and K. Gardner, personal communication). Elimination of PAS domain function by either method causes a marked increase in the interaction between hPASK and glycogen synthase (Fig. 4). It appears, therefore, that in addition to inhibiting kinase activity, the PAS domain also inhibits glycogen synthase binding by hPASK.
Fig. 4.
Interaction with glycogen synthase (GS) is inhibited by the hPASK PAS domain. HEK293 cells were transfected as indicated with CMV-based vectors encoding V5-tagged K1028R hPASK (behaves as wild-type in this assay) (lanes 1 and 3), LacZ (lane 2), hPASKΔN444 (lane 4), or hPASK LGY155–157EDI (lane 5) and either empty vector (lane 1) or glycogen synthase (lanes 2–5). After 24 h, the cells were harvested and subjected to immunoprecipitation (IP) with α-V5 antibody. Immunoprecipitates (A and C) and crude lysate (B) were analyzed by Western blotting with α-glycogen synthase antibody (A and B) or α-V5 antibody (C).
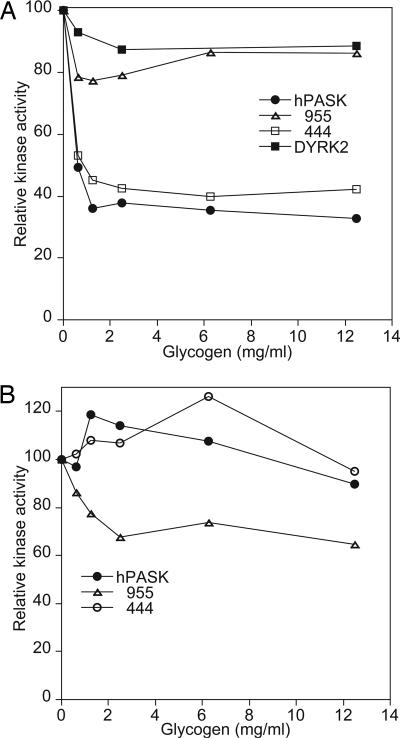
Glycogen Inhibits Glycogen Synthase Phosphorylation by hPASK. We noted that, although purified rabbit muscle glycogen synthase was an excellent substrate for hPASK, we could not achieve stoichiometric phosphorylation of recombinant muscle glycogen synthase expressed in E. coli (data not shown). Among other differences, the glycogen synthase purified from E. coli contains substantial glycogen, whereas the enzyme purified from rabbit muscle has minimal glycogen content. We hypothesized that this glycogen might be perturbing the ability of hPASK to phosphorylate glycogen synthase. To determine whether glycogen was inhibiting phosphorylation of recombinant glycogen synthase by hPASK, we titrated glycogen into the purified rabbit muscle enzyme preparation and measured the effect on phosphorylation. Addition of glycogen strongly inhibited the activity of hPASK toward rabbit muscle glycogen synthase (Fig. 5A) but had little effect on phosphorylation of the other known hPASK substrate, human UCK2 protein (Fig. 5B). The inhibition by glycogen occurred at low concentration (half-maximal effect at ≈35 μg/ml glycogen) and reached a plateau of ≈65% inhibition. Addition of higher concentrations of glycogen, up to >12 mg/ml, did not increase the extent of inhibition.
Glycogen synthase is known to bind glycogen, and we have been unable to detect an interaction between hPASK and glycogen. We reasoned, therefore, that the inhibitory effects of glycogen likely occur by virtue of its interaction with glycogen synthase. The simplest explanation would be that glycogen occludes the major phosphorylation site for hPASK, Ser-640. However, we found that recombinant DYRK2, which also phosphorylates Ser-640, was insensitive to the presence of glycogen (Fig. 4A). Furthermore, the Δ955 deletion mutant, which also predominantly phosphorylates Ser-640, is also insensitive to glycogen (Fig. 5A). In contrast, the Δ444 hPASK deletion mutant, which retains the midregion, is as sensitive to glycogen as the wild-type protein (Fig. 5A). In summary, glycogen inhibits glycogen synthase phosphorylation by hPASK, but not by occluding the principal phosphorylation site directly. Furthermore, only hPASK variants that contain an intact midregion and are thereby capable of stable glycogen synthase interaction are subject to this inhibition.
We noted that after 48 h of culture, the interaction observed between glycogen synthase and hPASK was greatly reduced and, at 72 h, was completely absent. When catalytically inactive glycogen synthase was expressed (point mutant P479Q) (30), the reduction in interaction with hPASK over time was not observed (data not shown). One interpretation of these data is that the interaction between hPASK and glycogen synthase was negatively regulated by glycogen. As time of culture increased, so did glycogen accumulation, resulting in decreased hPASK/glycogen synthase interaction. With the catalytically inactive glycogen synthase, no glycogen would be formed and the interaction between hPASK and glycogen synthase remained intact.
Discussion
We have identified glycogen synthase as an efficient hPASK phosphorylation substrate in vitro. We believe that the following evidence also supports a role for hPASK in directly phosphorylating glycogen synthase in vivo. First, the entire regulatory system is highly conserved. Glycogen synthase and the mechanism by which it is regulated is conserved from yeast to man (18, 31). PASK is similarly conserved, and genetic experiments have implicated PASK as a physiological regulator of glycogen synthase and glycogen accumulation in S. cerevisiae (20, 21). It is interesting to note that the exact site of PASK-dependent phosphorylation is similar but not identical in yeast and mammalian glycogen synthase. Mammalian PASK phosphorylates glycogen synthase at Ser-640, but yeast PASK phosphorylates glycogen synthase at the site analogous to Ser-644, four residues C-terminal (20). As will be discussed below, this finding suggests that the primary determinant of substrate specificity is not contained in the residues immediately surrounding the site of phosphorylation.
Second, hPASK phosphorylates glycogen synthase primarily at Ser-640, causing near complete inactivation. This site has been identified as one of the most important endogenous regulatory phosphorylation sites in mammalian glycogen synthase (16, 32). GSK-3 has long been implicated in the stepwise phosphorylation of four sites in the C terminus of glycogen synthase (10, 33, 34). Although GSK-3 is clearly a physiological kinase for the glycogen synthase C terminus, it has been demonstrated that GSK-3 is not required for Ser-640 phosphorylation in some cell types (11). We propose that hPASK is another Ser-640 kinase that is responsive to alternative stimuli. Unlike GSK-3, the activity of hPASK has been shown to be independent of insulin and probably regulated instead by a more direct metabolic signal (27).
Third, we have demonstrated that hPASK interacts specifically with glycogen synthase in cultured cells. The idea of the existence of such an interaction began when we observed that a hPASK mutant (Δ955) lacking the noncatalytic N terminus was unable to efficiently phosphorylate glycogen synthase. We speculated that there must be a region of the hPASK N terminus that acted to promote kinase activity, perhaps through direct interaction with glycogen synthase. Indeed, we found that the hPASK midregion (residues 444–955) is required for efficient phosphorylation of glycogen synthase in vitro and for interaction with glycogen synthase in cells. No function had previously been assigned to this region of hPASK. Although the hPASK midregion is required for efficient phosphorylation of glycogen synthase and another possible physiological substrate (UCK2), it is not required for the phosphorylation of generic, nonphysiological substrates, such as histones and synthetic peptides. We propose a model wherein the midregion of hPASK directly interacts with glycogen synthase, thereby targeting kinase activity to the substrate specifically. A similar substrate-targeting region has been discovered in many protein kinases (35–38).
Fourth, the interaction between the hPASK midregion and glycogen synthase is regulated. We have demonstrated that at least two factors play a major role in controlling the physical interaction between hPASK and glycogen synthase. The PAS domain of PAS kinase plays a negative role in regulating this interaction. If the PAS domain is deleted or disrupted, hPASK associates more stably with glycogen synthase. PAS domain function is usually controlled by the metabolic status of the host cell, as has been suggested for the PASK PAS domain (27). This observation raises the intriguing possibility that the hPASK–glycogen synthase interaction is regulated by the metabolic status of the cell, thereby enabling an additional layer of metabolic regulation of glycogen synthesis.
Glycogen negatively regulates the hPASK–glycogen synthase interaction, which would initially seem counterintuitive: Glycogen would thereby stimulate its own continued synthesis. It is possible, however, that this mechanism exists to spatially coordinate the synthesis of glycogen. It is becoming increasingly apparent that glycogen is synthesized in cells in a highly organized spatial pattern (39). Perhaps one function of hPASK is to maintain free, unlocalized glycogen synthase in a phosphorylated, inactive form until it is properly localized to an existing, properly organized glycogen particle. It is important to note that glycogen does not affect the ability of DYRK, another Ser-640 kinase, to phosphorylate glycogen synthase, nor does glycogen affect the ability of hPASK to phosphorylate other substrates, including UCK2, the efficient phosphorylation of which requires the hPASK midregion. These data strongly suggest that the hPASK midregion plays an important role in targeting hPASK catalytic activity to specific substrates within the cell.
In summary, we have shown that glycogen synthase is phosphorylated efficiently by hPASK in vitro. This phosphorylation event and interaction between the two proteins in cells depend on a region of hPASK between its regulatory PAS domain and catalytic serine/threonine kinase domain. This interaction is negatively regulated by PAS domain function and glycogen. hPASK has been recently implicated in glucose-sensing and glucose-responsive transcription in pancreatic β-cells. It is possible that glucose signaling by means of hPASK affects glycogen metabolism in vivo. If so, PASK would comprise a system for the metabolic control of glucose utilization and storage in mammalian cells.
Supplementary Material
Acknowledgments
We thank John Lawrence (University of Virginia, Charlottesville) for generously providing the rabbit muscle glycogen synthase antibody and Steven L. McKnight and Janet E. Lindsley for helpful discussions. This work was supported by a Sara and Frank McKnight Foundation Fellowship, the Searle Scholars Program (both to J.R.), and by National Institutes of Health Grants DK27221 (to P.J.R.) and DK036569 (to A.d.P.-R.).
Author contributions: W.A.W., P.J.R., and J.R. designed research; W.A.W., A.V.S., and J.R. performed research; A.V.S., B.P., and A.d.P.-R. contributed new reagents/analytic tools; W.A.W., P.J.R., and J.R. analyzed data; and W.A.W. and J.R. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: PASK, PAS kinase; hPASK, human PASK; G6P, glucose-6-phosphate; D-form, G6P-dependent form; I-form, G6P-independent form; DYRK, dual-specificity tyrosinephosphorylated and -regulated kinase; GSK-3; glycogen synthase kinase-3; UCK2, uridinecytidine kinase 2.
References
- 1.Roach, P. J., Skurat, A. V. & Harris, R. A. (2001) in The Endocrine Pancreas and Regulation of Metabolism, eds. Cherrington, A. D. & Jefferson, L. S. (Oxford Univ. Press, New York), pp. 609–647.
- 2.Bergstrom, J., Hermansen, L., Hultman, E. & Saltin, B. (1967) Acta Physiol. Scand. 71, 140–150. [DOI] [PubMed] [Google Scholar]
- 3.Cline, G. W., Rothman, D. L., Magnusson, I., Katz, L. D. & Shulman, G. I. (1994) J. Clin. Invest. 94, 2369–2376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shulman, G. I., Rothman, D. L., Jue, T., Stein, P., DeFronzo, R. A. & Shulman, R. G. (1990) N. Engl. J. Med. 322, 223–228. [DOI] [PubMed] [Google Scholar]
- 5.Cohen, P. (1982) Nature 296, 613–620. [DOI] [PubMed] [Google Scholar]
- 6.Roach, P. J. (1986) in The Enzymes, eds. Boyer, P. D. & Krebs, E. G. (Academic, Orlando, FL), Vol. 17, pp. 499–539. [Google Scholar]
- 7.Cohen, P. (1986) in The Enzymes, eds. Boyer, P. D. & Krebs, E. G. (Academic, Orlando, FL), Vol. 17, pp. 461–497. [Google Scholar]
- 8.Friedman, D. L. & Larner, J. (1963) Biochemistry 128, 669–675. [DOI] [PubMed] [Google Scholar]
- 9.Larner, J. (1990) Adv. Enzymol. Relat. Areas Mol. Biol. 63, 173–231. [DOI] [PubMed] [Google Scholar]
- 10.Roach, P. J. (1990) FASEB J. 4, 2961–2968. [PubMed] [Google Scholar]
- 11.Skurat, A. V., Wang, Y. & Roach, P. J. (1994) J. Biol. Chem. 269, 25534–25542. [PubMed] [Google Scholar]
- 12.Flotow, H. & Roach, P. J. (1989) J. Biol. Chem. 264, 9126–9128. [PubMed] [Google Scholar]
- 13.Nakielny, S., Campbell, D. G. & Cohen, P. (1991) Eur. J. Biochem. 199, 713–722. [DOI] [PubMed] [Google Scholar]
- 14.Fiol, C. J., Mahrenholz, A. M., Wang, Y., Roeske, R. W. & Roach, P. J. (1987) J. Biol. Chem. 262, 14042–14048. [PubMed] [Google Scholar]
- 15.Picton, C., Aitken, A., Bilham, T. & Cohen, P. (1982) Eur. J. Biochem. 124, 37–45. [DOI] [PubMed] [Google Scholar]
- 16.Skurat, A. V. & Roach, P. J. (1995) J. Biol. Chem. 270, 12491–12497. [DOI] [PubMed] [Google Scholar]
- 17.Skurat, A. V. & Dietrich, A. D. (2004) J. Biol. Chem. 279, 2490–2498. [DOI] [PubMed] [Google Scholar]
- 18.Hardy, T. A. & Roach, P. J. (1993) J. Biol. Chem. 268, 23799–23805. [PubMed] [Google Scholar]
- 19.Francois, J. & Parrou, J. L. (2001) FEMS Microbiol. Rev. 25, 125–145. [DOI] [PubMed] [Google Scholar]
- 20.Rutter, J., Probst, B. L. & McKnight, S. L. (2002) Cell 111, 17–28. [DOI] [PubMed] [Google Scholar]
- 21.Rutter, J., Michnoff, C. H., Harper, S. M., Gardner, K. H. & McKnight, S. L. (2001) Proc. Natl. Acad. Sci. USA 98, 8991–8996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hofer, T., Spielmann, P., Stengel, P., Stier, B., Katschinski, D. M., Desbaillets, I., Gassmann, M. & Wenger, R. H. (2001) Biochem. Biophys. Res. Commun. 288, 757–764. [DOI] [PubMed] [Google Scholar]
- 23.Amezcua, C. A., Harper, S. M., Rutter, J. & Gardner, K. H. (2002) Structure (Cambridge, U.K.) 10, 1349–1361. [DOI] [PubMed] [Google Scholar]
- 24.Taylor, B. L. & Zhulin, I. B. (1999) Microbiol. Mol. Biol. Rev. (Washington, DC) 63, 479–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gilles-Gonzalez, M. A. (2001) IUBMB Life 51, 165–173. [DOI] [PubMed] [Google Scholar]
- 26.Lindsley, J. E. & Rutter, J. (2004) Comp. Biochem. Physiol. B 139, 543–559. [DOI] [PubMed] [Google Scholar]
- 27.da Silva Xavier, G., Rutter, J. & Rutter, G. A. (2004) Proc. Natl. Acad. Sci. USA 101, 8319–8324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.DePaoli-Roach, A. A., Vilardo, P. G., Kim, J. H., Mavila, N., Vemuri, B. & Roach, P. J. (2003) Methods Enzymol. 366, 17–34. [DOI] [PubMed] [Google Scholar]
- 29.Roach, P. J., de Paoli-Roach, A. A. & Larner, J. (1978) J. Cyclic Nucleotide Res. 4, 245–257. [PubMed] [Google Scholar]
- 30.Orho, M., Bosshard, N. U., Buist, N. R., Gitzelmann, R., Aynsley-Green, A., Blumel, P., Gannon, M. C., Nuttall, F. Q. & Groop, L. C. (1998) J. Clin. Invest. 102, 507–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Farkas, I., Hardy, T. A., Goebl, M. G. & Roach, P. J. (1991) J. Biol. Chem. 266, 15602–15607. [PubMed] [Google Scholar]
- 32.Wang, Y. & Roach, P. J. (1993) J. Biol. Chem. 268, 23876–23880. [PubMed] [Google Scholar]
- 33.Picton, C., Woodgett, J., Hemmings, B. & Cohen, P. (1982) FEBS Lett. 150, 191–196. [DOI] [PubMed] [Google Scholar]
- 34.DePaoli-Roach, A. A., Ahmad, Z., Camici, M., Lawrence, J. C., Jr., & Roach, P. J. (1983) J. Biol. Chem. 258, 10702–10709. [PubMed] [Google Scholar]
- 35.Elia, A. E., Cantley, L. C. & Yaffe, M. B. (2003) Science 299, 1228–1231. [DOI] [PubMed] [Google Scholar]
- 36.Gao, T., Yatani, A., Dell'Acqua, M. L., Sako, H., Green, S. A., Dascal, N., Scott, J. D. & Hosey, M. M. (1997) Neuron 19, 185–196. [DOI] [PubMed] [Google Scholar]
- 37.Wilson, W. A., Mahrenholz, A. M. & Roach, P. J. (1999) Mol. Cell. Biol. 19, 7020–7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yedovitzky, M., Mochly-Rosen, D., Johnson, J. A., Gray, M. O., Ron, D., Abramovitch, E., Cerasi, E. & Nesher, R. (1997) J. Biol. Chem. 272, 1417–1420. [DOI] [PubMed] [Google Scholar]
- 39.Fernandez-Novell, J. M., Lopez-Iglesias, C., Ferrer, J. C. & Guinovart, J. J. (2002) FEBS Lett. 531, 222–228. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.