Abstract
Keratoconus is the most common corneal dystrophy that leads to severe visual impairment. Although the major etiological factors are genetic, the pathogenetic mechanism(s) is unknown. No medical treatments exist, and the only therapeutic approach is corneal transplantation. Recent data demonstrate the involvement of nerve growth factor (NGF) in trophism and corneal wound healing. In this study, we investigated alterations in the NGF pathway in keratoconus-affected corneas and found a total absence of the NGF-receptor TrkA (TrkANGFR) expression and a decreased expression of NGF and p75NTR. The absence of TrkANGFR expression was associated with a strong increase in the Sp3 repressor short isoform(s) and a lack of the Sp3 activator long isoform. Sp3 is a bifunctional transcription factor that has been reported to stimulate or repress the transcription of numerous genes. Indeed, we found that Sp3 short isoform(s) overexpression in cell culture results in a down-regulation of TrkANGFR expression. We suggest that an imbalance in Sp transcription-factor isoforms may play a role in controlling the NGF signaling, thus contributing to the pathogenesis of keratoconus. This mechanism for the transcriptional repression of the TrkANGFR gene can provide the platform for the development of a therapeutic strategy.
Keywords: cornea, nerve growth factor, transcription factor, corneal dystrophy, nerve growth factor's receptors
Keratoconus is characterized by a bilateral, noninflammatory progressive corneal ectasia, leading to impairment of visual acuity. It is the most common corneal dystrophy, with a prevalence of 8.8-54.4 per 100,000 and is the major cause of cornea transplantation in the Western world (1).
A genetic predisposition for keratoconus has been proposed, and multifactorial and Mendelian inheritance have been documented (1). The only identified molecular cause for keratoconus involves mutations in VSX1, a member of the Vsx1 group of vertebrate paired-like homeodomain transcription factors (2). Nevertheless, mutations in this gene are not specific to keratoconus, because they are also associated with posterior polymorphous dystrophy, another inherited but histopathologically and clinically dissimilar corneal dystrophy.
Previous reports have also shown that keratoconus-affected corneas have elevated levels of degradative enzymes and reduced levels of α1-proteinase inhibitor (α1-PI) and α2-macroglobulin (α2-M) (3). Moreover, Whitelock et al. (4) demonstrated a specific up-regulation of the Sp1 transcription factor in keratoconus-affected corneas. Some have proposed that an increase in Sp1 transcriptional activity is responsible for the reduced expression of α1-PI (5). Interestingly, Sacristan et al. (6) reported that Sp1 regulates expression of the NGF-receptor TrkA (TrkANGFR) through interactions with a cis-regulatory element in its promoter.
NGF and its signaling molecules have a crucial role in trophism and wound healing of the cornea. In normal conditions, all corneal cell types, including epithelium, endothelium, and keratocytes, produce and express NGF and its receptors TrkANGFR and p75NTR (7, 8). NGF has been shown to induce in vitro corneal epithelial-cell proliferation and differentiation and cornea wound healing in an experimental model of epithelial injury (9, 10). NGF-induced cornea recovery was also observed in patients affected by neurotrophic and autoimmune corneal ulcers, conditions unresponsive to any standard treatments (11, 12). NGF promotes corneal nerve function (13), and impairment of corneal innervation has been suggested to play a role in the pathogenesis of keratoconus (14, 15). In this study, a lack of TrkANGFR and a strong decrease in p75NTR and NGF levels were found in corneas of patients affected by keratoconus compared with healthy donors.
The Sp gene family of transcription factors has five members, referred to as Sp1-Sp5, which bind with similar affinity to GC-rich motifs (16). Whereas Sp1, Sp2, and Sp4 are transactivators, a number of laboratories have reported that Sp3 functions as a repressor of Sp1-mediated transcription (17). Sp3 was shown to encode three distinct gene products: a full-length protein (110 KDa), which is a transcription activator, and two isoforms (78 and 80 KDa), derived through internal translational initiation, which function as transcriptional repressors (18). Although the mechanism by which internally initiated Sp3 isoforms inhibit Sp1/Sp3-mediated transcription remains obscure, this means of regulation is thought to play an important role in cell-cycle- and signal-induced transcription (18). Because of the known up-regulation of Sp1 in keratoconus-affected corneas and its known ability to interact with the TrkANGFR promoter, we analyzed the expression levels of Sp3 isoforms in keratoconus corneas and found a strong increase in the Sp3 short protein(s) and an absence of the Sp3 long isoform. We also found that, when the Sp3 short isoform(s) was overexpressed in cell culture, it down-regulated TrkANGFR expression. These results strongly indicate that an imbalance in Sp transcription factors may be a key event in the inhibition of NGF signaling in keratoconus.
Materials and Methods
This study adhered to the tenets of the Declaration of Helsinki and was approved by the ethics committees of the University of Rome “Campus Bio-Medico.”
Full-thickness corneal buttons (central, 8.0 mm) were obtained at the time of penetrating keratoplasty: (i) ten corneas were obtained with advanced keratoconus, (ii) two corneas were obtained with keratoconus induced by refractive surgery, (iii) five corneas were obtained with bullous keratopathy, and (iv) three with Fuchs' dystrophy. In addition, ten normal central corneas were procured from the local eye bank (Banca degli Occhi, Rome) as a control group.
After surgical dissection, the corneas were processed for molecular and biochemical analysis, as described below. For in vitro studies, corneal fibroblasts were obtained from healthy corneas and HACAT cells from American Type Culture Collection (CRL-1435).
Semiquantitative RT-PCR. Human corneas were incubated at 55°C for 3 h in HIRT solution (10 mM Tris·HCl, pH 8/100 mM EDTA/0.5% SDS) containing proteinase K (20 mg/ml). After centrifugation at 16,000 × g, the supernatant was diluted 1:5 with Trizol, and total RNA was extracted according to the manufacturer's instructions (GIBCO/Invitrogen). PCR was performed by using the primers shown in Table 1. Step-cycle PCR was carried out to identify the logarithmic phase of amplification for all genes. Quantitation of PCR-amplified products was performed by using the FluorS imager (Bio-Rad) and the program imagequant to quantify the ethidium bromide signal.
Table 1. Primer sequences.
| Gene | Sequence |
|---|---|
| TrkA | Forward: 5′-TGGCTGATACTGGCATCTGCG-3′ |
| Reverse: 5′-AGCCGAGGA GTGAAATGGAAGG-3′ | |
| p75 | Forward: 5′-GGC ACC TCC AGA ACA AGA CCT C-3′ |
| Reverse: 5′-ACA GGG ATG AGG TTG TCG GTG-3′ | |
| NGF | Forward: 5′-CAGGACTCACAGGAGCAAGC-3′ |
| Reverse: 5′-GCCTTC CTGCTGAGCACACA-3′ | |
| H3 | Forward: 5′-AGA CTG CCC GCA AAT CGA CC-3′ |
| Reverse: 5′-CG CAC CAG ACG CTG GAA GG-3′ | |
| Sp3 | Forward: 5′-CCGGAATTCTGATGACTGCAGGCATTAATGCC-3′ |
| Reverse: 5′-CCGCTCGAGTTACTCCATTGTCTCATTTCC-3′ |
Protein Evaluation by Immunohistochemistry, Western Blot, and ELISA. Immunohistochemistry was performed by using rabbit anti-human TrkA antibody (2 μg/ml, Santa Cruz Biotechnology) according to a procedure published in ref. 19. Nonspecific binding was detected by substituting primary antibody with control rabbit IgG (Vector Laboratories).
For Western blot analysis, proteins were extracted in a solution containing Tris·HCl (pH 7.5) 150 mM NaCl, 1% Nonidet P-40, 2.5 glycerol, 0.1% SDS, and a mixture of protease inhibitors (Sigma). Samples were subjected to freeze and thaw several times, followed by steps of fast sonication. Equal amounts of proteins (50 μg) were separated by SDS/PAGE and transferred onto nitrocellulose membranes (Amersham Pharmacia).
Anti-TrkA (0.2 μg/ml), anti-Sp3 (1:1,000, Santa Cruz Biotechnology), and anti-β actin (1:10,000, Sigma) antibodies were used in TBS containing 5% milk (Sigma). Secondary antibodies, including the horseradish-peroxidase-conjugated affinity-purified donkey anti-rabbit IgG and conjugated affinity-purified donkey anti-mouse IgG, were used 1:10,000 in TBS with 5% milk (Jackson ImmunoResearch). Antibody binding was revealed by using the Super Signal West Pico Chemiluminescent substrate (Pierce). Quantitative analysis was performed by densitometry using the FluorS imager (Bio-Rad).
A two-site NGF-specific ELISA (sensitivity of 0.5 pg/ml) was performed according to the Weskamp and Otten standard procedure, with minor modifications as described in ref. 19.
Cell Culture and Transient Transfection. Human corneal keratocyte primary cultures were obtained from healthy corneas, following the standard procedure, in DMEM containing 10% heat-inactivated FBS, 2 mM glutamine, 100 units/ml penicillin, and 100 μg/ml streptomycin (Celbio, Euroclone, Milan) (19). Cells that grew from biopsies were trypsinized (Invitrogen) and cell purity assessed by the K3 epithelial-marker-exclusion test (AE5, 1:150, ICN). Subcultured corneal fibroblasts (third to fifth passages) were used for experiments. HACAT keratinocytes were cultured in RPMI medium 1640 supplemented with 10% FCS, 2 mM l-glutamine, 100 units/ml penicillin, and 100 units/ml streptomycin.
Human primary keratocytes at 70% density were transfected by electroporation according to the manufacturer's instructions (Amaxia, Amaxa Biosystems, Hannover, Germany) by using 3 μg of DNA. The cells were plated in a six-well plate on poly-d-lysine-coated glass coverslips and left to recover for 24 h. After 24 or 48 h, the cells were analyzed by immunofluorescence. HACAT keratinocytes were transfected by using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions.
Vector Design. The short Sp3 isoform was cloned from human corneal keratocytes by using the primers indicated in Table 1. The PCR-amplified fragment was gel purified (Qiagen, Milan) and used for ligation into the EcoRI-XhoI sites of the plasmid HA-pcDNA3 (a gift of Fabienne Hans, Institute Albert Bonniot, Grenoble, France). Expression of HA-Sp3 was confirmed by Western blot using anti-HA monoclonal antibody (1:1,000; Babco, Richmond, CA). Empty HA-vector, EGFP-N1 control vector (Clontec, Palo Alto, CA), and HA-Sp3 vector were amplified and DNA purified according to the manufacturer's instructions (Qiagen).
Immunofluorescence Microscopy. The cells were fixed in 2% paraformaldeyde, permeabilized, and incubated with anti-TrkA (0.4 μg/ml) (Santa Cruz Biotechnology) and anti-HA antibodies (1:500) overnight at 4°C. Thereafter, the cells were incubated with Cy3-conjugated affinity-purified donkey anti-rabbit IgG secondary antibody and Cy2-conjugated affinity-purified donkey anti-mouse IgG secondary antibody (1:250; Jackson ImmunoResearch). The cells were then treated with RNase I and stained with TOTO3 (Molecular Probes/Invitrogen). Images were collected with a Nikon C1 laser scanning confocal microscope, and the fluorescence intensity was analyzed with Nikon software.
Gene Reporter Assay. Human TrkA 5′-proximal promoter region (20) was obtained from genomic DNA by PCR amplification with the primers indicated in Table 1. The fragment produced was digested with MluI and BglII restriction enzymes and cloned into the corresponding sites of pGL3-basic reporter vector (Promega) to produce a TrkA promoter-reporter vector (pTrkA-Luc). All constructs were verified by sequencing.
HACAT cells were cotransfected with 2 μg of pTrkA-Luc and increasing amounts (100 ng-2 μg) of Sp3-pcDNA3 (or a corresponding amount of empty pcDNA3 vector) and 50 ng of empty pRL-TK vector. Twenty-four or 48 h after transfection, cells were harvested and lysed, and gene reporter assays were performed by using the Dual-Luciferase Reporter Assay system (Promega). Results were normalized for transfection efficiency with the values obtained for pRL-TK and reported as percent of luciferase activity (±SD) compared with transfections carried out in the absence of Sp3. Data are representative of at least three independent experiments performed in duplicate or triplicate.
Statistical Analysis. Throughout this article, measurements are expressed as mean ± SD. Each experiment was repeated at least three times. Statistical analysis was carried out by one-way ANOVA and results considered statistically significant at P < 0.05.
Results
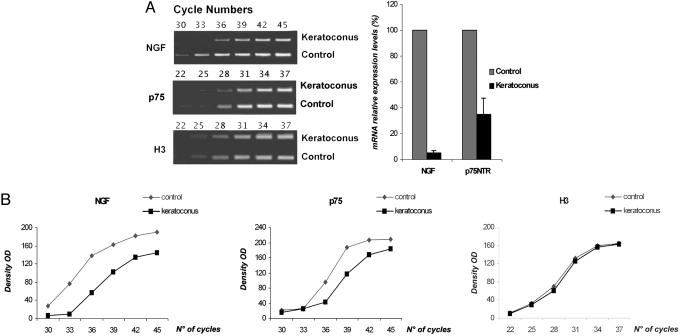
Alterations in the NGF Pathway in Keratoconus-Affected Corneas. In corneas affected by keratoconus, mRNA expression of NGF and its receptors, TrkANGFR and p75NTR, was analyzed by RT-PCR. TrkANGFR was not detected in keratoconus cornea after 50 cycles of PCR (Fig. 1, Keratoconus, lane 2) compared with control cornea (Fig. 1, control, lane 2). On the contrary, the positive control gene, histon H3, was amplified under the same number of cycles in both control and keratoconus corneas (Fig. 1, lanes 4). TrkANGFR mRNA was detected in other corneal diseases, such as Fuchs' dystrophy and bullous keratopathy (Fig. 1, Fuchs', lane 2 and bullous, lane 2), indicating a correlation between lack of TrkANGFR expression and keratoconus. Interestingly, TrkANGFR mRNA was detectable in circulating lymphocytes purified from the same keratoconus patients (Fig. 1, lymphocytes, lane 2), thus indicating that the lack of TrkANGFR was specific to corneal tissue in these patients.
Fig. 1.
Lack of TrkANGFR mRNA in corneas of keratoconus patients. RT-PCR analysis shows that lack of TrkANGFR mRNA is specific for keratoconus corneas (Keratoconus, lane 2). TrkANGFR mRNA is detectable in all evaluated corneas: normal (Control, lane 2), bullous keratopathy (Bullous, lane 2), Fuchs' distrophy (Fuchs, lane 2), and in peripheral lymphocytes obtained from the same keratoconus patients (Lymphocytes, lane 2). The band present in Keratoconus, lane 2 corresponds to primers. Lane 1, NGF RT-PCR; lane 2, TrkANGFR RT-PCR; lane 3, p75NTR RT-PCR; lane 4, H3 RT-PCR.
Analytical RT-PCR showed that both NGF (Fig. 1, lanes 1) and p75NTR (Fig. 1, lanes 3) mRNA was expressed in control, keratoconus, and other corneal diseases. However, semiquantitative RT-PCR demonstrated a 95% decrease of NGF mRNA and a 65% decrease of p75NTR mRNA in keratoconus samples compared with control corneas (Fig. 2A Right. Fig. 2 A Left illustrates that, in conditions where the housekeeping H3 gene amplification was comparable in control and keratoconus samples, higher number of cycles were necessary to amplify both p75NTR and NGF genes in keratoconus. Densitometric analysis for NGF, p75NTR, and H3 is shown in Fig. 2B. ELISA confirmed this significant decrease of NGF protein in keratoconus cornea (332.8 ± 85.4 pg/g; control = 763.8 ± 135.5 pg/g).
Fig. 2.
Decrease of NGF and p75NTR mRNA expression in keratoconus. (A) (Left) Semiquantitative RT-PCRs were run on ethidium-bromide-stained agarose gels. To identify the logarithmic phase of amplification for all genes, two sets of step-cycle PCRs were carried out. The first set was in a range of 22-37 cycles and revealed that H3 and p75NTR genes were within the linear dynamic range of amplification after 31 cycles. The second set was in a range of 30-45 cycles and revealed that NGF amplification was linear after 38 cycles. (Right) The histogram illustrates the percentage of mRNA relative expression for NGF and p75NTR. Means ± SD of three separate experiments are shown in the graph. (B) Cycle titration curves of RT-PCR amplification for the housekeeping gene H3, p75NTR, and NGF genes in keratoconus and control corneas.
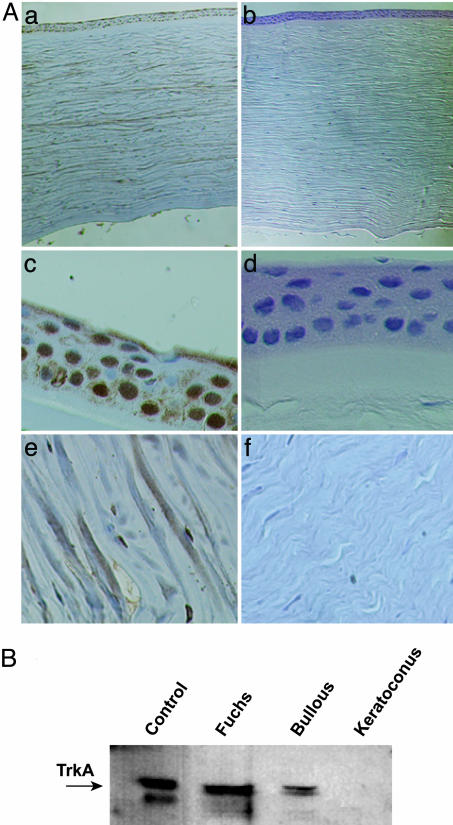
The absence of TrkANGFR protein in keratoconus was confirmed by immunohistochemistry and Western blot (Fig. 3 A and B). TrkANGFR protein was not detected by immunohistochemistry in keratoconus corneas or in corneal cells (epithelium, keratocytes, and endothelium) (Fig. 3A). Western blot analysis confirmed the absence of TrkANGFR protein in keratoconus and that the lack of this receptor was not observed in the other corneal diseases studied (Fuchs' and bullous) (Fig. 3B).
Fig. 3.
Lack of TrkANGFR protein expression in the cornea of keratoconus patients. (A) Representative immunohistochemical staining of TrkANGFR in control (a) and keratoconus (b) corneas. Higher magnification shows the lack of TrkANGFR in corneal cells, including epithelium (compare c with d) and keratocytes (compare e with f). Sections were counterstained with Harri's hematoxylin. (B) Western blot analysis of corneal protein extracts. The immunoreactive levels of TrkANGFR (arrow) are shown for control, Fuchs' dystrophy, bullous, and keratoconus corneas. In the first lane (Control), the lower band is a nonspecific immunoreactive band.
Different Expression Pattern of Sp3 Isoforms in Keratoconus. Because both mRNA and TrkANGFR protein are absent only in keratoconus disease, we hypothesized a transcriptional down-regulation of this gene in corneal tissue.
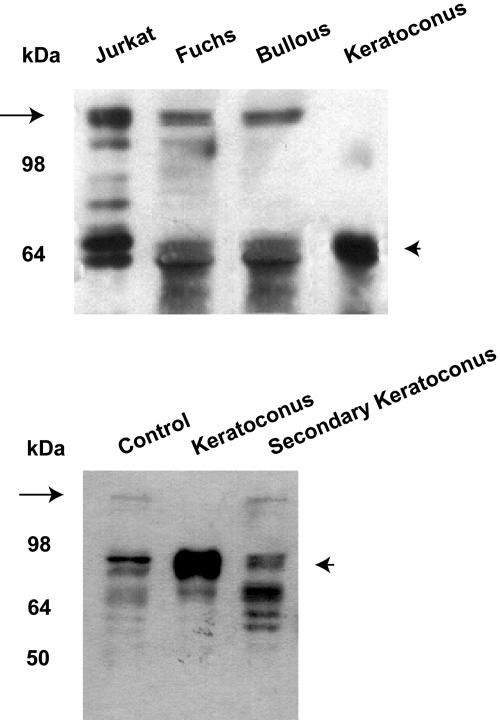
We first analyzed the human TrkANGFR minimal promoter (≈1,000 bp) (6) sequence isolated from the genome of patients with keratoconus, and no genetic mutations or polymorphisms that could explain the lack of expression of TrkANGFR have been identified. Because of the up-regulation of Sp1 in keratoconus-affected corneas and its possible interaction with the TrkANGFR gene, we investigated the expression profile of Sp3, the Sp1 transcriptional partner by Western blot analysis. Fig. 4 Upper shows the disappearance, only in keratoconus corneas, of the band corresponding to the long isoform of Sp3 (18) (Fig. 4, arrow) and a marked increase of the short isoform(s), identified as the repressor member(s) of the Sp family in ref. 18 (Fig. 4, arrowhead). Interestingly, keratoconus induced by refractive surgery (Fig. 4 Lower), maintained the same Sp3 expression profile of control cornea. By sequencing cDNAs isolated from keratoconus-affected corneas (n = 4), we confirmed that the disappearance of the long Sp3 isoform was not due to genetic mutations. Indeed, between the first ATG (long Sp3) and the internal ATG codons (short Sp3), we did not find any mutation resulting in a STOP codon that would affect translation only of the long Sp3 isoform.
Fig. 4.
Change in Sp3 pattern of expression in keratoconus cornea. Western blot, performed on protein extracts of corneas from Fuchs' dystrophy, bullous keratopathy, and keratoconus patients, shows an absence of Sp3 long isoform (arrow) and an overexpression of the Sp3 short isoform(s) (arrowhead) exclusively in keratoconus. Jurkat-cell extracts were used as a control group (Upper). Secondary keratoconus shows an expression profile similar to control cornea (Lower).
Overexpression of Sp3 Short Isoform(s) Down-Regulates TrkA Expression in Cell Culture. If TrkANGFR transcription is repressed by the internally initiated Sp3 short isoform(s), overexpression of these isoform(s) in cell culture should down-regulate the TrkA expression level.
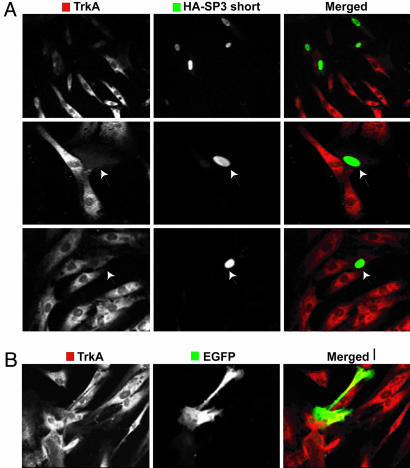
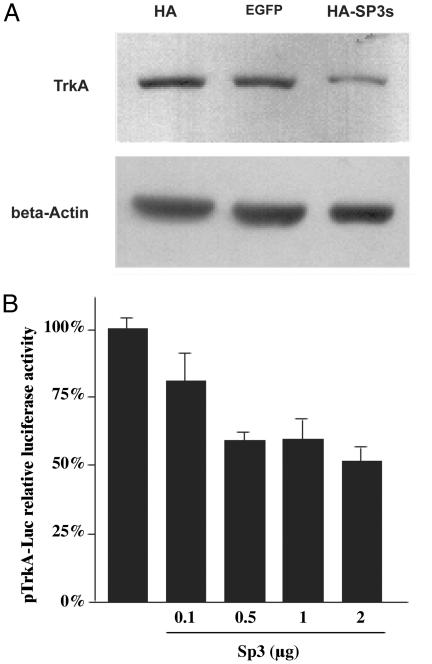
Therefore, we transfected primary corneal keratocytes with the short Sp3 isoform (HA-Sp3) (apparent molecular mass of 80 kDa), tagged at the N terminus with hemagglutinin (HA). Transfection efficiency was low, regardless of the vector used (empty HA, the reporter EGFP-N1, or the HA-Sp3 constructs). However, as shown in Fig. 5A, TrkANGFR fluorescence staining was 40% reduced in corneal keratocytes that overexpressed HA-Sp3 compared with EGFP-transfected cells (Fig. 5B). To better quantify this Sp3-short-isoform-mediated TrkANGFR down-regulation, a human HACAT cell line was made to overexpress the HA-Sp3 construct. Western blot analysis (Fig. 6A) showed a significant decrease of TrkANGFR expression (≈60%) in protein extracts of transfected HACAT cells when compared with cells transfected with the empty HA vector or the EGFP-N1 reporter vector.
Fig. 5.
Overexpression of Sp3 short isoform(s) down-regulates TrkANGFR expression in corneal keratocytes. (A) A panel of confocal images of cultured primary corneal keratocytes transfected with HA-Sp3 construct. Cells were double stained for TrkANGFR (red) and the HA-tag (green). Keratocytes that overexpress Sp3 short isoform(s) have nuclear staining (green) and reduced cytoplasmic TrkANGFR immunofluorescence. (B) Keratocytes that overexpress control plasmid (EGFP-N1 vector) do not have altered TrkANGFR immunostaining.
Fig. 6.
Sp3 down-regulates transcription from TrkANGFR proximal promoter. (A) Western blot of protein extracts of HACAT cells transfected with the HA-Sp3 construct. Protein levels of TrkANGFR are reduced only in extracts of cells that overexpress the HA-Sp3 short isoform(s) compared with cells that overexpress the empty HA vector or the control GFP vector (EGFP-N1). Protein loading was monitored by reprobing the nitrocellulose with β-actin antibody. (B) HACAT cells were cotransfected with pTrkA-Luc reporter construct and increasing amounts of HA-Sp3 vector. Luciferase activity was measured after 48 h. Results, which are representative of three independent experiments performed in triplicate, are reported as percent of luciferase activity (±SD) compared with the transfections carried out in the absence of HA-Sp3.
To further ascertain whether Sp3 short isoform(s) could down-regulate the expression of TrkANGFR, we produced a luciferase-reporter construct harboring the proximal promoter region of the TrkANGFR gene (pTrkA-Luc). We then used this construct to transiently transfect HACAT cells in the presence or absence of HA-Sp3. As shown in Fig. 6B, TrkANGFR promoter activity was progressively reduced when increasing amounts of HA-Sp3 were cotransfected with the pTrkA-Luc construct, further indicating an Sp3-mediated suppression of TrkANGFR gene transcription.
Discussion
The aim of this study was to identify the molecular basis of keratoconus, a disease lacking any medical treatment.
Molecular analyses of human corneas affected by keratoconus have led to the following conclusions: (i) keratoconus-affected corneas are characterized by a lack of TrkANGFR expression and a significant reduction in NGF and p75NTR expression, (ii) the absence of TrkANGFR is specific to keratoconus dystrophy, (iii) lack of TrkANGFR is associated with a strong increase in Sp3 short isoform(s) and a lack of Sp3 long isoform, and (iv) Sp3 short isoform(s), when overexpressed in cell culture, can down-regulate TrkANGFR expression.
NGF is known to play a crucial role in the maintenance of corneal trophism and in the equilibrium between cellular proliferation and differentiation (9, 10, 21). NGF affects in vitro and in vivo corneal epithelium and keratocytes function, promoting corneal wound healing in an experimental model and in patients affected by corneal ulcers (10-12). Several studies showed that knockout animals for the NGF receptor TrkANGFR develop corneal opacity and an impairment of corneal sensitive nerves (22, 23). Therefore, understanding the mechanisms responsible for TrkANGFR expression in corneal cells would provide important insight into the development of therapeutical strategies for the treatment of corneal diseases. In light of our results, keratoconus is a previously undescribed human disease characterized by a tissue-specific lack of TrkANGFR expression. Indeed, we have shown that the absence of TrkANGFR expression is specific to keratoconus dystrophy and limited to the cornea, because patients with keratoconus have circulating lymphocytes expressing TrkANGFR.
Congenital insensitivity to pain with anhidrosis (CIPA) is a disease characterized by a deficiency in TrkANGFR (24). Patients affected by CIPA have a highly complex syndrome that involves a wide number of tissues and organs, characterized by recurrent episodic fevers, anhidrosis, absence of reaction to noxious or painful stimuli, self-mutilating behavior, Ig deficiency, mental retardation, and severe visual impairment (25). The cornea-specific effects of keratoconus are in stark contrast to the devastating effects of CIPA, highlighting further the unique tissue-specific localization of TrkANGFR deficiency in keratoconus.
The complete absence of TrkANGFR expression in keratoconus cornea can be due to either mRNA degradation or repression of its transcription. Although we cannot exclude the possibility of a mechanism controlling TrkANGFR mRNA stability, dramatic alterations in the Sp3 protein expression profile indicate an Sp3-mediated down-regulation of TrkANGFR transcription. In fact, keratoconus corneas demonstrated an overexpression of short isoform(s) and a deficiency in long activator isoform. This imbalance may underlie the lack of TrkANGFR observed in keratoconus cornea. Indeed, cotransfection experiments using the luciferase assay have shown that the Sp3 short isoform(s) overexpression results in a significant down-regulation of TrkANGFR expression.
Furthermore, corneas from keratoconus induced by refractive surgery and other corneal diseases express normal TrkANGFR levels, and show a pattern of Sp3 protein expression similar to control corneas.
The basis for the altered profile of Sp3 expression in keratoconus remains unclear. From sequencing analyses, we can exclude the presence of a mutation resulting in a STOP codon in the cDNA of Sp3, particularly in the region between the initial ATG for the long isoform and the two internal translational sites. Recently, it has been suggested that, under many cellular conditions, including apoptosis, cellular stress, and viral infection, many proteins need to be synthesized via the internal ribosome entry site (IRES)-mediated mechanism (26). The switch from normal cap-dependent to the direct-internal-initiation mechanism (cap-independent initiation) would be an appealing explanation for the disappearance of Sp3 long isoform in keratoconus cornea.
The presence of an Sp3-mediated suppression of the TrkANGFR gene transcription in keratoconus cornea does not exclude the intervention or cooperation of other transcription factors in this disease. Whitelock et al. (4) analyzed a panel of transcription factors and found an enhanced level of Sp1 expression in corneal epithelium of patients affected by keratoconus compared with normal individuals. Interestingly, Sp1 is known to bind cis-elements in the promoter region of TrkANGFR, which contains several putative binding sites for Sp1/Sp3 factors (6, 27). Moreover, Gaudreault et al. (28) reported that alterations in Sp1 and Sp3 expression may trigger cells to express a set of keratins typical of the terminal differentiated state of corneal epithelial cells. We propose that TrkANGFR is another putative target gene, whose expression is sensitive to changes in the Sp family. An overexpression of the repressive isoform(s)of Sp3 could compete with the activator Sp1/Sp3 factors, leading to a shutting down of the human TrkANGFR gene in corneal cells. We have also analyzed the human TrkANGFR minimal promoter (≈1,000 bp) (6) sequence isolated from the genome of patients with keratoconus, and no genetic mutations or polymorphisms that could explain the lack of expression of TrkANGFR have been identified.
The magnitude of NGF signaling depends on the ratio of p75NTR and TrkANGFR activity (29, 30). It is intriguing that, in all patients with keratoconus, mRNA expression levels for NGF and p75NTR are altered, with a strong reduction (95%) in NGF mRNA. p75NTR, a member of the tumor necrosis factor receptor superfamily, is known to mediate apoptotic signals (31). Because TrkANGFR mediates survival of some neurons by silencing an ongoing p75NTR-mediated apoptotic signal (32), the absence of TrkANGFR and the presence of p75NTR might be responsible for increased apoptosis of corneal cells. Indeed, TUNEL analysis of corneas with keratoconus has shown the presence of diffuse stromal and epithelial apoptotic cell death (33).
In keratoconus, decreased corneal sensitivity, anatomic corneal nerve changes, and a progression of the disease after trigeminal-nerve injury have also been demonstrated (14, 15, 34). Several studies show that NGF regulates corneal nerves and, in particular, that TrkANGFR expression is essential for corneal sensitivity in mice (13, 23).
In conclusion, our study has shed light on molecular aspects of keratoconus, having identified that the imbalance in the Sp3 transcription factor isoforms induces an impairment of NGF signaling that merges multiple features of this disease. Absence of TrkANGFR and its regulation by the Sp family may well provide critical biochemical targets for future therapeutical strategies and drug design in keratoconus and other diseases characterized by a disregulation of NGF pathway.
Acknowledgments
We thank Dr. Alessio Cardinale for helpful comments and discussions. This work was supported, in part, by the Stem Cells National Program (Istituto Superiore di Sanità, Rome, Italy) and by the Italian Society of Ophthalmology.
Author contributions: A.L. and D.M. designed research; A.L., D.M., C.M., P.B., A.M.R., M.D.A., and P.R. performed research; A.M. contributed new reagents/analytic tools; M.C. analyzed data; and A.L., D.M., S.B., and E.G. wrote the paper.
Conflict of interest statement: No conflicts declared.
Abbreviations: HA, hemagglutinin; NGF, nerve growth factor; NGFR, nerve growth factor receptor.
References
- 1.Rabinowitz, Y. S. (1998) Surv. Ophthalmol. 42, 297-319. [DOI] [PubMed] [Google Scholar]
- 2.Heon, E., Greenberg, A., Kopp, K. K., Rootman, D., Vincent, A. L., Billingsley, G., Priston, M., Dorval, K. M., Chow, R. L., McInnes, R. R., et al. (2002) Hum. Mol. Genet. 11, 1029-1036. [DOI] [PubMed] [Google Scholar]
- 3.Zhou, L., Sawaguchi, S., Twining, S. S., Sugar, J., Feder, R. S. & Yue, B. Y. (1998) Invest. Ophthalmol. Vis. Sci. 39, 1117-1124. [PubMed] [Google Scholar]
- 4.Whitelock, R. B., Li, Y., Zhou, L. L., Sugar, J. & Yue, B. Y. (1997) Biochem. Biophys. Res. Commun. 235, 253-258. [DOI] [PubMed] [Google Scholar]
- 5.Maruyama, Y., Li, Y., Zhang, Y., Wang, X., Sugar, J. & Yue, B. Y. (2002) J. Cell. Biochem. 85, 482-489. [DOI] [PubMed] [Google Scholar]
- 6.Sacristan, M. P., de Diego, J. G., Bonilla, M. & Martin-Zanca, D. (1999) Oncogene 18, 5836-5842. [DOI] [PubMed] [Google Scholar]
- 7.Lambiase, A., Bonini, S., Micera, A., Rama, P. & Aloe, L. (1998) Invest. Ophthalmol. Vis. Sci. 39, 1272-1275. [PubMed] [Google Scholar]
- 8.You, L., Kruse, F. E. & Volcker, H. E. (2000) Invest. Ophthalmol. Vis. Sci. 41, 692-702. [PubMed] [Google Scholar]
- 9.Kruse, F. E. & Tseng, S. C. (1993) Invest. Ophthalmol. Vis. Sci. 34, 1963-1976. [PubMed] [Google Scholar]
- 10.Lambiase, A., Manni, L., Bonini, S., Rama, P., Micera, A. & Aloe, L. (2000) Invest. Ophthalmol. Vis. Sci. 41, 1063-1069. [PubMed] [Google Scholar]
- 11.Lambiase, A., Rama, P., Bonini, S., Caprioglio, G. & Aloe, L. (1998) N. Engl. J. Med. 338, 1174-1180. [DOI] [PubMed] [Google Scholar]
- 12.Lambiase, A., Bonini, S., Aloe, L. & Rama, P. (2000) Arch. Ophthalmol. 118, 1446-1449. [DOI] [PubMed] [Google Scholar]
- 13.Joo, M. J., Yuhan, K. R., Hyon, J. Y., Lai, H., Hose, S., Sinha, D. & O'Brien, T. P. (2004) Arch. Ophthalmol. 122, 1338-1341. [DOI] [PubMed] [Google Scholar]
- 14.Brookes, N. H., Loh, I. P., Clover, G. M., Poole, C. A. & Sherwin, T. (2003) Exp. Eye Res. 77, 515-524. [DOI] [PubMed] [Google Scholar]
- 15.Ruddle, J. B., Mackey, D. A. & Downie, N. A. (2003) Clin. Exp. Ophthalmol. 31, 363-365. [DOI] [PubMed] [Google Scholar]
- 16.Philipsen, S. & Suske, G. (1999) Nucleic Acids Res. 27, 2991-3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ghayor, C., Chadjichristos, C., Herrouin, J. F., Ala-Kokko, L., Suske, G., Pujol, J. P. & Galera, P. (2001) J. Biol. Chem. 276, 36881-36995. [DOI] [PubMed] [Google Scholar]
- 18.Kennett, S. B., Udvadia, A. J. & Horowitz, J. M. (1997) Nucleic Acids Res. 25, 3110-3117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Micera, A., Vigneti, E., Pickholtz, D., Reich, R., Pappo, O., Bonini, S., Maquart, F. X., Aloe, L. & Levi-Schaffer, F. (2001) Proc. Natl. Acad. Sci. USA 98, 6162-6167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chang, B. B., Persengiev, S. P., de Diego, J. G., Sacristan, M. P., Martin-Zanca, D. & Kilpatrick, D. L. (1998) J. Biol. Chem. 273, 39-44. [DOI] [PubMed] [Google Scholar]
- 21.Micera, A., Lambiase, A., Aloe, L., Bonini, S., Levi-Schaffer, F. & Bonini, S. (2004) Cytokine Growth Factor Rev. 15, 411-417. [DOI] [PubMed] [Google Scholar]
- 22.Smeyne, R. J., Klein, R., Schnapp, A., Long, L. K., Bryant, S., Lewin, A., Lira, S. A. & Barbacid, M. (1994) Nature 368, 246-249. [DOI] [PubMed] [Google Scholar]
- 23.de Castro, F., Silos-Santiago, I., Lopez de Armentia, M., Barbacid, M. & Belmonte, C. (1998) Eur. J. Neurosci. 10, 146-152. [DOI] [PubMed] [Google Scholar]
- 24.Indo, Y., Tsuruta, M., Hayashida, Y., Karim, M. A., Ohta, K., Kawano, T., Mitsubuchi, H., Tonoki, H., Awaya, Y. & Matsuda, I. (1996) Nat. Genet. 13, 485-488. [DOI] [PubMed] [Google Scholar]
- 25.Rosemberg, S., Marie, S. K. & Kliemann, S. (1994) Pediatr. Neurol. 11, 50-56. [DOI] [PubMed] [Google Scholar]
- 26.Jackson, R. J. (2000) in Translational Control of Gene Expression, eds. Sonenberg, N., Hershey, J. W. B. & Mathews, M. B. (Cold Spring Harbor Lab. Press, Cold Spring Harbor, NY), pp. 637-654.
- 27.Greco, A., Villa, R. & Pierotti, M. A. (1996) Oncogene 13, 2463-2466. [PubMed] [Google Scholar]
- 28.Gaudreault, M., Carrier, P., Larouche, K., Leclerc, S., Giasson, M., Germain, L. & Guerin, S. L. (2003) Invest. Ophthalmol. Vis. Sci. 44, 1447-1457. [DOI] [PubMed] [Google Scholar]
- 29.Yoon, S. O., Casaccia-Bonnefil, P., Carter, B. & Chao, M. V. (1998) J. Neurosci. 18, 3273-3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Casaccia-Bonnefil, P., Gu, C. & Chao, M. V. (1999) Adv. Exp. Med. Biol. 468, 275-282. [DOI] [PubMed] [Google Scholar]
- 31.Bamji, S. X., Majdan, M., Pozniak, C. D., Belliveau, D. J., Aloyz, R., Kohn, J., Causing, C. G. & Miller, F. D. (1998) J. Cell Biol. 140, 911-923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Majdan, M., Walsh, G. S., Aloyz, R. & Miller, F. D. (2001) J. Cell Biol. 155, 1275-1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kaldawy, R. M., Wagner, J., Ching, S. & Seigel, G. M. (2002) Cornea 21, 206-209. [DOI] [PubMed] [Google Scholar]
- 34.Dogru, M., Karakaya, H., Ozcetin, H., Erturk, H., Yucel, A., Ozmen, A., Baykara, M. & Tsubota, K. (2003) Ophthalmology 110, 1110-1118. [DOI] [PubMed] [Google Scholar]