Abstract
TRAF6 is an E3 ubiquitin ligase that mediates signaling from members of the tumor necrosis factor and Toll-like receptor superfamilies, including the p75 neurotrophin receptor. Recently, TRAF6 was shown to bind to another p75 cytoplasmic interactor, NRIF, and promote its nuclear localization. Here, we demonstrate that NRIF is a substrate for TRAF6-mediated K63 polyubiquitination and that this modification is necessary for its nuclear translocation. Activation of p75 resulted in NRIF polyubiquitination, association with TRAF6 and nuclear localization. NRIF was polyubiquitinated by TRAF6 in vitro and in cultured cells, and this was abrogated by mutation of K19 in the amino-terminus of NRIF. The K19R mutant NRIF displayed reduced TRAF6 association and neurotrophin-dependent nuclear localization. In neurons from traf6−/− mice, NRIF failed to enter the nucleus in response to p75 activation, and polyubiquitination and nuclear localization were attenuated in traf6−/− brain. Finally, unlike wild-type NRIF, the K19R NRIF failed to reconstitute p75-mediated apoptosis in nrif−/− neurons. These results reveal a unique mechanism of p75 signaling and a novel role for K63-linked ubiquitin chains.
Keywords: p75 receptor, polyubiquitination, neurotrophins, NRIF, TRAF6
Introduction
Members of the tumor necrosis factor (TNF) receptor and Toll-like receptor superfamilies mediate their effects primarily through the recruitment of signaling molecules in the TNF Receptor-Associated Factor (TRAF) family (Bradley and Pober, 2001). In mammals there are seven TRAFs known, based primarily on a region of homology in their carboxy-terminus, referred to as the TRAF/MATH domain, which is required for interaction with receptors, cytoplasmic interactors, and for TRAF homo- and heteromeric oligomerization (Dempsey et al, 2003). In addition, all but TRAF1 have RING and zinc-finger domains at their amino-terminus and, like many proteins containing a RING-finger domain, TRAF2 and 6 have been shown to function as E3 ubiquitin ligases (Sun and Chen, 2004). However, unlike the classical ubiquitin chain, which is polymerized by addition of ubiquitin moieties onto lysine 48 of ubiquitin and targets proteins to the proteasome, TRAF2 and 6 catalyze the formation of a unique chain linked through lysine 63, and, instead of degradation, the K63 polymer has been shown to mediate the activation of a protein kinase (Deng et al, 2000), facilitate DNA repair (Spence et al, 1995; Hofmann and Pickart, 1999) and regulate intracellular protein trafficking (Galan and Haguenauer-Tsapis, 1997). TRAF6 polyubiquitinates itself, resulting in the activation of transforming growth factor β-activated kinase (TAK1) through an unknown mechanism (Wang et al, 2001). TAK1 will then phosphorylate and stimulate the IκB kinase (IKK) complex, which subsequently activates the transcription factor NF-κB. TAK1 also phosphorylates MKK6 and 7, resulting in the activation of c-Jun N-terminal kinase (JNK) and p38 (Deng et al, 2000). In addition, TRAF6 can bind p62, a scaffolding protein containing an ubiquitin-associated (UBA) domain (Geetha and Wooten, 2002). P62 was originally identified by its interaction with atypical Protein Kinase C (aPKC) ζ (Puls et al, 1997), which also activates the IKK complex (Lallena et al, 1999), thereby providing another link from TRAF6 to NF-κB.
Recently, TRAF6 was shown to interact with another receptor-binding protein, the neurotrophin receptor interacting factor (NRIF) (Gentry et al, 2004), which was isolated based on its ability to bind the p75 neurotrophin receptor (Casademunt et al, 1999). The p75 receptor mediates a variety of biological effects, including the regulation of neurite outgrowth, myelination and cellular viability (Huang and Reichardt, 2003). In many cells, selective activation of the receptor leads to apoptosis, both in vitro and in vivo (Roux and Barker, 2002). For example, BDNF or the proform of NGF binding to p75 results in the death of perinatal sympathetic neurons (Bamji et al, 1998; Lee et al, 2001; Palmada et al, 2002), and deletion of the receptor in mice disrupts the naturally occurring neuronal loss during development (Bamji et al, 1998; Brennan et al, 1999; Frade and Barde, 1999).
Like many members of the TNF receptor superfamily, p75 can activate NF-κB and JNK and these signals are mediated by TRAF6 (Khursigara et al, 1999, 2001; Yeiser et al, 2004). The activation of JNK is known to be required for the receptor to induce cell death (Casaccia-Bonnefil et al, 1996; Yoon et al, 1998; Harrington et al, 2002); however, NRIF is also involved in this cell death signal, since the nrif−/− neurons are resistant to p75-induced apoptosis (Linggi et al, 2005). Moreover, ectopic expression of NRIF in sympathetic neurons causes cell death (Linggi et al, 2005). It is thought that NRIF affects neuronal survival by regulating gene transcription, since its amino-acid sequence includes five Kruppel-type zinc fingers, which are typically DNA-binding domains, and a sequence with homology to Kruppel-associated boxes, which are known to act as transcription repression domains (Margolin et al, 1994). Moreover, NRIF can localize to the nucleus (Gentry et al, 2004) and recombinant NRIF has been shown to bind DNA (Williams et al, 2004). The molecular mechanism by which NRIF translocates to the nucleus is not known; however, coexpression with TRAF6 in fibroblasts significantly increased its nuclear localization (Gentry et al, 2004), leading us to speculate that TRAF6 may control nuclear shuttling through ubiquitination. Here, we demonstrate that in response to p75 activation NRIF undergoes K63 polyubiquitination, associates with TRAF6 and p62, and translocates to the nucleus. In addition, NRIF is shown to be a target for TRAF6-mediated polyubiquitination based on in vitro and cell culture assays, and the polyubiquitination and nuclear translocation of NRIF is greatly reduced in neurons from traf6−/− mice. Finally, our data suggest that polyubiquitination of NRIF is required for p75 receptor-mediated apoptosis in sympathetic neurons.
Results
NRIF is ubiquitinated following ligand binding to the p75 receptor
Since NRIF and TRAF6 interact with each other (Gentry et al, 2004) as well as with p75 (Casademunt et al, 1999; Khursigara et al, 1999), and as TRAF6 is an ubiquitin ligase, we hypothesized that ligand binding to the receptor would promote the ubiquitination of NRIF. This was tested in PC12 cells using the NGF mutant Δ9/13 or BDNF, which can bind to p75 but not TrkA, thereby activating only the p75 signaling pathway (Hughes et al, 2001). The cells were treated with Δ9/13 NGF (50 ng/ml) or BDNF (100 ng/ml) for different times, lysed with SDS lysis buffer, NRIF immunoprecipitated and ubiquitination detected by Western blotting (Figure 1). NRIF was ubiquitinated shortly after addition of Δ9/13 NGF or BDNF, with maximal ubiquitination occurring within 10 min (Figures 1A and B). NGF did not induce ubiquitination of NRIF in PC12 cells (Figure 1C), thus indicating that NRIF is ubiquitinated in response to selective activation of the p75 receptor. Previous studies have shown that p75 interacts with TRAF6 (Khursigara et al, 1999), which in turn may bind to p62 (Sanz et al, 2000; Wooten et al, 2001). NRIF also associates with TRAF6 functionally to mediate p75 signaling (Gentry et al, 2004). Thus, we sought to investigate whether Δ9/13 NGF would drive the interaction of p62 and TRAF6 with NRIF. PC12 cells were stimulated with Δ9/13 NGF for 0, 5 and 15 min, followed by immunoprecipitation (IP) of NRIF, p62 or TRAF6 from lysates and Western blotting with antibody to NRIF, p62 and TRAF6. We observed that Δ9/13 NGF activation of p75 for 5 min resulted in a substantial increase in the interaction of both p62 and TRAF6 with NRIF (Figure 2), which is in concordance with the time for maximum ubiquitination of NRIF.
Figure 1.
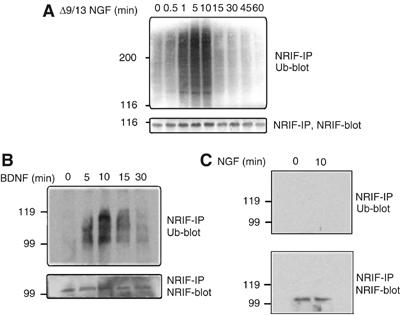
Activation of the p75 receptor induces the polyubiquitination of NRIF. PC12 cells were treated with 50 ng/ml of Δ9/13 mutant NGF (A), 100 ng/ml of BDNF (B) or 20 ng/ml NGF (C) for the indicated times; the cells were then lysed in SDS lysis buffer and the ubiquitination of NRIF was determined by IP of NRIF followed by immunoblotting with antiubiquitin (upper panels). The IP of NRIF on the same blot was verified by probing the blot with NRIF antibody (lower panels).
Figure 2.
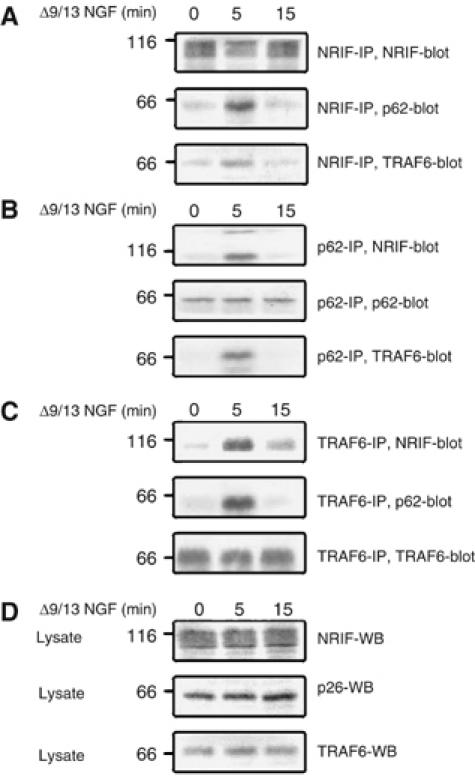
Activation of the p75 receptor increases NRIF association with TRAF6 and p62. PC12 cells were treated with 50 ng/ml of Δ9/13 mutant NGF for 0, 5, and 15 min, the cells were then lysed using Triton lysis buffer. NRIF (A), p62 (B) and TRAF6 (C) were immunoprecipitated and the precipitates were Western blotted for NRIF, p62 or TRAF6. (D) The same cell lysates (50 μg) were also immunoblotted for NRIF, p62 and TRAF6 to confirm expression.
TRAF6 attaches a lysine 63-linked polyubiquitin chain to NRIF
Since TRAF6 functions as an E3 ubiquitin ligase (Deng et al, 2000), we sought to determine whether it can polyubiquitinate NRIF. Therefore, Human embryonic kidney (HEK) 293 cells were transfected with NRIF, TRAF6 and ubiquitin constructs, and NRIF immunoprecipitated and Western blotted for ubiquitin. The coexpression of TRAF6 and NRIF resulted in NRIF polyubiquitination (Figure 3). A role for TRAF6 in modifying NRIF was further demonstrated by employing traf6 knockout (KO) mice. Wild-type (WT) and traf6 null mouse brain homogenates were immunoprecipitated with anti-NRIF and the ubiquitination status of NRIF examined by Western blotting. In contrast to lysates from traf6+/+ mice, in those from traf6−/− mice no ubiquitination of NRIF was detectable (Figure 4A). In addition to reducing NRIF ubiquitination, the absence of TRAF6 also diminished its interaction of p62 (Figure 4B). Altogether, these results suggest that TRAF6 mediates NRIF ubiquitination, which promotes the binding to p62.
Figure 3.
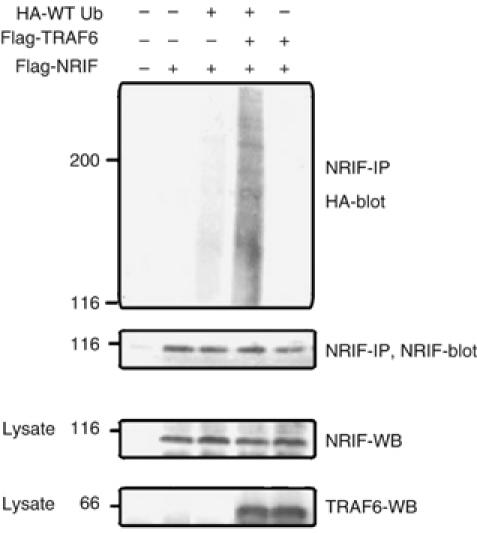
NRIF is polyubiquitinated by the E3 ubiquitin ligase TRAF6. HEK 293 cells were cotransfected with Flag-NRIF, Flag-TRAF6 or HA-ubiquitin, then lysed in SDS lysis buffer and the lysates were immunoprecipitated with anti-NRIF, followed by immunoblotting with an antibody to HA epitope to detect ubiquitin conjugates (upper panel), or an antibody to NRIF to verify the IP of NRIF (middle panel). The same cell lysates (50 μg) were immunoblotted with anti-NRIF and anti-TRAF6 to validate the expression of NRIF and TRAF6 (lower panels).
Figure 4.
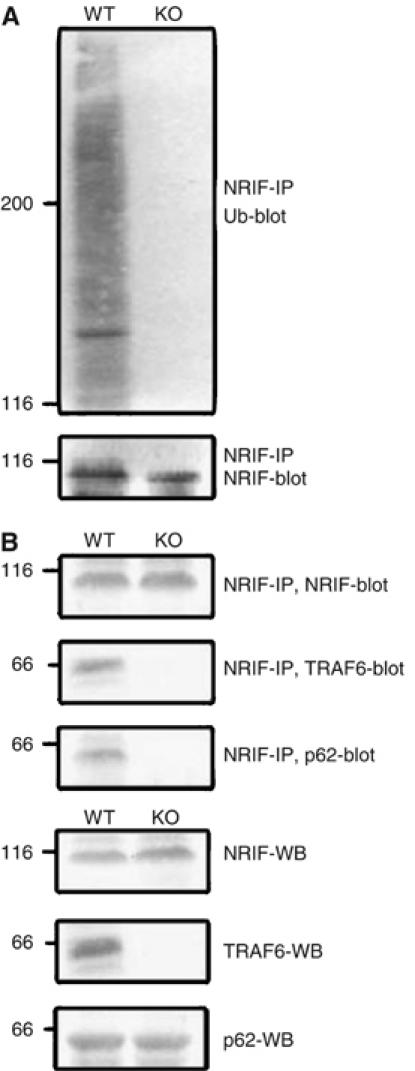
NRIF polyubiquitination is undetectable in traf6 null mice. (A) WT and traf6 KO mouse brain were homogenized in SDS lysis buffer, immunoprecipitated with anti-NRIF and immunoblotted with anti-ubiquitin (upper panel) or anti-NRIF (lower panel). (B) WT and traf6 KO mouse brains were homogenized in Triton lysis buffer, NRIF was immunoprecipitated and Western blotted with NRIF, TRAF6 or p62 antiserum to examine their interaction (upper panels). The same brain lysates were also Western blotted with anti-NRIF, TRAF6 and p62 to confirm the expression of these proteins (lower panels).
TRAF6 has been shown to ubiquitinate substrates through noncanonical lysine 63 chains (Deng et al, 2000); therefore, we proposed that such a polymer should be added to NRIF if TRAF6 was mediating this reaction. To test this hypothesis, HEK 293 cells were transfected with Flag-NRIF along with mutants of ubiquitin, where individual lysines had been changed to arginine: K29R, K48R and K63R, the cells were then lysed, NRIF immunoprecipitated and the covalent linkage of ubiquitin was examined by Western blotting with antibody to Flag. All ubiquitin constructs were expressed at similar levels (Figure 5A). Only the K63R ubiquitin mutant blocked the polyubiquitination of NRIF, whereas, in the presence of the K29R and K48R mutants, NRIF was polyubiquitinated to a degree comparable to that seen with WT ubiquitin. To further demonstrate that NRIF is K63 polyubiquitinated, we transfected PC12 cells with HA-tagged ubiquitin constructs, followed by treatment with Δ9/13 NGF. Therein, mutation of K63R blocked polyubiquitination of endogenous NRIF (Figure 5B) similar to results obtained in HEK cells (Figure 5A).
Figure 5.
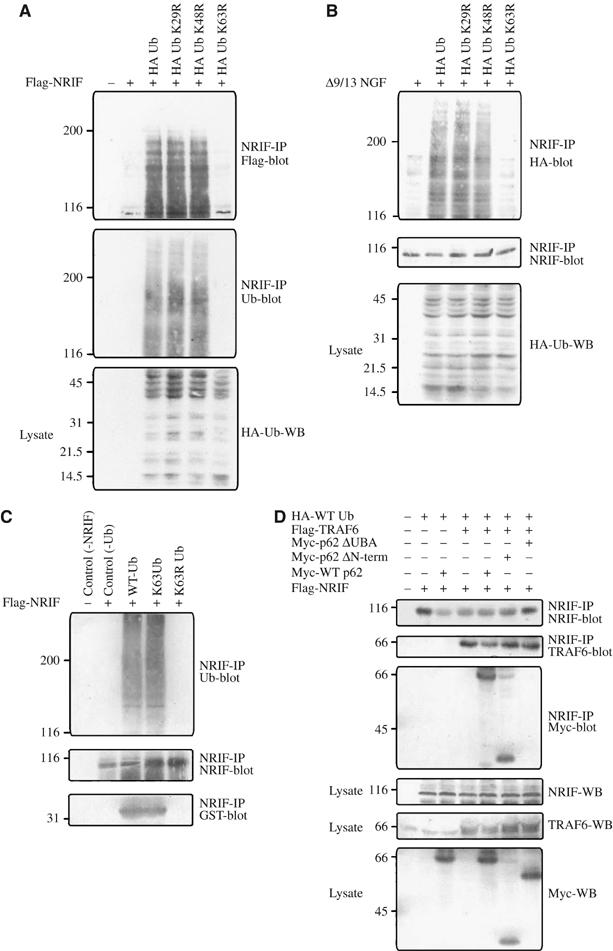
NRIF is K63-polyubiquitinated. (A) HEK cells were transfected with Flag-NRIF and either with HA-tagged WT ubiquitin, K29R, K48R or K63R point mutants of ubiquitin. The cells were then lysed in SDS lysis buffer, equal amounts of lysate were immunoprecipitated with NRIF antibody and immunoblotted with anti-Flag to detect NRIF (upper panel) or anti-ubiquitin (middle panel). The lysates were Western blotted with anti-HA to examine the expression of HA-tagged ubiquitin mutants (lower panel). (B) PC12 cells were transfected with HA-tagged WT, K29R, K48R or K63R ubiquitin mutants. The cells were treated with Δ9/13 NGF mutant for 5 min and lysed with SDS lysis buffer. NRIF was immunoprecipitated and Western blotted with anti-HA to examine the ubiquitination of NRIF (upper panel) or with anti-NRIF (middle panel) and the lysates were blotted with anti-HA (lower panel). (C) HEK 293 cells were transfected with Flag-NRIF, the cells lysed, then NRIF immunoprecipitated and the in vitro ubiquitination assay was carried out in the presence or absence of WT ubiquitin, a mutant with only K63 available for forming the ubiquitin chain (K63 Ub), or the K63R mutant ubiquitin along with E1, UbcH7 (E2) and TRAF6 (E3). This was followed by addition of 5 μg of GST-UBA in binding buffer to the above complex; the NRIF beads were then washed and subjected to Western blot analysis for ubiquitin, NRIF or GST (as indicated). Note that the GST-UBA only precipitated with the ubiquitinated form of NRIF. (D) HEK 293 cells were transfected with Flag-NRIF, myc-tagged WT p62, a p62 construct lacking the N-terminal 229 residues (p62-ΔN-term), or a p62 mutant lacking the UBA domain (p62-ΔUBA) along with Flag-TRAF6 or HA-Ub. Cells were then lysed in Triton lysis buffer, NRIF immunoprecipitated and the precipitates were immunoblotted with antibody to NRIF, TRAF6 or the myc-tag as indicated (upper panels). The lysates (50 μg) were subjected to Western blot analysis with anti-NRIF, anti-TRAF6 or anti-myc to verify the expression levels of NRIF, TRAF6 and p62 (lower panels).
To directly demonstrate that TRAF6 was able to mediate polyubiquitination of NRIF, an in vitro ubiquitination reaction was conducted. When the reaction was conducted with K63 ubiquitin, in which K63 is the only site that could be used for ubiquitin chain polymerization, we observed in vitro polyubiquitination of NRIF comparable to WT, but an absence of NRIF polyubiquitination was noted in reactions conducted with K63R mutant ubiquitin (Figure 5C).
p62 binds to polyubiquitinated NRIF through its UBA domain
In order to evaluate the role of ubiquitin in facilitating the interaction of NRIF with p62, an aliquot of the in vitro polyubiquitinated NRIF was allowed to interact with GST-UBA domain of p62. Only NRIF with the polyubiquitin modification coimmunoprecipitated with the UBA construct (Figure 5C). Similarly, in HEK 293 cells transfected with NRIF and p62 mutants, where the N-terminus had been deleted (amino acids 1–229) or where the C-terminal UBA domain was deleted (amino acids 384–440), only the constructs containing the UBA domain coimmunoprecipitated with NRIF (Figure 5D). In addition, the interaction between NRIF and p62 was TRAF6 dependent; no association was detected in the absence of TRAF6 (Figure 5D). These results suggest that the UBA domain of p62 can interact with NRIF only when NRIF is K63 polyubiquitinated by TRAF6.
TRAF6 ubiquitinates NRIF at lysine 19
The ubiquitination of proteins is often not restricted to a specific lysine; however, NRIF contains a lysine residue within a TRAF6 consensus-binding motif (PXEXXAr/Ac, where Ar is any aromatic residue and Ac is an acidic one (Ye et al, 2002)). The interaction between NRIF and TRAF6 was mapped to the N-terminal 161 amino acids on NRIF and within that region there is the sequence 14PHESEKF, which fits the consensus sequence except for a lysine insertion. Therefore, to test the possibility that this lysine is the ubiquitin acceptor site, we created a K19R mutant. When NRIF K19R was coexpressed with TRAF6 in HEK 293 cells, the ubiquitination of NRIF was significantly diminished, although not completely absent, suggesting that this is the primary lysine targeted by TRAF6 (Figure 6A). Mutating K19 also disrupted the interaction between NRIF and TRAF6 (Figure 6B).
Figure 6.
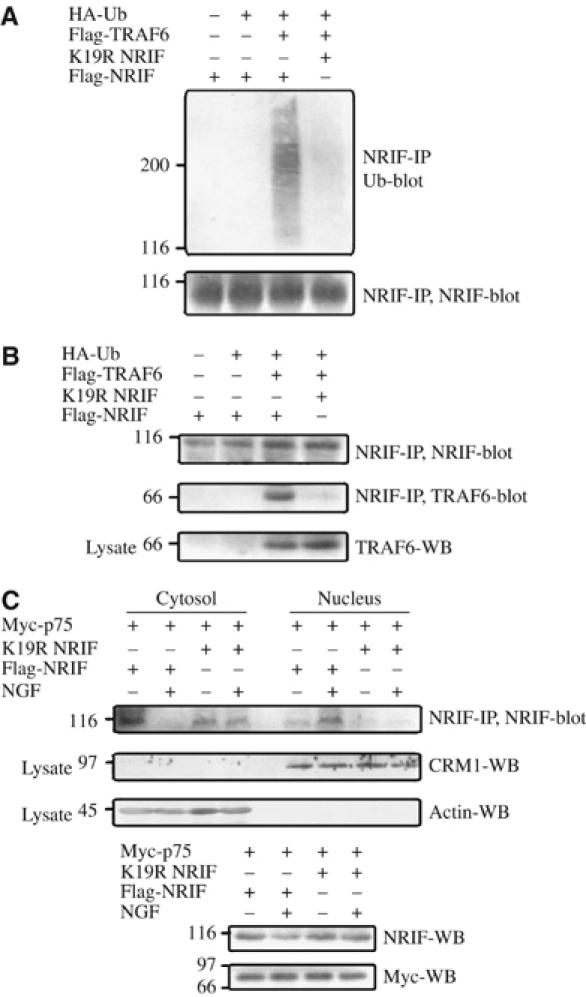
Lysine 19 is a polyubiquitination acceptor site in NRIF and is required for p75 receptor-mediated NRIF nuclear translocation. (A) HEK 293 cells were transfected with WT Flag-NRIF or its point mutant K19R along with Flag-TRAF6 and HA-tagged ubiquitin. The cells were then lysed with SDS lysis buffer and the extent of ubiquitination was determined by immunoprecipitating the lysate with anti-NRIF and Western blotting with anti-ubiquitin (upper panel) or anti-NRIF (lower panel). (B) HEK 293 cells transfected as in (A) were also lysed with Triton lysis buffer, NRIF immunoprecipitated and the precipitates immunoblotted with antisera to NRIF or TRAF6. The expression of TRAF6 was confirmed by Western blotting the lysate (lower panel). (C) HEK 293 cells were transfected with Flag-NRIF or K19R-NRIF along with myc-p75. The cells were then treated with 50 ng/ml NGF for 5 min, fractionated, and the cytosolic and nuclear fractions were immunoprecipitated with NRIF antibody and the precipitates subjected to Western blot analysis using an NRIF antibody. The nuclear and cytoplasmic fractions were immunoblotted for CRM1 and actin to confirm the purity of isolation of nuclear and cytosolic fractions (upper panels). Total lysates were subjected to Western blotting using antisera to NRIF or Myc to confirm the expression of NRIF and p75, respectively (lower panels).
Nuclear translocation of NRIF requires TRAF6-mediated polyubiquitination
We next considered what the physiological significance of TRAF6-mediated NRIF polyubiquitination might be. Previously, we observed that coexpression of TRAF6 enhanced the nuclear localization of NRIF in HEK 293 cells (Gentry et al, 2004); therefore, we hypothesized that the ubiquitination of NRIF was involved in targeting it to the nucleus. To test this possibility, HEK 293 cells were transfected with p75 along with WT NRIF or the K19R mutant NRIF, which prevented ubiquitination (Figure 6A). To avoid excess ubiquitination of NRIF induced by overexpression of TRAF6, we relied on the endogenous, albeit low, levels of this E3 ligase to ubiquitinate NRIF. Cytoplasmic and nuclear fractions were isolated and the localization of NRIF was determined by Western blotting. WT NRIF was localized to the nucleus in response to p75 receptor activation, whereas no K19R NRIF was detected in the nuclear fraction (Figure 6C). These results demonstrate that p75-induced nuclear localization of NRIF requires lysine 19, which is the site of ubiquitination.
To more directly address the need for the formation of the K63-linked chain of ubiquitin in this mobilization, we used the K63R ubiquitin mutant, which prevented polyubiquitination of NRIF (Figure 5). Expression of this mutant ubiquitin, along with p75 and NRIF, in 293 cells prevented NRIF nuclear localization in response to receptor activation (Figure 7A). Moreover, the K63R ubiquitin acted as a dominant negative in PC12 cells, preventing the shuttling of NRIF to the nucleus following Δ9/13 NGF treatment (Figure 7B).
Figure 7.
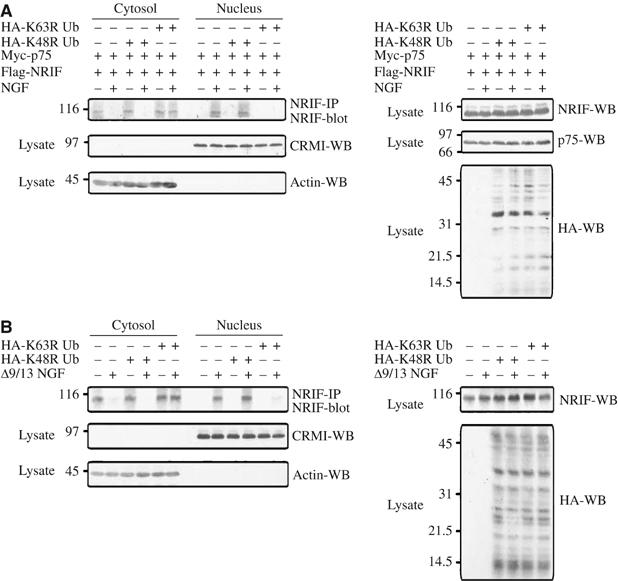
K63 polyubiquitinated NRIF is transported to the nucleus. (A) HEK 293 cells were transfected with Flag-NRIF, myc-tagged p75 along with HA-K48R or K63R ubiquitin mutants. The cells were then treated with 50 ng/ml NGF for 5 min, the cytosolic and nuclear fractions were immunoprecipitated with NRIF antibody and Western blotted using an NRIF antibody. The cytosolic and nuclear fractions were also immunoblotted for CRM1 and actin to validate the purity of the fractions (left panels). The total lysates were Western blotted using anti-NRIF, p75 or HA to confirm the expression of NRIF, p75 and ubiquitin constructs (right panels). (B) PC12 cells were transfected with the HA-tagged K48R or K63R ubiquitin mutant, then stimulated with Δ9/13 NGF (50 ng/ml) for 5 min, and cytosolic and nuclear fractions were isolated. NRIF was immunoprecipitated from these fractions and immunoblotted with anti-NRIF. The cytosolic and nuclear fractions were also immunoblotted for CRM1 and actin to validate the purity of the fractions (left panels). The expression of the NRIF and HA-tagged ubiquitin mutants was verified by Western blot with anti-NRIF and anti-HA (right panels).
Finally, to further link ubiquitination of NRIF with nuclear translocation in vivo, brains were isolated from traf6+/+ and −/− mice, fractionated and the amount of NRIF in the nuclear extract was determined. Although the expression of NRIF in total brain lysate from traf6+/+ and −/− mice was similar, its nuclear localization was significantly decreased in the tissue lacking TRAF6 relative to WT (Figure 8). We also observed that NRIF mobilization to the nucleus in response to p75 activation was abrogated in the absence of TRAF6. In sympathetic neurons from WT superior cervical ganglia, NRIF was observed in the nucleus after treatment with proNGF, which binds specifically to p75 (Lee et al, 2001); however, in the neurons from traf6−/− animals, NRIF remained cytoplasmic (Figure 9). Given that the ubiquitination of NRIF is dramatically reduced in traf6 null brain (Figure 4), these results suggest that polyubiquitination of NRIF by TRAF6 is required for its nuclear localization.
Figure 8.
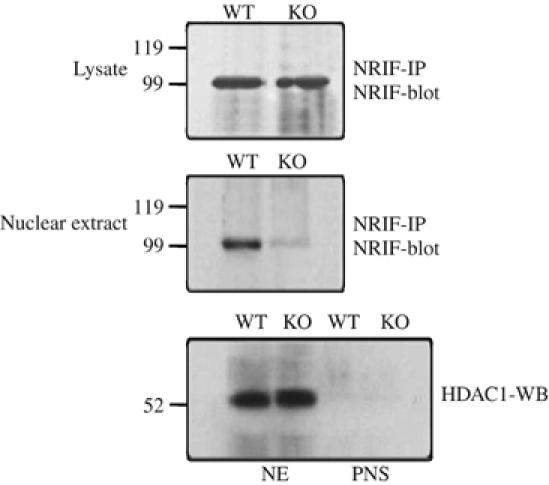
TRAF6 is necessary for NRIF nuclear localization in vivo. Whole brains were isolated from postnatal day 2 traf6 WT and KO mice, homogenized in RIPA buffer, then NRIF immunoprecipitated and the precipitates immunoblotted with NRIF antibody (upper panel). Nuclei were isolated from these tissues and NRIF immunoprecipitated. The precipitates were then subjected to Western blotting using an NRIF antibody (middle panel). Note the reduced presence of NRIF in traf6−/− nuclei relative to the WT. Nuclear extract (NE) and post nuclear supernatant (PNS) from WT and KO mouse brain were subjected to HDAC1 Western blot analysis to confirm the isolation of the nuclei, as well as serving as a loading control for the nuclear extracts (lower panel).
Figure 9.
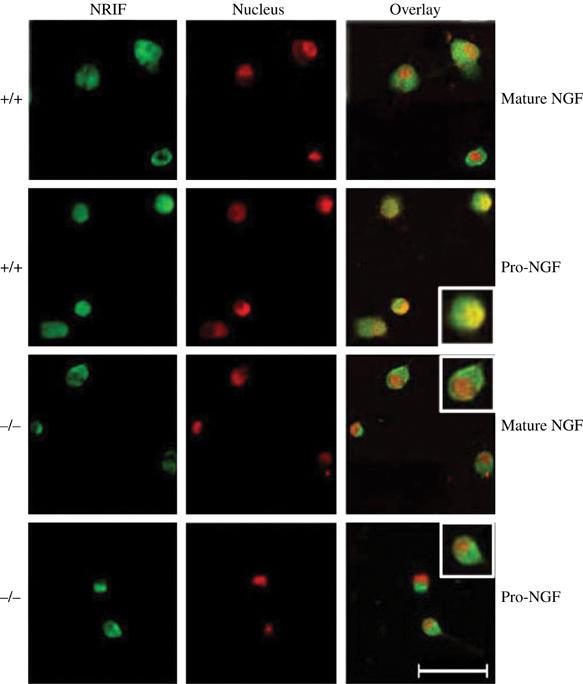
p75-mediated nuclear translocation of NRIF requires TRAF6. Sympathetic neurons from traf6+/+ and −/− mice were cultured directly in mature NGF (20 ng/ml) or proNGF (5 ng/ml) for 20 h, then the neurons were fixed and NRIF detected using an affinity-purified NRIF antibody revealed by Alexa 488-coupled secondary antibody (green). Nuclei were labeled with propidium iodide (red). The neurons were then imaged by confocal microscopy. Note the overlap of NRIF and nuclear staining in the presence of proNGF in traf6+/+ neurons (orange) but not in traf6−/− neurons. The insets depict individual neurons enlarged. Scale bar 50 μm.
Mutation of lysine 19 in NRIF prevents p75 receptor-mediated neuronal apoptosis
We recently demonstrated that nrif−/− sympathetic neurons are resistant to p75-induced apoptosis (Linggi et al, 2005). Given that NRIF is a DNA-binding protein, we hypothesized that its nuclear localization in response to receptor activation was necessary for the subsequent apoptosis. Since our results demonstrate that mutation of lysine 19 on NRIF prevented its ubiquitination and nuclear translocation, we used this mutant to test our hypothesis. WT NRIF or K19R mutant NRIF was transfected into nrif−/− sympathetic neurons and, 48 h later, p75 was activated by BDNF. As expected, expression of NRIF in the neurons induced some cell death under control conditions; however, the K19R mutant did not increase apoptosis above background. Most important, while the WT NRIF restored p75-mediated apoptosis in nrif−/− sympathetic neurons, the K19R mutant NRIF did not (Figure 10), suggesting that polyubiquitination and nuclear translocation of NRIF is required for p75 receptor-mediated neuronal apoptosis.
Figure 10.
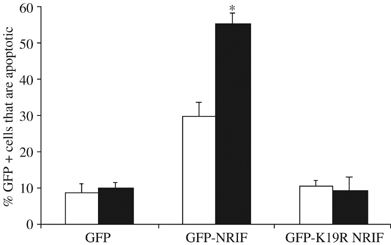
p75-mediated apoptosis can be restored in nrif−/− sympathetic neurons by expression of WT NRIF, but not K19R NRIF. Sympathetic neurons from nrif−/− mice were isolated, electroporated with GFP, GFP-NRIF, or GFP-K19R NRIF, and cultured in 20 ng/ml NGF for 2 days. The NGF was then removed, the neurons switched to media containing anti-NGF together with 12.5 mM KCl to promote survival and treated with (black bars) or without (empty bars) 100 ng/ml BDNF for 48 h. The cells were then fixed and GFP-positive neurons were evaluated for apoptotic nuclei by DAPI staining. Depicted are the mean±s.e.m. (n=3; *P<0.008).
Discussion
The p75 neurotrophin receptor is a multi-faceted signaling protein that regulates several key functions in the developing and injured mammalian nervous system, including neurite outgrowth and cellular survival (reviewed by Barker, 2004). One of the best characterized effects of p75 activation is the induction of apoptosis, which has been shown to require the intracellular associating proteins NRIF (Casademunt et al, 1999; Linggi et al, 2005) and TRAF6 (Yeiser et al, 2004). These two proteins are also required for the receptor to activate the stress kinase JNK, which is necessary for p75-mediated cell death. However, the ectopic expression of NRIF alone can induce apoptosis and this is independent of JNK activation, suggesting that this protein has a function independent of the kinase. Moreover, NRIF is a DNA-binding protein that can mobilize to the nucleus (Williams et al, 2004), thus raising the question as to how it would translocate from binding to a cell surface receptor into the nucleus. In this report, we demonstrate that ligand binding to p75 receptor stimulates NRIF polyubiquitination by the E3 ligase TRAF6 and this modification is necessary for the nuclear translocation of NRIF. Taken together, these results reveal a novel mechanism for regulating the nuclear shuttling of a transcription factor in response to an extracellular signal.
The addition of ubiquitin to proteins occurs through an enzymatic cascade; first an activating enzyme, E1, is charged with ubiquitin in an ATP-dependent process, then the ubiquitin is transferred to an E2-conjugating enzyme and, finally, it is attached to a substrate by an E3 ligase, which determines the target specificity (Pickart, 2001). Although protein ubiquitination has been best characterized as a mechanism for targeting proteins to the proteasome for degradation (Ciechanover and Ben-Saadon, 2004), recently it has become clear that this modification can also serve many other functions, largely by serving as a protein–protein interaction motif, similar to phosphorylation. Several cell plasma membrane proteins have been shown to internalize through a mechanism involving the attachment of monoubiquitin (Marmor and Yarden, 2004), which then recruits components of the internalization machinery such as Eps15 (de Melker et al, 2004). Ubiquitination can also modulate the activity of transcription factors, by recruiting coactivators or corepressors or by serving as a trigger for nuclear export (Muratani and Tansey, 2003; Shcherbik and Haines, 2004). In contrast, there are currently only a couple of examples of ubiquitin serving as a nuclear import signal. The nuclear translocation of the transcription factor NF-κB is indirectly controlled by ubiquitination. In response to various stimuli, ubiquitin is added to the endogenous inhibitor, IκB, which targets it for degradation, thereby revealing the hidden nuclear localization signal on NF-κB (Ghosh and Karin, 2002). A direct role for ubiquitin-targeting proteins to the nucleus has been shown for the E2-conjugating enzymes UbcM2, UbcH6 and UBE2E2; attachment of ubiquitin promotes their binding to importin-11, thereby facilitating nuclear translocation (Plafker et al, 2004). Similarly, ubiquitination causes the Saccharomyces cerevisiae transcription factors Mga2p and Spt23p to move to the nucleus, although the role of importins has not been determined in this case (Shcherbik and Haines, 2004). Our results demonstrate for the first time that ubiquitination of a mammalian transcription factor, NRIF, can directly induce its nuclear translocation.
Recently, ubiquitination has also been shown to promote the activation of a kinase, TAK1 (Wang et al, 2001). This kinase is activated by the E3 ligase TRAF6, but TAK1 is not the ubiquitination target, rather it is TRAF6 itself. This E3 polyubiquitinates itself by forming a chain linked through K63 on ubiquitin. This modification of TRAF6 results in the stimulation of the kinase through an unknown mechanism, ultimately resulting in the activation of NF-κB and JNK. Indeed, TRAF6 has been shown to mediate the activation of both pathways by p75 (Yeiser et al, 2004). TRAF6 also activates NF-κB by interacting with aPKC (Sanz et al, 2000), and this occurs through the scaffolding protein p62 (Wooten et al, 2001).
The K63 linkage of ubiquitin is different from the typical K48 polymer, which targets proteins for degradation through the proteasome. Recently, the solution structure of the K63 chain was shown to be substantially different from a K48 polymer (Varadan et al, 2004). The K63 chain, unlike K48, exists in a more open conformation that allows exposure of hydrophobic surfaces that could serve as an interface for binding to other proteins. Indeed, to date, the function of the K63-linked chain is proposed to mediate protein–protein interaction. Similar to polyubiquitination of TRAF6 facilitating the binding of TAK1, the proliferating cell nuclear antigen (PCNA) was shown to recruit specific DNA polymerases, required for error-free DNA repair, through the formation of a K63-linked polyubiquitin chain by the E3 ligase Rad5 (Hoege et al, 2002). Whether the polyubiquitin chain on NRIF promotes nuclear translocation through enhancing the binding of other proteins, such as importin-11, is not known.
Interestingly, one of the proteins that associated with NRIF in response to its ubiquitination was p62. While this protein was first identified by its interaction with aPKC, it has also been shown to activate transcription of an SV40 enhancer linked to a reporter gene, although it was unable to directly bind DNA, suggesting that it served as a coactivator (Rachubinski et al, 1999). We predict that NRIF functions as a transcription repressor since it contains KRAB domains, which typically function as repression domains able to recruit corepressors such as the NuRD complex (Schultz et al, 2001). Whether p62 would prevent repression and promote activation in the context of NRIF is unclear, but it is likely to modulate NRIF's transcriptional activity.
In summary, the data presented here demonstrate that TRAF6 polyubiquitinates NRIF and this modification is necessary for the translocation of NRIF into the nucleus. Although NRIF is the first example of ubiquitination directing a mammalian transcription factor to the nucleus, it is notable that the Toll-like receptors TLR7 and 9 induce interferon gene expression through the formation of a protein complex that includes TRAF6 and the transcription factor IRF-7 (Honda et al, 2004), suggesting that this mechanism may have broader applicability. We also found that NRIF ubiquitination and its nuclear mobilization occur in response to ligand binding to the p75 receptor. Given that TRAF6 and NRIF are required for p75-mediated apoptosis (Yeiser et al, 2004; Linggi et al, 2005), these results, taken together, suggest that activating the receptor causes NRIF to enter the nucleus and regulate the expression of genes that regulate neuronal survival/apoptosis. Future experiments will focus on identifying the target genes of NRIF.
Materials and methods
Cell culture
HEK 293 cells were maintained in high-glucose Dulbecco's modified Eagle's medium containing 10% fetal calf serum. PC12 cells were grown on plates coated with rat-tail collagen in DMEM containing 10% horse serum and 5% calf serum and antibiotics (50 U/ml penicillin and 50 μg/ml streptomycin). Sympathetic neurons from traf6+/+ and −/− mice at postnatal day 3–4 were isolated from the super cervical ganglia (SCG) as described by Palmada et al (2002). In brief, the neurons were dissociated with 0.25% trypsin and 0.3% collagenase for 30 min at 37°C, then plated on poly-L-ornithine- and laminin-coated four-well slides (Nalge Nunc International) containing 4.6 mM imidazole (used to elute pro- or mature NGF off the Ni beads), 5 ng/ml of pro-NGF or 20 ng/ml of NGF in Ultraculture medium (BioWhittaker) supplemented with 3% fetal calf serum (Gibco), 2 mM L-glutamine (Gibco).
HEK 293 cells were transfected employing the calcium phosphate method using a Mammalian Cell Transfection Kit (Specialty Media). The cells were lysed with Triton X-100 lysis buffer to detect protein–protein interactions (50 mM Tris–HCl, pH 7.5, 150 mM NaCl, 10 mM NaF, 0.5% Triton X-100, 1 mM Na3VO4, 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml leupeptin and aprotinin) or SDS lysis buffer to detect covalent interaction of ubiquitin and NRIF (Triton lysis buffer containing 1% SDS). Protein was estimated using bovine serum albumin (BSA) as a standard by Bradford procedure (Bio-Rad) for all samples except those containing SDS, which were estimated employing the DC assay (Bio-Rad).
Construction of NRIF and p62 mutants
The K19R-NRIF mutant was generated by site-directed mutagenesis using standard PCR methods. The primer 5′-CCCCATGAGTCTGTGAGGTTTGAAGATGTTTCT was used to amplify pcDNA3-NRIF, resulting in lysine 19 conversion to arginine. The complete sequence of the resulting mutant was confirmed. The p62 deletion mutants p62ΔN-term (lacking the amino-terminal 229 residues) and p62ΔUBA (lacking the carboxy-terminal 54 residues) were described previously (Seibenhener et al, 2004).
IP and Western blot analysis
A total of 1 mg of whole-cell lysate diluted in lysis buffer was incubated with 4 μg of primary antibody at 4°C for 3 h. The immunoprecipitates were collected with agarose-coupled secondary antibody for two more hours at 4°C and then washed three times with lysis buffer. Samples were boiled in SDS–PAGE sample buffer and resolved on 7.5 or 12% SDS–PAGE, transferred onto nitrocellulose membranes and analyzed by Western blotting with the appropriate antibodies.
For NRIF IP from brain, whole brain from traf6+/+ and −/− was isolated from postnatal day 4 mouse pups, lysed in RIPA lysis buffer (50 mM Tris–HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 0.1 mM phenylmethylsulfonyl fluoride, 10 μg/ml leupeptin, 2 μg/ml aprotinin) or SDS lysis buffer for detecting ubiquitin. Equal amounts of protein were immunoprecipitated with affinity-purified NRIF antibody (5 μl/0.5 ml) and protein A sepharose, separated by 10% SDS–PAGE and transferred to a nitrocellulose membrane and probed with NRIF antibody (1:2000 dilution) or anti-ubiquitin (1:500 dilution, SC-8017, Santa Cruz).
In vitro ubiquitination and GST-UBA-binding assay
HEK cells were transfected with Flag-tagged NRIF construct by the calcium phosphate method. The cells were lysed with SDS lysis buffer and 750 μg of the lysate was immunoprecipitated with antibody to NRIF and collected with agarose-coupled secondary antibody. The agarose beads containing the immunoprecipitated NRIF were ubiquitinated by the addition of 50 μl of reaction buffer (50 mM Tris, pH 7.5, 2.5 mM MgCl2, 2 mM DTT, 2 mM ATP) containing 100 ng E1, 200 ng UbcH7 (E2), along with 100 ng of TRAF6 (E3) and 5 μg of ubiquitin. Control samples without ubiquitin or TRAF6 were also included. Reactions were carried out by continuous shaking at 37°C for 2 h and then washed thrice with reaction buffer. The proteins were released by boiling for 2 min in SDS–PAGE sample buffer, electrophoresed on 7.5% SDS–PAGE and Western blotted with antiubiquitin and anti-NRIF. For the GST-UBA binding, the ubiquitinated NRIF captured on the agarose beads was incubated with 5 μg of p62 GST-UBA in binding buffer (20 mM Tris–HCl, pH 7.6, 50 mM NaCl, 0.1% Nonidet P-40, 0.5 M dithiothreitol and 1 mM phenylmethylsulfonyl fluoride) and allowed to interact at 37°C for 1 h. The agarose beads were washed three times with binding buffer and were boiled in sample buffer, resolved by 10% SDS–PAGE and Western blotted with antibody against ubiquitin or GST.
Cell fractionation
HEK cells were cotransfected with myc-tagged p75 and Flag-NRIF or K19R NRIF and, in some experiments, HA-K63R- or HA-K48R-ubiquitin, and, 24 h later, were stimulated with 50 ng/ml NGF for 5 min. PC12 cells were transfected with HA-K63R- or HA-K48R-ubiquitin using LipofectAMINE 2000 (Invitrogen) and, 24 h later, were treated with or without Δ9/13NGF. The cells were lysed in buffer A (10 mM HEPES, pH 7.9, 10 mM KCl, 0.1 mM EDTA, 0.1 mM EGTA, 0.4% Nonidet P-40, 1 mM dithiothreitol, 0.1 mM phenylmethylsulfonyl fluoride and 10 μg/ml aprotinin with 1 mM Na3VO4). The cell suspension was incubated on ice for 10 min, then the nuclei were pelleted at 750 g for 5 min. The supernatant was saved as the cytosol fraction. The nuclear pellet was further lysed in TLB (20 mM Tris, pH 7.5, 137 mM NaCl, 2 mM EDTA, 1% Triton X-100, 25 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride, and 10 μg/ml aprotinin and 1 mM Na3VO4), sonicated and centrifuged. This was then followed by IP of NRIF in both cytosol and nuclear fractions and Western blotting for NRIF to detect changes in its localization.
Isolation of nuclear extract from brain
Whole brains from traf6+/+ and −/− were isolated from postnatal day 2 mouse pups and nuclear extract prepared as described by Dignam et al (1983). In brief, the tissue was homogenized in buffer A (10 mM HEPES, pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and leupeptin), centrifuged at 1000 g for 10 min to pellet nuclei, and the nuclear pellet was resuspended in buffer C (20 mM HEPES, pH 7.9, 0.84 M NaCl, 1.5 mM MgCl2, 0.4 mM EDTA, 0.5 mM DTT and 0.5 mM phenylmethylsulfonyl fluoride, 10 μg/ml aprotinin, and leupeptin) to extract protein. This nuclear extract was then cleared by centrifuging 14 000 g for 20 min and NRIF immunoprecipitated as described above.
Immunostaining
At 20 h after plating in the proNGF, NGF or imidazole, the neurons were fixed in 4% paraformaldehyde, permeabilized with 0.1% sodium citrate and 0.1% Triton X-100, blocked with 10% goat serum in PT, and incubated overnight with the affinity-purified NRIF antibody (1:100 dilution), followed by incubation with anti-rabbit Alexa 488. Nuclei were visualized by propidium iodide and images were acquired using a confocal laser-imaging system (LSM 510; Carl Zeiss MicroImaging, Inc.) at × 400.
Transfection of primary neurons
SCGs from nrif−/− animals were isolated at postnatal day 2–4 and sympathetic neurons were dissociated with 0.25% trypsin and 0.3% collagenase for 30 min at 37°C. The non-neuronal cells were removed with a 1-h preplating on uncoated Falcon plate (Becton Dickinson). Then, the cell suspensions were transfected with GFP, GFP-NRIF or GFP-K19R NRIF by electroporation using program O-03 on an Amaxa Nucleofactor device. Neurons were then cultured as above with 20 ng/ml NGF and, 2 days later, the NGF was removed by washing the cultures in Ultraculture medium lacking NGF, containing 0.1 μg/ml anti-NGF (Chemicon International). Then, neurons were switched to media containing anti-NGF together with 12.5 mM KCl to promote survival with or without 100 ng/ml BDNF. After 48 h, the cells were fixed in 4% paraformaldehyde and GFP-positive neurons were counted by DAPI staining of nuclei. In each case at least 200 neurons were counted.
Acknowledgments
We are grateful to Barbara Hempstead and colleagues for providing the proNGF and to Kenneth Neet for providing Δ9/13 NGF. This work was supported by grants from the NIH NINDS NS38220 (BDC), NS33661 (MWW), and the Southeast Affiliate of the American Heart Association (TG).
References
- Bamji SX, Majdan M, Pozniak CD, Belliveau DJ, Aloyz RK, Causing CG, Miller FD (1998) The p75 neurotrophin receptor mediates neuronal apoptosis and is essential for naturally occurring sympathetic neuron death. J Cell Biol 140: 911–923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barker PA (2004) p75NTR is positively promiscuous: novel partners and new insights. Neuron 42: 529–533 [DOI] [PubMed] [Google Scholar]
- Bradley JR, Pober JS (2001) Tumor necrosis factor receptor-associated factors (TRAFs). Oncogene 20: 6482–6491 [DOI] [PubMed] [Google Scholar]
- Brennan C, Rivas-Plata K, Landis SC (1999) The p75 neurotrophin receptor influences NT-3 responsiveness of sympathetic neurons in vivo. Nat Neurosci 2: 699–705 [DOI] [PubMed] [Google Scholar]
- Casaccia-Bonnefil P, Carter BD, Dobrowsky RT, Chao MV (1996) Death of oligodendrocytes mediated by the interaction of nerve growth factor with its receptor p75. Nature 383: 716–719 [DOI] [PubMed] [Google Scholar]
- Casademunt E, Carter BD, Benzel I, Frade JM, Dechant G, Barde YA (1999) The zinc finger protein NRIF interacts with the neurotrophin receptor p75(NTR) and participates in programmed cell death. EMBO J 18: 6050–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciechanover A, Ben-Saadon R (2004) N-terminal ubiquitination: more protein substrates join in. Trends Cell Biol 14: 103–106 [DOI] [PubMed] [Google Scholar]
- de Melker AA, van der Horst G, Borst J (2004) Ubiquitin ligase activity of c-Cbl guides the epidermal growth factor receptor into clathrin-coated pits by two distinct modes of Eps15 recruitment. J Biol Chem 279: 55465–55473 [DOI] [PubMed] [Google Scholar]
- Dempsey PW, Doyle SE, He JQ, Cheng G (2003) The signaling adaptors and pathways activated by TNF superfamily. Cytokine Growth Factor Rev 14: 193–209 [DOI] [PubMed] [Google Scholar]
- Deng L, Wang C, Spencer E, Yang L, Braun A, You J, Slaughter C, Pickart C, Chen ZJ (2000) Activation of the IkappaB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103: 351–361 [DOI] [PubMed] [Google Scholar]
- Dignam JD, Lebovitz RM, Roeder RG (1983) Accurate transcription initiation by RNA polymerase II in a soluble extract from isolated mammalian nuclei. Nucleic Acid Res 11: 1475–1489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frade JM, Barde YA (1999) Genetic evidence for cell death mediated by nerve growth factor and the neurotrophin receptor p75 in the developing mouse retina and spinal cord. Development 126: 683–690 [DOI] [PubMed] [Google Scholar]
- Galan JM, Haguenauer-Tsapis R (1997) Ubiquitin lys63 is involved in ubiquitination of a yeast plasma membrane protein. EMBO J 16: 5847–5854 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geetha T, Wooten MW (2002) Structure and functional properties of the ubiquitin binding protein p62. FEBS Lett 512: 19–24 [DOI] [PubMed] [Google Scholar]
- Gentry JJ, Rutkoski NJ, Burke TL, Carter BD (2004) A functional interaction between the p75 neurotrophin receptor interacting factors, TRAF6 and NRIF. J Biol Chem 279: 16646–16656 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Karin M (2002) Missing pieces in the NF-kappaB puzzle. Cell 109: S81–S96 [DOI] [PubMed] [Google Scholar]
- Harrington AW, Kim JY, Yoon SO (2002) Activation of Rac GTPase by p75 is necessary for c-jun N-terminal kinase-mediated apoptosis. J Neurosci 22: 156–166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature 419: 135–141 [DOI] [PubMed] [Google Scholar]
- Hofmann RM, Pickart CM (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96: 645–653 [DOI] [PubMed] [Google Scholar]
- Honda K, Yanai H, Mizutani T, Negishi H, Shimada N, Suzuki N, Ohba Y, Takaoka A, Yeh WC, Taniguchi T (2004) Role of a transductional-transcriptional processor complex involving MyD88 and IRF-7 in Toll-like receptor signaling. Proc Natl Acad Sci USA 101: 15416–15421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EJ, Reichardt LF (2003) Trk receptors: roles in neuronal signal transduction. Annu Rev Biochem 72: 609–642 [DOI] [PubMed] [Google Scholar]
- Hughes AL, Messineo-Jones D, Lad SP, Neet NE (2001) Distinction between differentiation, cell cycle, and apoptosis signals in PC12 cells by the nerve growth factor mutant delta9/13, which is selective for the p75 neurotrophin receptor. J Neurosci Res 63: 10–19 [DOI] [PubMed] [Google Scholar]
- Khursigara G, Bertin J, Yano H, Moffett H, DiStefano PS, Chao MV (2001) A prosurvival function for the p75 receptor death domain mediated via the caspase recruitment domain receptor-interacting protein 2. J Neurosci 21: 5854–5863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khursigara G, Orlinick JR, Chao MV (1999) Association of the p75 neurotrophin receptor with TRAF6. J Biol Chem 274: 2597–2600 [DOI] [PubMed] [Google Scholar]
- Lallena MJ, Diaz-Meco MT, Bren G, Paya CV, Moscat J (1999) Activation of IkappaB kinase beta by protein kinase C isoforms. Mol Cell Biol 19: 2180–2188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R, Kermani P, Teng KK, Hempstead BL (2001) Regulation of cell survival by secreted proneurotrophins. Science 294: 1945–1948 [DOI] [PubMed] [Google Scholar]
- Linggi MS, Burke TL, Williams BB, Harrington A, Kraemer R, Hempstead BL, Yoon SO Carter BD (2005) NRIF is an essential mediator of apoptotic signaling by the p75 neurotrophin receptor. J Biol Chem 280: 13801–13808 [DOI] [PubMed] [Google Scholar]
- Margolin JF, Friedman JR, Meyer WK, Vissing H, Thiesen HJ, Rauscher FJ III (1994) Kruppel-associated boxes are potent transcriptional repression domains. Proc Natl Acad Sci USA 91: 4509–4513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marmor MD, Yarden Y (2004) Role of protein ubiquitylation in regulating endocytosis of receptor tyrosine kinases. Oncogene 23: 2057–2070 [DOI] [PubMed] [Google Scholar]
- Muratani M, Tansey WP (2003) How the ubiquitin–proteasome system controls transcription. Nat Rev Mol Cell Biol 4: 192–201 [DOI] [PubMed] [Google Scholar]
- Palmada M, Kanwal S, Rutkoski NJ, Gufstafson-Brown C, Johnson RS, Wisdom R, Carter BD (2002) c-jun is essential for sympathetic neuronal death induced by NGF withdrawal but not by p75 activation. J Cell Biol 158: 453–461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70: 503–533 [DOI] [PubMed] [Google Scholar]
- Plafker SM, Plafker KS, Weissman AM, Macara IG (2004) Ubiquitin charging of human class III ubiquitin-conjugating enzymes triggers their nuclear import. J Cell Biol 167: 649–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puls A, Schmidt S, Grawe F, Stabel S (1997) Interaction of protein kinase C zeta with ZIP, a novel protein kinase C-binding protein. Proc Natl Acad Sci USA 94: 6191–6196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rachubinski RA, Marcus SL, Capone JP (1999) The p56 (lck)-interacting protein p62 stimulates transcription via the SV40 enhancer. J Biol Chem 274: 18278–18284 [DOI] [PubMed] [Google Scholar]
- Roux PP, Barker PA (2002) Neurotrophin signaling through the p75 neurotrophin receptor. Prog Neurobiol 67: 203–233 [DOI] [PubMed] [Google Scholar]
- Sanz L, Diaz-Meco MT, Nakano H, Moscat J (2000) The atypical PKC-interacting protein p62 channels NF-kappaB activation by the IL-1–TRAF6 pathway. EMBO J 19: 1576–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz DC, Friedman JR, Rauscher FJ III (2001) Targeting histone deacetylase complexes via KRAB-zinc finger proteins: the PHD and bromodomains of KAP-1 form a cooperative unit that recruits a novel isoform of the Mi-2alpha subunit of NuRD. Genes Dev 15: 428–443 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seibenhener ML, Babu JR, Geetha T, Wong HC, Krishna NR, Wooten MW (2004) Sequestosome 1/p62 is a polyubiquitin chain binding protein involved in ubiquitin proteasome degradation. Mol Cell Biol 24: 8055–8068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shcherbik N, Haines DS (2004) Ub on the move. J Cell Biochem 93: 11–19 [DOI] [PubMed] [Google Scholar]
- Spence J, Sadis S, Haas AL, Finley D (1995) A ubiquitin mutant with specific defects in DNA repair and multiubiquitination. Mol Cell Biol 15: 1265–1273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun L, Chen ZJ (2004) The novel functions of ubiquitination in signaling. Curr Opin Cell Biol 16: 119–126 [DOI] [PubMed] [Google Scholar]
- Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D (2004) Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem 279: 7055–7063 [DOI] [PubMed] [Google Scholar]
- Wang C, Deng L, Hong M, Akkaraju GR, Inoue J, Chen ZJ (2001) TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412: 346–351 [DOI] [PubMed] [Google Scholar]
- Williams BB, Gentry JJ, Rutkotski NJ, Linggi MS, Carter BD (2004) The p75 interacting factor NRIF functions as a transcriptional repressor (abstract). Soc Neurosci: Program No. 726.21 [Google Scholar]
- Wooten MW, Seibenhener ML, Mamidipudi V, Diaz-Meco MT, Barker PA, Moscat J (2001) The atypical protein kinase C-interacting protein p62 is a scaffold for NF-kappaB activation by nerve growth factor. J Biol Chem 276: 7709–7712 [DOI] [PubMed] [Google Scholar]
- Ye H, Arron JR, Lamothe B, Cirilli M, Kobayashi T, Shevde NK, Segal D, Dzivenu OK, Vologodskaia M, Yim M, Du K, Singh S, Pike JW, Darnay BG, Choi Y, Wu H (2002) Distinct molecular mechanism for initiating TRAF6 signalling. Nature 418: 443–447 [DOI] [PubMed] [Google Scholar]
- Yeiser EC, Rutkoski NJ, Naito A, Inoue J, Carter BD (2004) Neurotrophin signaling through the p75 receptor is deficient in traf6−/− mice. J Neurosci 24: 10521–10529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon SO, Casaccia-Bonnefil P, Carter B, Chao MV (1998) Competitive signaling between TrkA and p75 nerve growth factor receptors determines cell survival. J Neurosci 18: 3273–3281 [DOI] [PMC free article] [PubMed] [Google Scholar]


