Abstract
The pre-B-cell receptor (pre-BCR), composed of Ig heavy and surrogate light chain (SLC), signals pre-BII-cell proliferative expansion. We have investigated whether the pre-BCR also signals downregulation of the SLC genes (VpreB and λ5), thereby limiting this expansion. We demonstrate that, as BM cells progress from the pre-BI to large pre-BII-cell stage, there is a shift from bi- to mono-allelic λ5 transcription, while the second allele is silenced in small pre-BII cells. A VpreB1-promoter-driven transgene shows the same pattern, therefore suggesting that VpreB1 is similarly regulated and thereby defines the promoter as a target for transcriptional silencing. Analyses of pre-BCR-deficient mice show a temporal delay in λ5 downregulation, thereby demonstrating that the pre-BCR is essential for monoallelic silencing at the large pre-BII-cell stage. Our data also suggest that SLP-65 is one of the signaling components important for this process. Furthermore, the VpreB1/λ5 alleles undergo dynamic changes with respect to nuclear positioning and heterochromatin association, thereby providing a possible mechanism for their transcriptional silencing.
Keywords: B-cell development, heterochromatin, pre-B-cell receptor, SLP-65, surrogate light chain
Introduction
Early in B-cell development, functional VDJH rearrangement of the IgH locus results in the expression of a μH chain (μHC) which, together with surrogate light chain (SLC), forms the precursor-B-cell receptor (pre-BCR) (Pillai and Baltimore, 1987; Karasuyama et al, 1990; Tsubata and Reth, 1990). It has been proposed that expression of the pre-BCR serves as a checkpoint to monitor functional IgH recombination and signals proliferative expansion of pre-BII cells, resulting in the establishment of a broad repertoire of antigen-binding Ig molecules (Melchers, 2005). Indeed, pre-BII-cell expansion is prevented in mice lacking either the transmembrane region of μHC, the SLC components VpreB or λ5, or the entire SLC (Kitamura et al, 1991, 1992; Mundt et al, 2001; Shimizu et al, 2002). While the pre-BCR promotes pre-BII-cell proliferation, in vitro studies suggest that the signaling molecules SLP-65 (BLNK/BASH) and btk act by limiting this proliferation (Middendorp et al, 2002; Flemming et al, 2003). In mice lacking these signaling molecules, B-cell development is impaired at the pre-B-cell stage (Hendriks and Middendorp, 2004). These cells express unusually high levels of pre-BCR and the enhanced in vitro proliferative capacity requires expression of the pre-BCR and IL7R (Flemming et al, 2003). Furthermore, SLP-65 and btk act as tumor suppressors since mice lacking these molecules develop pre-B-cell tumors (Flemming et al, 2003; Hayashi et al, 2003; Jumaa et al, 2003; Kersseboom et al, 2003).
It has been proposed that an additional function of the pre-BCR is to signal downregulation of the genes encoding the SLC, that is, VpreB and λ5 (Grawunder et al, 1995; Dul et al, 1996; Melchers, 2005), although this remains controversial (Stephan et al, 2001). Self-limiting expression of the pre-BCR would provide a means of regulating the extent of pre-BCR-induced cellular expansion. The pre-BCR has also been implicated in downregulating components of the recombinase machinery, such as the recombination-activating genes (RAG1 and 2) and terminal deoxynucleotidyl transferase (TdT) (Grawunder et al, 1995). This would contribute to the mechanism ensuring heavy-chain allelic exclusion. However, more recent evidence suggest that downregulation of Rag and TdT is initiated by μHC, in a pre-BCR-like complex lacking SLC (Galler et al, 2004). Therefore downregulation of the SLC genes may also be regulated by μHC in a pre-BCR-like complex rather than by the conventional pre-BCR.
Here, we have investigated whether the pre-BCR signals downregulation of SLC gene expression. Our results suggest that the pre-BCR, via the tumor suppressor SLP-65, induces silencing of SLC gene transcription. Furthermore, our data suggest that silencing of the VpreB1/λ5 locus is initiated by transient association with heterochromatin and maintained by relocation of both alleles at the nuclear periphery.
Results
The pre-BCR is expressed on transitional pre-BI and large pre-BII cells
To determine whether the pre-BCR limits its own expression by downregulating SLC gene transcription, it was essential first to determine at which stages during B-cell development SLC, that is, VpreB and λ5, and the pre-BCR are expressed (Figure 1A). As both are difficult to detect on the surface of primary B-lineage cells, we used a technique that enhances detection. As shown in Figure 1B, while 88% of the pre-BI population expressed SLC, only 15% expressed the pre-BCR on the cell surface. These proportions were confirmed by staining for cytoplasmic SLC versus μHC (Figure 1C); 83% stained positive for SLC, with 13% also expressing μHC. Both these populations expressed similar levels of SLC. We have termed the μHC+ subpopulation as transitional pre-BI cells (Martensson et al, 2002). The pre-BII population consists of large, cycling and small resting cells in the ratio of ∼1:4 (Rolink et al, 1994). Of the pre-BII population, 18 and 12% were positive for SLC and pre-BCR, respectively (Figure 1D). Cytoplasmic staining demonstrated that only large pre-BII cells expressed SLC, whereas both large and small cells, as expected, stained positive for μHC (Figure 1E). Thus, the pre-BCR is expressed on transitional pre-BI and large pre-BII cells. Furthermore, although the SLC level is lower in the large pre-BII cells, that of the pre-BCR is relatively similar.
Figure 1.
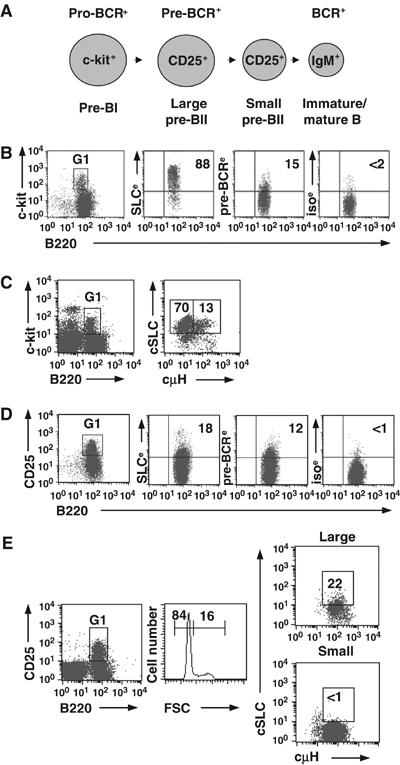
Pro- and pre-BCR expression in ex vivo mouse BM B cells. (A) B-cell development showing expression of the pro-BCR (SLC with glycoproteins), pre-BCR (SLC with μHC) and BCR (μHC with IgL chain). Large cells are cycling and small cells are noncycling. Cell surface markers used are also shown. All cells are B220+CD19+. (B) CD19-enriched BM cells were gated (G1) on pre-BI cells and analyzed for surface expression of SLC, pre-BCR or isotype control, (e) indicates that the EAS kit was used. As the levels of λ5 (shown) and VpreB were similar, λ5+ cells represent SLC+ cells. (C) BM cells were gated (G1) on pro-/pre-BI cells and analyzed for intracellular expression of SLC and μHC. (D) As in (B), except that cells were gated (G1) on pre-BII cells. (E) As in (C), except that cells were gated (G1) on pre-BII cells and size. Representative results from at least three experiments.
The λ5 gene is still transcribed at the large pre-BII-cell stage
After determining the stages at which the pre-BCR is expressed, we investigated the transcriptional status of the λ5 gene during BM B-cell development. To directly measure transcription rather than mRNA steady-state levels, the cells were analyzed using RNA fluorescent in situ hybridization (FISH). This technique has the advantage of detecting primary transcripts within individual nuclei. A single-stranded DNA probe, specific for λ5 primary transcripts, was used (Figure 2A). A probe specific for CD45 was used as a control, since this gene is biallelically transcribed (Skok et al, 2001) and expressed in all B-lineage cells up to the plasma cell stage.
Figure 2.
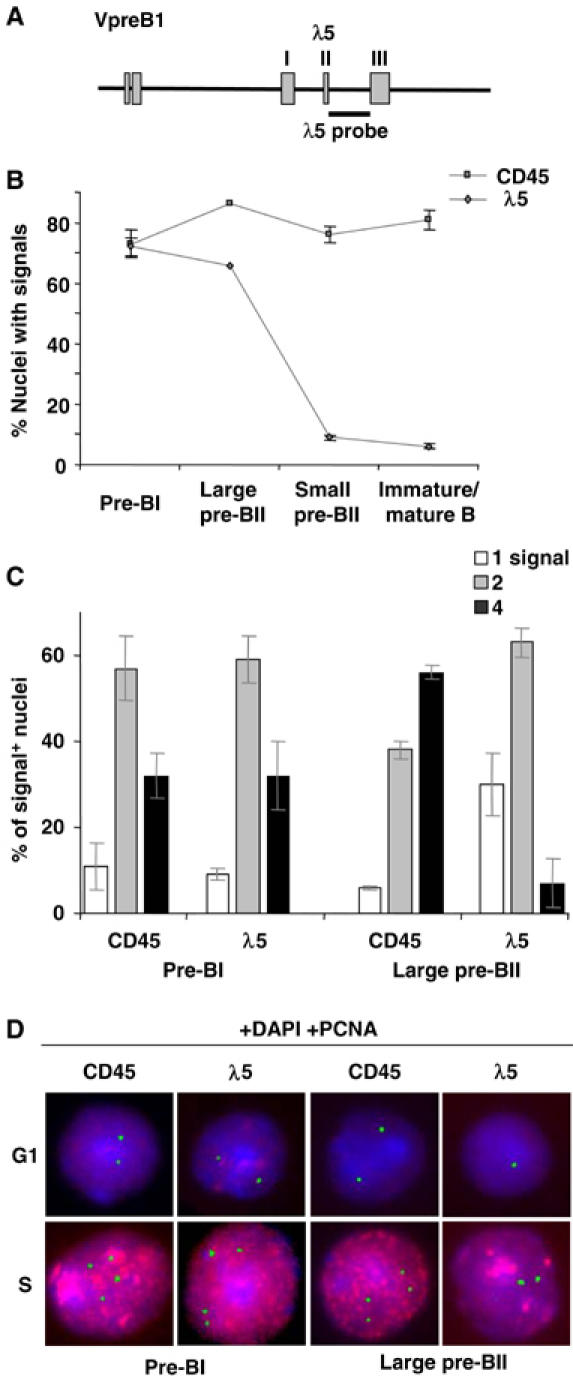
Transcription patterns of the λ5 and CD45 genes during B-cell development. (A) Schematic diagram of the VpreB1-λ5 locus. The position of the λ5 probe is indicated. (B) RNA FISH. Percentages (±s.e.m.) of nuclei with CD45 or λ5 signals in sorted BM cells from indicated stages. (C) Percentages (±s.d.) of signal-positive nuclei containing either one, two or four signals at the pre-BI and large pre-BII stages. A total of >900 nuclei were counted for each stage (>3 experiments). (D) Typical images of nuclei hybridized with either the CD45 or λ5 probe (green), in combination with an antibody recognizing PCNA (red), distinguishing S-phase nuclei. DNA was counterstained with DAPI (blue) to verify nuclear integrity. For CD45, typically two (G1 phase) or four (S phase) foci were observed in both pre-BI and pre-BII cells. While two (G1) or four (S) λ5 foci were also observed in pre-BI cells, only one (G1) or two (S) were observed in large pre-BII cells.
Initially, the various stages of BM B-cell development were analyzed (Supplementary data 1). If expression of the pre-BCR turns off the SLC genes, we postulated that λ5 should be transcriptionally silent in large pre-BII cells. As shown in Figure 2B, the proportion of nuclei containing CD45 signals remained constant (75–85%) at all stages. At the pre-BI stage, λ5 signals were detected in 70–75% of nuclei. Unexpectedly, the proportion of nuclei containing λ5 foci (∼65%) at the large pre-BII stage was similar to that of the pre-BI stage. In contrast, at the small pre-BII and immature B-cell stages, the proportion of nuclei with λ5 signals was greatly reduced and close to background levels. This demonstrates therefore that the λ5 gene is transcribed in the majority of pre-BI and large pre-BII cells.
λ5 transcription is biallelic in pre-BI but monoallelic in large pre-BII cells
As the RNA FISH technique allows detection of transcription at individual alleles, a more detailed analysis of the number of λ5 and CD45 transcription sites in individual nuclei was carried out (Figure 2 and Supplementary data 2). This revealed that the vast majority (∼90%) of signal-positive pre-BI nuclei contained either two or four CD45 or λ5 RNA foci, with very few containing only one (Figure 2C). It is likely that four signals represent transcription from replicated alleles, as 25–45% of pre-BI cells are in S/G2/M of the cell cycle (Karasuyama et al, 1994; Rolink et al, 1994). To determine whether cells containing four foci were in fact in the S phase of the cell cycle, RNA FISH was performed together with proliferating cell nuclear antigen (PCNA) staining to specifically identify replicating nuclei (Morris and Mathews, 1989). These analyses revealed that the majority of pre-BI nuclei with either four CD45 or λ5 signals were PCNA-positive and therefore in S phase (Figure 2D). In contrast, the majority of nuclei containing two signals were PCNA-negative and therefore either noncycling, or cells not yet in S phase. The few PCNA-negative nuclei with four CD45 or λ5 signals were large and probably in early G2 phase. Thus, at the pre-BI stage, transcription of CD45 and λ5 was detected on both alleles in cells in G1- and on replicated alleles in S-phase cells. Furthermore, the transcription pattern of CD45 and λ5 was similar in these cells. We therefore conclude that the λ5 gene is biallelically transcribed at the pre-BI-cell stage.
Thereafter, we analyzed large pre-BII cells (Figure 2, Supplementary data 2). The vast majority (∼95%) of signal-positive nuclei contained two or four CD45 signals, with most of the latter also being PCNA+ (Figure 2C and D). The proportion of nuclei with four CD45 foci was greater in the large pre-BII (∼55%) compared to that in the pre-BI (∼25%) population, in agreement with 65–70% of large pre-BII cells residing in S/G2/M of the cell cycle (Rolink et al, 1994). Surprisingly, and in contrast to the CD45 transcription pattern, the vast majority (>95%) of large pre-BII nuclei contained either one or two λ5 foci with most of the latter being PCNA+ (Figure 2C and D). In pre-BI nuclei the proportion with four signals was similar for both genes, whereas in large pre-BII nuclei the proportion with four CD45 and two λ5 foci was similar, about 55 and 60%, respectively. Since the large pre-BII population consists of cycling cells, these data demonstrate that at this stage the predominant transcription pattern of CD45 is biallelic, whereas that of λ5 is monoallelic. At the small pre-BII- and immature B-cell stages, while most nuclei contained two CD45 signals, as would be expected for resting cells, λ5 signals were observed in <10% of nuclei. Thus, silencing of both λ5 alleles has occurred by the small pre-BII stage and both alleles remain inactive at the subsequent immature B-cell stage. Furthermore, the observed switch from bi- to monoallelic λ5 transcription follows cell surface expression of the pre-BCR, whereas silencing of the second λ5 allele is coincident with differentiation and cell cycle exit.
Silencing of λ5 transcription also follows induction of pre-BCR expression in vitro
To investigate more directly whether pre-BCR expression results in downregulation of SLC gene transcription, we utilized a transgenic (TG) Ig-tTA/tet-μ/Rag2−/− (tet-μH) mouse model in which expression of a TG μHC can be experimentally manipulated (Hess et al, 2001). The in vitro induction of μHC in pre-BI cells from the tet-μH mice causes the cells to proliferate in a pre-BCR-dependent fashion and this is followed by differentiation into small, resting cells.
CD19+ BM cells from tet-μH mice (treated with tetracycline) were cultured in vitro for 4 days in the absence of tetracycline to allow re-expression of μHC (Figure 3 and Supplementary data 3). FACS analysis showed that, on days 0–2, SLC was expressed on the surface of the majority (70–80%) of cells, whereas, after days 3 and 4 in culture, <5% of cells expressed SLC (Figure 3A). At day 1, induction of the TG was apparent with ∼40% of the cells being surface μHC+, which increased to 63% by day 2. Forward scatter analysis showed that on days 0 and 1 ∼45% of the cells were large and at day 2 this proportion had increased to ∼58%. However, by day 4, the vast majority (95%) of cells had become small, resting cells.
Figure 3.
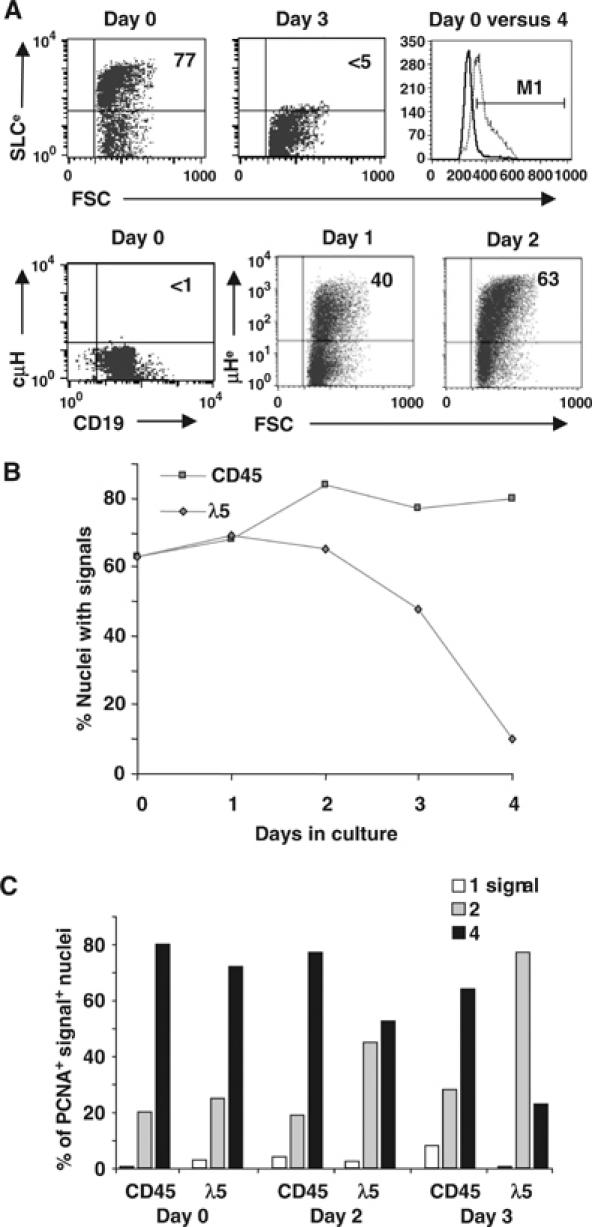
Transcription pattern of the λ5 gene after induction of pre-BCR expression. (A) FACS analysis of CD19-enriched BM pre-BI cells from tet-μH mice before and at the indicated times following the in vitro induction of μHC expression. The following is shown: surface expression of SLC and μHC using EAS (e) and cytoplasmic μH expression; cell size at days 0 and 4 (dotted and filled lines, respectively, in histogram). (B) RNA FISH on the cells in (A) using either CD45 or λ5 probes daily following induction of TG μHC, that is, pre-BCR, expression. Percentages of nuclei with signals are shown. (C) Percentages of PCNA+ signal-positive nuclei containing one, two or four signals at the indicated times. The data from one representative experiment (out of three) are shown. A total of >900 nuclei were counted for each time point.
RNA FISH analysis showed that the number of nuclei with CD45 and λ5 signals was similar (65–70%) on days 0 and 1 (Figure 3B and Supplementary data 3). With time, the proportion of CD45-positive nuclei increased to ∼80%. In contrast, by day 2, the proportion of λ5-positive nuclei had not increased and by day 4 this had fallen to 10%. Thus, by the time that most of the in vitro cultured cells had differentiated into small, resting cells, the majority had silenced λ5 transcription. Thereafter, the number of signals in individual nuclei was determined (Figure 3C and Supplementary data 3). As observed in ex vivo cells, transcription was also detected from replicated alleles in the cultured cells, which was confirmed using PCNA. The proportion of PCNA-positive nuclei containing four CD45 foci was similar (∼80%) at days 0–2, with a slight decrease (∼65%) at day 3. The proportion of PCNA-positive nuclei containing four λ5 foci was similar (∼75%) to that of CD45 at days 0 and 1. However, at day 2, and even more so at day 3, the percentage with four λ5 foci was greatly reduced (55 and 20%, respectively), while that with two increased. By day 4, the few nuclei transcribing λ5 did so from only one allele. Thus, also in these in vitro cultures, there is a switch from bi- to monoallelic λ5 transcription following pre-BCR expression, while differentiation and cell cycle exit is accompanied by silencing of the second λ5 allele.
VpreB1 shows the same transcription pattern as λ5
The asynchronous silencing of the two λ5 alleles prompted us to investigate whether this pattern is unique for λ5 or whether other genes are regulated in the same fashion, with an obvious candidate being VpreB. In mice, there are two genes, VpreB1 and VpreB2, which are 97% identical (Kudo and Melchers, 1987). VpreB1 is located 4.5 kb upstream of λ5 and VpreB2 ∼1 Mb downstream. Analysis of pre-BI cells, using two different VpreB probes, revealed nuclei containing between two and eight signals (data not shown), perhaps due to a difference in the transcription of the two genes (Dul et al, 1996), making interpretation of the results difficult. We therefore took advantage of a TG mouse line (−214VpreB1-HuCD122), in which expression of the reporter gene is under the control of the VpreB1 promoter (Licence et al, 2003). As reporter gene expression mimics that of the endogenous VpreB1 gene during B-cell development, this was used as a marker for VpreB1. Mice homozygous for the transgene were analyzed by RNA FISH, using a probe (TG) which detects primary transcripts from the transgene and λ5 as a control. As previously observed for λ5 and CD45, transcription of the transgene was also detected from replicated alleles. As shown in Figure 4, the majority (90%) of signal-positive pre-BI nuclei contained either two or four TG signals, with a ratio similar to that of λ5. At the large pre-BII stage, the majority of signal-positive nuclei contained either one or two TG signals, again with a ratio similar to that of λ5. At the small pre-BII stage, the transgene was not transcribed, with <5% of nuclei containing signals (data not shown). This demonstrates that the transcription pattern of the VpreB1 promoter-driven transgene is similar to that of λ5, and that there is a switch from bi- to monoallelic transcription at the large pre-BII stage and silencing of the second allele in small pre-BII cells. Thus, asynchronous allelic silencing is not unique to the λ5 gene, but is also observed for VpreB1. Furthermore, these results demonstrate that the 350-bp VpreB1 promoter region is sufficient to ensure appropriate mono- and biallelic silencing.
Figure 4.
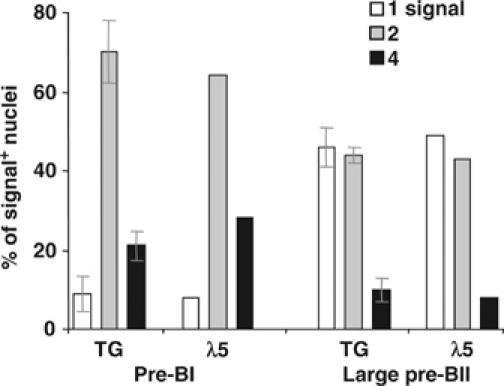
Transcription pattern of the −214VpreB1-HuCD122 transgene during B-cell development. RNA FISH analysis of ex vivo BM pre-BI and large pre-BII cells from −214VpreB1-HuCD122+/+ TG mice, in which the reporter gene is driven by the VpreB1 promoter. The percentages of signal-positive nuclei with one, two or four TG or λ5 signals are shown. For the TG, a total of >200 nuclei was counted for each stage (two experiments).
Pre-BCR expression per se is not sufficient to silence λ5 transcription
The observed switch from bi- to monoallelic SLC gene transcription takes place in ex vivo large pre-BII cells and at day 2 after induction of the pre-BCR in vitro. However, the pre-BCR is already expressed on transitional pre-BI cells (Figure 1B) and at day 1 in the in vitro culture system (Figure 3A). It was therefore unclear whether pre-BCR expression per se was sufficient to signal downregulation of SLC gene transcription. As transitional pre-BI cells only represent 10–15% of the total pre-BI population, a switch to monoallelic SLC gene transcription in this subpopulation might not have been detected and this was therefore investigated. In ex vivo transitional pre-BI cells, the majority of signal-positive nuclei contained either two (∼75%) or four (∼20%) CD45 or λ5 signals (Figure 5A). In in vitro cultured transitional pre-BI cells, the pattern of λ5 transcription was similar to that of the total pre-BI-cell population, with most nuclei containing either two (∼70%) or four (25–30%) signals (Figure 5B). From this we conclude that the λ5 gene is biallelically transcribed in transitional pre-BI cells and that pre-BCR expression per se is not sufficient to induce monoallelic λ5 transcription.
Figure 5.
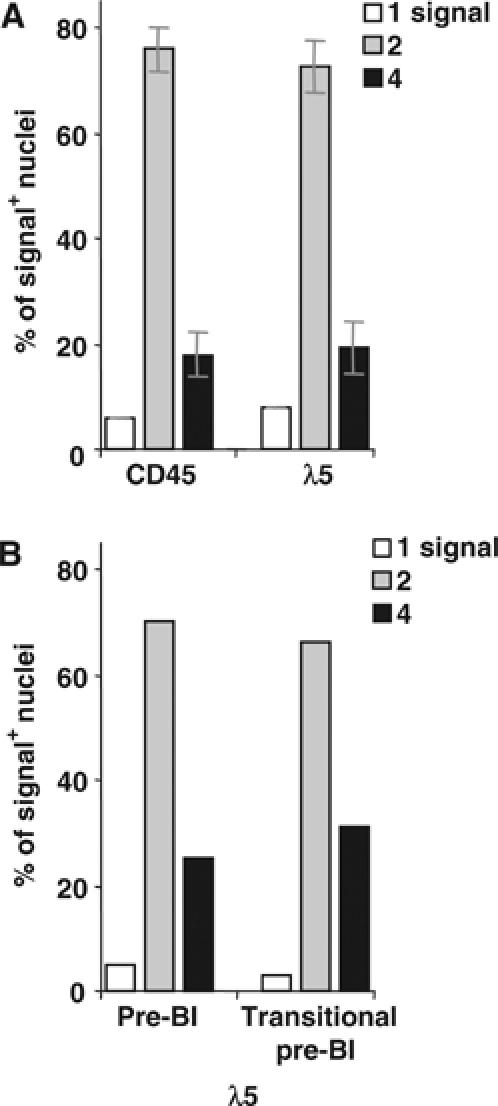
RNA FISH analysis of transitional pre-BI cells from normal mice. (A) Sorted ex vivo BM transitional pre-BI cells. (B) In vitro cultured transitional pre-BI cells. Total pre-BI cells before culture were included for comparison. The percentages of signal-positive nuclei with one, two or four signals are shown using the indicated probes. The results from (A) two experiments (±s.d.) and (B) one experiment are shown. A total of >300 nuclei were counted for each population.
Silencing of λ5 transcription is temporally delayed in pre-BCR-deficient mice
As pre-BCR expression per se did not induce silencing of the λ5 gene, it may be that another receptor complex is responsible for this signal. To investigate this, the transcriptional status of the λ5 gene was analyzed in mice lacking a functional pre-BCR. For these experiments, VpreB1−/−VpreB2−/− (VpreB−/−) mice (Mundt et al, 2001), in which the λ5 gene is intact, were used. Although the number of pre-BII cells is reduced (>20-fold) in these mice (Mundt et al, 2001), a pre-BII-cell population (CD25+cμH+) is still present (Figure 6A). We postulated that if the pre-BCR initiates the switch from bi- to monoallelic λ5 transcription, pre-BII cells from these mice should transcribe both alleles. Alternatively, if another receptor is responsible, λ5 transcription should be monoallelic. In VpreB−/− mice, the majority (>90%) of signal-positive pre-BI nuclei contained either two or four λ5 or CD45 signals (Figure 6B). In pre-BII cells, the majority (>80%) of nuclei contained two CD45 signals and ∼10% contained four (Figure 6B). In agreement with this and, as expected, the majority of pre-BII cells were PCNA− (data not shown). Analysis of nuclei with λ5 signals revealed a pattern that was similar to that of CD45, with >70% of nuclei containing two signals and ∼15% four. Thus, the pre-BCR is required to silence the λ5 gene at the pre-BII-cell stage. Furthermore, as the majority (∼90%) of pre-BII cells in VpreB−/− mice are resting, these results suggest that cell cycle exit per se is not sufficient to silence the λ5 gene. To investigate whether λ5 transcription is maintained, IgM+ splenic B cells from VpreB−/− and wt mice were analyzed. In both these populations, the majority (>90%) of signal-positive nuclei contained two CD45 foci, while <5% were observed for λ5 (data not shown). Thus, by the mature B-cell stage, the λ5 gene is also inactive in mutant mice. This demonstrates therefore that silencing of the λ5 gene is temporally delayed in pre-BCR-deficient mice.
Figure 6.
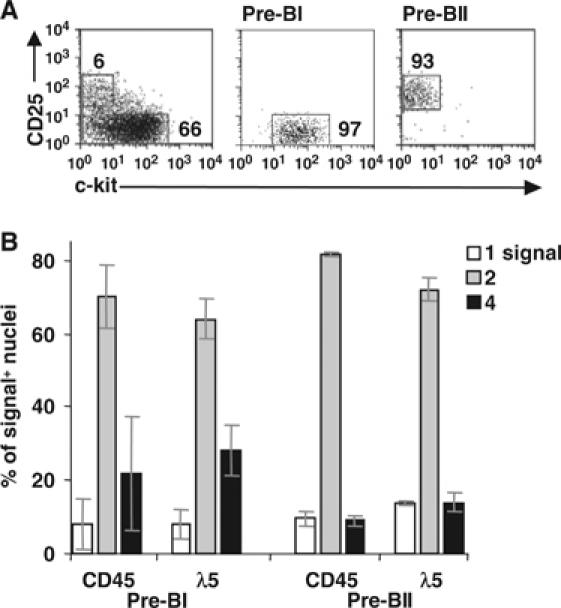
Analysis of pre-BCR-deficient pre-BI and pre-BII cells. (A) CD19-enriched BM cells from VpreB−/− mice were sorted as pre-BI and pre-BII cells. (B) RNA FISH analysis of the cells in (A). The percentages (±s.d.) of signal-positive nuclei with one, two or four signals using the indicated probes are shown. A total of >600 nuclei were counted for each population (at least two experiments).
Biallelic λ5 transcription in pre-BCR-expressing cells in the absence of SLP-65
While the pre-BCR promotes pre-BII-cell proliferation, the signaling molecule SLP-65 (BLNK/BASH) may be involved in controlling this proliferation by downregulating pre-BCR expression and inducing pre-B-cell differentiation (Flemming et al, 2003). These cells express unusually high levels of pre-BCR and show enhanced pre-BCR-dependent in vitro proliferative capacity. To investigate whether the high level of pre-BCR results from an inability to downregulate the SLC genes, two cell lines derived from SLP-65−/− mice (Flemming et al, 2003) were analyzed. FACS analysis of these cells confirmed the high pre-BCR levels (Supplementary data 4). RNA FISH analysis demonstrated that similar proportions (∼70%) of nuclei contained CD45 and λ5 signals (data not shown). Using either probe, the vast majority (>90%) of nuclei contained either two or four foci (Figure 7A). Furthermore, the ratio of two versus four signals was similar for the probes. Thus, the λ5 gene is biallelically transcribed in SLP-65−/− pre-B-cell lines.
Figure 7.
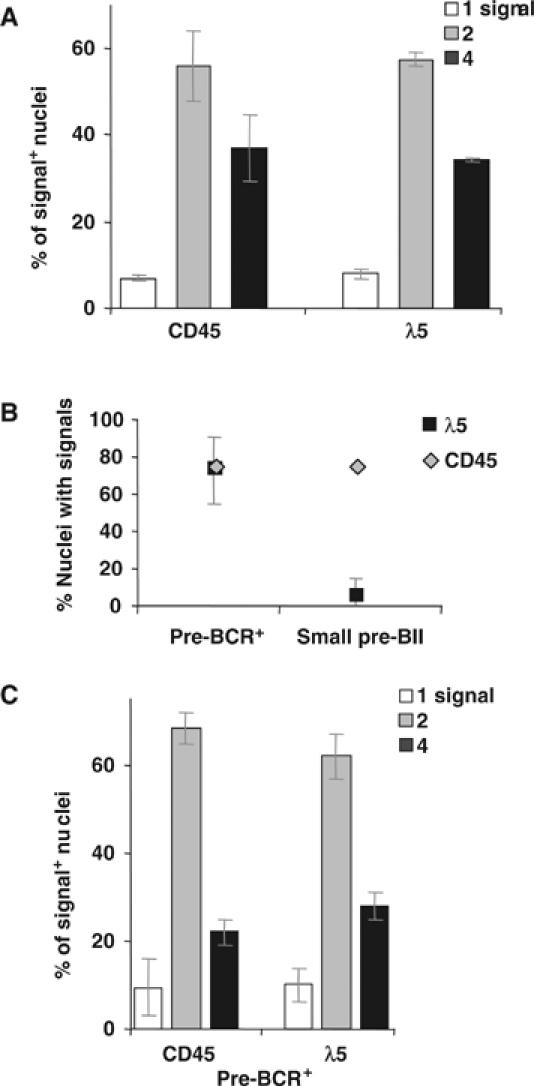
RNA FISH analysis of pre-B cells lacking SLP-65. (A) The percentages (±s.e.m.) of signal-positive nuclei with one, two or four signals using the indicated probes are shown for the SLP-65−/− cell line, 26.5. (B) Percentages (±s.d.) of nuclei with CD45 or λ5 signals are shown for pre-BCR+ and small pre-BII cells from SLP-65−/− mice. (C) Percentages (±s.d.) of signal-positive nuclei containing one, two or four signals in pre-BCR+ cells. (A–C) A total of >300 nuclei were counted (at least two experiments).
It is possible that leukemic clones predominate during culture and that the above lines do not represent the situation in vivo. Although pre-BCR-dependent in vitro proliferation is increased in SLP-65−/− pre-B cells, the situation in vivo is not fully clear (Hayashi et al, 2003). Therefore, to more directly determine whether SLP-65 is important for downregulation of λ5 transcription, ex vivo BM cells from mutant mice were analyzed. In agreement with previous results (Flemming et al, 2003; Hayashi et al, 2003), a pre-BCR+ cell population, most of which were c-kit−CD43+CD25– and, in addition, a pre-BII population, of which >90% were small cells, were detected (Supplementary data 4). RNA FISH analysis of these two cell populations revealed similar (75–85%) proportions of nuclei with CD45 signals (Figure 7B). In contrast, λ5 signals were observed in ∼80% of pre-BCR+ but in <5% of small pre-BII cells. Thus, silencing of both λ5 alleles also takes place in mutant small pre-BII cells. In pre-BCR+ cells, the majority of nuclei contained two (∼70%) or four (∼20%) CD45 or λ5 signals (Figure 7C). Most nuclei with four signals were PCNA+, in agreement with a proportion of cells in S phase. Analysis of small pre-BII nuclei showed that >80% contained two and only ∼5% four CD45 signals, with the vast majority of nuclei being PCNA−, as expected in a noncycling cell population. Thus, in contrast to wt mice, both λ5 alleles are transcribed in mutant pre-BCR+ cells. This therefore suggests that SLP-65 is important for silencing SLC gene transcription.
The position of λ5 alleles alters in relation to γ-satellite DNA during B-cell development
The unusual asynchronous silencing of the two VpreB1/λ5 alleles prompted us to investigate the mechanism by which this is achieved. Previous studies have implicated centromeric recruitment in transcriptional silencing (Brown et al, 1997, 1999; Skok et al, 2001). In order to ascertain whether this process is involved in silencing the VpreB1/λ5 locus, 3D-FISH was carried out to examine the position of VpreB1/λ5 alleles relative to centromeric clusters in sorted BM B-cell populations (Figure 8A). The experiments were performed as described previously (Roldan et al, 2005) using a γ-satellite probe to detect centromeric clusters in conjunction with a VpreB1/λ5 locus probe (termed λ5).
Figure 8.
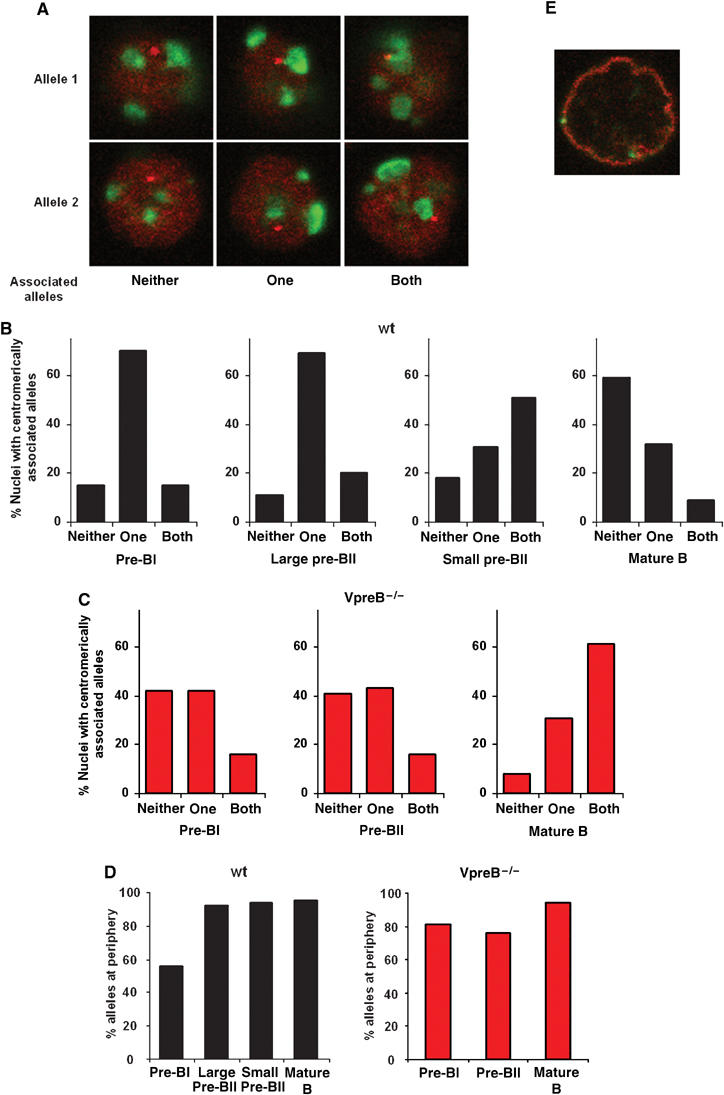
DNA FISH analysis of BM and splenic B cells from wt and pre-BCR-deficient mice. (A) Confocal section images of nuclei after DNA FISH are shown, combining the λ5 locus probe (red) with a probe detecting γ-satellite DNA (green). Images demonstrate association between a single, both or neither λ5 allele and γ-satellite DNA. Alleles 1 and 2 indicate the image plane where the respective allele is observed. (B) Percentages of nuclei with a single, both or neither λ5 allele associated with γ-satellite DNA in the indicated cell populations from wt mice. Association was scored when an allele was in apparent contact with γ-satellite foci. (C) As in (B), except that cells were from VpreB−/− mice. (D) Percentages of alleles located at the nuclear periphery in the indicated cell populations. Cell populations were from wt (left) and VpreB−/− (right) mice. (E) Immuno-DNA FISH analysis of wt splenic B cells using the λ5 locus probe (green) and staining with antibodies specific to the amino- and carboxy-terminal portions of lamin B (red).
The pattern of colocalization between λ5 alleles and γ-satellite DNA in pre-BI and large pre-BII nuclei was relatively similar, with a single allele colocalized in the majority (∼70%) of nuclei (Figure 8B and Supplementary data 5). In contrast, at the small pre-BII stage, where both λ5 alleles are transcriptionally inactive, half of the cells have both λ5 alleles positioned at centromeric clusters. In mature, resting splenic B cells, more than half (60%) of the nuclei have both λ5 alleles positioned away from centromeric regions. These data indicate that, during transcriptional silencing, λ5 alleles undergo dynamic changes in nuclear location with respect to centromeric heterochromatin, suggesting that recruitment of λ5 to these regions may be important for silencing. Recruitment of λ5 alleles to heterochromatic domains is transient and by the mature B-cell stage maintenance of the silent state appears to be independent of continued association with these domains.
As shown above, transcriptional silencing of λ5 is delayed in pre-BCR-deficient mice and not observed until the mature B-cell stage. Therefore, if centromeric recruitment is involved in transcriptional silencing, the pattern of association between λ5 alleles and heterochromatin should be different in these mice. Analysis of VpreB−/− mice showed a similar pattern of centromeric association in the pre-BI and pre-BII-cell populations (Figure 8C and Supplementary data 5), both of which show biallelic λ5 transcription. The proportion of nuclei with neither λ5 allele associated with γ-satellite DNA was higher than in wt mice (40% compared to 10–15%), while the proportion of cells with one allele associated was much lower (∼40% compared to 70%). In mature splenic B cells from VpreB−/− mice, most (60%) nuclei had both alleles positioned at centromeric clusters. This is in contrast to wt mature splenic B cells where most (∼60%) nuclei have both alleles positioned away from heterochromatic regions. The positioning of λ5 alleles in the mutant splenic B cells is therefore reminiscent of the pattern observed in wt small pre-BII cells. This is noteworthy, since these populations represent the first respective stage at which transcriptional silencing of both λ5 alleles is observed. These data suggest that silencing both in wt and pre-BCR-deficient mice is achieved by repositioning of λ5 alleles to centromeric clusters; however, in the mutant mice, transcriptional silencing and repositioning are both delayed.
Relocation of λ5 alleles in relation to heterochromatin may be required to initiate and maintain transcriptional repression
Subnuclear compartmentalization of genes at the nuclear periphery has been shown to be a way of regulating gene expression (Kosak et al, 2002; Fuxa et al, 2004). To assess whether association with the nuclear periphery has a role in regulating expression of the VpreB1/λ5 locus, we examined the positioning of λ5 alleles relative to this subcompartment.
In the pre-BI fraction from wt mice, about half of the alleles were peripheral. In contrast, the vast majority (∼95%) of alleles were located at the nuclear periphery in both the large and small pre-BII cells as well as in mature, splenic B cells (Figure 8D, left, and Supplementary data 5). These data are supported by a more detailed analysis of nuclei from resting, splenic B cells in which both λ5 alleles localized to the inner nuclear membrane, as revealed by lamin B staining (Figure 8E). The analyses on centromeric repositioning of λ5 alleles did not show any differences between the pre-BI and large pre-BII cells. This is in contrast to the results examining the location of λ5 relative to the nuclear periphery. Here, there is a difference between the two developmental stages, at a time when there is a change from bi- to monoallelic transcription. Thus, in addition to centromeric recruitment, localization to the nuclear periphery may be important in initiating silencing of the VpreB1/λ5 locus. Furthermore, once silenced, relocation at the nuclear periphery away from heterochromatin may have a role in maintaining the silent state of the VpreB1/λ5 locus.
To investigate this further, we examined the position of λ5 alleles relative to the nuclear periphery in nuclei from VpreB−/− mice (Figure 8D, right, and Supplementary data 5). In both the pre-BI and pre-BII-cell populations, ∼80% of λ5 alleles were at this location, a proportion that increased to ∼95% in the mature B-cell fraction. As the percentage of λ5 alleles located at the nuclear periphery was ∼95% from the large pre-BII stage and onwards in wt mice, this shows that, in the mutant mice, this process is first completed at the mature B-cell stage. Positioning at the nuclear periphery may therefore contribute to the initiation of transcriptional silencing, but only in conjunction with centromeric recruitment, as this process alone is insufficient to initiate silencing of λ5 transcription. Furthermore, in mutant mice transcriptional silencing, repositioning of λ5 alleles to centromeric clusters and the nuclear periphery are delayed until the mature B-cell stage.
Discussion
Our results provide evidence to suggest that the pre-BCR mediates transcriptional silencing of the VpreB1/λ5 locus in wt mice. This process is initiated in large pre-BII cells and followed by complete silencing in small pre-BII cells. However, pre-BCR expression per se is not sufficient, as biallelic transcription is observed in transitional pre-BI cells. This may therefore indicate that either the pre-BCR acts together with another receptor or, alternatively, that the pre-BCR is not signaling competent until the large pre-BII stage. The pre-BCR is necessary for inactivation of the SLC genes in pre-BII cells. However, silencing still takes place in pre-BCR-deficient mice although it is temporally delayed, being first observed at the mature B-cell stage. Downregulation at this stage in mutant mice may be due to a signal mediated by the BCR. However, as judged by the positioning of the VpreB1/λ5 locus in respect to nuclear subcompartmentalization, this signal is insufficient to fully compensate for that of the pre-BCR.
Our results suggest that association of λ5 alleles to centromeric heterochromatin is part of the mechanism of transcriptional silencing, a finding supported by studies in B cells (Brown et al, 1997, 1999). However, as shown here, recruitment in itself is not sufficient to repress λ5 transcription; association is also observed in wt pre-BI cells where both alleles are active. Initiation of silencing also requires the positioning of both alleles at the nuclear periphery. The association of one allele with centromeric clusters observed in wt pre-BI cells may indicate that the allele is in a poised state, such that when the appropriate pre-BCR signal is delivered silencing takes place immediately. Somewhat unexpectedly however, pre-BI cells from pre-BCR-deficient mice did not show the same pattern of nuclear compartmentalization of λ5 alleles as wt pre-BI cells. This may therefore indicate a role for the pro-BCR at this stage (Ohnishi et al, 2000) or, alternatively, this could be the result of the block in B-cell development observed in the mutant mice. Here, the pre-BI population is 2–3-fold enriched (Mundt et al, 2001) and, in addition, 20–25% of cells express μHC compared to 10–15% in wt mice (Galler et al, 2004). Thus, it may be that only a proportion of mutant pre-BI cells are equivalent to wt pre-BI cells, but these are masked by the remaining cells.
In non-B-lineage cells the λ5 5′ regulatory region is hypermethylated, whereas in pre-B and immature B cells it is hypomethylated (Sabbattini et al, 2001). This indicates therefore that methylation may play a role in the tissue- but not stage-specific expression of the SLC genes. Therefore, in the B-lineage, association with heterochromatin domains may play a role in transcriptional silencing, where repression is facilitated by proteins residing in these domains (Brown et al, 1997). Such candidate proteins are Ikaros and Aiolos (Georgopoulos et al, 1992; Hahm et al, 1994; Morgan et al, 1997), for which DNA-binding sites exist in both the VpreB and λ5 promoters (Martensson and Ceredig, 2000; Sabbattini and Dillon, 2005). Furthermore, both Ikaros and Aiolos mRNA levels are upregulated in pre-B cells (Morgan et al, 1997), when initial silencing of the VpreB1/λ5 locus is taking place. Support for a role for these factors comes from analyses of a λ5-driven transgene (Sabbattini et al, 2001). Here, mutation of one of the Ikaros-binding sites allows expression of the mutated, but not wt, transgene in a fraction of activated B cells. Furthermore, the binding sites for Ikaros in the SLC gene promoters overlap with those of other factors, for example, EBF, which is crucial for activation of these genes (Mårtensson and Mårtensson, 1997; Sigvardsson et al, 1997; Persson et al, 1998; O'Riordan and Grosschedl, 1999). If these are mutually exclusive, which may well be the case (Mårtensson and Mårtensson, 1997; Sabbattini et al, 2001), one possibility is that relocation to centromeric clusters facilitates binding of the repressive factor. Additionally, Ikaros interacts with histone modification components, which is of interest since histone modifications indicative of an active state have been observed across the VpreB1/λ5 locus in pre-B but not mature B cells (Jenuwein and Allis, 2001; Georgopoulos, 2002; Szutorisz et al, 2005). Taken together with our results, this supports a model in which subnuclear compartmentalization, heterochromatin and its residing Ikaros and Aiolos complexes play an essential role in regulating SLC gene transcription.
As shown here, SLP-65-deficient pre-BCR+ pre-B cells transcribe both λ5 alleles, suggesting that the pre-BCR-mediated signal for silencing the SLC genes is SLP-65-dependent. The presence of small pre-BII cells in which λ5 is inactive suggests that other signaling molecules downstream of the pre-BCR can partially compensate. As there is a complete block in B-cell development at the pre-BCR+ stage in mice lacking both SLP-65 and btk (Jumaa et al, 2001), btk would be one such candidate, while other potential candidates are LAT and SLP-76 (Hendriks and Middendorp, 2004; Jumaa et al, 2005). Although SLP-65-deficient pre-B cells show enhanced in vitro proliferative capacity (Flemming et al, 2003), ex vivo pre-B cells exhibit decreased proliferation (Hayashi et al, 2003). This is in agreement with our observation that the proportion of PCNA+ nuclei with four signals in ex vivo pre-BCR+ cells from SLP-65−/− mice is lower than that observed in wt large pre-BII cells, suggesting that the loss of SLP-65 results in diminished proliferative capacity in vivo. Nevertheless, although diminished, control of this pre-BCR-driven proliferation is important, since a proportion (∼1/15) of SLP-65-deficient mice develop pre-B-cell tumors (Flemming et al, 2003; Jumaa et al, 2003), whereas SLP-65/λ5 double-deficient mice do not (Jumaa et al, 2005).
The level of SLC expressed in pre-BI and transitional pre-BI cells is similar, whereas that in large pre-BII cells is lower. This therefore correlates with monoallelic transcription of the SLC genes. Furthermore, comparison of protein levels in heterozygous and homozygous −214VpreB1-HuCD122 mice demonstrates that the mean fluorescence intensity of HuCD122 reporter protein is ∼2-fold higher in homozygous mice (Supplementary data 6), demonstrating a direct correlation between VpreB1 promoter-driven transcription and protein levels. Unexpectedly however, surface pre-BCR levels, although very low, are similar in transitional pre-BI and large pre-BII cells and therefore do not reflect SLC levels. If monoallelic transcription of the SLC genes at the pre-BII-cell stage directly serves as a means of limiting the extent of pre-BCR-driven proliferative expansion by reducing SLC protein levels, we would expect an effect on the pre-BII population in haplo-deficient λ5-, VpreB- and SLC-targeted mice. However, none has been reported (Kitamura et al, 1992; Mundt et al, 2001; Shimizu et al, 2002). This may be explained by our recent observation that the SLC and pre-BCR levels at the large pre-BII-cell stage are similar in mice with either only one or two VpreB1 alleles or only one or two VpreB2 alleles, even though the SLC levels at the pre-BI stage are different (Mundt et al, 2005). Therefore, it may be that progression to the large pre-BII stage requires a defined surface pre-BCR level. If so, monoallelic transcription in itself would have no direct effect on pre-BCR levels but could serve as preparation for cessation of SLC expression, ultimately leading to cell cycle exit.
Materials and methods
Mice
For wt, C57BL/6 mice were used (typically 5 weeks). The Ig-tTA/tet-μ/Rag2−/− (tet-μH), −214VpreB1-HuCD122 (line F133), VpreB−/− and SLP-65−/− mice (12–14 weeks) have been described previously (Jumaa et al, 1999; Hess et al, 2001; Mundt et al, 2001; Licence et al, 2003). All animal experiments were performed in accordance with the current UK guidelines under project licences 80/1501, 1736.
FACS analysis and sorting of mouse BM and splenic B cells
BM and spleen cells were isolated using conventional techniques. Antibodies used have been described previously (Mundt et al, 2001). Where possible, propidium iodide was included to exclude dead cells. Expression of SLC or the pre-BCR was analyzed using the LM34 (λ5), VP245 (VpreB) and SL156 antibodies (Karasuyama et al, 1993; Winkler et al, 1995). An enzymatic amplification staining kit was used (EAS, Flow-Amp) to analyze cell surface SLC and pre-BCR expression in Figure 1. For details of sorting protocols, see Supplementary data 1.
Cell cultures
Enrichment of CD19+ BM cells from tet-μH mice and the culture system (using irradiated stromal cells) have been described previously (Hess et al, 2001). BM pre-BI cells were sorted and then cultured as above, including IL7. SLP65−/− pre-B cells (clones 26.5 and El) were cultured in the presence of IL7 (Flemming et al, 2003).
Probes
The λ5 intron 2 (1.3 kb) and CD45 cDNA fragments were amplified by PCR using the following primers: λ5 5′-GTAAGTGGTTCTCATGGTCTCA-3′; 5′-CTGCGCAGATAGAGAAGGAGAT-3′ and CD45 (Skok et al, 2001), and cloned into appropriate vectors. A human β-globin intron 2 probe (Gribnau et al, 1998) was used to detect the transgene. Probes were prepared and labeled with digoxygenin-dUTP as described previously (Chakalova et al, 2004).
RNA FISH
RNA FISH was performed as described previously (Gribnau et al, 2000), except that the hybridization solution contained 50% formamide. The anti-PCNA (Santa Cruz Biotechnology) antibody was visualized using Texas-Red-labeled goat anti-mouse IgG (Jackson ImmunoResearch). Images were examined under an Olympus BX4 white field epifluorescence microscope, under oil using an × 100 objective and appropriate filters. For each probe, typically 100–150 nuclei were counted per slide. For the total numbers per probe, see figure legends.
DNA FISH
DNA FISH and immunostaining (using antiserum to lamin B) were carried out as described previously (Kosak et al, 2002; Roldan et al, 2005). A probe specific for γ-satellite DNA labeled with dUTP-FITC and a BAC clone (RP24–166A3, BAC PAC Resources) for VpreB1/λ5, labeled with dUTP-Cy3 were used. Cells were analyzed by confocal microscopy on a Leica SP2 AOBS (Acousta Optical Beam Splitter). Optical sections were collected with sections separated by a distance of 0.3 μm. Only cells with two λ5 signals were evaluated.
Supplementary Material
Parker et al, Suppl 1
Parker et al, Suppl 2
Parker et al, Suppl 3
Parker et al, Suppl 4
Parker et al, Suppl 5
Parker et al, Suppl 6
Acknowledgments
We thank C Ayling for genotyping mice and D Alexander and M Turner for reading the manuscript. This work was funded by the Biotechnology and Biological Sciences Research Council (LE grant 202/C15831, MP, GM, SL, I-LM, LC, CO, PF, CA), the Deutsche Forschungsgemeinschaft (DFG WI 1183/3 and GK 592, GG, TW and SFB 620, HJ) and a Wellcome Trust University Award (JS). We have no conflicting financial interests.
References
- Brown KE, Baxter J, Graf D, Merkenschlager M, Fisher AG (1999) Dynamic repositioning of genes in the nucleus of lymphocytes preparing for cell division. Mol Cell 3: 207–217 [DOI] [PubMed] [Google Scholar]
- Brown KE, Guest SS, Smale ST, Hahm K, Merkenschlager M, Fisher AG (1997) Association of transcriptionally silent genes with Ikaros complexes at centromeric heterochromatin. Cell 91: 845–854 [DOI] [PubMed] [Google Scholar]
- Chakalova L, Carter D, Fraser P (2004) RNA fluorescence in situ hybridization tagging and recovery of associated proteins to analyze in vivo chromatin interactions. Methods Enzymol 375: 479–493 [DOI] [PubMed] [Google Scholar]
- Dul JL, Argon Y, Winkler T, ten Boekel E, Melchers F, Martensson IL (1996) The murine VpreB1 and VpreB2 genes both encode a protein of the surrogate light chain and are co-expressed during B cell development. Eur J Immunol 26: 906–913 [DOI] [PubMed] [Google Scholar]
- Flemming A, Brummer T, Reth M, Jumaa H (2003) The adaptor protein SLP-65 acts as a tumor suppressor that limits pre-B cell expansion. Nat Immunol 4: 38–43 [DOI] [PubMed] [Google Scholar]
- Fuxa M, Skok J, Souabni A, Salvagiotto G, Roldan E, Busslinger M (2004) Pax5 induces V-to-DJ rearrangements and locus contraction of the immunoglobulin heavy-chain gene. Genes Dev 18: 411–422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galler GR, Mundt C, Parker M, Pelanda R, Martensson I-L, Winkler TH (2004) Surface μ heavy chain signals down-regulation of the V(D)J-recombinase machinery in the absence of surrogate light chain components. J Exp Med 199: 1523–1532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos K (2002) Haematopoietic cell-fate decisions, chromatin regulation and ikaros. Nat Rev Immunol 2: 162–174 [DOI] [PubMed] [Google Scholar]
- Georgopoulos K, Moore DD, Derfler B (1992) Ikaros, an early lymphoid-specific transcription factor and a putative mediator for T cell commitment. Science 258: 808–812 [DOI] [PubMed] [Google Scholar]
- Grawunder U, Leu TM, Schatz DG, Werner A, Rolink AG, Melchers F, Winkler TH (1995) Down-regulation of RAG1 and RAG2 gene expression in preB cells after functional immunoglobulin heavy chain rearrangement. Immunity 3: 601–608 [DOI] [PubMed] [Google Scholar]
- Gribnau J, de Boer E, Trimborn T, Wijgerde M, Milot E, Grosveld F, Fraser P (1998) Chromatin interaction mechanism of transcriptional control in vivo. EMBO J 17: 6020–6027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gribnau J, Diderich K, Pruzina S, Calzolari R, Fraser P (2000) Intergenic transcription and developmental remodeling of chromatin subdomains in the human beta-globin locus. Mol Cell 5: 377–386 [DOI] [PubMed] [Google Scholar]
- Hahm K, Ernst P, Lo K, Kim GS, Turck C, Smale ST (1994) The lymphoid transcription factor LyF-1 is encoded by specific, alternatively spliced mRNAs derived from the Ikaros gene. Mol Cell Biol 14: 7111–7123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi K, Yamamoto M, Nojima T, Goitsuka R, Kitamura D (2003) Distinct signaling requirements for Dmu selection, IgH allelic exclusion, pre-B cell transition, and tumor suppression in B cell progenitors. Immunity 18: 825–836 [DOI] [PubMed] [Google Scholar]
- Hendriks RW, Middendorp S (2004) The pre-BCR checkpoint as a cell-autonomous proliferation switch. Trends Immunol 25: 249–256 [DOI] [PubMed] [Google Scholar]
- Hess J, Werner A, Wirth T, Melchers F, Jack HM, Winkler TH (2001) Induction of pre-B cell proliferation after de novo synthesis of the pre-B cell receptor. Proc Natl Acad Sci USA 98: 1745–1750 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenuwein T, Allis CD (2001) Translating the histone code. Science 293: 1074–1080 [DOI] [PubMed] [Google Scholar]
- Jumaa H, Bossaller L, Portugal K, Storch B, Lotz M, Flemming A, Schrappe M, Postila V, Riikonen P, Pelkonen J, Niemeyer CM, Reth M (2003) Deficiency of the adaptor SLP-65 in pre-B-cell acute lymphoblastic leukaemia. Nature 423: 452–456 [DOI] [PubMed] [Google Scholar]
- Jumaa H, Hendriks RW, Reth M (2005) B cell signaling and tumorigenesis. Annu Rev Immunol 23: 415–445 [DOI] [PubMed] [Google Scholar]
- Jumaa H, Mitterer M, Reth M, Nielsen PJ (2001) The absence of SLP65 and Btk blocks B cell development at the preB cell receptor-positive stage. Eur J Immunol 31: 2164–2169 [DOI] [PubMed] [Google Scholar]
- Jumaa H, Wollscheid B, Mitterer M, Wienands J, Reth M, Nielsen PJ (1999) Abnormal development and function of B lymphocytes in mice deficient for the signaling adaptor protein SLP-65. Immunity 11: 547–554 [DOI] [PubMed] [Google Scholar]
- Karasuyama H, Kudo A, Melchers F (1990) The proteins encoded by the VpreB and lambda 5 pre-B cell-specific genes can associate with each other and with mu heavy chain. J Exp Med 172: 969–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasuyama H, Rolink A, Melchers F (1993) A complex of glycoproteins is associated with VpreB/lambda 5 surrogate light chain on the surface of mu heavy chain-negative early precursor B cell lines. J Exp Med 178: 469–478 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karasuyama H, Rolink A, Shinkai Y, Young F, Alt FW, Melchers F (1994) The expression of Vpre-B/lambda 5 surrogate light chain in early bone marrow precursor B cells of normal and B cell-deficient mutant mice. Cell 77: 133–143 [DOI] [PubMed] [Google Scholar]
- Kersseboom R, Middendorp S, Dingjan GM, Dahlenborg K, Reth M, Jumaa H, Hendriks RW (2003) Bruton's tyrosine kinase cooperates with the B cell linker protein SLP-65 as a tumor suppressor in pre-B cells. J Exp Med 198: 91–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura D, Kudo A, Schaal S, Muller W, Melchers F, Rajewsky K (1992) A critical role of lambda 5 protein in B cell development. Cell 69: 823–831 [DOI] [PubMed] [Google Scholar]
- Kitamura D, Roes J, Kuhn R, Rajewsky K (1991) A B cell-deficient mouse by targeted disruption of the membrane exon of the immunoglobulin mu chain gene. Nature 350: 423–426 [DOI] [PubMed] [Google Scholar]
- Kosak ST, Skok JA, Medina KL, Riblet R, Le Beau MM, Fisher AG, Singh H (2002) Subnuclear compartmentalization of immunoglobulin loci during lymphocyte development. Science 296: 158–162 [DOI] [PubMed] [Google Scholar]
- Kudo A, Melchers F (1987) A second gene, VpreB in the lambda 5 locus of the mouse, which appears to be selectively expressed in pre-B lymphocytes. EMBO J 6: 2267–2272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licence S, Persson C, Mundt C, Martensson IL (2003) The VpreB1 enhancer drives developmental stage-specific gene expression in vivo. Eur J Immunol 33: 1117–1126 [DOI] [PubMed] [Google Scholar]
- Mårtensson A, Mårtensson I-L (1997) Early B cell factor binds to a site critical for lambda5 core enhancer activity. Eur J Immunol 27: 315–320 [DOI] [PubMed] [Google Scholar]
- Mårtensson I-L, Ceredig R (2000) Review article: role of the surrogate light chain and the pre-B-cell receptor in mouse B-cell development. Immunology 101: 435–441 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mårtensson I-L, Rolink A, Melchers F, Mundt C, Licence S, Shimizu T (2002) The pre-B cell receptor and its role in proliferation and Ig heavy chain allelic exclusion. Semin Immunol 14: 335–342 [DOI] [PubMed] [Google Scholar]
- Melchers F (2005) The pre-B-cell receptor: selector of fitting immunoglobulin heavy chains for the B-cell repertoire. Nat Rev Immunol 5: 578–584 [DOI] [PubMed] [Google Scholar]
- Middendorp S, Dingjan GM, Hendriks RW (2002) Impaired precursor B cell differentiation in Bruton's tyrosine kinase-deficient mice. J Immunol 168: 2695–2703 [DOI] [PubMed] [Google Scholar]
- Morgan B, Sun L, Avitahl N, Andrikopoulos K, Ikeda T, Gonzales E, Wu P, Neben S, Georgopoulos K (1997) Aiolos, a lymphoid restricted transcription factor that interacts with Ikaros to regulate lymphocyte differentiation. EMBO J 16: 2004–2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morris GF, Mathews MB (1989) Regulation of proliferating cell nuclear antigen during the cell cycle. J Biol Chem 264: 13856–13864 [PubMed] [Google Scholar]
- Mundt C, Licence S, Maxwell G, Mårtensson I-L (2005) The expression level of VpreB1, but not VpreB2, is sufficient to support normal B cell development. Int Immunol (in press) [DOI] [PubMed] [Google Scholar]
- Mundt C, Licence S, Shimizu T, Melchers F, Martensson IL (2001) Loss of precursor B cell expansion but not allelic exclusion in VpreB1/VpreB2 double-deficient mice. J Exp Med 193: 435–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi K, Shimizu T, Karasuyama H, Melchers F (2000) The identification of a nonclassical cadherin expressed during B cell development and its interaction with surrogate light chain. J Biol Chem 275: 31134–31144 [DOI] [PubMed] [Google Scholar]
- O'Riordan M, Grosschedl R (1999) Coordinate regulation of B cell differentiation by the transcription factors EBF and E2A. Immunity 11: 21–31 [DOI] [PubMed] [Google Scholar]
- Persson C, Martensson A, Martensson IL (1998) Identification of a tissue- and differentiation stage-specific enhancer of the VpreB1 gene. Eur J Immunol 28: 787–798 [DOI] [PubMed] [Google Scholar]
- Pillai S, Baltimore D (1987) Formation of disulphide-linked mu 2 omega 2 tetramers in pre-B cells by the 18K omega-immunoglobulin light chain. Nature 329: 172–174 [DOI] [PubMed] [Google Scholar]
- Roldan E, Fuxa M, Chong W, Martinez D, Novatchkova M, Busslinger M, Skok JA (2005) Locos ‘decontraction' and centromeric recruitment contribute to allelic exclusion of the immunoglobin heavy-chain gene. Nat Immunol 6: 31–41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolink A, Grawunder U, Winkler TH, Karasuyama H, Melchers F (1994) IL-2 receptor alpha chain (CD25, TAC) expression defines a crucial stage in pre-B cell development. Int Immunol 6: 1257–1264 [DOI] [PubMed] [Google Scholar]
- Sabbattini P, Dillon N (2005) The lambda5-VpreB1 locus—a model system for studying gene regulation during early B cell development. Semin Immunol 17: 121–127 [DOI] [PubMed] [Google Scholar]
- Sabbattini P, Lundgren M, Georgiou A, Chow C, Warnes G, Dillon N (2001) Binding of Ikaros to the lambda5 promoter silences transcription through a mechanism that does not require heterochromatin formation. EMBO J 20: 2812–2822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu T, Mundt C, Licence S, Melchers F, Martensson IL (2002) VpreB1/VpreB2/lambda 5 triple-deficient mice show impaired B cell development but functional allelic exclusion of the IgH locus. J Immunol 168: 6286–6293 [DOI] [PubMed] [Google Scholar]
- Sigvardsson M, O'Riordan M, Grosschedl R (1997) EBF and E47 collaborate to induce expression of the endogenous immunoglobulin surrogate light chain genes. Immunity 7: 25–36 [DOI] [PubMed] [Google Scholar]
- Skok JA, Brown KE, Azuara V, Caparros ML, Baxter J, Takacs K, Dillon N, Gray D, Perry RP, Merkenschlager M, Fisher AG (2001) Nonequivalent nuclear location of immunoglobulin alleles in B lymphocytes. Nat Immunol 2: 848–854 [DOI] [PubMed] [Google Scholar]
- Stephan RP, Elgavish E, Karasuyama H, Kubagawa H, Cooper MD (2001) Analysis of VpreB expression during B lineage differentiation in lambda5-deficient mice. J Immunol 167: 3734–3739 [DOI] [PubMed] [Google Scholar]
- Szutorisz H, Canzonetta C, Georgiou A, Chow CM, Tora L, Dillon N (2005) Formation of an active tissue-specific chromatin domain initiated by epigenetic marking at the embryonic stem cell stage. Mol Cell Biol 25: 1804–1820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsubata T, Reth M (1990) The products of pre-B cell-specific genes (lambda 5 and VpreB) and the immunoglobulin mu chain form a complex that is transported onto the cell surface. J Exp Med 172: 973–976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winkler TH, Rolink A, Melchers F, Karasuyama H (1995) Precursor B cells of mouse bone marrow express two different complexes with the surrogate light chain on the surface. Eur J Immunol 25: 446–450 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Parker et al, Suppl 1
Parker et al, Suppl 2
Parker et al, Suppl 3
Parker et al, Suppl 4
Parker et al, Suppl 5
Parker et al, Suppl 6


