Abstract
TGF-β-activated kinase 1 (TAK1), a member of the MAPKKK family, is thought to be a key modulator of the inducible transcription factors NF-κB and AP-1 and, therefore, plays a crucial role in regulating the genes that mediate inflammation. Although in vitro biochemical studies have revealed the existence of a TAK1 complex, which includes TAK1 and the adapter proteins TAB1 and TAB2, it remains unclear which members of this complex are essential for signaling. To analyze the function of TAK1 in vivo, we have deleted the Tak1 gene in mice, with the resulting phenotype being early embryonic lethality. Using embryonic fibroblasts lacking TAK1, TAB1, or TAB2, we have found that TNFR1, IL-1R, TLR3, and TLR4-mediated NF-κB and AP-1 activation are severely impaired in Tak1m/m cells, but they are normal in Tab1-/- and Tab2-/- cells. In addition, Tak1m/m cells are highly sensitive to TNF-induced apoptosis. TAK1 mediates IKK activation in TNF-α and IL-1 signaling pathways, where it functions downstream of RIP1–TRAF2 and MyD88–IRAK1–TRAF6, respectively. However, TAK1 is not required for NF-κB activation through the alternative pathway following LT-β signaling. In the TGF-β signaling pathway, TAK1 deletion leads to impaired NF-κB and c-Jun N-terminal kinase (JNK) activation without impacting Smad2 activation or TGF-β-induced gene expression. Therefore, our studies suggests that TAK1 acts as an upstream activating kinase for IKKβ and JNK, but not IKKα, revealing an unexpectedly specific role of TAK1 in inflammatory signaling pathways.
Keywords: JNK, NF-κB, TAB1, TAB2, TAK1, TGF-β
The NF-κB and c-Jun N-terminal kinase (JNK) pathways play critical roles in the immune response and protect cells from apoptosis induced by various stressors and inflammatory cytokines. Due to their roles in various disease states and their potential as therapeutic targets, the signaling pathways activated by the inflammatory cytokines TNF-α and IL-1 have been intensely studied. TNF-α induces trimerization of its cognate receptor (TNFR1) and binding of the adaptor proteins TRADD, TRAF2, TRAF5, and RIP1, which leads to recruitment of the IKK complex and JNK to the receptor complex (Song et al. 1997). Activated IKK triggers degradation of IκBα and subsequent translocation of NF-κB to the nucleus, leading to transcription of a subset of TNF-regulated genes (Hayden and Ghosh 2004). Once activated, JNK translocates to the nucleus, where it regulates the activity of the AP-1 transcription factor complex (c-Jun and ATF-2) (Chen and Goeddel 2002). In the IL-1 signaling pathway, interaction of IL-1 with its cognate receptor (IL-1R) and IL-1R accessory protein (IL-1Racp) forms an activated receptor complex that recruits the adaptor proteins MyD88, IRAK1, IRAK4, and TRAF6. Phosphorylation and degradation of IRAK1 induces ubiquitination of TRAF6 and release of the adapter protein complex into the cytoplasm to activate downstream kinases, including IKK and JNK (Deng et al. 2000; Takaesu et al. 2001; Jiang et al. 2002).
TGF-β-activated kinase 1 (TAK1), a member of the MAPKKK family, was originally identified as a key regulator of MAP kinase activation in TGF-β-induced signaling pathways (Yamaguchi et al. 1995). During Xenopus embryonic development, TAK1 is involved in mesoderm induction and patterning mediated by bone morphogenetic protein (BMP), a TGF-β family ligand (Shibuya et al. 1998). Recent studies have reported that Drosophila TAK1 is required for both JNK and NF-κB (Relish) activation in response to immune challenge by gram-negative bacteria infection and that IMD signaling is impaired in TAK1-/- flies (Vidal et al. 2001; Boutros et al. 2002; Silverman et al. 2003; Park et al. 2004). In mammals, in vitro and overexpression studies suggest TAK1 is involved in TNFR1 and IL-1R/TLR-mediated signaling pathways upstream of IKK and JNK/p38 MAP kinases (Takaesu et al. 2003). Two mammalian TAK1 adaptor proteins, TAB1 and TAB2, were isolated as TAK1-interacting proteins by yeast two-hybrid screening. TAB1 interacts constitutively with TAK1 and induces TAK1 kinase activity when overexpressed (Shibuya et al. 1996). Following IL-1 stimulation, TAB2 translocates from the cell membrane to the cytosol and links TAK1 with TRAF6, thereby mediating TAK1 activation (Takaesu et al. 2000). In vitro biochemical studies have demonstrated that TRIKA1 (a complex of Ubc13 and Uev1A) and TRIKA2 (the complex of TAK1, TAB1, and TAB2) are key signal-transducing complexes that activate IKK and MKK6 in a manner dependent upon ubiquitination of TRAF6 (Wang et al. 2001). However, the physiological roles of mammalian TAK1, TAB1, and TAB2 in inflammatory signaling in vivo remain to be definitively established. TAB1-deficient mouse embryonic fibroblasts (MEFs) have been generated; however, TAB1 function has not been tested in TNFR1 and IL-1R/TLR-mediated signaling (Komatsu et al. 2002). Surprisingly, TAB2-deficient MEFs are capable of activating IKK and JNK normally in response to TNF-α or IL-1 (Sanjo et al. 2003). Thus, available genetic evidence has not yet recapitulated the predicted role for the TAK1 complex in mammalian cells in signaling to NF-κB and AP-1.
Therefore, to definitively establish the function of TAK1 in vivo, we have generated mutant Tak1m/m mice, which have an embryonic lethal phenotype that is distinct from that of TAB1 and TAB2-deficient mice. To circumvent the difficulty posed by this early lethality, we have investigated signaling to JNK and NF-κBinembryonic fibroblasts derived from these mice. Unlike Tab1-/- and Tab2-/- cells, Tak1m/m cells exhibit dramatically impaired NF-κB and JNK activation through TNFR1, IL-1R, and TLR3, and are highly sensitive to TNF-α-induced apoptosis. TAK1, but not TAB1 or TAB2, is an essential signal transducer from the RIP–TRAF2 and MyD88–IRAK1–TRAF6 complexes to the IKK complex. Although TAK1 is essential for TLR4-induced AP-1 activation, the requirement for TAK1 in NF-κB activation by LPS appears less complete. However, TAK1 is not required for the induction of p100 processing or NF-κB activation by LT-β, suggesting differential requirements for activation of the alternative IKKα-mediated NF-κB pathway. In Tak1m/m cells, Smad2 activation and gene expression following TGF-β are normal, whereas NF-κB and JNK activation are impaired. Therefore, this in vivo analysis of the mammalian TAK1 complex establishes the specific requirement for TAK1, but not TAB1 or TAB2, as an upstream activator of IKKβ and JNK in multiple signaling pathways to AP-1 and NF-κB.
Results
Generation of Tak1 mutant embryos
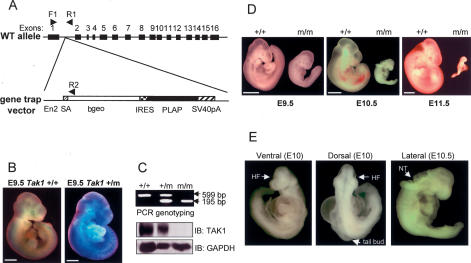
To explore the biological function of TAK1 in vivo, we generated TAK1-deficient (Tak1m/m) mice by microinjecting gene trap-mutated Tak1 embryonic stem cells (ES) into blastocysts of C57BL/6 mice (Fig. 1A; Stryke et al. 2003; Austin et al. 2004; Skarnes et al. 2004). To confirm the generation of embryos from the gene trap-mutated Tak1 ES cells, β-galactosidase expression was analyzed with embryonic day 9.5 (E9.5) wild-type (Tak1+/+) and heterozygous embryos (Tak1+/m) (Fig. 1B). PCR genotyping demonstrated that β-galactosidase was expressed in heterozygous (Tak1+/m), but not wild-type embryos (Tak1+/+). Immunoblotting analysis with anti-TAK1 antibody confirmed the expression of TAK1 in wild-type (Tak1+/+) and heterozygous (Tak1+/m), but not homozygous mutant (Tak1m/m) embryos (Fig. 1C).
Figure 1.
Targeted disruption of the Tak1 gene. (A) Schematic drawing of the Tak1 gene-trapping strategy. The position of primers (F1, R1, and R2) used in PCR genotyping are indicated by arrowheads. (En2) engrailed 2 gene; (βgeo) a fusion protein between β-galactosidase and neomycin phosphotransferase; (IRES) internal ribosomal entry site; (PLAP) human placental alkaline phosphatase; (SV40 pA) SV40 polyadenylation signal. (B) β-galactosidase staining of E9.5 wild-type (Tak1+/+) and heterozygous embryos (Tak1+/m). Bar, 100 μm. (C) PCR genotyping and immunoblotting analysis of TAK1 in E10 wild-type (+/+), heterozygous (+/m), and homozygous (m/m) mutant embryos. The size of PCR fragments is 599 base pairs (bp) and 195 bp for wild-type and insertional mutant alleles, respectively. (D) Lateral view of the wild type (+/+) and Tak1 mutant (m/m) embryos at E9.5, E10.5, and E11.5. (E) Ventral (left) and Dorsal (middle) view of E10 Tak1 mutant embryos and lateral view of E10.5 Tak1 mutant embryos (right). Whole embryos were used for PCR genotyping. (HF) Head fold; (NT) neural tube.
Abnormal development of neural tube in Tak1 mutant embryos
We next examined the genotypes of newborn offspring following intercrossing of Tak1 heterozygous parents. No homozygous mutant mice (Tak1m/m) were scored, suggesting that disruption of the Tak1 gene causes an embryonic lethality (Table 1). All of the embryos between E8.5 and E9.5 were scored according to Mendelian expectations, whereas at E11.5 all homozygous mutant embryos were dead and being resorbed, and empty deciduas were frequently observed (Table 1). Thus, these results suggest that Tak1 mutant embryos begin to die around E10. Most wild-type embryos and mutant littermates did not exhibit any gross morphological difference until E9.0, after which the mutant embryos displayed severe developmental abnormalities in the neural tube (Fig. 1D). At E9.5, most of the mutant embryos were smaller than wild-type and heterozygous embryos, and at E10 and E10.5, the head fold and neural tube were wavy and not closed in the mutant embryos (Fig. 1E). After E10.5, the embryonic structures were resorbed in the Tak1 mutants. Interestingly, the mutant phenotype of Tak1 embryos is substantially different from Tab1 (abnormal cardiovascular and pulmonary morphogenesis) and Tab2 (liver degeneration and apoptosis) mutant embryos (Table 2; Komatsu et al. 2002; Sanjo et al. 2003). The phenotypes of the mutant embryos suggest that the TAK1 complex does not only function as a single unit but, instead, that each component has a distinct role during development.
Table 1.
Genotype of offspring derived from Tak1 heterozygous parents
| Tak1+/+ | Tak1+/- | Tak1-/- | |
|---|---|---|---|
| E8.5 | 27 | 52 | 22 |
| E9.5 | 17 | 34 | 13 |
| E10.5 | 13 | 28 | 9 (3)a |
| E11.5 | 16 | 38 | 5 (5) |
| E12.5 | 21 | 53 | 1 (1) |
| E13.5 | 15 | 37 | 0 |
| Newborn | 46 | 81 | 0 |
The number of dead embryos is indicated in parentheses.
Table 2.
Comparison of phenotypes of Tak1, Tab1, and Tab2 mutant embryos
| Tak1-/- | Tab1-/-b | Tab2-/-c |
|---|---|---|
| Ubiquitous expression at E14.5 (strong expression in nervous system, kidney, gut, testis, liver, lung, and pancreas)a | Ubiquitous expression through embryogenesis | Ubiquitous expression (strong expression in heart and liver) |
| Embryonic lethality around E10 | Embryonic lethality at late stage of gestation | Embryonic lethality around E12.5 |
| Neural fold dysmorphogenesis | Cardiovascular and lung dysmorphogenesis | Liver degeneration and apoptosis |
TAK1, but not TAB1 or TAB2, is required for NF-κB and JNK activation in TNF-α and IL-1 signaling pathways
Despite numerous attempts, we were unable to culture embryonic fibroblasts from TAK1m/m embryos to obtain sufficient cells for study. Therefore, to analyze the function of TAK1 in inflammatory signaling pathways, we generated immortalized embryonic fibroblasts derived from E9.75 wild-type (Tak1+/+), heterozygous (Tak1+/m), and homozygous mutant (Takm/m) embryos by stably transfecting linearized SV40 large T antigen plasmid (Kobayashi et al. 2005). Immunoblotting analysis with anti-TAK1 antibody showed that endogenous TAK1 is expressed in Tak1+/+ and Tak1+/m cells, but not Tak1m/m cells (data not shown).
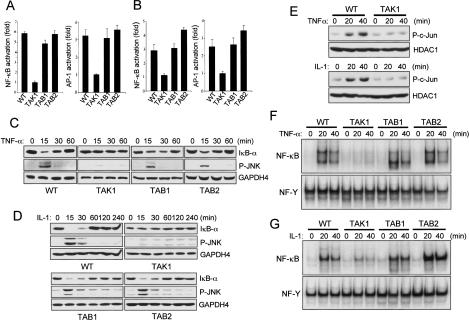
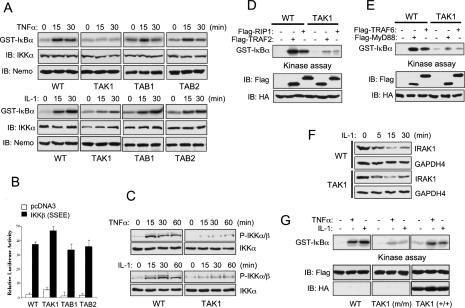
To address the hypothesis that the TAK1 complex, including TAK1, TAB1, and TAB2 is required for NF-κB and JNK activation in response to inflammatory cytokines, we tested the requirement for each member of the TAK1 complex in TNF-α and IL-1 signaling pathways by luciferase analysis. Cells were transfected with NF-κB and AP-1-responsive luciferase reporters and stimulated with TNF-α or IL-1 (Fig. 2A,B). Following activation by TNF-α and IL-1, Tab1-/- and Tab2-/- cells exhibit wild-type or greater levels of NF-κB and AP-1, whereas both NF-κB and AP-1 activation were significantly decreased in Tak1m/m cells. This is consistent with in vitro experiments that indicate overexpression of kinase-inactive TAK1 mutant (TAK1-DN) inhibited both TNF-α and IL-1-induced NF-κB activation in a dose-dependent manner (Supplementary Fig. S1A,B). Interestingly, Tab2-/- cells show a small, but reproducible increase in signaling to both AP-1 and NF-κB. We also examined TNF-α and IL-1-induced NF-κB and JNK activation by immunoblotting with antibodies specific for IκBα and phospho-JNK1/2, respectively (Fig. 2C,D). In wild-type, Tab1-/-, and Tab2-/- cells, IκBα degradation and JNK phosphorylation occurs normally and peaks at 15 min in response to TNF-α and IL-1. In contrast, in the absence of TAK1 IκBα degradation and JNK phosphorylation following TNF-α and IL-1 stimulation is markedly decreased. Furthermore, phosphorylation and nuclear translocation of c-Jun following TNF-α or IL-1 stimulation occurs normally in wild-type cells, whereas Tak1m/m cells exhibit impaired c-Jun activation (Fig. 2E). We next used electrophoretic mobility gel-shift assay (EMSA) to analyze NF-κB DNA-binding activity following TNF-α and IL-1 stimulation (Fig. 2 F,G). Wild-type, Tab1-/-, and Tab2-/- cells exhibited similar NF-κB DNA-binding activities in response to TNF-α and IL-1, whereas in Tak1m/m cells, TNF-α-induced binding activity was completely abolished and IL-1-induced activity was partially reduced. In agreement with the luciferase analysis, Tab2-/- cells showed higher IL-1-induced NF-κB DNA-binding activity than wild-type or Tab1-/- cells, implying that TAB2 negatively regulates NF-κB activation. Therefore, these results provide genetic evidence of a requirement for TAK1, but not TAB1 or TAB2, in NF-κB and JNK activation in the TNF-α and IL-1 signaling pathways.
Figure 2.
TAK1 deficiency inhibits NF-κB and JNK activation by TNF-α and IL-1. (A,B) Wild-type (WT), Tak1m/m (TAK1), Tab1-/- (TAB1), and Tab2-/- (TAB2) cells were transfected with either pBIIx-luc (left) or AP1-luc (right) reporter together with Renilla luciferase vector. Twenty-four hours after transfection, cells were treated with TNF-α (10 ng/mL; A) or IL-1 (10 ng/mL; B) for 5 h and then analyzed for luciferase activity. Results are expressed as the fold induction in luciferase activity relative to that of untreated cells. Error bars indicate standard deviation. (C,D) IκB-α degradation and JNK phosphorylation by TNF-α (C) or IL-1 (D) were analyzed by immunoblotting with antibodies specific for IκB-α and phospho-JNK, respectively. Immunoblotting with anti-GAPDH4 antibody was performed as a loading control. (E) Cells were treated with TNF-α (10 ng/mL; top) or IL-1 (10 ng/mL; bottom) for the indicated time, nuclear extracts were prepared and immunoblotted with anti-phosho-c-Jun antibody. Immunoblotting with anti-HDAC1 antibody was performed as a loading control. (F,G) Cells were treated with TNF-α (10 ng/mL; E) or IL-1 (10 ng/mL; F) for the indicated times and nuclear extract were prepared. NF-κB DNA-binding activity was analyzed by EMSA.
TAK1, but not TAB1 or TAB2, is required for NF-κB and JNK activation in TLR-mediated signaling pathways
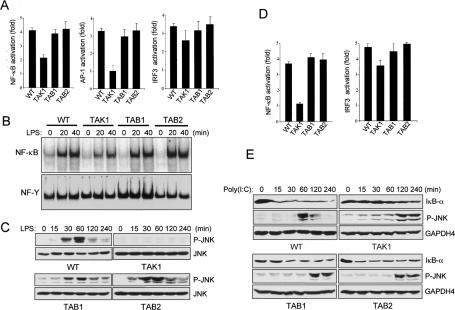
Previous studies have reported that TAK1 plays a critical role in regulating the activation of multiple protein kinases during LPS signaling (Lee et al. 2000). We therefore examined the function of the TAK1 complex in TLR4-mediated signaling. Since all of the cells were weakly responsive to LPS, we cotransfected CD4/TLR4, a dominant active TLR4 chimera, together with either NF-κBor AP-1-responsive luciferase reporter constructs. Using this system, wild-type, Tab1-/-, and Tab2-/- cells exhibit similar degrees of NF-κB and AP-1 activation, while in Tak1m/m cells, NF-κB activation was partially decreased and AP-1 activation was markedly decreased (Fig. 3A). In contrast, IRF3 activation remains largely intact in wild-type, Tak1m/m, Tab1-/-, and Tab2-/- cells. Therefore, these results indicate that in TLR4-mediated signaling, TAK1, but not TAB1 or TAB2, is required for AP-1 activation, but not for IRF3 activation. Intriguingly, our in vitro experiment showed that overexpression of kinaseinactive TAK1 mutant (TAK1-DN) inhibited IRF3 activation by CD4/TLR4 in a dose-dependent manner. This implies that overexpression of TAK1-DN may induce nonspecific inhibition of TLR4-induced IRF3 activation (Supplementary Fig. S1C,D). We next analyzed LPS-induced NF-κB DNA-binding activity by using electrophoretic mobility gel-shift assay. As shown in Figure 3B, similar binding activities were observed in wild-type, Tab1-/-, and Tab2-/- cells in response to LPS. However, in contrast to TNF-α and IL-1 signaling, Tak1m/m cells exhibited only a modest decrease in DNA-binding activity in response to LPS. To examine the function of the TAK1 complex in LPS-induced JNK activation, we performed immunoblotting with anti-phospho-JNK1/2 antibody (Fig. 3C). JNK phosphorylation following LPS stimulation occurs normally in wild-type, Tab1-/-, and Tab2-/- cells, while it is severely impaired in Tak1m/m cells. Therefore, these results suggest that while TAK1, but not TAB1 or TAB2, is required for JNK activation in a TLR4-mediated signaling pathway, it is only partially required for NF-κB activation in response to LPS.
Figure 3.
TLR-mediated NF-κB and JNK activation is impaired in Tak1m/m cells. (A) Wild type, Tak1m/m, Tab1-/-, and Tab2-/- cells were cotransfected with CD4/TLR4 and Renilla luciferase vectors together with either pBIIx-luc (left), AP1-luc (middle), or IRF3-luc (right) reporters. Twenty-four hours after transfection, cells were lysed and analyzed for luciferase activity. Results are expressed as fold change in luciferase activity relative to untreated cells. (B) Cells were treated with LPS (1 μg/mL) for the indicated times, nuclear extracts were prepared, and NF-κB DNA-binding activity was analyzed by EMSA. (C) LPS-induced JNK phosphorylation was analyzed by immunoblotting with antiphospho-JNK antibody. Immunoblotting with anti-GAPDH4 antibody was performed to control for gel loading. (D) Cells were transfected with pBIIx-luc (left) or IRF3-luc (right) reporters together with Renilla luciferase vector. Twenty-four hours after transfection, cells were treated with poly(I:C) (5 μg/mL) and luciferase activity was analyzed as in A. (E) Poly(I:C)-induced I-κB-α degradation and JNK phosphorylation were analyzed by immunoblotting with anti-I-κB-α and antiphospho-JNK antibodies, respectively. Immunoblotting with anti-GAPDH4 antibody was performed to control for gel loading.
To assess whether this partial dependence on TAK1 in LPS signaling is due to the divergent nature of signaling downstream of TLR4—through both MyD88-dependent and independent pathways using the adapters TRIF and TRAM—we examined the role of TAK1 in signaling from a TLR that has a more linear path of activation to NF-κB. Ligation of TLR3 by poly(I:C) leads to TAK1 activation through TRIF–TRAF6 (Sato et al. 2003), followed by NF-κB activation (Jiang et al. 2003). However, TAK1 activation is not required for IRF3 activation through TLR3 (Jiang et al. 2004). We therefore analyzed TLR3-mediated NF-κB and IRF3 activation using the luciferase-reporter assays (Fig. 3D). Wild-type, Tab1-/-, and Tab2-/- cells normally induced NF-κB and IRF3 activation in response to poly(I:C); however, in Tak1m/m cells, NF-κB activation following poly(I:C) stimulation was significantly decreased, although IRF3 activation was largely intact. This is consistent with in vitro experiments that indicate overexpression of TAK1-DN inhibits poly(I:C)-induced NF-κB activation in a dose-dependent manner, but not IRF3 activation (Supplementary Fig. S1E,F). Therefore, these results indicate that TAK1, but not TAB1 or TAB2 is required for TLR3-mediated NF-κB activation, but not IRF3 activation. We next used immunoblotting analysis with antibodies specific for IκBα and phospho-JNK1/2 to analyze poly(I:C)-induced NF-κB and JNK activation, respectively (Fig. 3E). In wild-type, Tab1-/-, and Tab2-/- cells, IκBα degradation occurs after 15 min post-induction with poly(I:C), however, degradation is reduced and significantly delayed in Tak1m/m cells. Therefore, these results suggest that TAK1, but not TAB1 or TAB2 functions in early NF-κB activation through TLR3. Surprisingly, Tak1m/m, Tab1-/-, and Tab2-/- cells all exhibited delayed JNK phosphorylation in response to poly(I:C). Therefore, these results suggest that in TLR3-mediated signaling pathways, TAK1, but not TAB1 or TAB2, is required for NF-κB activation, but not for JNK activation and IRF3 activation. Taken together, TAK1 plays a differential role in signaling from TLR3 and TLR4 that probably reflects the complexity of the pathways that emanate from these receptors.
Tak1m/m cells are highly sensitive to TNF-α-induced apoptosis
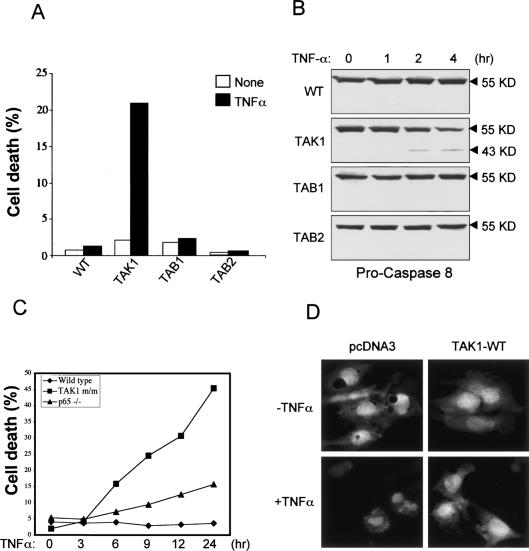
Activation of NF-κB following TNF-α stimulation protects cells from TNF-α-induced apoptosis (Beg and Baltimore 1996; Van Antwerp et al. 1996). In p65, IKKβ and NEMO/IKKγ-deficient MEFs, defective NF-κB activation increases their sensitivity to TNF-α-induced apoptosis (Beg et al. 1995; Tanaka et al. 1999; Rudolph et al. 2000). To examine whether loss of each member of the TAK1 complex affects TNF-α-induced apoptosis, we treated cells with TNF-α for 8 h, and analyzed cell death by using Annexin-V and PI staining (Fig. 4A). In agreement with our analysis of NF-κB activation, TNF-α alone did not induce any apoptosis in wild-type, Tab1-/-, and Tab2-/- cells, whereas Tak1m/m cells were highly sensitive to TNF-α-induced apoptosis. Consistent with this result, immunoblotting analysis with anti-Caspase 8 antibody showed that TNF-α induced cleavage of pro-Caspase 8 (55 KD) to active Caspase-8 (43 KD) in Tak1m/m cells, but not wild-type, Tab1-/-, and Tab2-/- cells (Fig. 4B). Therefore, these results indicate that TNF-α induces apoptosis in Tak1m/m cells through Caspase 8 activation. To examine the kinetics of TNF-α-induced apoptosis in Tak1m/m cells, we treated wild-type, p65-/-, and Tak1-/- cells with TNF-α and analyzed cell death by using Annexin-V and PI staining at multiple time points. As shown in Figure 4C, wild-type cells are resistant to TNF-α-induced apoptosis, whereas both p65-/- and Tak1-/- cells are highly sensitive to TNF-α-induced apoptosis. Intriguingly, Tak1m/m cells exhibited higher sensitivity to TNF-α-induced apoptosis than p65-/- cells. Recent studies have reported that TAK1 is essential for regulation of IAP-mediated JNK1 activation, which plays a role in protecting cells from TNF-α-induced apoptosis (Sanna et al. 2002). In addition, the JNK/JunD pathway transduces a survival signal in collaboration with NF-κB following TNFα stimulation. In the absence of NF-κB activation, however, the JNK pathway mediates pro-apoptotic signaling in response to TNFα (Lamb et al. 2003). Therefore, our results suggest that TAK1 deletion increases sensitivity to TNF-α-induced apoptosis due to defects of both NF-κB and JNK activation. We next tested whether exogenous expression of TAK1 proteins can rescue the mutant phenotypes of Tak1m/m cells (Fig. 4D). Cells were cotransfected with GFP together with either pcDNA3 or wild-type TAK1 (TAK1-WT), and treated with TNF-α for 4 h. As expected, Tak1m/m cells expressing pcDNA3 undergo TNF-α-induced apoptosis; however, cells expressing exogenous TAK1 proteins are resistant to apoptosis. Thus, exogenous TAK1 expression restored resistance to TNF-α-induced apoptosis in Tak1m/m cells.
Figure 4.
Tak1m/m cells are highly sensitive to TNF-α-induced apoptosis. (A) Wild-type (WT), Tak1m/m, Tab1-/-, and Tab2-/- cells were treated with TNF-α (10 ng/mL) for 8 h. Apoptosis was analyzed by staining with FITC-Annexin V and propidium iodide (PI) staining. Data given is the percentage of cells that stained positive with both PI and Annexin V. (B) Cells were stimulated with TNF-α for the indicated times, lysed, and immunoblotted with anti-caspase 8 antibody. Immunoblotting with anti-GAPDH4 antibody was performed to control for gel loading. (C) Wild-type, p65-/- and Tak1m/m cells were treated with TNF-α (10 ng/mL) for the indicated times. Apoptosis was analyzed by FITC-Annexin V and propidium iodide (PI) staining as in A. (D) Cells were transfected with the GFP expression vector together with pcDNA3, or HA-tagged wild-type TAK1 (TAK1-WT). Twenty-four hours after transfection, cells were treated with TNF-α (10 ng/mL) for 4 h and analyzed by fluorescent microscopy.
TAK1 functions upstream of the IKK complex and downstream of RIP1–TRAF2 and MyD88–IRAK1–TRAF6 in TNFα and IL-1/TLR signaling pathways
Activation of the IKK complex is essential for NF-κB activation by various inflammatory cytokines (Hayden and Ghosh 2004). We therefore analyzed TNF-α and IL-1-induced IKK activation using in vitro kinase assays (Fig. 5A). In wild-type, Tab1-/-, and Tab2-/- cells, the IKK complex was activated normally with peak activity at 15 min post-induction of TNF-α, whereas IKK complex activation was severely impaired in Tak1m/m cells. In wild-type, Tab1-/-, and Tab2-/- cells, IL-1-induced IKK complex activation occurs normally and peaks at 30 min post-induction; however, IKK complex activation was moderately decreased in Tak1m/m cells. Consistent with these results, luciferase analysis with NF-κB-responsive reporters showed that wild-type, Tak1m/m, Tab1-/-, and Tab2-/- cells all exhibited a similar degree of NF-κB activation by overexpression of IKKβ-SSEE, a dominant active mutant (Fig. 5B). Therefore, these results indicate that TAK1 functions upstream of the IKK complex in NF-κB activation by TNF-α or IL-1, and that disruption of the Tak1 gene does not affect signaling to NF-κB downstream of the IKK complex. We next examined the requirement for TAK1 in phosphorylation of the IKK complex by TNF-α or IL-1 (Fig. 5C). Immunoblotting analysis with anti-phospho-IKKα/β antibody showed that phosphorylation of IKKα/β following TNF-α or IL-1 stimulation occurs normally and peaks at 15–30 min in wild-type cells, whereas in Tak1m/m cells, TNF-α-induced phosphorylation was significantly decreased and IL-1-induced phosphorylation was modestly decreased. Hence, these results suggest that TAK1 plays a differential role in regulating activation of the IKK complex in TNF-α and IL-1 signaling pathways. Taken together, TAK1 functions upstream of the IKK complex, where TAK1 mediates phosphorylation and activation of the IKK complex in TNF-α and IL-1 signaling pathways.
Figure 5.
TAK1 is required for TNF- and IL-1-induced IKK activation. (A) Wild-type (WT), Tak1m/m, Tab1-/-, and Tab2-/- cells were treated with TNF-α (10 ng/mL; top) or IL-1 (10 ng/mL; bottom) for the indicated times. IKK activity was analyzed by in vitro kinase assay using a GST-IκB-α substrate. Protein input was analyzed by immunoblotting analysis with antibodies specific for IKKα and Nemo. (B) Cells were transfected with pcDNA3 or IKKβ (SSEE) together with pBIIx-luc and Renilla luciferase vectors. NF-κB activity was analyzed by luciferase assay and is expressed as fold change compared with untreated control. (C) Wild-type (WT) and Tak1m/m cells were treated with TNF-α (10 ng/mL; top) or IL-1 (10 ng/mL; bottom) for the indicated times, and then immunoblotted with antiphospho-IKKα/β antibody. Immunoblotting analysis with anti-IKKα antibody was performed as a control. (D) Cells were transfected with HA-IKKβ together with either pcDNA3, Flag-tagged TRAF2, or RIP1 expression vector. Cell were harvested 24 h after transfection, and cells were immunoprecipitated with anti-HA antibody and protein A agarose. IKKβ kinase activity was analyzed by in vitro kinase assay as in A.(E) Cells were transfected with HA-IKKβ expression vector together with either pcDNA3, Flag-tagged TRAF6, or MyD88 expression vector. Cells were harvested 24 h after transfection and IKKβ was immunoprecipitated with anti-HA antibody and protein A agarose. IKKβ activity was analyzed by in vitro kinase assay. (F) Cells were treated with IL-1 (10 ng/mL) for the indicated times, and then immunoblotted with anti-IRAK1 antibody. Immunoblotting with anti-GAPDH4 antibody was performed to control for gel loading. (G) Wild-type and Tak1m/m cells were transfected with Flag-IKKβ expression vector together with pcDNA3, or HA-tagged wild-type TAK1 (HA-TAK1-WT). Cells were stimulated 24 h later with TNF-α (10 ng/L) or IL-1 (10 ng/mL) for 20 min, lysed, and immunoprecipitated IKKβ kinase activity was analyzed by in vitro kinase assay.
In the TNF-α signaling pathway, RIP1 and TRAF2 are required for IKK complex activation and recruitment of the IKK complex to TNFRI receptor complex, respectively (Devin et al. 2000; Chen and Goeddel 2002). To test whether TAK1 is required for TRAF2 or RIP1-mediated IKK complex activation, wild-type and Tak1m/m cells were transfected with HA-tagged IKKβ together with pcDNA3, Flag-tagged TRAF2, or RIP1 (Fig. 5D). In vitro kinase assays showed that wild-type cells exhibited IKK complex activation in the presence of TRAF2 or RIP1, whereas TRAF2 or RIP1-induced IKK complex activation was impaired in Tak1m/m cells. Thus, this result indicates that TAK1 is required for signaling from TRAF2 and RIP1 to the IKK complex. Given that MyD88, IRAK1/4, and TRAF6 are key regulators in IL-1R- and TLR signaling pathways (Takeda and Akira 2005), we next tested MyD88 or TRAF6-mediated activation of the IKK complex in wild-type and Tak1m/m cells (Fig. 5E). Cells were transfected with HA-tagged IKKβ together with pcDNA3, Flag-tagged MyD88, or TRAF6, and the IKK complex activity was analyzed by in vitro kinase assay. As expected, expression of MyD88 or TRAF6 induced activation of the IKK complex in wild-type cells, whereas Tak1m/m cells exhibited an impaired activation of the IKK complex. In addition, immunoblotting analysis with anti-IRAK1 antibody showed that IL-1-induced IRAK1 degradation occurs normally in both wild-type and Tak1m/m cells (Fig. 5F). Thus, these results indicate that in the IL-1 signaling pathway, TAK1 is required for signaling from MyD88 or TRAF6 to the IKK complex, but not for IRAK degradation. Therefore, these results suggest that TAK1 plays a role in mediating IKK complex activation from TRAF2–RIP1 and MyD88–TRAF6–IRAK1 in TNFR1 and IL-1R/TLR-mediated signaling pathways, respectively.
To examine whether exogenous expression of TAK1 proteins can restore IKK activity in Tak1m/m cells, wild-type and Tak1m/m cells were transfected with Flag-tagged IKKβ together with pcDNA3 or HA-tagged wild-type TAK1 (Fig. 5G). In vitro kinase assays showed that Tak1m/m cells expressing exogenous TAK1 proteins exhibited IKK complex activation in response to TNF-α or IL-1, whereas IKK complex activation was impaired in the cells expressing pcDNA3. Thus, exogenous TAK1 expression restored activity of the IKK complex in TNF-α or IL-1 signaling pathways. Taken together, it appears that TAK1 functions upstream of IKK complex as an IKK kinase, and downstream of TRAF2–RIP1 and MyD88–TRAF6–IRAK1. Hence, TAK1 mediates IKK complex activation from the receptor complex, including TRAF2–RIP1 and MyD88–TRAF6–IRAK1 in TNF-α and IL-1-induced signaling pathways, respectively.
Since ERK and p38 MAP kinases are essential downstream molecules in TNFR1 and IL-1R/TLR-mediated signaling pathways (Baud and Karin 2001; Takeda and Akira 2005), we analyzed the kinetics of ERK and p38 MAP kinase activation by immunoblotting with antibodies specific for phospho-ERK1/2 and phospho-p38 MAP kinases (Supplementary Fig. S2A,B). Wild-type and Tak1m/m cells both exhibited phosphorylation of ERK and p38 MAP kinases in response to TNF-α, although phosphorylation of p38 MAP kinase is modestly decreased in Tak1m/m cells. In contrast, in Tak1m/m cells, ERK phosphorylation was delayed and p38 phosphorylation was severely impaired following IL-1 stimulation, relative to wild-type cells in which ERK and p38 phosphorylation occurs normally. Therefore, these results suggest that TAK1 plays a differential role in regulating activation of ERK and p38 MAP kinases in TNF-α and IL-1-induced signaling pathways.
TAK1 is not required for alternative NF-κB activation in LT-β signaling pathway
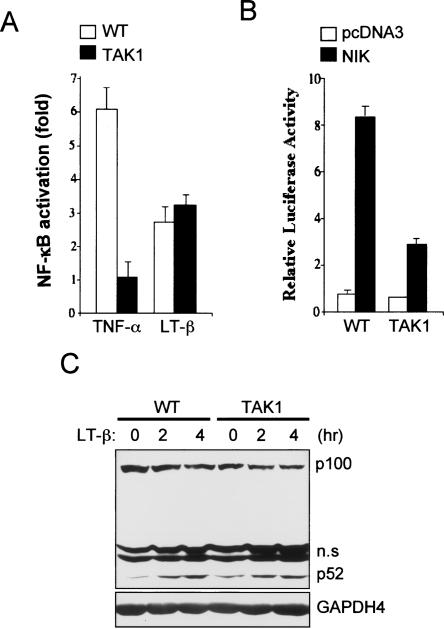
In the alternative NF-κB activation pathway, IKKα homodimers are activated via NF-κB-inducing kinase (NIK) in response to cytokines such as LT-β, BAFF, and CD40 (Dejardin et al. 2002; Derudder et al. 2003). Activated IKKα, in turn, phosphorylates the NF-κB p100 protein and induces its processing to the mature p52 subunit (Xiao et al. 2004). Finally, p52-containing heterodimers, notably p52:RelB, translocate into the nucleus and drive expression of a subset of NF-κB target genes (Xiao et al. 2001; Bonizzi and Karin, 2004; Hayden and Ghosh 2004). To examine the role of TAK1 in the alternative NF-κB pathway, we treated wild-type and Tak1m/m cells with TNF-α or LT-β, and analyzed NF-κB-responsive gene expression using a luciferase-reporter assay (Fig. 6A). As expected, TNF-α-induced NF-κB activation occurs normally in wild-type cells, but is completely abolished in Tak1m/m cells. In contrast, wild-type and Tak1m/m cells exhibited a similar degree of NF-κB activation in response to LT-β, indicating that LT-β-induced, NF-κB activation is normal in the absence of TAK1. NF-κB activation by NIK overexpression was partially decreased in Tak1m/m cells, relative to wild-type cells (Fig. 6B), which is consistent with previous reports that NIK overexpression strongly induces both classical and alternative NF-κB activation pathways (Malinin et al. 1997; Woronicz et al. 1997). These results, therefore, suggest that in the LT-β signaling pathway, TAK1 mediates signaling of NIK to the classical NF-κB activation pathway, but is not involved in activation of the alternative NF-κB pathway. To further test this hypothesis, we treated cells with LT-β at multiple time points and immunoblotted with the anti-p100 antibody (Fig. 6C). Wild-type and Tak1m/m cells both exhibited a similar degree of induction of p100 processing to p52 subunit in response to LT-β, indicating that LT-β-induced p100 processing is normal in Tak1m/m cells. Therefore, TAK1 is not involved in activation of IKKα in the alternative NF-κB pathway.
Figure 6.
LT-β-induced NF-κB activation is normal in Tak1m/m cells. (A) Wild-type (WT) and Tak1m/m cells were transfected with pBIIx-luc reporter along with Renilla luciferase vector. Twenty-four hours after transfection, cells were treated with TNF-α (10 ng/mL) or anti-LT-βR antibody (2 μg/mL) for 5 or 12 h, respectively, and then analyzed for luciferase activity. Results are expressed as the fold change in luciferase activity relative to untreated cells. (B) Cells were transfected with pcDNA3 or NIK expression vector along with pBIIx-luc reporter and Renilla luciferase vector. NF-κB activity was analyzed by luciferase assay. (C) Cells were stimulated with anti-LT-βR antibody (2 μg/mL) for the indicated times, and then immunoblotted with anti-p100 antibody. Protein amount was analyzed by immunoblotting with anti-GAPDH4 antibody.
Role of TAK1 complex in TGF-β signaling pathway
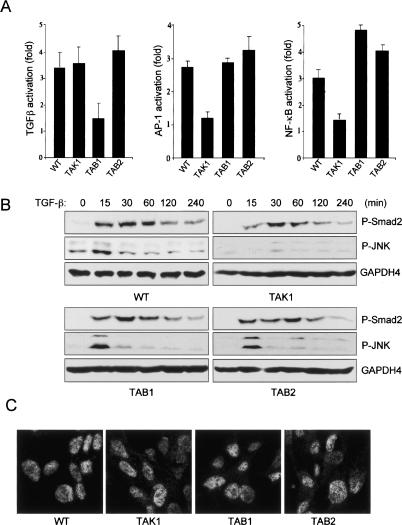
TAK1, originally identified as a key regulator in TGF-β signaling, is believed to be involved in mesoderm induction and patterning through BMP signaling during Xenopus early development (Yamaguchi et al. 1995; Shibuya et al. 1998). To examine the role of each member of the TAK1 complex in TGF-β signaling, we analyzed TGF-β, NF-κB, and AP-1-responsive gene induction in response to TGF-β signaling by using luciferase reporters (Fig. 7A). TGF-β-induced NF-κB and AP-1 activation were impaired in the absence of TAK1, although NF-κB and AP-1 activation following TGF-β stimulation occurs normally in wild-type, Tab1-/-, and Tab2-/- cells. Consistent with these results, JNK phosphorylation was completely abolished in Tak1m/m cells relative to wild-type, Tab1-/-, and Tab2-/- cells in which the phosphorylation occurs normally (Fig. 7B). Intriguingly, wild-type, Tak1m/m, and Tab2-/- cells all exhibit normal gene expression in response to TGF-β, whereas gene expression was severely impaired in Tab1-/- cells. Thus, these results indicate that in TGF-β-induced signaling pathways, TAK1 is required for NF-κB and JNK activation, but not for gene induction, TAB1 is required for gene induction but not for NF-κB and JNK activation, and TAB2 is not required for gene induction or activation of NF-κB and JNK. This implies that TAK1 and TAB1, but not TAB2 play differential roles in TGFβ signaling pathways. We next examined TGFβ-induced Smad2 activation by immunblotting and immunostaining with antiphospho-Smad2 antibody (Fig. 7B,C). Loss of TAK1, TAB1, or TAB2 does not affect phosphorylation or nuclear translocation of Smad2 following TGF-β stimulation. This indicates that the TAK1 complex is not required for TGF-β-induced Smad2 activation—the classical TGF-β signaling pathway. Taken together, these results suggest that each member of the TAK1 complex plays different roles in TGF-β signaling pathways. TAB1 is required for efficient induction of a TGFβ-responsive reporters, TAK1 is required for TGFβ-induced NF-κB, and AP-1 activation and deficiency in TAB2 does not appear to affect TGFβ signaling.
Figure 7.
Role of TAK1 complex in TGF-β signaling pathway. (A) Wild-type, Tak1m/m, Tab1-/-, and Tab2-/- cells were transfected with the either 3TP-lux (left), AP-1-luc (middle), or pBIIxluc (right) reporter construct together with Renilla luciferase vector. Twenty-four hours after transfection, cells were treated with TGF-β (1 ng/mL) for 24 h and then analyzed for luciferase activity. Results are expressed as the fold induction in luciferase activity relative to that of untreated cells. (B) Smad2 and JNK phosphorylation by TGF-β (1 ng/mL) were analyzed by immunoblotting with antiphospho-Smad2 antibody and antiphospho-JNK antibody, respectively. Immunoblotting analysis with anti-GAPDH4 antibody was performed as a control. (C) Cells were stimulated with TGF-β (1 ng/mL) for 20 min, stained with antiphospho-Smad2 antibody, and analyzed by immunofluorescence microscopy.
Discussion
The inducible transcription factors AP-1 and NF-κB are key mediators of multiple aspects of the immune and inflammatory response and are activated upon stimulation with proinflammatory cytokines, microbial products, and various cell stressors. Multiple receptors, most notably the Toll/IL-1 and TNF receptor families, have been shown to activate both AP-1 and NF-κB, and activation of the IKK complex and JNK have been considered the points of convergence in signaling pathways leading to NF-κB and AP-1. Recently, however, the protein kinase TAK1 has emerged as a potential regulator of both these pathways, acting upstream of IKK and JNK as both an IKK kinase (IKKK) and JNK kinase kinase (JNKKK). Initial support for this hypothesis came from a variety of in vitro experiments that suggested that the interaction of the TAK1 complex (TAK1, TAB1, and TAB2) with ubiquitinated TRAF family members led to TAK1 activation, followed by activation of IKK and JNK. Although the biochemical evidence for such a role for the TAK1 complex appeared persuasive, there is almost no definitive genetic evidence supporting the hypothesis that TAK1, TAB1, and TAB2 are activated through a ubiquitination-dependent mechanism, and that the TAK1 complex plays such an essential role in AP-1 and NF-κB signaling pathways in mammalian cells.
Therefore, to investigate the biological role of TAK1, we generated TAK1-deficient mice by using gene trap mutated Tak1 ES (Fig. 1A; Stryke et al. 2003; Austin et al. 2004; Skarnes et al. 2004). Tak1 homozygous mutant embryos died around E10 and exhibited abnormal development of the neural tube, suggesting that TAK1 is critical for early embryonic development (Fig. 1D,E). The phenotype of Tak1 embryos is strikingly different from that of Tab1 and Tab2-deficient embryos, since Tab1 mutant embryos die at a late stage of gestation due to abnormal cardiovascular and lung morphogenesis, while Tab2 mutant embryos die around E12.5 due to liver degeneration and apoptosis (Komatsu et al. 2002; Sanjo et al. 2003). These results further emphasize that TAK1 has one or more unique functions that are independent of the adaptor proteins, TAB1 and TAB2.
In the NF-κB activation pathway, TAK1 interacts with TRAF2 or TRAF6, and IKKα/β following TNF-α or IL-1 stimulation, and reduction of TAK1 through RNA interference inhibits IL-1 and TNF-α-induced IKK activation (Takaesu et al. 2003). In the JNK activation pathway, TAK1 interacts directly with JNK1, and expression of a kinase-inactive TAK1 inhibits TNF-α-induced JNK activation (Sanna et al. 2002). Although primary Tak1m/m embryonic fibroblasts could not be cultured, we were able to generate SV40 large T antigen-transformed embryonic fibroblasts for analyzing signaling in the absence of TAK1 (Kobayashi et al. 2005). We have found that loss of TAK1 leads to defects in NF-κB and JNK-responsive reporter gene expression, NF-κB DNA-binding activity, IκBα degradation, IKK complex activation, and JNK phosphorylation in TNF-α signaling pathways. In contrast, NF-κB and JNK activation following TNF-α stimulation was largely intact in Tab1-/- and Tab2-/- cells. Thus, these results indicate that TAK1, but not TAB1 or TAB2, is required for NF-κB and JNK activation in TNF-α signaling pathways. In addition, in vitro kinase assays in Tak1m/m cells demonstrate that TAK1 is necessary for signaling from RIP1 and TRAF2 to the IKK complex (Fig. 5). Thus, TAK1 is a key functional intermediate between the recruitment of TRAF2–RIP1 to TNFR1 (Devin et al. 2000; Chen and Goeddel 2002) and activation of the IKK complex. It is well established that NF-κB and JNK activation plays a role in protecting cells from TNF-α-induced apoptosis (Beg and Baltimore 1996; Van Antwerp et al. 1996; Lamb et al. 2003). Consequently, Tak1m/m cells were highly sensitive to apoptosis and display elevated levels of caspase 8 activation relative to wild-type, Tab1-/-, and Tab2-/- cells (Fig. 4). As expected, exogenous TAK1 expression could rescue the defect of Tak1m/m cells in TNF-α-induced apoptosis. In contrast to previous in vitro studies that have suggested that the TAK1 adaptor proteins TAB1 and TAB2 are critical for TAK1 activation and TNF-α signaling, our results suggest that only TAK1 is required for TNF-α-induced IKK activation and JNK phosphorylation leading to NF-κB and JNK activation in vivo.
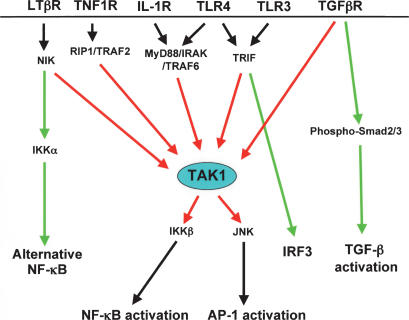
In vitro studies have reported that the MyD88–IRAK1–IRAK4–TRAF6 complex regulates TAK1 activation through TAB1 and TAB2, thereby leading to IKK complex and MKK6 activation in response to IL-1 (Wang et al. 2001; Jiang et al. 2002). However, when we examined the role of TAK1, TAB1, and TAB2 in the IL-1 signaling pathway, only TAK1 deficiency abolished IL-1-induced NF-κB and JNK activation. Intriguingly, NF-κB DNA-binding activity and IKK complex activity following IL-1 stimulation were not completely inhibited in Tak1m/m cells, although no NF-κB-responsive reporter gene expression was observed following TNF-α and IL-1 stimulation. One likely explanation for this discrepancy is that the poor transfection efficiency of embryonic fibroblasts precludes using luciferase analysis to discriminate between the complete loss following TNFα and near-complete loss of NF-κB activation following IL-1 that is detected by biochemical analyses. In vivo, these results suggest that although TAK1 is crucial for both IL-1 and TNF-α-induced JNK activation, the requirement for TAK1 in IL-1-induced IKK complex and NF-κB activation is less complete. In vitro kinase assays indicate that the requirement for TAK1 function is positioned between IRAK–MyD88–TRAF6 and activation of the IKK complex (Fig. 5). Considered together with the result that TAK1 deficiency caused delayed ERK activation and impaired p38 activation in response to IL-1 (Supplementary Fig. S2), TAK1 may function as a branching signal transducer from the MyD88–IRAK1–TRAF6 complex to the NF-κB, JNK, ERK, and p38 MAP kinase pathways (Fig. 8).
Figure 8.
Schematic model showing role of TAK1 complex in inflammatory signaling pathways. Red lines indicate TAK1-dependent signaling pathways, while green lines indicate TAK1-independent pathways.
Previous studies have demonstrated that TAK1 is required for IKK complex and JNK activation in LPS-induced signaling (Lee et al. 2000). However, we find that while TAK1 deletion abolishes JNK-responsive reporter gene expression, NF-κB DNA-binding affinity and reporter gene expression are only partially reduced (Fig. 3A–C). Alternatively, when TAK1 activity is disrupted by DN-TAK1 overexpression, consistent with previously published results (Lee et al. 2000), we find that NF-κB activation by TLR4 is completely abrogated (Supplementary Fig. S1C). NF-κB and JNK activation following LPS stimulation is normal in Tab1-/- and Tab2-/- cells; thus TAK1, but not TAB1 or TAB2, is required for LPS-induced JNK activation. These results imply that TAK1 mediates signaling of MyD88–IRAK–TRAF6 complex to NF-κB activation, while TRAM-mediated NF-κB activation is altogether independent of TAK1. Previous observations that TAK1 is involved in all TLR4 signaling pathways to NF-κB likely resulted from nonphysiological effects induced by overexpression of DN-TAK1 constructs. Consistent with this hypothesis, while Tak1m/m cells exhibit normal LPS-induced IRF3 activation, which is dependent on TRIF, overexpression of DN-TAK1 completely inhibits both NF-κB and IRF3 activation. Our analysis of TLR3-induced NF-κB activation further supports a selective role for TAK1 in TLR signaling. Following poly(I:C) stimulation, TAK1, but not TAB1 or TAB2, is required for NF-κB activation, but not for IRF3 activation. Thus TAK1, but not the adapter proteins TAB1 and TAB2, appears to play a vital role in inflammatory signaling to both AP-1 and the classical NF-κB pathway.
In recent years, two NF-κB activation pathways have been recognized that play distinct roles in the regulation of immune responses in mammals (Bonizzi and Karin 2004; Hayden and Ghosh 2004). In the classical pathway, diverse stimuli, such as proinflammatory cytokines TNF-α and IL-1, pathogen-associated molecular patterns (PAMPs), and antigen receptors (i.e., TcR and BcR) induce phosphorylation and degradation of IκBs in an IKKβ and NEMO-dependent manner. The so-called alternative pathway is instead dependent on IKKα. Cytokines, such as LT-β, BAFF, and CD40 activate IKKα via the protein kinase NIK, and the activated IKKα in turn induces processing of NF-κB2/p100 precursor to mature p52, followed by translocation of p52 containing complexes to the nucleus (Senftleben et al. 2001). We found that TAK1 deletion does not affect NF-κB reporter gene expression or the processing of p100 to p52 following LT-β stimulation. Interestingly, NF-κB reporter gene expression following NIK overexpression, which activates both IKKα and IKKβ (Woronicz et al. 1997), was moderately decreased in Tak1m/m cells. These results raise the intriguing possibility that TAK1 may be necessary for the IKKβ activation that occurs upon NIK overexpression, but that TAK1 is not required for NIK-mediated IKKα activation and p100 processing in LT-β signaling. Furthermore, these results highlight a distinct requirement for the activation of alternative and classical NF-κB, upstream of the IKK complex itself, and suggest that TAK1 and NIK may occupy analogous positions as IKKKs in these two signaling pathways.
It has been reported that TAK1 is involved in TGF-β-induced signaling of Xenopus and mammalian cells (Shibuya et al. 1996; Yamaguchi et al. 1995). To test requirements for each member of the TAK1 complex in TGF-β signaling, we analyzed luciferase activities by using NF-κB, JNK, and TGF-β-responsive reporters. Intriguingly, TAB1 is required for efficient induction of TGFβ-responsive reporters, while TAK1 is required for TGFβ-induced NF-κB and AP-1 activation, and deficiency in TAB2 does not appear to affect TGFβ signaling. However, the TAK1 complex, including TAK1, TAB1, and TAB2 is not required for Smad2 activation in TGFβ signaling. Therefore, these results suggest that each member of the TAK1 complex plays different roles in regulating NF-κB, JNK, and Smad2 activation in TGF-β signaling pathways (Fig. 8). Whether the whole TAK1 complex is required for TGF-β-induced signaling remains to be determined by generating cells lacking TAK1 together with TAB1 and TAB2.
A surprising conclusion from our studies is the lack of any effect on TNFα, IL-1, LPS, and poly(I:C) signaling in cells lacking TAB1 or TAB2. At present, the early lethality of the individual knock-outs has prevented analysis of signaling in cells lacking both TAB1 and TAB2. In addition, the recent description of human TAB3 as a TAB2 homolog has further complicated the genetic analysis of the role of TAB proteins. The murine TAB3 gene has not yet been cloned, although efforts are underway to knock-out this gene. A recent study has reported that down-regulation of both TAB2 and TAB3 using RNA interference was necessary to observe inhibition of IKK and JNK activation by TNF-α and IL-1, suggesting that TAB2 and TAB3 may play a redundant role in these signaling pathways (Cheung et al. 2003; Ishitani et al. 2003). Therefore, it is clear that additional analysis will be necessary to determine the exact role that TAB proteins play in regulation of TAK1 activity. However, the different developmental phenotypes observed in TAK1, TAB1, and TAB2 knock-outs strongly suggest that the three proteins do not always function together as a complex. Remarkably, only the lack of TAB1 leads to a dramatic reduction of TGF-β-induced gene-expression, once again strongly hinting at a unique role for TAB1 that does not depend on either TAK1 or the other TAB-proteins. Our results also raise questions about the widely discussed model of ubiquitination-dependent activation of TAK1, where TAB1 is believed to play an essential role. While we have been unable to genetically address the role of ubiquitination in TAK1 regulation, it does appear that additional studies will be necessary to fully elucidate the details of how the different TAB proteins might regulate TAK1.
Materials and methods
Cells and antibodies
HEK 293 (human kidney embryonic cells) and COS1 (simian fibroblast cells) were purchased from ATCC. HEK 293, COS1, and immortalized MEF cells were cultured in DMEM medium (GIBCO) containing 7% FBS, 2 mM L-glutamine, 100 unit/mL of penicillin, and 100 μg/mL streptomycin and 5 × 10-5 M β-mercaptoethanol. Antibodies used were anti-TAK1 (Ninomiya-Tsuji et al. 1999), antiphospho-IKKα/β, antiphospho-c-Jun, antiphospho-Erk1/2, antiphospho-JNK1/2, anti-JNK1/2, antiphospho-p38, and antiphospho-Smad2 (Cell Signaling), anti-IκBα (Santa Cruz, C21), and anti-GAPDH4 (Research Diagnostics). Cells were treated with poly(I:C) and LPS (Sigma), anti-LT-βR (Alexis Biochemicals), and rhTNF-α, mTGF-β, and mIL-1β (R&D Systems) as indicated.
Generation of Tak1m/m mice
Tak1 gene trap ES clone (XB444) was obtained from Dr. Williams Skarnes (BayGenomics) (Stryke et al. 2003; Skarnes et al. 2004). The Tak1 ES clones were injected into C57BL/6 blastocysts and transferred to ICR females. Male chimeric mice were mated with C57BL/6 females, resulting in transmission of the inserted allele to the germline. Positive mice were interbred and maintained on a mixed 129 × C57BL/6 background.
Generation of MEFs
E9.75 embryos from timed breeding of Tak1-/- mice were dissected free of maternal tissues and Reichert's membrane, washed with PBS, and incubated with 0.1% collagenase (Sigma) and Trypsin-EDTA (GIBCO) for 30 min at 37°C. The cell suspension was plated in 12-well plates containing DMEM supplemented with 10% fetal bovine serum. Tak1m/m MEFs were identified by immunoblotting with TAK1 antibody or PCR using extraembryonic tissues. MEFs were stably transfected with linearlized SV40 Large T antigen plasmid by using FuGENE 6.
Immunofluorescence
For immunofluorescence analysis, cells were grown on Alcian Blue-coated coverslips, treated with TGF-β (1 ng/mL) for 20 min, washed with PBS, fixed with 4% paraformaldehyde for 30 min at room temperature, and permeabilized with permeabilization buffer (0.05% saponin, 1% FBS, 10 mM HEPES, 10 mM glycine in PBS at pH 7.5) for 30 min at room temperature. Cells were incubated with antiphospho-Smad2 antibody for 30 min at room temperature, followed by incubation with goat anti-rabbit-FITC under identical conditions.
TNF-induced apoptosis
MEF cells (3 × 105) were seeded in a 6-cm Petri dish 24 h prior to TNF-α stimulation at the indicated concentration for the time. Cells were harvested in PBS and incubated with Annexin V-FITC and propidium iodide (BD Pharmingen) according to manufacturer's instructions and analyzed by flow cytometer.
Luciferase reporter assay
MEF cells (2.5 × 105) grown on 12-well plates were transiently transfected using FuGENE 6 with the NF-κB-dependent reporter construct pBIIx-luc together with the Renilla luciferase vector (Promega). Total DNA concentration in each experiment was maintained by adding the appropriate empty vector to the DNA mixture. Twenty-four hours after transfection, cells were lysed, and luciferase activity was measured using the dual luciferase assay kit (Promega). The levels of transfected proteins in lysates were examined by immunoblotting with appropriate epitope tag-specific antibodies.
In vitro kinase assay
For in vitro kinases assays, 1 × 107 MEF cells grown on a 10-cm Petri dish were transiently transfected with various DNAs using the FuGENE 6 reagent as described above. Forty-eight hours after transfection, the cells were lysed in TNT lysis buffer for 30 min. Alternatively, the cells were either untreated or treated with TNF (10 ng/mL) and IL-1 (10 ng/mL) and then lysed in TNT lysis buffer. Protein amount in each lysate was determined by using a Bio-Rad protein assay kit (Bio-Rad) and then normalized among the samples. Protein from lysates was immunoprecipitated using either anti-Nemo antibody and protein G-Sepharose or anti-Flag (M2)-coupled agarose beads overnight at 4°C, and the precipitates were washed in TNT lysis buffer and then kinase buffer (20 mM HEPES at pH 7.5, 20 mM MgCl2, 1 mM EDTA, 2 mM NaF, 2 mM-glycerophosphate, 1 mM dithiothreitol, 10 μM ATP). Precipitates were incubated for 15 min at 37°C in kinase buffer containing appropriate GST-fused substrate proteins and 10 μCi of [γ32P]ATP (Amersham Biosciences). The substrate was then precipitated using glutathioneagarose (Amersham Biosciences), resolved by SDS-PAGE, and phosphorylated proteins were visualized by autoradiography.
Galactosidase staining of embryos
β-Galactosidase staining was performed as previously described (Lawson et al. 1999).
Acknowledgments
We thank Williams Skarnes (BayGenomics) for providing Tak1m/m ES cells. M.S.H. was supported by NIH/National Institute of General Medical Sciences Medical Scientist Training Grant GM07205. This work was supported by a grant from NIH (R37-AI33443).
Article published online ahead of print. Article and publication date are at http://www.genesdev.org/cgi/doi/10.1101/gad.1360605.
Supplemental material is available at http://www.genesdev.org.
Corresponding author.
E-MAIL Sankar.ghosh@yale.edu; FAX (203) 737-1764.
References
- Austin C.P., Battey, J.F., Bradley, A., Bucan, M., Capecchi, M., Collins, F.S., Dove, W.F., Duyk, G., Dymecki, S., Eppig, J.T., et al. 2004. The knockout mouse project. Nat. Genet. 36: 921–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baud V. and Karin, M. 2001. Signal transduction by tumor necrosis factor and its relatives. Trends Cell. Biol. 11: 372–377. [DOI] [PubMed] [Google Scholar]
- Beg A.A. and Baltimore, D. 1996. An essential role for NF-κBin preventing TNF-α-induced cell death. Science 274: 782–784. [DOI] [PubMed] [Google Scholar]
- Beg A.A., Sha, W.C., Bronson, R.T., Ghosh, S., and Baltimore, D. 1995. Embryonic lethality and liver degeneration in mice lacking the RelA component of NF-κ B. Nature 376: 167–170. [DOI] [PubMed] [Google Scholar]
- Bonizzi G. and Karin, M. 2004. The two NF-κB activation pathways and their role in innate and adaptive immunity. Trends. Immunol. 25: 280–288. [DOI] [PubMed] [Google Scholar]
- Boutros M., Agaisse, H., and Perrimon, M. 2002. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell 3: 711–722. [DOI] [PubMed] [Google Scholar]
- Chen G. and Goeddel, D.V. 2002. TNF-R1 signaling: A beautiful pathway. Science 296: 1634–1635. [DOI] [PubMed] [Google Scholar]
- Cheung P.C., Campbell, D.G., Nebreda, A.R., and Cohen, P. 2003. Feedback control of the protein kinase TAK1 by SAPK2a/p38α. EMBO J. 22: 5793–5805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dejardin E., Dorin, N.M., Delhase, M., Hass, E., Cao, Y., Makris, C., Li, Z-W., Karin, M., Ware, C.F., and Green, D.R. 2002. The lymphotoxin-β receptor induces different patterns of gene expression via two NF-κB pathways. Immunity 17: 525–535. [DOI] [PubMed] [Google Scholar]
- Deng L., Wang, C., Spencer, E., Yang, L., Braun, A., You, J., Slaughter, C., Pickart, C., and Chen, Z.J. 2000. Activation of the IκB kinase complex by TRAF6 requires a dimeric ubiquitin-conjugating enzyme complex and a unique polyubiquitin chain. Cell 103: 351–361. [DOI] [PubMed] [Google Scholar]
- Derudder E., Dejardin, E., Pritchard, L.L., Green, D.R., Korner, M., and Baud, V. 2003. RelB/p50 dimers are differentially regulated by tumor necrosis factor-α and lymphotoxin-β receptor activation. J. Biol. Chem. 278: 23278–23284. [DOI] [PubMed] [Google Scholar]
- Devin A., Cook, A., Lin, Y., Rodriguez, Y., Kelliher, M., and Liu, Z-G. 2000. The distinct roles of TRAF2 and RIP in IKK activation by TNFR1: TRAF2 recruits IKK to TNF-R1 while RIP mediates IKK activation. Immunity 12: 419–429. [DOI] [PubMed] [Google Scholar]
- Hayden M.S. and Ghosh, S. 2004. Signaling to NF-κB. Genes & Dev. 18: 2195–2224. [DOI] [PubMed] [Google Scholar]
- Ishitani T., Takaesu, G., Ninomiya-Tsuji, J., Shibuya, H., Gaynor, R.B., and Matsumoto, K. 2003. Role of the TAB2-related protein TAB3 in IL-1 and TNF signaling. EMBO J. 22: 6277–6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jadrich J.L., O'Connor, M.B., and Coucouvanis, E. 2003. Expression of TAK1, a mediator of TGF-β and BMP signaling, during mouse embryonic development. Gene Expr. Patterns 3: 131–134. [DOI] [PubMed] [Google Scholar]
- Jiang Z., Ninomiya-Tsuji, J., Qina, Y., Matsumoto, K., and Li, X. 2002. Interleukin-1 (IL-1) receptor-associated kinase-dependent IL-1-induced signaling complexes phopsphorylate TAK1 and TAB2 at the plasma membrane and activate TAK1 in the cytosol. Mol. Cell. Biol. 22: 7158–7167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang Z., Zamanian-Daryoush, M., Nie, H., Silva, A.M., Williams, B.R.G., and Li, X. 2003. Poly(dI-dC)-induced toll-like receptor 3 (TLR3)-mediated activation of NF-κB and MAP kinase is through an interleukin-1 receptor-associated kinase (IRAK)-independent pathway employing the signaling components TLR3–TRAF6–TAK1–TAB2–PKR. J. Biol. Chem. 278: 16713–16719. [DOI] [PubMed] [Google Scholar]
- Jiang Z., Mak, T.W., Sen, G., and Li, X. 2004. Toll-like receptor 3-mediated activation of NF-κB and IRF3 diverges at Toll-IL-1 receptor domain-containing adapter inducing IFN-β. Proc. Natl. Acad. Sci. 101: 3533–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi H., Tanaka, N., Asao, H., Miura, S., Kyuuma, M., Semura, K., Ishii, N., and Sugamura, K. 2005. Hrs, a mammalian master molecule in vesicular transport and protein sorting, suppresses the degradation of ESCRT proteins signal transducing adaptor molecule 1 and 2. J. Biol. Chem. 280: 10468–10477. [DOI] [PubMed] [Google Scholar]
- Komatsu Y., Shibuya, H., Takeda, N., Ninomiya-Tsuji, J., Yasui, T., Miyado, K., Sekimoto, T., Ueno, N., Matsumoto, K., and Yamada, G. 2002. Targeted disruption of the Tab1 gene causes embryonic lethality and defects in cardiovascular and lung morphogenesis. Mech. Dev. 119: 239–249. [DOI] [PubMed] [Google Scholar]
- Lamb J.A., Ventura, J-J., Hess, P., Flavell, R.A., and Davis, R.J. 2003. JunD mediates survival signaling by the JNK signal transduction pathway. Mol. Cell 11: 1479–1489. [DOI] [PubMed] [Google Scholar]
- Lawson K.A., Dunn, N.R., Roelen, B.A.J., Zeinstra, L.M., Davis, A.M., Wright, C.V.E., Korving, J., and Hogan, B.L. 1999. Bmp4 is required for the generation of primordial germ cells in the mouse embryo. Proc. Natl. Acad. Sci. 13: 424–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Mira-Arbibe, L., and Ulevitch, R.J. 2000. TAK1 regulates multiple protein kinase cascades activated by bacterial lipopoly saccharide. J. Leukoc. Biol. 68: 909–915. [PubMed] [Google Scholar]
- Malinin N.L., Boldin, M.P., Kovalenko, A.V., and Wallach, D. 1997. MAP3K-realted kinase involved in NF-κ B induction by TNF, CD95 and IL-1. Nature 385: 540–544. [DOI] [PubMed] [Google Scholar]
- Ninomiya-Tsuji J., Kishimoto, K., Hiyama, A., Inoue, J., Cao, Z., and Matsumoto, K. 1999. The kinase TAK1 can activate the NIK-IκB as well as the MAP kinase cascade in the IL-1 signalling pathway. Nature 398: 252–256. [DOI] [PubMed] [Google Scholar]
- Park J.M., Brady, H., Ruocco, M.G., Sun, H., Williams, D., Lee, S.J., Kato Jr., T., Richards, N., Chan, K., Mercurio, F., et al. 2004. Targeting of TAK1 by the NF-κB protein Relish regulates the JNK-mediated immune response in Drosophila. Genes & Dev. 18: 584–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudolph D., Yeh, W.C., Wakeham, A., Rudolph, B., Nallainathan, D., Potter, J., Elia, A.J., and Mak, T.J, 2000. Severe liver degeneration and lack of NF-κB activation in NEFMO/IKKγ-deficient mice. Genes & Dev. 14: 854–862. [PMC free article] [PubMed] [Google Scholar]
- Sanjo H., Takeda, K., Tsujimura, T., Ninomiya-Tsuji, J., Matsumoto, K., and Akira, S. 2003. TAB2 is essential for prevention of apoptosis in fetal liver but not for interleukin-1 signaling. Mol. Cell. Biol. 23: 1231–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanna M.G., Correia, J.S., Ducrey, O., Lee, J., Nomoto, K., Schrantz, N., Deveraux, Q.L., and Ulevitch, R.J. 2002. IAP suppression of apoptosis involves distinct mechanisms: The TAK1/JNK1 signaling cascade and caspase inhibition. Mol. Cell. Biol. 22: 1754–1766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sato S., Sugiyama, M., Yamamoto, M., Watanabe, Y., Kawai, T., Takeda, K., and Akira, S. 2003. Toll/IL-1 receptor domain-containing adaptor inducing IFN-β (TRIF) associates with TNF receptor-associated factor 6 and TANK-binding kinase 1, and activates two distinct transcription factors, NF-κ B and IFN-regulatory factor-3, in the Toll-like receptor signaling. J. Immunol. 171: 4304–4310. [DOI] [PubMed] [Google Scholar]
- Senftleben U., Cao, Y., Xiao, G., Greten, F.R., Krahn, G., Bonnizi, G., Chen, Y., Hu, Y., Fong, A., Sun, S.C., et al. 2001. Activation by IKKα of a second, evolutionary conserved, NF-κ B signaling pathway. Science 293: 1495–1499. [DOI] [PubMed] [Google Scholar]
- Shibuya H., Yamaguchi, K., Shirakabe, K., Tonegawa, A., Gotoh, Y., Ueno, N., Irie, K., Nishida, E., and Matsumoto, K. 1996. TAB1: An activator of the TAK1 MAPKKK in TGF-signal transduction. Science 272: 1179–1182. [DOI] [PubMed] [Google Scholar]
- Shibuya H., Iwata, H., Masuyama, N., Gotoh, Y., Yamaguchi, K., Irie, K., Matsumoto, K., Nishida, E., and Ueno, N. 1998. Role of TAK1 and TAB1 in BMP signaling in early Xenopus development. EMBO J. 17: 1019–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman N., Zhou, R., Erlich, R.L., Hunter, M., Bernstein, E., Schneider, D., and Maniatis, T. 2003. Immune activation of NF-B and JNK requires Drosophila TAK1. J. Biol. Chem 278: 48928–48934. [DOI] [PubMed] [Google Scholar]
- Skarnes W.C., von Melchner, H., Wurst, W., Hicks, G., Nord, R.S., Cox, T., Young, S.G., Ruiz, P., Soriano, P., Tessier-Lavigne, M., et al. 2004. A public gene trap resource for mouse functional genomics. Nat. Genet. 36: 543–544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song H.Y., Regnier, C.H., Kirschning, C.J., Goeddel, D.V., and Rothe, M. 1997. Tumor necrosis factor (TNF)-mediated kinase cascades: Bifurcation of nuclear factor-B and c-jun N-terminal kinase (JNK/SAPK) pathways at TNF receptor-associated factor 2. Proc. Natl. Acad. Sci. 94: 9792–9796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stryke D., Kawamoto, M., Huang, C.C., Johns, S.J., King, L.A., Harper, C.A., Meng, E.C., Lee, R.E., Yee, A., L'Italien, L., et al. 2003. BayGenomics: A resource of insertional mutations in mouse embryonic stem cells. Nucleic Acids Res. 31: 278–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaesu G., Kishida, S., Hiyama, A., Yamaguchi, K., Shibuya, H., Irie, K., Ninomiya-Tsuji, J., and Matsumoto, K. 2000. TAB2, a novel adaptor protein, mediates activation of TAK1 MAPKKK by linking TAK1 to TRAF6 in the IL-1 signal transduction pathway. Mol. Cell 5: 649–658. [DOI] [PubMed] [Google Scholar]
- Takaesu G., Ninomiya-Tsuji, J., Kishida, S., Li, X., Stark, G.R., and Matsumoto, K. 2001. Interleukin-1 (IL-1) receptor-associated kinase leads to activation of TAK1 by inducing TAB2 translocation in the IL-1 signaling pathway. Mol. Cell. Biol. 21: 2475–2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takaesu G., Surabhi, R.M., Park, K.J., Ninomiya-Tsuji, J., Matsumoto, K., and Gaynor, R.B. 2003. TAK1 is critical for IκB kinase-mediated activation of the NF-B pathway. J. Mol. Biol. 326: 105–115. [DOI] [PubMed] [Google Scholar]
- Takeda K. and Akira, S. 2005. Toll-like receptors in innate immunity. Int. Immunol. 17: 1–14. [DOI] [PubMed] [Google Scholar]
- Tanaka M., Fuentes, M.E., Yamaguchi, K., Durnin, M.H., Dalrymple, S.A., Hardy, K.L., and Goeddel, D.V. 1999. Embryonic lethality, liver degeneration, and impaired NF-κ B activation in IKK-β-deficient mice. Immunity 10: 421–429. [DOI] [PubMed] [Google Scholar]
- Van Antwerp D.J., Martin, S.J., Kafri, T., Green, D.R., and Verma, I.M. 1996. Suppression of TNF-α-induced apoptosis by NF-κB. Science 274: 787–789. [DOI] [PubMed] [Google Scholar]
- Vidal S., Khush, R.S., Leulier, F., Tzou, P., Nakamura, M., and Lemaitre, B. 2001. Mutations in the Drosophila dTAK1 gene reveal a conserved function for MAPKKKs in the control of rel/NFB-dependent innate immune responses. Genes & Dev. 15: 1900–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Deng, L., Hong, M,. Akkaraju, G.R., Inoue, J., and Chen, Z.J. 2001. TAK1 is a ubiquitin-dependent kinase of MKK and IKK. Nature 412: 346–351. [DOI] [PubMed] [Google Scholar]
- Woronicz J.D., Gao, X., Cao, Z., Rothe, M., and Goeddel, D.V. 1997. IκB kinase-β: NF-κB activation and complex formation with IκB kinase-α and NIK. Science 278: 866–869. [DOI] [PubMed] [Google Scholar]
- Xiao G., Harhaj, E.W., and Sun, S-C. 2001. NF-κB-inducing kinase regulates the processing of NF-κB2 p100. Mol. Cell 7: 401–409. [DOI] [PubMed] [Google Scholar]
- Xiao G, Fong, A., and Sun, S.C. 2004. Induction of p100 processing by NF-κB-inducing kinase involves docking IκB kinase α (IKKα) to p100 and IKKα-mediated phosphorylation. J. Biol. Chem. 279: 30099–30105. [DOI] [PubMed] [Google Scholar]
- Yamaguchi K., Shirakabe, K., Shibuya, H., Irie, K., Oishi, I., Ueno, N., Taniguchi, T., Nishida, E., and Matsumoto, K. 1995. Identification of a member of the MAPKKK family as a potential mediator of TGF-β signal transduction. Science 270: 2008–2011. [DOI] [PubMed] [Google Scholar]