Abstract
Synaptonemal complex (SC) assembly must occur between correctly paired homologous chromosomes to promote formation of chiasmata. Here, we identify the Caenorhabditis elegans HORMA-domain protein HTP-1 as a key player in coordinating establishment of homolog pairing and synapsis in C. elegans and provide evidence that checkpoint-like mechanisms couple these early meiotic prophase events. htp-1 mutants are defective in the establishment of pairing, but in contrast with the pairing-defective chk-2 mutant, SC assembly is not inhibited and generalized nonhomologous synapsis occurs. Extensive nonhomologous synapsis in htp-1; chk-2 double mutants indicates that HTP-1 is required for the inhibition of SC assembly observed in chk-2 gonads. htp-1 mutants show a decreased abundance of nuclei exhibiting a polarized organization that normally accompanies establishment of pairing; analysis of htp-1; syp-2 double mutants suggests that HTP-1 is needed to prevent premature exit from this polarized nuclear organization and that this exit stops homology search. Further, based on experiments monitoring the formation of recombination intermediates and crossover products, we suggest that htp-1 mutants are defective in preventing the use of sister chromatids as recombination partners. We propose a model in which HTP-1 functions to establish or maintain multiple constraints that operate to ensure coordination of events leading to chiasma formation.
Keywords: Homolog pairing, synapsis, meiosis, recombination, nuclear reorganization, HTP-1
Sexually reproducing organisms carry two copies of each chromosome, one received from each parent. These two copies, known as homologs, are separated during gametogenesis so that gametes carry a single copy of each chromosome; the original chromosome number is then restored following fertilization. Meiosis is the specialized cell division program that accomplishes separation of homologs, and defects in this process can result either in the production of aneuploid gametes or in arrest of the meiotic cycle and consequent sterility. To achieve correct homolog segregation during meiosis, chromosomes typically undergo the following events: initial recognition of homologs among all chromosomes present in the nucleus; complete alignment of the homologs along their lengths—a process that in most organisms culminates with the assembly of a proteinaceous structure, the synaptonemal complex (SC), between the aligned homologs; and formation of chiasmata, which are cytologically visible manifestations of interhomolog crossover recombination events that hold the homologs together until they are separated during the first meiotic division (Page and Hawley 2003).
Pairing (i.e., initial recognition and alignment) of homologous chromosomes during meiosis coincides with a reorganization of chromatin that includes both changes in the structure of chromosomes and movements of chromosomes within the nucleus (Zickler and Kleckner 1998). In many organisms, this reorganization includes a polarization of nuclear components involving clustering of chromosomes toward one side of the nucleus and/or clustering of telomeres to a small region of the nuclear envelope (Scherthan 2001; Harper et al. 2004). Mutants in which telomere clustering is impaired (e.g., fission yeast taz-1, budding yeast ndj-1, and maize pam-1) display delayed or incomplete homolog pairing and defects in downstream events including homolog segregation, suggesting that this organization facilitates productive interhomolog interactions (Chua and Roeder 1997; Conrad et al. 1997; Cooper et al. 1998; Nimmo et al. 1998; Golubovskaya et al. 2002). In Caenorhabditis elegans, the clustering of chromosomes in the nucleus during early prophase appears to be mechanistically linked to the establishment of homolog pairing, since mutations in either chk-2 or him-3 that prevent chromosome clustering also result in almost complete elimination of homolog pairing (MacQueen and Villeneuve 2001; Couteau et al. 2004).
Dispersal of chromosomes from the highly polarized nuclear organization observed in early prophase occurs as the SC is installed (synapsis) between homologs. Completion of synapsis defines onset of the pachytene stage of meiotic prophase, when the SC is seen as a tripartite structure composed of two axial/lateral elements (one along each homolog), plus a central region that glues together the lateral elements (von Wettstein et al. 1984). Although the role of the SC central region is not fully understood, it is thought that most crossovers are completed within the context of the SC. Moreover, mutants that lack SC central region components show a reduction in the number of crossovers ranging from 50% to 70% in budding yeast to almost complete elimination in C. elegans and Drosophila (Sym and Roeder 1994; Page and Hawley 2001; MacQueen et al. 2002; Colaiacovo et al. 2003). Crossovers are initiated at the DNA level by the deliberate creation of DNA double-strand breaks (DSBs), which in order to be productive for chiasma formation must be repaired using the homolog instead of the sister chromatid as a repair template. Proteins associated with the lateral elements of the SC in yeast, C. elegans, and Drosophila have been proposed to play a role either in blocking the use of sister chromatids as recombination templates or in directly promoting the use of the homolog, thereby promoting chiasma formation (Schwacha and Kleckner 1997; Couteau et al. 2004; Wan et al. 2004; Webber et al. 2004). Therefore, different SC components appear to play several distinct roles to ensure that DSBs are successfully matured into the chiasmata that allow homolog segregation.
Although the location of the SC between aligned homologs could suggest an involvement of the SC in homolog pairing, studies in mutants that lack components of the central region of the SC in yeast and C. elegans show that establishment of homolog pairing is independent of synapsis but that a mature SC is needed to maintain intimate homolog alignment (Sym et al. 1993; MacQueen et al. 2002). In fact, pairing and synapsis can be uncoupled to such an extent that SC is formed between nonhomologous chromosomes. For example, haploid plants and haploid yeast strains undergoing meiosis can achieve widespread SC formation despite the absence of homologs (Gillies 1974; Loidl et al. 1991; Santos et al. 1994). Nonhomologous synapsis is also observed in several yeast meiotic mutants that undergo delayed synapsis (Zickler and Kleckner 1999). These observations not only show that the SC is indifferent to homology but also that the central region of the SC has the inherent tendency to synapse pairs of chromosomal axes until maximal synapsis is achieved (Loidl 1990). Therefore, SC assembly must be actively regulated so that it is restricted to occur in a productive manner, linking axes of properly aligned homologous chromosomes.
In the present study, we investigate the coordination of homolog pairing and synapsis during meiosis in C. elegans. This analysis focuses on the roles of HTP-1, one of four C. elegans members of a meiosis-enriched HORMA-domain protein family that also includes the axial/lateral element component HIM-3. Our studies demonstrate a requirement for HTP-1 in the establishment of homolog pairing, the prevention of nonhomologous synapsis, and the conversion of DSBs into interhomolog crossovers. Moreover, we provide evidence for two monitoring mechanisms that coordinate early prophase events, one that couples SC polymerization with successful homolog recognition, and one that couples release from chromosome clustering and termination of homology search with stabilization of pairing through synapsis. We discuss our results in the context of a model in which the diverse phenotypic defects seen in htp-1 mutants are explained as a premature release from constraints that are imposed during meiotic prophase to ensure chiasma formation between homologous chromosomes, implicating HTP-1 as a key player in establishment and/or maintenance of these constraints.
Results
Mutations affecting him-3 paralog htp-1
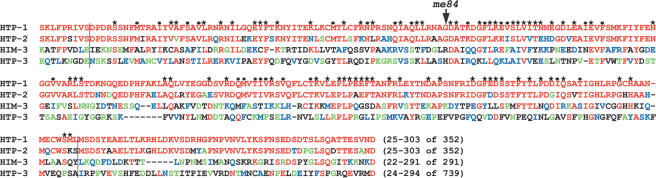
We isolated the me84 mutant following EMS mutagenesis in a screen for mutants with chromosomes that lacked chiasmata at diakinesis, the last stage of meiotic prophase (see Materials and Methods). Mapping and sequence analysis identified me84 as a mutant allele of htp-1/F41H10.10, which encodes a paralog of HIM-3, a meiosis-specific component of chromosome axes needed for chromosome pairing, synapsis, and recombination (Zetka et al. 1999; Couteau et al. 2004). HTP-1 and HIM-3 are homologs of yeast Hop1 (Hollingsworth et al. 1990), Arabidiosis ASY-1 (Caryl et al. 2000; Armstrong et al. 2002), and rice PAIR-2 (Nonomura et al. 2004), also implicated in meiotic chromosome function. Members of this protein family are characterized by the presence of a HORMA domain, a conserved structural motif also found in Mad2, a spindle assembly checkpoint component (Aravind and Koonin 1998). Missense mutations affecting residues in the HORMA domain have been shown to disrupt HIM-3 function, demonstrating the importance of this domain in meiosis (Couteau et al. 2004). The htp-1(me84) allele contains a missense mutation that results in a glycine-to-glutamate substitution at residue 97 of the HTP-1 protein; this position is immediately adjacent to a pair of residues (DA) that is highly conserved among meiosis-enriched HORMA domain proteins (Fig. 1; data not shown). htp-1(me84) is likely to be a severe loss-of-function or null allele based on its phenotypic similarities to two deletion alleles generated by the C. elegans Gene Knockout Consortium that are predicted to be molecular null alleles. htp-1(gk174) lacks the whole coding region, and htp-1(gk150) is missing 730 bp of the 1059-bp coding region (including most of the HORMA domain).
Figure 1.
Alignment of the C. elegans HIM-3 homologs: For each protein, the residues included and the total length are indicated in brackets. Characters above the HTP-1 sequence indicate residues that match (*) or are similar to (•) the HORMA domain model from the Conserved Domain Database (CDD) (Marchler-Bauer et al. 2005). Residues are colored using HTP-1 as the reference sequence. Red indicates identity with HTP-1, blue indicates conservative substitutions, and green indicates semiconservative substitutions. The arrow marks the position of the substitution (G to E) caused by the me84 mutation. Vertical black lines delimit the HORMA domain.
C. elegans has a total of four him-3 paralogs: him-3, htp-1, htp-2, and htp-3. Both him-3 and htp-3 are substantially diverged from htp-1 (26.6% and 20.6% identity at the amino acid level) and have clear orthologs in Caenorhabditis briggsae and Caenorhabditis remanei. htp-1 and htp-2 are extremely similar to each other (82% identity at amino acid level and 90% nucleotide identity in the coding region), and RNAi experiments would target both genes simultaneously. Multiple members of this subfamily are also found in C. briggsae (two) and C. remanei (three), but clear orthologous relationships are difficult to discern among subfamily members, and htp-1 and htp-2 are the most closely related gene pair, suggesting that multiple duplications (and possibly deletions) have occurred in this subfamily during Caenorhabditis evolution. Despite the high similarity between htp-1 and htp-2, the severe defects in meiotic prophase described below for htp-1 mutants indicate that an intact htp-2 gene cannot functionally compensate for loss of htp-1 activity. Thus isolation of htp-1 mutant alleles enabled investigation of meiotic functions that are either specific to htp-1 or that involve partially redundant roles of htp-1 and htp-2 for which htp-2 alone is insufficient.
htp-1 mutants load SC components but display nonhomologous synapsis
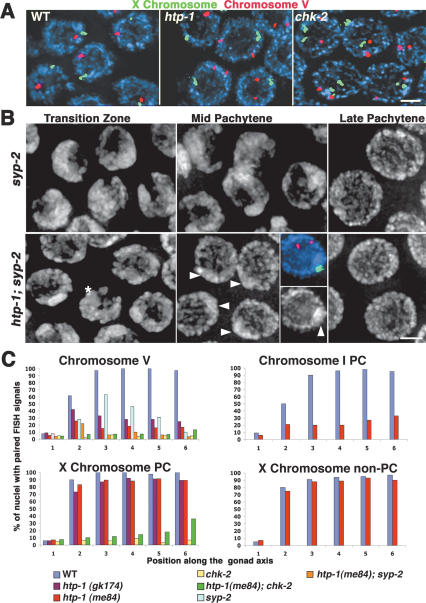
Pairing and synapsis of homologous chromosomes culminate at the pachytene stage with the homologs aligned along their entire lengths and the SC located between them. This alignment is revealed in wild-type gonads stained with DAPI by the presence of parallel tracks of DAPI-stained chromatin (Fig. 2A). In contrast, chk-2- and him-3-null mutants, which are defective in the establishment of pairing, display unaligned single DAPI tracks at this stage (Fig. 2C; MacQueen and Villeneuve 2001; Couteau et al. 2004). Pachytene-region nuclei in htp-1 mutants appear superficially quite similar to wild type, with parallel tracks of DAPI-stained chromatin suggesting extensive synapsis between chromosomes (Fig. 2B). Formation of abundant SC in htp-1 mutants is confirmed by immunostaining of htp-1 gonads (Figs. 2, 3A, 4B) with antibodies against lateral element component HIM-3 and central region components SYP-1 (MacQueen et al. 2002) and SYP-2 (Colaiacovo et al. 2003). As in wild type, HIM-3 forms linear structures along chromatin tracks, suggesting that all chromosomes have an axial element; furthermore, SYP-1 is present between parallel DNA tracks and colocalizes with HIM-3 (Fig. 2D–G). Thus, whereas HIM-3 is required for loading of SC central region components (Colaiacovo et al. 2003), HTP-1 is not.
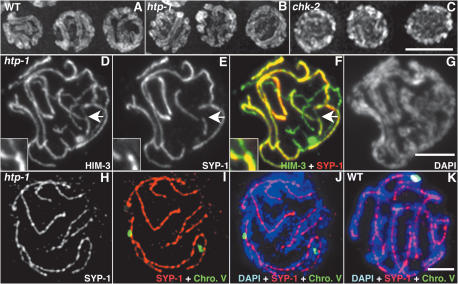
Figure 2.
htp-1 mutants exhibit nonhomologous synapsis. (A–C) DAPI-stained nuclei from the pachytene region of whole-mount gonads. Wild-type and htp-1 mutant nuclei show the presence of parallel DAPI-stained chromatin tracks, which are not observed in chk-2 nuclei. (D–G) Squash preparation of htp-1(me84) pachytene nucleus labeled with α-HIM-3 antibodies, α-SYP-1 antibodies, and DAPI. Thick HIM-3 lines in D correspond to regions stained with SYP-1 in E and F, demonstrating synapsis of chromosome axes; DAPI shows that regions of overlap of HIM-3 and SYP-1 are flanked by parallel tracks of DAPI (G). The overlay of HIM-3 and SYP-1 also shows that some axis segments are unsynapsed. The region indicated by the arrow is magnified in the inset and shows a pairing partner switch. (H–J) Squash preparation of htp-1(me84) pachytene nucleus stained with α-SYP-1 antibodies, hybridized with a FISH probe for the 5S rDNA locus on chromosome V, and counterstained with DAPI. FISH signals are separated but are associated with two different α-SYP-1 signals that are located between parallel tracks of DAPI-stained chromatin, demonstrating nonhomologous synapsis. (K) Squash preparation of wild-type pachytene nucleus; both FISH signals are colocalized and associated with a single α-SYP-1 track. Bars, 5 μm.
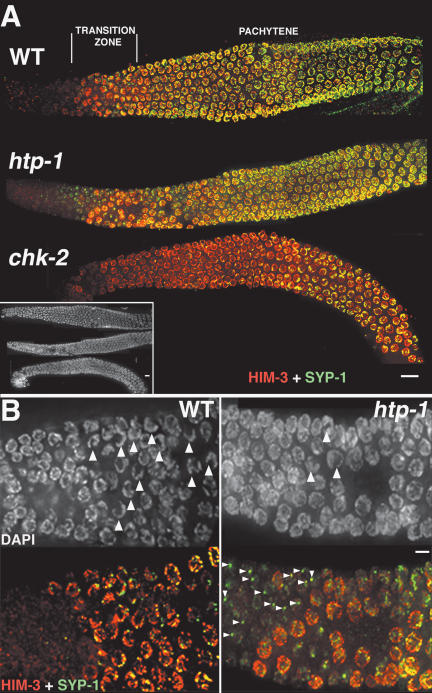
Figure 3.
Loading of SC components in htp-1, chk-2, and wild-type (WT) gonads. (A) Whole gonads stained with α-HIM-3 and α-SYP-1 antibodies. The three gonads are aligned according to the DAPI image (inset); the positions of the transition zone and the pachytene region are indicated in reference to wild type. HIM-3 loads in the normal position in both htp-1 and chk-2 mutant gonads. SYP-1 appears to be loaded at roughly wild-type levels in the pachytene region of htp-1(me84) mutant gonads, but loading of SYP-1 is substantially delayed and restricted in chk-2 mutants. (B) High-magnification image of the transition zone region of the gonad. In wild-type (WT) gonads, DAPI staining reveals the presence of numerous nuclei with clustered chromosomes (arrowheads), while in htp-1 mutant gonads, nuclei with clustered chromosomes are very infrequent. In wild-type gonads, SYP-1 signals are only detected in nuclei that have accumulated substantial amounts of HIM-3, while in htp-1 mutant gonads, SYP-1 is detected as bright aggregates (arrowheads) before HIM-3 is present on the chromosomes. All images are projections from 3D data stacks designed to include the whole depth of the nuclei shown. Bars: A, 10 μm; B, 5 μm.
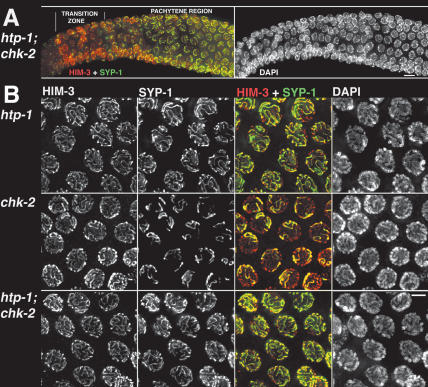
Figure 4.
HTP-1 prevents SYP-1 loading when homolog pairing fails. (A) htp-1; chk-2 double-mutant gonad showing nuclei from early to mid prophase stained with α-HIM-3 and α-SYP-1 antibodies. As in the htp-1 mutant, and in contrast with the chk-2 single mutant, bright SYP-1 foci are present before HIM-3 is detected on chromosomes, and extensive loading of SYP-1 is seen in the early–mid pachytene region. DAPI staining reveals the absence of nuclei displaying chromosome clustering in htp-1; chk-2 double-mutant gonads. (B) High-magnification images of late-pachytene region nuclei. Both the htp-1 mutant and the htp-1; chk-2 double mutant display almost complete colocalization of HIM-3 and SYP-1, showing that most chromosomal regions are synapsed; in contrast, only a small subset of chromosome axes are associated with SYP-1 in the chk-2 mutant. All images are projections from 3D data stacks designed to include the whole depth of the nuclei shown. Bars: A, 10 μm; B, 5 μm.
However, closer examination of pachytene region nuclei in htp-1 mutants revealed several abnormalities. First, whereas in wild-type pachytene nuclei, SYP-1 is colocalized with HIM-3 along the full lengths of parallel-aligned chromosome pairs, in the mid-pachytene region of htp-1 mutant gonads, some segments of α-HIM-3-stained chromosomal axes are not stained by α-SYP-1, indicating that they are not synapsed (Fig. 2F); however, by the late-pachytene region, almost all axes appear fully synapsed (Fig. 4B). Moreover, we detected instances of pairing partner switches in which two intimately aligned, SYP-1-associated chromosome axes became separated and then each one aligned and synapsed with a different partner (inset in Fig. 2D–F).
Second, fluorescence in situ hybridization (FISH) analysis revealed that pairing of homologous chromosomes is substantially impaired but not eliminated in htp-1 mutant gonads (Figs. 2I,J, 5A). We performed quantitative analysis of pairing by assessing the frequency of paired FISH signals in six zones along the distal–proximal axis of the gonad (Fig. 5C). In wild-type gonads, zone 1 corresponds to premeiotic nuclei in which chromosomes are unpaired, zone 2 is composed mostly of nuclei in the transition zone where nuclei are entering meiotic prophase, and zones 3–6 represent early- to late-pachytene stages. For an interstitial region of chromosome V, the frequency of paired signals in the pachytene region of htp-1 mutant gonads (zones 3–6) never arose above 33%; this contrasts both with the essentially complete pairing detected in wild type (>94%) and the absence of pairing in the chk-2 mutant (in which pairing remained at premeiotic levels) (Fig. 5C; MacQueen and Villeneuve 2001). htp-1 mutants also exhibited low levels of pairing, albeit above premeiotic levels, for a probe targeting the right end of chromosome I (which contains a cis-acting domain termed the pairing center [PC] that confers local stabilization of homolog pairing even in the absence of synapsis) (MacQueen et al. 2002) as well as for probes targeting regions of chromosomes II and IV (Fig. 5C; data not shown). In agreement with these observations, Couteau and Zetka (2005) also report a severe decrease in pairing of autosomes in htp-1 mutants; the higher levels of residual pairing detected in our study may reflect differences in the ages of the animals scored.
Figure 5.
htp-1 mutants are defective in the establishment of homolog pairing. (A) Pachytene nuclei from whole-mount gonads hybridized with probes for the 5S rDNA locus on chromosome V (red) and for the X-chromosome PC end (green). FISH signals for both probes are paired in all wild-type nuclei; in the htp-1 mutant, signals for the X-chromosome probe are paired, while signals for chromosome V are unpaired in most nuclei; in the chk-2 mutant, lack of pairing for both probes is seen in all nuclei. (B) High-magnification images of DAPI-stained nuclei. The top panels show maintenance of clustered chromosomes in the mid-pachytene region of the syp-2 mutant, followed by eventual dispersal of chromosomes at the very end of the pachytene region. The bottom panels show the corresponding regions of an htp-1; syp-2 gonad. The transition zone region contains very few nuclei with chromosome clustering (asterisk), and by the early-pachytene region, all nuclei show dispersed chromosomes. A DAPI-bright region is evident in midpachytene region nuclei (arrowheads); the inset shows a pachytene nucleus hybridized with probes for chromosomes V and X, demonstrating that the X chromosomes are associated and coincide with the DAPI-bright region visible at this stage. Bars, 2 μm. (C) Graphs displaying quantitation of pairing levels for different chromosome regions in wild-type and mutant gonads. Numbers along the X-axis correspond to different zones along the length of the gonad; zone 1 contains premeiotic nuclei, zone 2 is composed mostly of nuclei in the transition zone, and zones 3–6 are early to late pachytene. The Y-axis indicates the percent of nuclei in each zone that displayed paired FISH signals.
The fact that autosomes are frequently unpaired despite extensive SC formation implies that synapsis is taking place between nonhomologous chromosomes. Simultaneous labeling of nuclei with α-SYP-1 antibodies, DAPI, and FISH confirmed that htp-1 mutants do, indeed, exhibit extensive nonhomologous synapsis. Not only were chromosome V FISH signals unpaired most of the time, but they were also usually associated with long α-SYP-1 stretches that were flanked on both sides by parallel aligned DAPI-stained chromatin tracks, demonstrating nonhomologous synapsis of chromosome V (Fig. 2H–J).
In addition to extensive nonhomologous synapsis in the pachytene region of htp-1 mutant gonads, other abnormalities are also evident in the transition zone region of the gonad, where nuclei are entering meiotic prophase. In wild-type gonads, nuclei in the transition zone display a characteristic clustering of the chromatin toward one side of the nucleus, with the nucleolus situated at the opposite side. Initial loading of meiotic chromosome axis components also occurs within the transition zone, followed by loading of SC central region proteins (MacQueen et al. 2002). In htp-1 mutants, very few nuclei exhibit the polarized nuclear organization characteristic of the transition zone (Fig. 3B). Furthermore, the early pattern of α-SYP-1 signals relative to α-HIM-3 signals is perturbed in htp-1 mutants. In wild-type gonads, nuclei that have not yet accumulated substantial chromosomal HIM-3 rarely show α-SYP-1 signals (Fig. 3B). In htp-1 mutants, in contrast, a single, bright focus of α-SYP-1 staining, which may correspond to aggregation of SYP-1 protein, is observed in or adjacent to numerous nuclei that have not yet accumulated chromosomal HIM-3 (Fig. 3B). Once HIM-3 is loaded on chromosomes, the bright α-SYP-1 foci disappear and SYP-1 starts forming linear structures similar to those observed in wild type (Fig. 3B). Both the aggregates of α-SYP-1 signal and the reduced incidence of nuclei with clustered chromosomes suggest that HTP-1 plays an important role in early steps of meiotic prophase.
The X chromosomes pair correctly in htp-1 mutants
C. elegans hermaphrodites possess five pairs of autosomes plus a pair of X chromosomes, all of which are correctly paired at pachytene in wild-type gonads. The lack of CHK-2 or HIM-3 results in global pairing failure, indicating that these two proteins are essential for pairing of all chromosomes (MacQueen and Villeneuve 2001; Couteau et al. 2004). However, in mutants carrying weak alleles of him-3, the X chromosomes achieve high levels of pairing, while pairing of autosomes is severely impaired, suggesting that X-chromosome pairing is more robust against certain perturbations of the meiotic pairing program (Couteau et al. 2004; Nabeshima et al. 2004). In htp-1 mutant gonads, despite the widespread pairing defect observed for the autosomes, the X chromosomes achieved nearly wild-type levels of pairing with normal kinetics (Fig. 5A,C). We assessed pairing using probes targeting both the PC end (in both me84 and gk174 mutants) and the non-PC end (in the me84 mutant) of the X chromosome. Both ends achieved almost 100% pairing in the pachytene region (Fig. 5C), suggesting that X chromosomes are correctly synapsed along their whole lengths and are therefore not involved in nonhomologous synapsis with the autosomes.
The ability of the X chromosomes to pair in the htp-1 mutants suggests either that HTP-1 does not function in X-chromosome pairing, or that HTP-1 and HTP-2 may play redundant roles. By performing htp-2 RNAi in an htp-1 mutant, Couteau and Zetka (2005) have revealed roles for HTP-2 in promoting both stabilization of X-chromosome pairing and SC assembly; however, these studies could not distinguish between an exclusive role for HTP-2 or redundant roles for HTP-1 and HTP-2 in these processes.
Pairing defects in htp-1 mutants are not solely a consequence of indiscriminate synapsis
Extensive synapsis between nonhomologous chromosomes raised the possibility that the pairing defect might be an indirect consequence of indiscriminate assembly of SC. This hypothesis is testable, as mutants lacking SC central region proteins SYP-1 or SYP-2 are successful in initial establishment of homolog pairing, but are defective in stabilizing these homolog interactions (MacQueen et al. 2002; Colaiacovo et al. 2003). Thus, in time-course FISH experiments, syp-1 and syp-2 mutants achieve peak levels of pairing that are substantially higher than the autosomal pairing levels observed in htp-1 mutants. Therefore, if the pairing defect in htp-1 mutants were solely a consequence of the indiscriminate loading of SYP-1 and SYP-2, initial pairing should be restored to syp-2 mutant levels in htp-1; syp-2 double mutants. This was not observed. Instead, whereas the syp-2 mutant achieved peak pairing levels of 65% for the 5S locus in zone 3, the htp-1; syp-2 double mutant exhibited only 10% paired signals in the corresponding region of the gonad (Fig. 5C). This finding indicates that the pairing defect in htp-1 mutants is not simply a secondary consequence of inappropriate installation of SC central region components SYP-1 and SYP-2 and indicates that HTP-1 plays an active role in initial establishment of homolog pairing.
Furthermore, similar to htp-1 gonads, htp-1; syp-2 double-mutant gonads also contain very few nuclei with the clustered chromosome configuration that is characteristic of the wild-type transition zone (and coincides with the establishment of chromosome pairing) (Fig. 5B). This is strikingly different from syp-2 and syp-1 mutant gonads in which chromosome clustering persists in nuclei throughout most of the pachytene region, with chromosomes becoming dispersed only at the very end of the pachytene region (Fig. 5B; MacQueen et al. 2002; Colaiacovo et al. 2003). Instead, chromosomes are already dispersed in the mid-pachytene region of htp-1; syp-2 gonads, and DAPI staining reveals unpaired chromatin tracks similar to those seen in the late-pachytene region of the syp-2 single mutant (Fig. 5B). A bright chromatin mass is present in most mid-pachytene region nuclei, which FISH analysis reveals as the X-chromosome pair (Fig. 5B). Suppression of the syp-2 “extended transition zone” phenotype in the htp-1; syp-2 double mutant suggests that HTP-1 plays a role in inhibiting dispersal of chromosomes from the clustered chromosome configuration.
In htp-1 mutants, chromosomes V and I showed a fairly stable level of residual homolog pairing from the early-meiotic prophase until the late-pachytene region of the gonad (zones 2–6 in Fig. 5C). In contrast, htp-1; syp-2 double-mutant gonads show a marked drop in detectable pairing of chromosome V after zone 2 (Fig. 5C). This result suggests that the residual levels of pairing observed for the autosomes in htp-1 mutants are, in fact, stabilized through synapsis.
HTP-1 plays an active role in inhibiting SC assembly between nonhomologous chromosomes
Previous studies have shown that initial establishment of homolog pairing is not dependent on the installation of the central region of the SC between homologous chromosome axes (MacQueen et al. 2002). Furthermore, recent data show that assembly of the SC central region can also take place between nonhomologous chromosome axes (Couteau et al. 2004; Nabeshima et al. 2004; this work). This raises the question of whether nonhomologous synapsis is a default outcome that will occur when chromosome axes containing HIM-3 are present but initial homolog pairing fails. We tested this possibility by examining localization of SYP-1 in chk-2 mutants, previously shown to be profoundly defective for establishment of homolog pairing but competent for loading HIM-3 (MacQueen and Villeneuve 2002). Costaining with α-HIM-3 and α-SYP-1 antibodies revealed that loading of SYP-1 is severely delayed and reduced in chk-2 gonads compared with the htp-1 mutant. In the region of the gonad corresponding to the transition zone, where initial SYP-1 loading takes place in wild type, no SYP-1 signal is detected in chk-2 gonads (Fig. 3A). Most nuclei in the early–mid-pachytene region of chk-2 gonads display one or a few bright foci of α-SYP-1 staining and by the mid- to late-pachytene region, most nuclei contain a few robust SYP-1 stretches along only a subset of chromosomes; these SYP-1 stretches colocalize with thick HIM-3 stretches, suggesting that the SYP-1 stretches represent instances where two chromosome axes are linked together by SC (Figs. 3A, 4B).
The late and limited assembly of the SC central region in chk-2 gonads suggests that there may be a mechanism to prevent SC formation between nonhomologous chromosomes when initial pairing fails. The fact that htp-1 mutants, in contrast, assemble nearly wild-type levels of SC central region components (Figs. 3A, 4B) even though homolog pairing is impaired led us to hypothesize that HTP-1 might be part of a mechanism that coordinates establishment of pairing with SC assembly by preventing loading of central region components between nonhomologous axes. We tested this hypothesis by analyzing loading of both HIM-3 and SYP-1 in htp-1; chk-2 double mutants. The blockage to SYP-1 loading observed in chk-2 mutants is not observed in the htp-1; chk-2 double mutant, and by mid–late pachytene, most chromosome axes are synapsed as in the htp-1 single mutants (Fig. 4B). However, the extent of SYP-1 loading in early–mid pachytene in htp-1; chk-2 gonads appears lower than in the htp-1 single mutant, suggesting that the double mutant is somewhat slower to achieve maximal synapsis (Fig. 4A). As in the chk-2 single mutant, nuclei with clustered chromosomes are completely absent in the htp-1; chk-2 double-mutant (Fig. 4A), and FISH analysis shows that pairing is severely defective for both the autosomes and the X chromosomes (Fig. 5C). Therefore, synapsis must be taking place between nonhomologous chromosomes. These results implicate HTP-1 as part of an active mechanism that prevents loading of SYP-1 between nonhomologous chromosomes.
Although X-chromosome pairing levels were quite low over much of the assayed region of the gonads in htp-1; chk-2 worms (comparable to the chk-2 single mutant), the level of paired signals increased significantly by the late-pachytene region to 36% in zone 6 (Fig. 5C). As this late rise in pairing levels coincides with the peak in SYP-1 loading (Fig. 4), we infer that ongoing installation of the SC central region can succeed in stabilizing associations between chromosomes even at late time points.
DSBs are created and repaired in htp-1 mutants
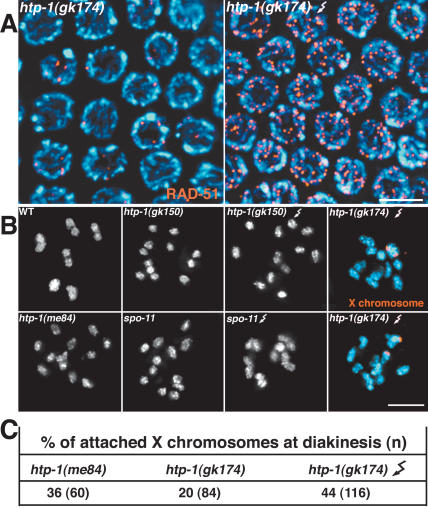
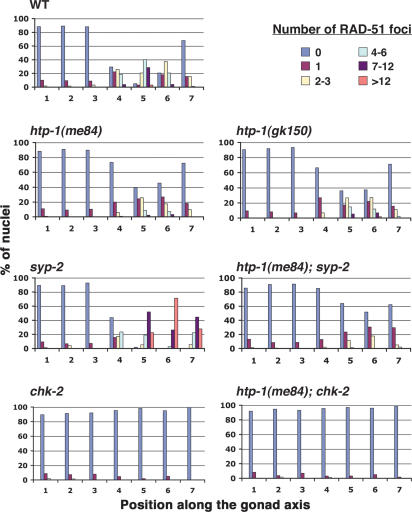
In wild-type gonads, the occurrence of crossover recombination events becomes evident at the diakinesis stage, when the homologs appear linked together by chiasmata (Fig. 8B, below). In contrast, diakinesis-stage oocytes in htp-1 mutants contain mostly unattached univalent chromosomes, indicative of a severe crossover deficit (Fig. 8B; see below). To study the nature of this crossover deficiency, we monitored the progression of meiotic recombination by analyzing dynamics of foci stained with an antibody against the DNA strand exchange protein RAD-51 (Fig. 6). We quantified numbers of foci in seven zones along the length of the gonad (zones 1 and 2 contain premeiotic nuclei, zone 3 contains mostly transition zone nuclei, and zones 4–7 represent early to late pachytene). In wild type, numbers of RAD-51 foci rose substantially in zone 4, peaked in zone 5, and diminished in zones 6 and 7, reflecting creation of DSBs and progression of recombination/repair beyond the strand exchange step. In htp-1 mutants, the overall number of RAD-51 foci in zones 4–7 was substantially reduced compared with wild type. Whereas the average number of foci per nucleus in zone 5 of wild type was 5.5, the corresponding zone in htp-1(me84) and (gk150) mutant gonads had averages of only 1.5 and 2 foci, respectively. Furthermore, whereas nuclei lacking RAD-51 foci were infrequent (5%) in zone 5 of control gonads, they comprised 36%–40% of nuclei in zone 5 of htp-1 mutant gonads. Couteau and Zetka (2005) similarly found an overall reduction of RAD-51 foci in htp-1 mutants; the higher numbers of foci detected in htp-1 mutants in our study likely reflect differences in the antibody staining procedures, as evidenced by the fact that we also detected substantially higher numbers of RAD-51 foci in our wild-type controls.
Figure 8.
γ-Irradiation of htp-1 mutants does not fully rescue chiasma formation between the X chromosomes. (A) α-RAD-51 immunostaining of mid-pachytene region nuclei from an untreated htp-1 mutant worm (left) and an htp-1 worm treated with 5000 rads of γ-irradiation and fixed 1 h later (right). (B) Each panel shows the DAPI-stained chromosomes (white or blue) from a single diakinesis-stage oocyte nucleus of the indicated genotype. Crooked arrows indicate nuclei from worms that had been exposed to 5000 rads of γ-irradiation and then fixed and stained 18 h later. Orange in the right panels corresponds to FISH signals used to identify the X chromosomes. Wild-type (WT) nuclei show six bivalents, while htp-1 and spo-11 mutants typically show 12 unattached chromosomes (univalents). Following γ-irradiation, chiasma formation is efficiently restored in spo-11 mutants (six bivalents present) but not in htp-1 mutants. Bars, 5 μm. (C) The table shows quantitation of diakinesis nuclei showing chiasmata between the X chromosomes in htp-1 mutants.
Figure 6.
Quantitative time-course analysis of RAD-51 foci. Graphs depicting quantitation of RAD-51 foci in gonads of the indicated genotypes. Numbers along the X-axes indicate position along the length of the gonad (zones 1 and 2 contain premeiotic nuclei, zone 3 contains mostly nuclei in the transition zone, and zones 4–7 represent early- to late-pachytene nuclei). Bars represent the percentages of nuclei in a given zone with the numbers of RAD-51 foci indicated by the color code.
Although levels of RAD-51 foci were reduced in the htp-1 mutants, they were substantially higher than the levels we observed in chk-2 mutants (which were previously reported to lack RAD-51 foci [Alpi et al. 2003]), or in the htp-1; chk-2 double mutants (<0.03 foci per nucleus in zone 5 for both genotypes). Moreover, the dynamics of appearance and disappearance of RAD-51 foci in the htp-1 mutants paralleled those observed in wild-type gonads: Foci increased in abundance around zone 4 and were mostly gone by zone 7 (Fig. 6). This pattern suggests that meiotic DSBs are being created and repaired in htp-1 mutants, albeit without forming crossovers. The overall reduced levels of RAD-51 foci in htp-1 mutants could reflect a decrease in the number of DSBs formed and/or an altered pathway of DSB repair.
An htp-1 mutation suppresses the accumulation of RAD-51 foci associated with asynapsis and heterosynapsis
In syp-1 and syp-2 mutants, recombination is initiated, but RAD-51 foci persist and accumulate at high levels at mid/late pachytene (Colaiacovo et al. 2003). Since the homologs are not tightly juxtaposed in the pachytene region of these mutants due to the lack of SC, it is likely that homologs are unavailable as templates for DSB repair. Nevertheless, RAD-51 foci start reducing in number at late pachytene in syp-2 gonads (zone 7 in Fig. 6) and are gone at diakinesis, suggesting eventual repair of DSB by a sister-chromatid-dependent pathway (Colaiacovo et al. 2003). The accumulation of RAD-51 foci observed in the pachytene region of syp-2 gonads contrasts with the timely disappearance of RAD-51 foci in htp-1 mutant gonads, where most homologous regions are not in proximity because of generalized nonhomologous synapsis. We found that htp-1 mutations (both me84 and gk150) suppress the elevated and persistent RAD-51 foci levels of the syp-2 mutant (Fig. 6; data not shown). In fact, htp-1; syp-2 double mutants exhibited even lower levels of RAD-51 foci than were seen in htp-1 single mutants alone (an average of 0.5 foci per nucleus in zone 5 compared with 1.5 in the htp-1 single mutant).
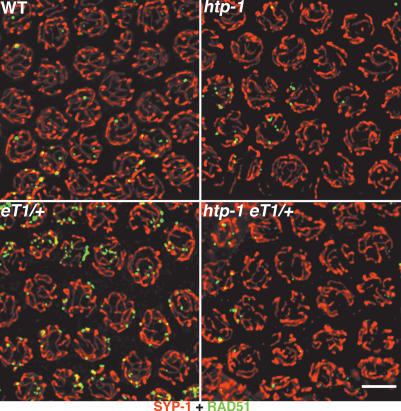
RAD-51 foci have also been seen to accumulate in late-pachytene nuclei in wild-type worms heterozygous for reciprocal translocations (Fig. 7; Alpi et al. 2003). This persistence/accumulation of RAD-51 foci is presumably a consequence of heterosynapsis between translocated chromosome segments and their normal sequence counterparts (MacQueen et al. 2005), and may reflect a delay in the repair of DSBs in heterosynapsed regions and/or continued DSB formation. We found that the htp-1 mutation suppresses accumulation of RAD-51 foci in reciprocal translocation heterozygotes (Fig. 7), as might be expected based on the fact that RAD-51 foci do not accumulate in htp-1 mutants with normal karyotype despite extensive nonhomologous synapsis. These results are consistent with the idea that htp-1 mutants may use an alternate pathway for DSBs repair that avoids RAD-51 accumulation.
Figure 7.
An htp-1 mutation suppresses the accumulation of RAD-51 foci associated with translocation heterozygosity. Late-pachytene region nuclei stained with α-SYP-1 and α-RAD-51 antibodies. In the wild type and htp-1 mutant, most nuclei in this region contain few or no RAD-51 foci; in contrast, in otherwise wild-type worms heterozygous for the reciprocal translocation eT1 (eT1/+), multiple RAD-51 foci are present in most nuclei in this region of the gonad. In contrast, most nuclei in the corresponding region of an htp-1; eT1/+ mutant gonad contain few or no RAD-51 foci. Images are projections from a 3D data stack designed to include the whole depth of the nuclei shown. Bar, 5 μm.
Homologous synapsis of X chromosomes does not ensure crossing over and chiasma formation
The presence of univalents at diakinesis is generally reflective of failure in crossing over, but in mutants that also show a defect in pairing and/or synapsis it is usually not possible to discern if the recombination defect is simply a secondary consequence of the pairing/synapsis defect. The fact that the X chromosomes appear fully paired and synapsed in the htp-1 mutant provided an opportunity to assess whether HTP-1 has a role in recombination beyond its roles in pairing and regulation of synapsis. We analyzed the frequency of crossing over on the X chromosomes, finding that the recombination frequency between two markers near opposite ends of the chromosome was reduced in htp-1 to 30% of the wild-type level (Table 1). Furthermore, we used FISH at the diakinesis stage to assess whether the X chromosomes were attached (indicating chiasma formation) or present as univalents. X chromosomes were present as univalents in 80% of htp-1(gk174) and 64% of htp-1(me84) oocytes (Fig. 8). Thus in htp-1 mutants, the X chromosomes suffer a substantial decrease in crossovers that is not readily accounted for by pairing and synapsis defects, suggesting a separable role for HTP-1 during meiotic recombination.
Table 1.
Reduction of crossover recombination in htp-1 mutants
| Genotype | Recombinant progeny | Total progeny | Map distance (cM)a |
|---|---|---|---|
| +/(htp-1 or +); dpy-3 unc-3/++ | 453 hermaphrodites | 1590 | 35 |
| htp-1/htp-1; dpy-3 unc-3/++ | 21 hermaphrodites | 233 | 11.6 |
| 29 males | 160 |
Map distances were calculated as in Kelly et al. (2000).
DSB are not limiting for X-chromosome crossing over in htp-1 mutants
It is possible that the reduction in RAD-51 foci observed in htp-1 mutants is due to a decrease in the number of DSBs being formed, and this, in turn, could explain the reduced recombination on the X chromosomes. We therefore sought to test whether DSBs were limiting for crossing over on the X chromosomes in htp-1 mutants by testing whether DSBs induced by γ-irradiation could rescue the crossover deficit in htp-1 mutants. We observed only a modest increase in the percentage of attached X chromosomes (from 20% to 44%) following irradiation of htp-1 worms; using the same γ-irradiation treatment, we successfully induced formation of chiasmata for 97% of chromosome pairs in control spo-11 mutant worms (which lack the DSB-forming enzyme and completely lack chiasmata) (Fig. 8). These results suggest that even if htp-1 mutants do form fewer DSBs than wild type, this defect alone cannot account for the decrease in crossover formation on the X chromosomes. They also suggest that HTP-1 has a function during recombination that is exerted after DSB formation.
Discussion
The analysis reported here has revealed requirements for HTP-1 in numerous aspects of the meiotic prophase program, including establishment of homolog pairing, regulation of SC assembly, and conversion of DSBs to inter-homolog crossovers. Furthermore, our data provide evidence for the existence of two previously unknown monitoring mechanisms that coordinate early prophase events, one that monitors whether homologs have successfully recognized their partners, and a second that monitors whether homolog interactions have been stabilized by the SC.
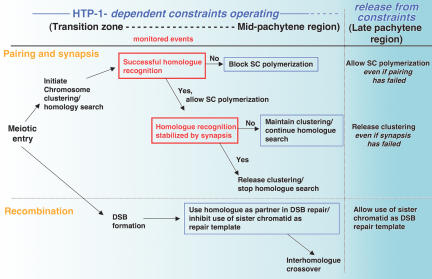
Below we discuss our results in the context of a model (Fig. 9) in which the proposed monitoring mechanisms impose constraints that operate during early meiotic prophase to (1) inhibit SC polymerization until homolog recognition is achieved, and (2) maintain chromosome clustering until homolog interactions are stabilized by the SC. We use the term “constraint” to refer to a restriction that limits the conditions under which a given process (e.g., SC polymerization, chromosome dispersal, homologous recombination) is allowed to occur. We show that most of the apparently diverse defects in htp-1 mutants can be interpreted as an absence of or premature release from early prophase constraints governing key aspects of the meiotic program, suggesting a requirement for HTP-1 in establishment or maintenance of these constraints.
Figure 9.
Model in which HTP-1-dependent constraints govern key aspects of the meiotic program. We propose that HTP-1 functions to establish or maintain constraints that operate during early prophase to (1) couple SC polymerization with successful homolog recognition, (2) couple release from chromosome clustering and termination of homology search with stabilization of pairing through synapsis, and (3) inhibit the use of sister chromatids as recombination partners. Whereas during wild-type meiosis the conditions for proceeding with synapsis and chromosome dispersal are usually met early within the domain in which the proposed constraints operate, analysis of mutants provides evidence for continued operation of these constraints through the mid-pachytene region of the gonad. We further propose that these constraints are lifted in the late-pachytene region of the gonad. Red boxes indicate monitored events; blue boxes indicate functions that require HTP-1. See text for further discussion.
Coordination of homolog pairing and SC assembly
Homolog pairing is a key defining event of meiotic prophase; while other meiotic events show some variability among organisms, correct pairing is in all cases critical for proper homolog segregation. In most organisms, initial pairing interactions are followed by SC polymerization, which renders homologs intimately associated along their lengths. However, the occurrence of nonhomologous synapsis is a well-established phenomenon. For example, haploid plants display extensive synapsis despite the fact that each chromosome is present as a single copy (Gillies 1974), suggesting that the SC per se cannot distinguish homologs from nonhomologous chromosomes. Therefore, homolog recognition and SC assembly must be exquisitely coordinated to ensure SC assembly only between homologs; this coordination is clearly lost in htp-1 mutants. We postulate that successful homolog recognition is monitored by a mechanism that inhibits SC assembly when homolog pairing has not occurred, thereby avoiding heterologous SC installation. Evidence for such inhibition is seen in the pairing-defective chk-2 mutant, in which HIM-3 is loaded onto chromosomes at the onset of meiotic prophase but SYP-1 loading is greatly impaired during early–mid pachytene. However, gradual accumulation of chromosomal SYP-1 in the late-pachytene region in chk-2 gonads indicates that SYP-1 is present and capable of loading onto chromosomes. We propose that the delayed onset of SYP-1 loading reflects an active inhibition of SC polymerization, triggered by pairing failure, which is eventually released at late pachytene. Furthermore, we propose that HTP-1 is required to establish or maintain this inhibition, as homolog pairing also fails in htp-1; chk-2 double mutants, but SC polymerization is not inhibited.
How might HTP-1 prevent improper loading of SC central region components? One possibility is suggested by our observation of strong nonchromosomal α-SYP-1 foci in htp-1 gonads in the region where nuclei have not yet accumulated substantial chromosomal HIM-3. This observation suggests that HTP-1 may prevent SC assembly by sequestering SYP-1 and/or SYP-2 in a context that prevents them from aggregating or assembling on chromosomes. HTP-1 might accomplish this by acting as a molecular mimic of HIM-3 (which is required for SYP-1 and SYP-2 assembly onto chromosomes), or by preventing the proposed association between SYP-1 and SYP-2. Alternatively, HTP-1 might act by preventing accumulation of SYP-1 or SYP-2, possibly by promoting their degradation. Under either scenario, it is necessary to postulate that a mechanism that senses the success of homolog recognition would lead to release from HTP-1-mediated inhibition of SC assembly.
Homology search and the duration of chromosome clustering
In most organisms, chromosome pairing coincides with a major reorganization of chromatin within the nucleus. In C. elegans, the onset of pairing coincides with a nuclear reorganization in which chromatin is clustered toward one side of the nucleus. By the time chromosomes are dispersed from this clustered stage in wild-type gonads, all chromosomal regions show almost 100% pairing (MacQueen et al. 2002; this work), suggesting that homology search is completed during the clustered stage. Furthermore, a mechanistic link between clustering and establishment of pairing is suggested by the fact that the known mutations that eliminate clustering (chk-2, him-3-null allele) also eliminate pairing (MacQueen et al. 2001; Couteau et al. 2004). In contrast to chk-2 and him-3 mutants, htp-1 mutants do not lack nuclei with clustered chromatin, but instead show a reduction in their number. There are at least two possible explanations for this reduction: (1) A subset of nuclei fail to initiate chromosome clustering, or (2) nuclei spend less time in the clustered configuration. Homolog pairing in htp-1 mutants peaks in the region of the gonad where nuclei enter meiosis, which is also the zone that contains the few nuclei with clustered chromatin. After this zone, levels of pairing decrease slightly and then remain stable throughout the pachytene region, suggesting that homolog searching has stopped. In contrast, in syp-1 and syp-2 mutants, chromatin clustering is maintained until the late-pachytene region, and pairing levels both peak later and remain higher than in htp-1 mutants until the late-pachytene region. The correlation between the high levels of pairing and maintenance of clustering in syp-2 mutants and the low levels of pairing and reduced number of nuclei with clustered chromosomes in htp-1 mutants further strengthens the notion that pairing and chromosome clustering are mechanistically coupled. We can explain the pairing phenotypes of the htp-1 and syp-2 single mutants and the htp-1; syp-2 double mutant by postulating that homology searching takes place only while nuclei remain in the clustered state and that exit from this state effectively terminates the homology search. This scenario suggests that reduced levels of pairing in htp-1 mutants may reflect premature exit from a state of active homology search.
Dispersal of chromosomes from the clustered configuration is a regulated process
The identification of mutants that fail to achieve telomere bouquet formation in budding yeast (ndj1) and maize (pam1) or chromosome clustering in C. elegans (chk-2) indicates that the acquisition of these highly polarized nuclear organizations is actively regulated. However, much less is known about what triggers exit from this polarized state. Two hypothesis have been proposed to explain chromosome dispersal in C. elegans (MacQueen et al. 2002). The first proposes that physical forces created by SC morphogenesis are sufficient to disperse the chromosomes; the second proposes that dispersal is controlled by a signal dependent on synapsis progression. Our studies suggest that exit from the clustered stage does not depend on SC morphogenesis per se, since htp-1;syp-2 double mutants, which lack SC, do not accumulate clustered nuclei throughout the pachytene region as observed in syp-2 mutants. Instead, our results support that idea that chromosome dispersal is regulated by a signaling mechanism that monitors stabilization of homolog interactions by the SC. Under this scenario, HTP-1 could be involved in the generation of an inhibitory signal that blocks chromosome dispersal, in the maintenance of this inhibition, or both. Chromosomes do eventually disperse in syp-2 mutants, suggesting that the constraint imposed by this monitoring mechanism (i.e., making chromosome dispersal dependent on stabilization of homolog pairing by the SC) is ultimately lifted at late pachytene.
Such a mechanism may monitor SC assembly at specific subregions of each homolog pair rather than throughout the whole length of the homologs. Attractive candidate regions to fulfill this role are the PCs, cis-acting chromosome domains that play dual roles to ensure synapsis between homologous chromosomes (McKim et al. 1988, 1993; Herman and Kari 1989; Villeneuve 1994; MacQueen et al. 2002, 2005). PCs are found near one end of each of the C. elegans chromosomes and serve both to promote local stabilization of homologous interactions independently of the SC and to promote SC assembly. Furthermore, multiple lines of evidence support the view that SC polymerization is nucleated in the vicinity of the PCs and proceeds in a highly processive directional fashion along the length of a chromosome pair (Nabeshima et al. 2004; MacQueen et al. 2005). Thus, we postulate that PCs may also serve as chromosomal regions where stabilization of homolog pairing by nascent SC is monitored to allow chromosome dispersal. Under this model, once homolog interactions at PCs are stabilized by the SC, production of an inhibitory signal would cease, triggering exit from the clustered configuration and ending the homology search. Our hypothesis is in agreement with current models that also favor a signal-regulated exit from the telomere bouquet in budding yeast and Sordaria (Trelles-Sticken et al. 1999; Storlazzi et al. 2003). In these cases it has been proposed that bouquet exit is triggered by progression of DSB-dependent recombination-related interactions. In C. elegans, however, recombination progression is unlikely to be the relevant monitored event, as spo-11 is not required either for normal entry into or exit from the clustered stage and is also not required for the persistence of chromosome clustering seen in syp-1 and syp-2 mutants (Dernburg et al. 1998; MacQueen et al. 2002; Colaiacovo et al. 2003).
Pairing of the X chromosomes
In contrast with the autosomes, pairing of the X chromosomes is remarkably unaffected in htp-1 mutants. Recent studies have shown that partial loss-of-function him-3 mutants in which clustering of chromatin is not eliminated also show a similar pairing phenotype (Couteau et al. 2004; Nabeshima et al. 2004). One possible explanation for the preferential pairing of the X chromosomes is that coincident with the onset of chromatin clustering, the X chromosomes somehow become spatially separated from the autosomes. In fact, despite lack of SC in htp-1; syp-2 double-mutant gonads, the X chromosomes appear to be associated with each other throughout most of meiotic prophase, while most autosomes remain unassociated with their homologs. This observation of SC-independent colocalization of X chromosomes in adjacent territories is reminiscent of the situation during meiotic prophase in Drosophila males, where in the absence of SC, chromosome pairs are sorted into distinct chromosome territories within meiotic prophase nuclei (Vazquez et al. 2002). It is possible that in worms, a similar mechanism separates the X chromosomes from the autosomes soon after entering meiosis, thereby creating a first wave of chromosome sorting in the nucleus.
Role of HTP-1 during meiotic recombination
The severe defect in crossover formation observed in htp-1 mutants could be an expected consequence of generalized pairing failure; however, this defect is also observed for the X chromosomes, which appear correctly paired and synapsed, implying a role for HTP-1 in meiotic recombination per se. The fact that RAD-51 foci both appear (albeit in reduced numbers) and disappear and that chromosome fragments are not observed at diakinesis suggests that DSBs are being made and repaired in htp-1 mutants, but repair is occurring by a pathway that does not yield interhomolog crossovers. Since most chromosomal regions are nonhomologously synapsed, we infer that most DSBs are being repaired in a context in which homologs are not available as repair templates. One obvious possibility is that htp-1 mutants are capable of using sister chromatids instead of the homologs as repair templates.
During meiosis, the choice of the homolog as the preferred recombination partner is an actively promoted event, and the use of sister chromatids as repair templates is actively inhibited (Schwacha and Kleckner 1997; Wan et al. 2004; Webber et al. 2004); this constraint on recombination partner choice is indicated in Figure 9. Under circumstances in which homologs are not available, however, DSBs are eventually repaired, presumably by using sister chromatids as templates. For example, following induction of meiosis in haploid yeast, where the sister chromatid is the only homologous partner available, DSBs are formed and repaired (de Massy et al. 1994); however, DSB disappearance is substantially delayed compared with diploid strains. Similarly, in C. elegans syp-1 and syp-2 mutants, where the homologs are not in close parallel alignment, RAD-51 foci form and eventually disappear, but disappearance is substantially delayed relative to wild type (Colaiacovo et al. 2003; this work). In both of these examples, repair appears to occur with delayed kinetics, suggesting that a barrier to sister-directed repair is operating under these circumstances until late prophase, when this constraint is ultimately lifted.
In contrast to the above examples of delayed DSB repair, in some mutants with impaired sister chromatid cohesion or axial element structure (C. elegans him-3 null and Drosophila ord mutants), RAD-51 or γ-H2Av (a marker for DSBs) foci disappear without a delay (Couteau et al. 2004; Webber et al. 2004). DSB repair seems to be taking place between sister chromatids in these mutants, implying that normally developed axes are important for establishing or maintaining the barrier to sister-chromatid repair. Based on the timing of disappearance of RAD-51 foci in htp-1 and htp-1; syp-2 mutants, we propose that HTP-1 is similarly required to establish or maintain the barrier that inhibits use of sisters as repair templates during early prophase.
In htp-1; syp-2 double mutants, there is a significant reduction in RAD-51 foci compared with htp-1 single mutants. This reduction could reflect a decrease in the number of DSBs formed; however, we suggest the alternative possibility that this reduction may reflect a contribution of SC central region proteins to inhibition of sister-directed repair. A possible role for the SC central region in inhibiting sister-directed repair was also suggested by the recent work of Nabeshima et al. (2004).
We note that in syp-2 mutant gonads, the number of RAD-51 foci begins to diminish in the late-pachytene region—which is also the region where nuclei finally exit from the clustered chromosome configuration. This observation suggests a coincident removal of two constraints at late pachytene—allowing both use of sister chromatids as repair templates and release from chromosome clustering despite synapsis failure—perhaps reflecting a coordinated developmental transition at this stage of meiotic prophase. Furthermore, the proposed lifting of early prophase constraints would take place in parallel with cessation of new DSB formation. Thus, if htp-1 mutants suffer a premature release from constraints that should be in place until late pachytene, they may also suffer premature imposition of a late-prophase state characterized by loss of competence for DSB formation (Reddy and Villeneuve 2004); this scenario could account for the reduced numbers of RAD-51 foci in htp-1 mutants. Since induction of DSBs by γ-irradiation is not sufficient to ensure crossover formation between the correctly paired X chromosomes, however, a premature loss of DSB competence alone can not account for the reduction of crossovers observed in htp-1 mutants. Instead, we suggest that htp-1 nuclei at early meiotic prophase have acquired a developmental state that mimics the late-pachytene stage in wild-type gonads, both in inhibiting DSB formation and in allowing repair of DSBs using sister chromatids as repair templates.
Are pairing and synapsis coordinated by checkpoint-like mechanisms?
We have proposed roles for HTP-1 in the operation of multiple constraints that together ensure a successful outcome of meiotic prophase, that is, the formation of chiasmata between homologous chromosomes. Two of these constraints involve monitoring mechanisms that make progression of subsequent events (SC polymerization and release from chromosome clustering) contingent upon successful completion of the monitored events (homolog recognition and stabilization of recognition through synapsis). There are obvious parallels between the proposed couplings and the operation of the spindle assembly checkpoint, which monitors attachment of kinetochores to spindle microtubules and prevents the onset of anaphase until all chromosomes achieve bipolar attachment (for review, see Nasmyth 2005). Moreover, a central player in the signaling process that couples detection of unattached kinetochores with inhibition of the anaphase promoting complex is Mad2, which is structurally homologous to HTP-1. Both proteins are composed mostly of a motif known as the HORMA domain, which accounts for 97% of the residues of Mad2 and 62% of HTP-1. Mad2 exists in two conformational states, and its function requires both Mad2 protein that is localized at the kinetochore, to trigger the signal, and a cytoplasmatic fraction that amplifies the signal and blocks activators of anaphase (De Antoni et al. 2005). We can envision similar modes of action for HTP-1 in the coordination of pairing and synapsis. For example, HTP-1 may interact with chromosome domains where homolog pairing and/or synapsis are monitored and may also exist in a nucleoplasmic or cytoplasmic form that is able to transduce a “wait synapsis” or “wait dispersal” signal. As discussed above, the “wait synapsis” command might be executed either by promoting degradation of SYP-1 and SYP-2 or by sequestering them in a soluble form; good candidates for the target(s) of the “wait dispersal” signal await discovery. Lack of HTP-1 would result in unregulated SC assembly and premature release from clustering in a manner analogous to how Mad2 depletion results in premature onset of anaphase (Gorbsky et al. 1998).
We note that previous work identified a role for Hop1, the sole Saccharomyces cerevisiae member of the meiosis-enriched HORMA domain protein family, in the DNA-damage-checkpoint-mediated pachytene arrest that is triggered by intermediate blocks in the recombination pathway (Bailis et al. 2000; Roeder and Bailis 2000; Woltering et al. 2000). An interaction between Hop1 and axial element component Red1 is required for function of the checkpoint, and it has been proposed that recombination intermediates need to be processed in the presence of these proteins to be recognized by checkpoint activators (Woltering et al. 2000). A similar pachytene checkpoint operates in C. elegans, inducing apoptosis in late pachytene in response to unrepaired recombination intermediates (Gartner et al. 2000). The early-prophase checkpoint-like mechanisms proposed here are distinct from this pachytene checkpoint in that they operate independently of recombination.
Recombination independence is a hallmark of the early meiotic prophase program in C. elegans (Dernburg et al. 1998): Both homolog pairing and synapsis take place normally in the absence of recombination. Regulatory mechanisms coupling pairing and synapsis may have evolved in conjunction with the mechanisms that allowed successful homolog synapsis in the absence of recombination. The evolutionary events that uncoupled synapsis initiation from recombination and consolidated features to solidify pairing partner choice and to nucleate synapsis to a single domain on each chromosome may have necessitated the development of checkpoint-like mechanisms to coordinate these events. The expansion of the htp-1/him-3 gene family in Caenorhabditis may have provided a means to accomplish this goal using existing protein modules well suited for the task.
Materials and methods
Genetics
All C. elegans strains were cultured at 20°C under standard conditions as described in Brenner (1974). The wild-type background used was Bristol N2. The following mutations and chromosome rearrangements were used (Dernburg et al. 1998; MacQueen and Villeneuve 2001; Colaiacovo et al. 2003; this work): LG IV: htp-1(me84, gk174, gk150), spo-11(ok79), dpy-13(e184) unc-24(e138), nT1 [unc-?(n754) let-?(m435)] (IV, V), nT1[unc-?(n754) let-? qIs50](IV;V).LGV: syp-2(ok307), chk-2(me64) rol-9(sc148), eT1(III;V). LG X: dpy-3(e27), unc-3(e151).
The me84 mutation was generated by ethyl methanesulfonate (EMS) and identified in a “green chromosomes screen” (Nabeshima et al. 2004). Briefly, worms homozygous for the integrated transgene ruIs32 expressing a histone H2B::GFP fusion protein under control of a germline promoter (Praitis et al. 2001) were mutagenized with EMS and allowed to produce F1 progeny. F2 progeny from individual F1 were scored under the microscope for presence of univalents at diakinesis. Using the method of Wicks et al. (2001), me84 was mapped between SNP markers in F36H12 and B0273 on chromosome IV, a region containing htp-1(F41H10.10). Sequencing of F41H10.10 in me84 mutants confirmed the presence of a single base change in the predicted coding region. gk150 and gk174 deletion alleles of htp-1 were generated by the C. elegans Gene Knockout Consortium.
Recombination analysis was performed as in Kelly et al. (2000). The recombination frequency (p) for the control was calculated as p =1 – (1 – 2R)1/2, where R is the frequency of phenotypically recombinant progeny (Brenner 1974). The recombination frequency calculated for htp-1 was a weighted average of the recombination frequency calculated from scoring hermaphrodite (XX) progeny and that calculated from scoring male (XO) progeny (where p = R): [phermaphrodite(2 × no. of hermaphrodites) + pmale(no. of males)]/(total no. of X chromosomes sampled). Map distances in centimorgans = 100 × p.
FISH
Probes: DNA from the following YACs was amplified as described in Zalevsky et al. (1999): Y51E12 (X, left), Y68A3 (X, right), Y48E9 (I, right), Y25B10 (II, left), and Y40H8 (IV, right). The 5S rDNA was amplified as described in Dernburg et al. (1998). Following amplification, DNA was labeled with d(UTP) digoxigenin or d(UTP) biotin using the Nick Translation kit from Roche.
Dissection and fixation: Gonads from 18–20-h post-L4 adults were dissected in egg buffer (118 mM NaCl, 48 mM KCl2, 2mM CaCl2, 2 mM MgCl2, 5 mM HEPES at pH 7.4), permeabilized for 2 min with 0.06% Triton and then fixed for 2 min in 3.7% paraformaldehyde. Slides were frozen in liquid nitrogen and then stored in methanol at –20°C. Before hybridization, slides were incubated in 50% formamide 2× SSC for 4 h at 37°C. Hybridization was carried out at 93°C for 3 min. Post-hybridization washes were carried out in 50% formamide 2× SSC at 37°C for 30 min. Slides were then washed in 2× SSC, blocked with 0.5% BSA in 2× SSC, and incubated with α-biotin and/or α-digoxigenin antibodies labeled with FITC or Cy3 for 2 h at room temperature. Finally, slides were counterstained with DAPI and mounted with Vectashield. Time-course analysis of pairing was carried out as described in MacQueen and Villeneuve (2001). The average numbers of nuclei scored per zone (probes: Y51E12, 5S rDNA, Y48E9, Y68A3) for wild type, htp-1(me84), htp-1(gk174), htp-1(me84); chk-2, and htp-1(me84); syp-2 were zone 1, n = 67; zone 2, n = 78; zone 3, n = 83; zone 4, n = 88; zone 5, n = 72; zone 6, n = 69. For syp-2 and chk-2 mutants (probes 5S rDNA and Y51E12), between 42 and 52 nuclei were scored per zone.
γ-Irradiation and quantification of attached X chromosomes at diakinesis
Adult worms (24 and 48 h post-L4) were treated with a dose of 5000 rads of γ-rays from a 137Cs source and then fixed 18 h after irradiation. Gonad dissection and FISH were carried out as described above.
Immunostaining
Gonads from 20-h post-L4 adults were dissected in egg buffer containing 0.1% Tween and immediately fixed in 1% paraformaldehyde for 5 min. Slides were frozen in liquid nitrogen, then immersed for 1 min in methanol at –20°C and transferred to PBST (1× PBS, 0.1% Tween). Blocking in 0.5% BSA in PBST was carried out for 1 h. Primary antibodies were incubated overnight at room temperature using the following dilutions: α-HIM-3 (1:100), α-SYP-1 (1:50), α-RAD-51 (1:200). Following three washes in PBST, secondary antibodies were added (Alexa 488 α-rabbit and Alexa 555 α-guinea pig). Slides were then counterstained with DAPI and mounted using Vectashield. Time-course analysis of RAD-51 foci was performed as described in Colaiacovo et al. (2003), except that we divided the gonad into seven zones. The average numbers of nuclei scored per zone for wild type, htp-1(me84), htp-1(gk150), and htp-1(me84); syp-2 were zone 1, n = 138; zone 2, n = 149; zone 3, n = 140; zone 4, n = 127; zone 5, n = 101; zone 6, n = 88; zone 7, n = 75. For chk-2, htp-1; chk-2, and syp-2: zone 1, n = 64; zone 2, n = 66; zone 3, n = 59; zone 4, n = 64; zone 5, n = 58; zone 6, n = 50; zone 7, n = 45.
Squash preparations were prepared as described in Colaiacovo et al. (2003). For antibody staining, squash preparations were processed in the same way as whole-mount gonads. For simultaneous DNA in situ hybridization and antibody staining, squashes were incubated with α-SYP-1 antibodies as described above and then fixed in 3.7% paraformaldehyde in PBST before they were hybridized with the 5S rDNA probe.
All images (FISH and immunostaining) were acquired as stacks of optical sections with an interval of 0.2 μm using a Delta Vision deconvolution microscopy system.
Acknowledgments
We thank the Caenorhabditis Genetics Center and the Caenorhabditis Gene Knockout Consortium for strains and M. Zetka for the HIM-3 antibody. We also thank F. Couteau and M. Zetka for communicating unpublished results and helpful discussions and members of the Villeneuve laboratory for criticalreading of the manuscript. This work was supported by NIH grant R01GM53804 to A.M.V., and by an individual postdoctoral fellowship from the Human Frontiers Science Program to E.M.-P.
Article and publication are at http://www.genesdev.org/cgi/doi/10.1101/gad.1338505.
References
- Alpi A., Pasierbek, P., Gartner, A., and Loidl, J. 2003. Genetic and cytological characterization of the recombination protein RAD-51 in Caenorhabditis elegans. Chromosoma 112: 6–16. [DOI] [PubMed] [Google Scholar]
- Aravind L. and Koonin, E.V. 1998. The HORMA domain: A common structural denominator in mitotic checkpoints, chromosome synapsis and DNA repair. Trends Biochem. Sci. 23: 284–286. [DOI] [PubMed] [Google Scholar]
- Armstrong S.J., Caryl, A.P., Jones, G.H., and Franklin, F.C. 2002. Asy1, a protein required for meiotic chromosome synapsis, localizes to axis-associated chromatin in Arabidopsis and Brassica. J. Cell Sci. 115: 3645–3655. [DOI] [PubMed] [Google Scholar]
- Bailis J.M., Smith, A.V., and Roeder, G.S. 2000. Bypass of a meiotic checkpoint by overproduction of meiotic chromosomal proteins. Mol. Cell Biol. 20: 4838–4848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S. 1974. The genetics of Caenorhabditis elegans. Genetics 77: 71–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caryl A.P., Armstrong, S.J., Jones, G.H., and Franklin, F.C. 2000. A homologue of the yeast HOP1 gene is inactivated in the Arabidopsis meiotic mutant asy1. Chromosoma 109: 62–71. [DOI] [PubMed] [Google Scholar]
- Chua P.R. and Roeder, G.S. 1997. Tam1, a telomere-associated meiotic protein, functions in chromosome synapsis and crossover interference. Genes & Dev. 11: 1786–1800. [DOI] [PubMed] [Google Scholar]
- Colaiacovo M.P., MacQueen, A.J., Martinez-Perez, E., McDonald, K., Adamo, A., La Volpe, A., and Villeneuve, A.M. 2003. Synaptonemal complex assembly in C. elegans is dispensable for loading strand-exchange proteins but critical for proper completion of recombination. Dev. Cell. 5: 463–474. [DOI] [PubMed] [Google Scholar]
- Conrad M.N., Dominguez, A.M., and Dresser, M.E. 1997. Ndj1p, a meiotic telomere protein required for normal chromosome synapsis and segregation in yeast. Science 276: 1252–1255. [DOI] [PubMed] [Google Scholar]
- Cooper J.P., Watanabe, Y., and Nurse, P. 1998. Fission yeast Taz1 protein is required for meiotic telomere clustering and recombination. Nature 392: 753–754. [DOI] [PubMed] [Google Scholar]
- Couteau F. and Zetka, M. 2005. HTP-1 coordinates synaptonemal complex assembly with homolog alignment during meiosis in C. elegans. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Couteau F., Nabeshima, K., Villeneuve, A., and Zetka, M. 2004. A component of C. elegans meiotic chromosome axes at the interface of homolog alignment, synapsis, nuclear reorganization, and recombination. Curr. Biol. 14: 585–592. [DOI] [PubMed] [Google Scholar]
- De Antoni A., Pearson, C.G., Cimini, D., Canman, J.C., Sala, V., Nezi, L., Mapelli, M., Sironi, L., Faretta, M., Salmon, E.D., et al. 2005. The Mad1/Mad2 complex as a template for Mad2 activation in the spindle assembly checkpoint. Curr. Biol. 15: 214–225. [DOI] [PubMed] [Google Scholar]
- de Massy B., Baudat, F., and Nicolas, A. 1994. Initiation of recombination in Saccharomyces cerevisiae haploid meiosis. Proc. Natl. Acad. Sci. 91: 11929–11933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dernburg A.F., McDonald, K., Moulder, G., Barstead, R., Dresser, M., and Villeneuve, A.M. 1998. Meiotic recombination in C. elegans initiates by a conserved mechanism and is dispensable for homologous chromosome synapsis. Cell 94: 387–398. [DOI] [PubMed] [Google Scholar]
- Gartner A., Milstein, S., Ahmed, S., Hodgkin, J., and Hengartner, M.O. 2000. A conserved checkpoint pathway mediates DNA damage-induced apoptosis and cell cycle arrest in C. elegans. Mol. Cell 5: 435–443. [DOI] [PubMed] [Google Scholar]
- Gillies C.B. 1974. The nature and extent of synaptonemal complex formation in haploid barley. Chromosoma 48: 441–453. [Google Scholar]
- Golubovskaya I.N., Harper, L.C., Pawlowski, W.P., Schichnes, D., and Cande, W.Z. 2002. The pam1 gene is required for meiotic bouquet formation and efficient homologous synapsis in maize (Zea mays L.). Genetics 162: 1979–1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorbsky G.J., Chen, R.H., and Murray, A.W. 1998. Microinjection of antibody to Mad2 protein into mammalian cells in mitosis induces premature anaphase. J. Cell Biol. 141: 1193–1205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper L., Golubovskaya, I., and Cande, W.Z. 2004. A bouquet of chromosomes. J. Cell Sci. 117: 4025–4032. [DOI] [PubMed] [Google Scholar]
- Herman R.K. and Kari, C.K. 1989. Recombination between small X chromosome duplications and the X chromosome in Caenorhabditis elegans. Genetics 121: 723–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollingsworth N.M., Goetsch, L., and Byers, B. 1990. The HOP1 gene encodes a meiosis-specific component of yeast chromosomes. Cell 61: 73–84. [DOI] [PubMed] [Google Scholar]
- Kelly K.O., Dernburg, A.F., Stanfield, G.M., and Villeneuve, A.M. 2000. Caenorhabditis elegans msh-5 is required for both normal and radiation-induced meiotic crossing over but not for completion of meiosis. Genetics 156: 617–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loidl J. 1990. Coming to grips with a complex matter. Chromosoma 100: 289–292. [DOI] [PubMed] [Google Scholar]
- Loidl J., Nairz, K., and Klein F. 1991. Meiotic chromosome synapsis in a haploid yeast. Chromosoma 100: 221–228. [DOI] [PubMed] [Google Scholar]
- MacQueen A.J. and Villeneuve, A.M. 2001. Nuclear reorganization and homologous chromosome pairing during meiotic prophase require C. elegans chk-2. Genes & Dev. 15: 1674–1687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Colaiacovo, M.P., McDonald, K., and Villeneuve, A.M. 2002. Synapsis-dependent and -independent mechanisms stabilize homolog pairing during meiotic prophase in C. elegans. Genes & Dev. 16: 2428–2442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacQueen A.J., Phillips, C.M., Bhalla, N., Weiser, P., Villeneuve, A.M., and Dernburg, A.F. 2005. Chromosome sites play dual roles to establish homologous synapsis during meiosis in C. elegans. Cell (in press). [DOI] [PMC free article] [PubMed]
- Marchler-Bauer A., Anderson, J.B., Cherukuri, P.F., DeWeese-Scott, C., Geer, L.Y., Gwadz, M., He, S., Hurwitz, D.I., Jackson, J.D., Ke, Z., et al. 2005. CDD: A Conserved Domain Database for protein classification. Nucleic Acids Res. 33: D192–D196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K.S., Howell, A.M., and Rose, A.M. 1988. The effects of translocations on recombination frequency in Caenorhabditis elegans. Genetics 120: 987–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKim K.S., Peters, K., and Rose, A.M. 1993. Two types of sites required for meiotic chromosome pairing in Caenorhabditis elegans. Genetics 134: 749–768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nabeshima K., Villeneuve, A.M., and Hillers, K.J. 2004. Chromosome-wide regulation of meiotic crossover formation in Caenorhabditis elegans requires properly assembled chromosome axes. Genetics 168: 1275–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasmyth K. 2005. How do so few control so many? Cell 120: 739–746. [DOI] [PubMed] [Google Scholar]
- Nimmo E.R., Pidoux, A.L., Perry, P.E., and Allshire, R.C. 1998. Defective meiosis in telomere-silencing mutants of Schizosaccharomyces pombe. Nature 392: 825–828. [DOI] [PubMed] [Google Scholar]
- Nonomura K.I., Nakano, M., Murata, K., Miyoshi, K., Eiguchi, M., Miyao, A., Hirochika, H., and Kurata, N. 2004. An insertional mutation in the rice PAIR2 gene, the ortholog of Arabidopsis ASY1, results in a defect in homologous chromosome pairing during meiosis. Mol. Genet. Genomics 271: 121–129. [DOI] [PubMed] [Google Scholar]
- Page S.L. and Hawley, R.S. 2001. c(3)G encodes a Drosophila synaptonemal complex protein. Genes & Dev. 15: 3130–3143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ____. 2003. Chromosome choreography: The meiotic ballet. Science 301: 785–789. [DOI] [PubMed] [Google Scholar]
- Praitis V., Casey, E., Collar, D., and Austin, J. 2001. Creation of low-copy integrated transgenic lines in Caenorhabditis elegans. Genetics 157: 1217–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy K.C. and Villeneuve, A.M. 2004. C. elegans HIM-17 links chromatin modification and competence for initiation of meiotic recombination. Cell 118: 439–452. [DOI] [PubMed] [Google Scholar]
- Roeder G.S. and Bailis, J.M. 2000. The pachytene checkpoint. Trends Genet. 16: 395–403. [DOI] [PubMed] [Google Scholar]
- Santos J.L., Jimenez, M.M., and Diez M. 1994. Meiosis in haploid rye: Extensive synapsis and low chiasma frequency. Heredity 73: 580–588. [Google Scholar]
- Scherthan H. 2001. A bouquet makes ends meet. Nat. Rev. Mol. Cell. Biol. 2001 2: 621–627. [DOI] [PubMed] [Google Scholar]
- Schwacha A. and Kleckner, N. 1997. Interhomolog bias during meiotic recombination: Meiotic functions promote a highly differentiated interhomolog-only pathway. Cell 90: 1123–1135. [DOI] [PubMed] [Google Scholar]
- Storlazzi A., Tesse, S., Gargano, S., James, F., Kleckner, N., and Zickler, D. 2003. Meiotic double-strand breaks at the interface of chromosome movement, chromosome remodeling, and reductional division. Genes & Dev. 17: 2675–2687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sym M. and Roeder, G.S. 1994. Crossover interference is abolished in the absence of a synaptonemal complex protein. Cell 79: 283–292. [DOI] [PubMed] [Google Scholar]
- Sym M., Engebrecht, J.A., and Roeder, G.S. 1993. ZIP1 is a synaptonemal complex protein required for meiotic chromosome synapsis. Cell 72: 365–378. [DOI] [PubMed] [Google Scholar]
- Trelles-Sticken E., Loidl, J., and Scherthan, H. 1999. Bouquet formation in budding yeast: Initiation of recombination is not required for meiotic telomere clustering. J. Cell Sci. 112: 651–658. [DOI] [PubMed] [Google Scholar]
- Vazquez J., Belmont, A.S., and Sedat J.W. 2002. The dynamics of homologous chromosome pairing during male Drosophila meiosis. Curr. Biol. 12: 1473–1483. [DOI] [PubMed] [Google Scholar]
- Villeneuve A.M. 1994. A cis-acting locus that promotes crossing over between X chromosomes in Caenorhabditis elegans. Genetics 136: 887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Wettstein D., Rasmussen, S.W., and Holm, P.B. 1984. The synaptonemal complex in genetic segregation. Annu. Rev. Genet. 18: 331–413. [DOI] [PubMed] [Google Scholar]
- Wan L., de los Santos, T., Zhang, C., Shokat, K., and Holling-sworth, N.M. 2004. Mek1 kinase activity functions downstream of RED1 in the regulation of meiotic double strand break repair in budding yeast. Mol. Biol. Cell 15: 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Webber H.A., Howard, L., and Bickel, S.E. 2004. The cohesion protein ORD is required for homologue bias during meiotic recombination. J. Cell Biol. 164: 819–829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicks S.R., Yeh, R.T., Gish, W.R., Waterston, R.H., and Plasterk, R.H. 2001. Rapid gene mapping in Caenorhabditis elegans using a high density polymorphism map. Nat. Genet. 28: 160–164. [DOI] [PubMed] [Google Scholar]
- Woltering D., Baumgartner, B., Bagchi, S., Larkin, B., Loidl, J., de los Santos, T., and Hollingsworth, N.M. 2000. Meiotic segregation, synapsis, and recombination checkpoint functions require physical interaction between the chromosomal proteins Red1p and Hop1p. Mol. Cell Biol. 20: 6646–6658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zalevsky J., MacQueen, A.J., Duffy, J.B., Kemphues, K.J., and Villeneuve, A.M. 1999. Crossing over during Caenorhabditis elegans meiosis requires a conserved MutS-based pathway that is partially dispensable in budding yeast. Genetics 153: 1271–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zetka M.C., Kawasaki, I., Strome, S., and Muller, F. 1999. Synapsis and chiasma formation in Caenorhabditis elegans require HIM-3, a meiotic chromosome core component that functions in chromosome segregation. Genes & Dev. 13: 2258–2270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zickler D. and Kleckner, N. 1998. The leptotene–zygotene transition of meiosis. Annu. Rev. Genet. 32: 619–697. [DOI] [PubMed] [Google Scholar]
- ____. 1999. Meiotic chromosomes: Integrating structure and function. Annu. Rev. Genet. 33: 603–754. [DOI] [PubMed] [Google Scholar]