Abstract
Current methodologies that monitor immune responses rely on invasive techniques that sample tissues at a given point in time. New technologies are needed to elucidate the temporal patterns of immune responses and the spatial distribution of immune cells on a whole-body scale. We describe a noninvasive, quantitative, and tomographic approach to visualize a primary anti-tumor immune response by using positron emission tomography (PET). Bone marrow chimeric mice were generated by engraftment of hematopoietic stem and progenitor cells transduced with a trifusion reporter gene encoding synthetic Renilla luciferase (hRluc), EGFP, and Herpes virus thymidine kinase (sr39TK). Mice were challenged with the Moloney murine sarcoma and leukemia virus complex (M-MSV/M-MuLV), and the induced immune response was monitored by using PET. Hematopoietic cells were visualized by using 9-[4-[18F]fluoro-3-(hydroxymethyl)butyl]guanine ([18F]FHBG), a radioactive substrate specific for the sr39TK PET reporter protein. Immune cell localization and expansion were seen at the tumor and draining lymph nodes (DLNs). 2-[18F]fluoro-2-deoxy-d-glucose ([18F]FDG), which is sequestered in metabolically active cells, was used to follow tumor growth and regression. Elevated glucose metabolism was also seen in activated lymphocytes in the DLNs by using the [18F]FDG probe. When M-MSV/M-MuLV-challenged mice were treated with the immunosuppressive drug dexamethasone, activation and expansion of immune cell populations in the DLNs could no longer be detected with PET imaging. The method we describe can be used to kinetically measure the induction and therapeutic modulations of cell-mediated immune responses.
Keywords: immunosuppressive therapy, lymphocytes
During an anti-tumor immune response, innate and adaptive immunity synergize through cellular events that create regional variations in immune cell populations throughout the body (1). Immunotherapy protocols in cancer treatment are directed at enhancing a range of immune processes, including antigen presentation, activation of specific T cells, and localization of immune effectors to tumors. Methods used to determine the efficacy of treatment often rely on invasive techniques, such as histologic evidence of lymphocyte infiltration (2). Single time points are unable to reflect contextual influences of living organs, variations throughout the body, and the plasticity of the immune system (3).
Noninvasive imaging of reporter gene expression in living subjects can be accomplished by using small animal optical imaging, positron emission tomography (PET), and magnetic resonance imaging approaches with distinct reporter genes compatible with each imaging modality (4). Each of these imaging techniques and their respective reporter genes have unique advantages and limitations. Optical bioluminescence imaging (BLI) uses an enzymatic reaction between a luciferase enzyme and its substrate, luciferin, to produce visible light detected by a highly sensitive cooled charge-coupled device (CCD) camera. The optical imaging strategies have the advantages of being relatively low cost and high-throughput, but they are limited by their nontomographic nature, lack of fine spatial resolution, and inability to accommodate up to larger animals and humans.
PET is a highly sensitive detection method that provides three-dimensional information within the whole body (5). By using molecules labeled with positron-emitting radioisotopes in trace quantities, one can measure rates of biochemical processes in tissues. 2-[18F]Fluoro-2-deoxy-d-glucose ([18F]FDG), a glucose analog that accumulates within metabolically active cells when phosphorylated by hexokinase, has been commonly used as a diagnostic tool in clinical practice to assess neural and cardiac function and tumor progression (5). 9-[4-[18F]fluoro-3-(hydroxymethyl)butyl-]guanine ([18F]FHBG), an acycloguanosine substrate, is preferentially phosphorylated and trapped within cells by the protein encoded by a mutant herpes simplex virus type 1 thymidine kinase reporter gene (sr39tk) (6). Advances in detector design have led to the development of micro-PET devices that allow accurate measurements in rodents (7).
Past studies employing imaging modalities to follow immune cell trafficking have relied on the introduction of PET or bioluminescence reporter genes into cells ex vivo. By using BLI, antigen-specific lymphocyte populations have been shown to traffic to sites of inflammation in tumor (lymphoma) and autoimmune (experimental autoimmune encephalomyelitis and collagen-induced arthritis) animal models (8). We and others have used PET imaging to monitor adoptively transferred T cells as they migrate to, and accumulate at, tumors (9, 10). These adoptive transfer methods have been based on memory immune responses using vaccinated or transgenic mice.
In the present study, we generated bone marrow (BM) chimeric mice reconstituted with hematopoietic stem and progenitor cells transduced with the trifusion reporter gene (hrl-egfp-tk) (11). The reporter was used for concomitant multimodality imaging by using flow cytometry, BLI, and PET. A strongly immunogenic, nonmetastasizing retrovirally induced rhabdomyosarcoma [M-MSV/M-MuLV (Moloney murine sarcoma virus/Moloney murine leukemia virus)] was used to initiate a potent anti-tumor immune response (12). MSV is a replication-defective, acutely transforming retrovirus carried with helper activity provided by M-MuLV, which encodes the gag, pol, and env components that are necessary for cell infection and replication (13). The tumor was chosen for its short latency period (7-10 days) and rapid regression (2-3 wk) after the induction of a strong immune reaction in immunocompetent adult mice. [18F]FHBG and [18F]FDG PET imaging were used to sequentially follow the localization of immune cell populations relative to tumor progression in the same animal. Transduced hematopoietic cells were seen at the tumor and draining lymph nodes (DLNs). Alterations of the immune response after therapeutic manipulation with an immunosuppressive drug, dexamethasone (DEX), were also detected with PET. Such approaches to visualize immune cell expansion and activation can be used for the evaluation and development of immunotherapies for cancer and other diseases.
Materials and Methods
Animals. Immunocompetent C57BL/6 mice and immunodeficient CB-17SCID/SCIDmice were bred and maintained according to the guidelines of the Department of Laboratory Animal Medicine (DLAM) at the University of California, Los Angeles. All animal studies were carried out by using protocols that had been approved by DLAM.
Trifusion Lentivirus Production. M-MSV/M-MuLV stock was prepared as described (9). The lentiviral trifusion reporter construct (pFUG-TF) is composed of a synthetic Renilla luciferase (hrl), enhanced green fluorescent protein (egfp), and a truncated version of sr39tk as a PET reporter (ttk) driven by a ubiquitin promoter (11, 14). Lentivirus was prepared as described (15). Concentrated virus was titered on Baf3 cells. Infectivity was determined by GFP expression as analyzed on FACSCanto (Becton Dickinson).
Generation of BM Chimeras. Six- to 10-week-old C57BL/6 mice were injected i.v. with 150 mg/kg 5-fluorouracil (APP). BM was harvested 4 days after treatment and cultured with IL-3 (6 ng/ml), IL-6 (10 ng/ml), stem cell factor (SCF) (100 ng/ml), and 5% WeHI as growth factors. After 48 h, the cells were infected with pFUG-TF lentivirus [≈10 multiplicity of infection (moi)] and 1.6 μg/ml of polybrene under spin conditions (2,500 rpm, 90 min, 30°C, Beckman CS-6R centrifuge) and then incubated overnight at 37°C. Four- to 8-week-old C57BL/6 mice were lethally irradiated (1,200 rad) before i.v. injection of 1-2 × 106 transduced BM cells on the same day.
Optical Bioluminescence. Shaved mice were anesthetized and injected with 5-10 μg of coelenterazine (Biotium, Hayward, CA, dissolved in PBS) i.v. Each animal was then placed in a Xenogen-IVIS optical imaging system (Xenogen, Alameda, CA), and whole body images were obtained and analyzed as described (16). For organ imaging, organs of interest were harvested, incubated with coelenterazine, and immediately imaged. Values are expressed in photons·s-1·cm-2·steradian-1.
In Vitro Enzymatic Analysis. MACS beads (Miltenyi Biotec, Auburn, CA) were used to positively select for B220+, CD4+, CD8+, CD11b+, and Ter-119+ cells. Sorting for c-kit+Thy1.2lolin-/loSca-1+ hematopoietic stem cells (HSCs) has been described (17). Renilla luciferase assays were performed with Renilla Luciferase Assay Reagents (E2810, Promega) and an LMax II384 luminometer (Molecular Devices).
Generation and Treatment of Tumor-Bearing Chimeras. Chimeras were challenged i.m. in the right triceps with M-MSV/M-MuLV in a volume of 100 μl. Chimeras in the treatment group were given 10 mg/kg DEX daily by i.p. injection, starting 5-7 days before tumor challenge.
Micro-PET Imaging. Animals were anesthetized, injected i.p. with [18F]FDG or i.v. with [18F]FHBG, and scanned by using a FOCUS micro-PET scanner (Concorde Microsystems, Knoxville, TN) as described (18). Images were reconstructed from the scanner data by using an iterative maximum a posteriori algorithm (19). Quantification was performed by drawing regions of interest (ROIs) and taking the ratio of the %ID/g (percent injected dose per gram of tissue) of the ROI over the %ID/g of a contralateral region of the animal. For PET imaging of organs, animals were euthanized after tracer uptake, and organs of interest were harvested and imaged.
Immunohistochemistry. Tumors were harvested, embedded, and stained as described (9). Sections were stained with primary hamster αCD3 [145-2C11, 2.5 μg/ml, American Type Culture Collection (ATCC)], biotinylated rat αCD45 (104, 5 μg/ml, Becton Dickinson), biotinylated rat αCD11b (M1/70, 5 μg/ml, ATCC), or biotinylated rat αGR-1 (8C5, 5 μg/ml, ATCC). Peroxidaseconjugated StreptABComplex/HRP (K0377, DAKO) was used as a secondary reagent for the biotinylated antibodies.
Results
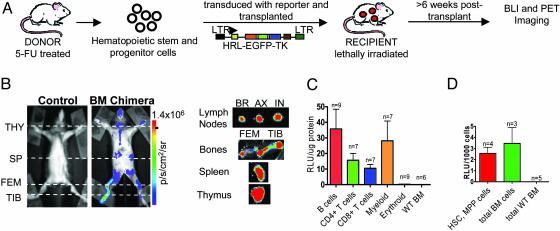
Generation of BM Chimeric Mice with Multimodality Reporter Genes. A trifusion lentiviral reporter construct was used to stably introduce reporter genes into hematopoietic stem and progenitor cells. The reporter construct (pUbi-hrl-egfp-tk) comprises three genetically fused reporter components (Renilla luciferase, EGFP, and mutated thymidine kinase) driven by a ubiquitin promoter (11, 14). To generate BM chimeric mice with reporter gene expression, stem and progenitor hematopoietic cells were enriched from the BM, transduced with trifusion lentivirus, and transplanted into lethally irradiated recipient mice (Fig. 1A). The reporter genes were not toxic to the transduced cells, which retained their ability to rescue the hematopoietic cell compartment of lethally irradiated recipients.
Fig. 1.
Trifusion reporter is stably expressed in the hematopoietic cell compartment of BM chimeras. (A) Schematic of the generation of BM chimeric mice. The trifusion reporter construct is composed of a ubiquitin promoter driving expression of an optical bioluminescence reporter gene Renilla luciferase (hrl), a green fluorescent protein marker (egfp), and a PET reporter mutant herpes simplex virus type 1 thymidine kinase (tk). (B) (Left) Renilla luciferase expression in control and BM chimeric mice was imaged by using a charge-coupled device (CCD) camera. (Right) Organs were imaged ex vivo to assess tissue origin of bioluminescence signal. THY, thymus; SP, spleen; FEM, femurus; TIB, tibia; BR, brachial; AX, axillary; IN, inguinal. p/s/cm2/sr, photons·s-1·cm-2·steridian-1. The results are representative of 40 BM chimeric mice. (C) B cells (B220+), T cells (CD4+ and CD8+), myeloid cells (CD11b+), and erythroid cells (Ter119+) were purified from lymphoid organs of chimeras. (D) HSCs and multipotent progenitor (MPP) (c-kit+Thy1.2-/loLin-Sca-1+) cells were sorted by FACS. (C and D) Cells were lysed, and Renilla luciferase activity was assayed. BM cells from C57BL/6 mice were used as a control. RLU, relative light units.
Successful reconstitution of reporter-expressing hematopoietic cells was determined by BLI of chimeras 6-8 wk posttransplant by using a cooled charge-coupled device (CCD) system. Bioluminescence signal could be detected at areas corresponding to the thymus, spleen, femur, and tibia by whole body BLI, indicating the presence of Renilla luciferase-expressing hematopoietic cells (Fig. 1B). Organs were imaged ex vivo to assess the tissue origin of the signals observed. Both primary (bones and thymus) and secondary (spleen and lymph nodes) lymphoid organs had strong luminescence signals (Fig. 1B). The bioluminescence signal observed in individual lymph nodes imaged ex vivo was undetectable in the whole-body image. This discrepancy can be explained by the attenuation of the signal by a number of variables, such as tissue depth and skin and hair pigmentation, as well as the positioning of the animal during imaging.
To confirm reporter gene expression in both differentiated and primitive hematopoietic cell types, cell populations were purified, and luciferase expression was analyzed in vitro. T and B cells (CD4+, CD8+, and B220+) were isolated from the spleen and thymus, myeloid cells (CD11b+) from BM, and mature erythroid cells (Ter119+) from peripheral blood. Luciferase activity was detected in lymphoid and myeloid cell populations, confirming multilineage reconstitution by transduced hematopoietic stem and progenitor cells (Fig. 1C). Erythrocyte precursors (Ter119+CD45+) isolated from BM did not have luciferase activity (data not shown).
Primitive hematopoietic cells were purified to confirm that early, repopulating cells had reporter gene expression. The c-kit+, Thy1.2lo, lineage marker-/lo, Sca-1+ population (KTLS), which represents a primitive subset including self-renewing long-term HSC (LT-HSC), short-term HSC (ST-HSC) with restricted self-renewing potential, and non-self-renewing multipotent progenitor (MPP), was sorted by FACS (17). KTLS and unsorted BM cells from chimeras had comparable levels of luciferase activity, indicating that stem and progenitor hematopoietic cells were successfully transduced with the trifusion lentivirus (Fig. 1D).
[18F]FDG Imaging of Metabolically Active Cells in BM Chimeric and CB-17SCID/SCID Tumor-Bearing Mice. The immunological process we chose to image was the rejection of a strongly immunogenic, well localized, and nonmetastasizing rhabdomyosarcoma. The rejection of the tumor is characterized by massive expansion of T lymphocytes, granulocytes, and macrophages at the tumor lesion, DLNs, and spleen. Rejection is mediated by CD8+ cytolytic T cells, which recognize peptides from the gag and env proteins of M-MuLV, requires help from CD4+ T cells, and depends on presentation by H-2 Db alleles (20). When BM chimeric mice were challenged and imaged with BLI, the only signal detected corresponded to the tumor site (data not shown). The limited efficiency of light transmission through tissue and skin and spatial resolution (≈5 mm) of BLI made it difficult to discern between signals emitted from the tumor and DLNs.
Micro-PET imaging, which has a spatial resolution of <1.3 mm, lacks attenuation, and offers excellent tomographic images, was used to image a localized immune response at the tumor and DLNs. Two radiolabeled PET substrates, [18F]FDG and [18F]FHBG probes, were used to differentially monitor tumor progression and immune cell localization, respectively. Sequential imaging with [18F]FDG and [18F]FHBG probes was performed on alternating days within the same animal.
[18F]FDG imaging, which detects highly metabolically active cells, was used to monitor tumor progression. [18F]FDG is phosphorylated by hexokinase to form [18F]FDG-6-phosphate and subsequently trapped in the cell. [18F]FDG PET scans of healthy normal mice typically have substrate accumulation in the brain, heart, kidneys, and bladder.
BM chimeric mice were challenged and imaged with the [18F]FDG probe on day 13. High-definition whole-body mouse images were generated by algebraic reconstruction of collected three-dimensional emission data (19). A three-dimensional computed tomography (CT) image of the basic skeletal structure of the mouse demonstrates the plane where the coronal, transverse, and sagittal images were taken (Fig. 2A). PET images are displayed as a measure of %ID/g in a false color scale, with red indicating the highest amount of radioactivity present at that location. The three-dimensional data were analyzed by dividing the image into 64 adjacent planes from the ventral to dorsal side of the mouse (Fig. 5, which is published as supporting information on the PNAS web site). [18F]FDG signal was observed in the tumor, brain, heart, kidneys, and bladder (Fig. 2B). Interestingly, [18F]FDG retention was also observed at the tumor DLNs, suggesting the presence of highly metabolically active cells (Fig. 2B). Analysis of histological sections confirmed that the [18F]FDG signal was not due to tumor metastasis (data not shown) but rather to the presence of active lymphocytes and other immune cells. Accumulation of [18F]FDG was also observed in the spleen, possibly indicative of metabolically active immune cell populations (Fig. 2B).
Fig. 2.
Whole-body [18F]FDG PET imaging of metabolically active tumor cells and immune cells. (A) Whole-body tomographic computed tomography (CT) image of untreated mouse. Coronal, transverse, and sagittal planes are shown. BM chimeric mice (B) and CB-17SCID/SCID mice (C) were injected i.p. with [18F]FDG on day 13 post tumor challenge and imaged as described in Materials and Methods. Whole-body coronal, transverse, and sagittal PET sections (1.6-mm thickness) are shown. TUM, tumor; H, heart; BL, bladder; SP, spleen; KID, kidney.
Immunodeficient CB-17SCID/SCID mice, which lack T and B cells, were challenged with M-MSV/M-MuLV and imaged with the [18F]FDG probe to determine whether lymphocytes were required for the DLN signal observed. [18F]FDG imaging detected only tumor growth, and no DLN signal was seen (Fig. 2C). This result indicates that [18F]FDG sequestration in DLNs requires the presence of an intact adaptive immune system.
[18F]FHBG Imaging of Transduced Immune Cells Localized at the Tumor and DLNs. The [18F]FHBG probe, which is phosphorylated and trapped by cells expressing the PET reporter gene (tk), was used to follow transduced hematopoietic cells. Mice were serially imaged between day 6 and day 23 after tumor induction (Fig. 3A). PET images presented are single coronal sections at the plane of the tumor and DLNs (Fig. 3B). [18F]FHBG retention was detected in the tumor and DLNs between days 8-14 post tumor challenge (Fig. 3B). This observation is representative of a total of 10 animals performed in three separate experiments. Transduced immune cells were found to localize and expand in the tumor and DLNs at the height of the anti-tumor immune response around day 10.
Fig. 3.
Alterations in immune cell localization and activation with or without DEX treatment can be detected by [18F]FHBG and [18F]FDG PET imaging. (A) Schematic diagram of imaging schedule and DEX treatment. Mice were injected i.v with [18F]FHBG and imaged on day 8, 10, and 14 (B) and i.p with [18F]FDG and imaged on day 9, 13, and 15 (F). The coronal section displayed is centered at the tumor (TUM) and DLNs (1.6-mm thickness). (B and F) Shown are untreated mice (No DEX) (Left) and DEX-treated mice (DEX) (Right). Representative of a total of 10 animals performed in three separate experiments. ROIs were drawn to quantify [18F]FHBG (C) and [18F]FDG (G) uptake in the TUM and DLN on different days post tumor challenge. The quantification of [18F]FHBG and [18F]FDG signals was calculated as a ratio of %ID/g at region of interest to the %ID/g at a background region. Immediately after whole-body imaging, each organ was isolated day 14 post challenge to assess organ size to the [18F]FHBG (D) and [18F]FDG (H) signal. (H) Naive unchallenged C57BL/6 mouse lymph nodes were used as a control. Representative of three animals from each group. CON MUS, contralateral muscle; CON LNs, contralateral LNs. (E) Picture of the DLNs and CON LNs from untreated (No DEX) and DEX-treated (DEX) mice.
The kinetics of tumor progression and immune cell expansion over days 6-21 post tumor challenge was quantified. ROIs were drawn, quantified, and represented as %ID/g tissue. %ID/g denotes the amount of tracer accumulated in a ROI normalized to the total amount of tracer and to the mass of the tissue analyzed. [18F]FHBG retention in the tumor and DLNs peaked around day 10, representing the maximum number of immune cells present (Fig. 3C). The mean PET signal (%ID/g) was 1.47 ± 0.15 on day 8, 2.60 ± 0.27 on day 10 for the tumor, P < 0.05; and 1.42 ± 0.25 on day 8, 2.53 ± 0.33 on day 10 for the DLNs, P < 0.05. It is important to note that each BM chimeric mouse had slightly different kinetics of tumor progression and immune response. Nevertheless, a general cellular growth and expansion followed by regression at both the tumor and DLNs were observed. Data are representative of four animals.
Treatment with an Immunomodulatory Drug Resulted in a Selective Loss of [18F]FHBG Accumulation in the DLNs. An immunomodulatory drug was used to test whether therapeutic manipulations that altered immune function and cellular composition could be detected by PET imaging. DEX, a potent immunosuppressive glucocorticoid, was used to modify the anti-tumor immune response. DEX has been previously shown to reduce the lymphocyte numbers in circulation by the redistribution of cells to lymph nodes, induction of apoptosis and growth suppression of thymocytes, and inhibition of numerous cytokines required for the maintenance and activation of immune cells (21).
Mice chronically treated with DEX had an ≈3-fold decrease of lymphocyte counts in the circulating blood (data not shown). However, DEX-treated mice were still able to reject the tumor with an ≈6-day delay compared with untreated mice. The dose of DEX administered may have resulted in only a partial inhibition of CD4+ and CD8+ T cells. Accumulation of [18F]F-HBG in DLNs was not seen in DEX-treated mice, indicating a reduction of immune cell expansion. Surprisingly, transduced immune cells were detected at the tumor between days 8-14 (Fig. 3B). Immune cells were still able to infiltrate and expand at the tumor in DEX-treated mice. [18F]FHBG uptake at the tumor of DEX-treated mice was quantified. The peak of immune cell infiltration was between days 10 and 11 (1.48 ± 0.24%ID/g on day 8, 3.60 ± 0.63%ID/g on day 10-11, P < 0.05) (Fig. 3C). Quantification of [18F]FHBG accumulation in DLNs was undetectable and close to background levels (data not shown). There was no difference in mean PET signal between DLN ROIs on day 8 (1.46 ± 0.24%ID/g) and day 10 images (1.40 ± 0.37%ID/g), P > 0.05.
Lymphoid organs were isolated and imaged on the PET scanner ex vivo to independently assess organ size to the PET signal. On days 13-15 post tumor challenge, mice were injected with [18F]FHBG and euthanized, and organs were harvested and imaged. Tumor lesions in untreated and DEX-treated animals accumulated 2- to 3-fold more substrate compared with the contralateral muscle, representing the infiltration and expansion of immune cells at the tumor site (Fig. 3D). Similarly, in untreated mice, a significantly larger number of immune cells were present in the tumor DLNs compared with the contralateral lymph nodes (LNs) (Fig. 3D). This increase correlated with a 3- to 5-fold expansion of the size of tumor DLNs after induction of the cellular immune response (Fig. 3E). In DEX-treated mice, there was a lack of [18F]FHBG uptake in the tumor DLNs, which correlated with small LN size (Fig. 3 D and E). Data of ex vivo imaged organs was representative of three animals in each group. PET DLN signals were reflective of LN size and the state of lymphocyte activation, proliferation, and expansion.
[18F]FDG Imaging of Tumor Progression and Lymphocyte Activation. BM chimeric mice were challenged and serially imaged with the [18F]FDG probe. Based on the [18F]FDG signal, the tumor was seen to peak on day 9 and completely regress by day 15 (3.72 ± 0.45%ID/g on day 9, 1.51 ± 0.22%ID/g on day 15-17, P < 0.05) (Fig. 3 F and G). Activated immune cells were observed at the tumor DLNs between days 13 and 15 (1.68 ± 0.24%ID/g on day 7-8, 5.05 ± 0.69%ID/g on day 13-15, P < 0.05) (Fig. 3 F and G). Data are representative of a total of 10 animals in three separate experiments. Tumor DLN were shown to accumulate 2- to 6-fold more [18F]FDG than the contralateral nodes when organs were imaged ex vivo, correlating with a 3- to 5-fold increase of DLN size (Fig. 3H).
Lymph nodes from naive C57BL/6 mice were imaged to demonstrate the baseline level of glucose uptake without tumor challenge. The intensity of [18F]FDG LN signal of naive mice was at a similar background level as the contralateral LN of challenged mice (Fig. 3H). There is a clear difference in lymphocyte glucose metabolism between naive and activated state.
[18F]FDG imaging was also carried out in DEX-treated mice to monitor tumor progression and the effects of DEX on lymphocyte activation in DLNs. On day 14 post tumor challenge, [18F]FDG uptake was still observed at the tumor site, indicating the presence of tumor cells (Fig. 3F). DEX-treated mice had delayed tumor rejection compared with untreated mice. ROI quantification of [18F]FDG sequestered at the tumor site demonstrated that the highest level of tumor cell glucose metabolism occurred around day 14 (2.60 ± 0.54%ID/g on day 7, 6.82 ± 0.98%ID/g on day 13-15, P < 0.05) (Fig. 3G). [18F]FDG uptake was undetectable on day 21, indicating complete tumor eradication. PET imaging of organs ex vivo on day 14 showed a 3- to 10-fold more [18F]FDG accumulation at the tumor compared with the contralateral muscle in both DEX-treated and untreated mice (Fig. 3H). [18F]FDG signal was not detected at the DLNs of DEX-treated mice (1.44 ± 0.12%ID/g on day 7, 1.18 ± 0.12%ID/g on day 13-15, P > 0.05).
Histological Analysis of Tumor Sections Detects Myeloid Cell Populations in DEX-Treated Tumors. Previous studies on the M-MSV/M-MuLV-induced sarcomas have established that CD4+ T cells recruit large numbers of macrophages that lead to the recruitment and expansion of CD8+ T cells (20). The [18F]FHBG uptake observed in DEX-treated lesions may reflect the accumulation of DEX-resistant cell types, such as macrophages, granulocytes, or natural killer (NK) cells. Histological analysis was performed to characterize the immune cell populations in the tumor microenvironment. Tumors were harvested for histological analysis on the day when the highest level of [18F]FHBG PET signal at the tumor was seen. The presence of reporter-transduced myeloid and T cells at the tumor lesion of untreated chimeras was confirmed by staining sections with a pan-myeloid marker (α-CD11b) and pan-T marker (α-CD3) (Fig. 4). In DEX-treated lesions, a similar number of myeloid cells (α-CD11b) and a reduced number of T cells (α-CD3) were seen (Fig. 4). The data suggest that myeloid cells were primarily responsible for the [18F]FHBG uptake at the tumor in DEX-treated mice.
Fig. 4.
Histological analysis of tumor sections detects myeloid cell populations in DEX-treated tumors. Shown are stainings of tumor sections from untreated (No DEX) and DEX-treated (DEX) animals with α-CD3 and α-CD11b. (Magnification: ×200.)
Discussion
Many of the problems in evaluating immunotherapy protocols stem from the lack of effective tools to follow the extent and duration of responses to treatment. In this study, we have demonstrated a strategy to monitor a specific primary immune response against a tumor challenge. BM chimeric mice were successfully reconstituted with hematopoietic stem and progenitor cells expressing the trifusion reporter gene. Through the use of micro-PET imaging, we were able to serially visualize immune cell localization, expansion, and activation in the tumor and DLNs.
Estimating the number of cells at the tumor site based on the PET signal is essential to determine the sensitivity and applicability of this technique to other tumor or autoimmune models. The correlation of [18F]FHBG PET signal to cell numbers of MSCV-sr39TK-transduced primary T cells injected intra-tumorally was previously investigated by Su et al. (22). Their data showed a linear regression of 0.2%ID/g signal per 106 cells. By using these calculations, we estimated the highest number of immune cells present at the tumor site at day 10 to be ≈5-10 × 106 cells in a 0.3-ml volume. The estimation of the limit of detection of [18F]FHBG uptake was ≈1 × 106 cells in a 0.3-ml volume. It should be noted that the accuracy of these estimations could be influenced by multiple factors. For example, we might underestimate the actual cell numbers, given the weaker ubiquitin promoter used in our study, compared with the MSCV promoter used by Su et al. (22). Furthermore, differences in lentiviral and retroviral (22) constructs and titer should be considered in regard to the number of integrated copies of the reporter constructs, as well as the sites of integration. Another factor that may influence thymidine kinase (TK) activity is the activation status of the cells. Splenocytes from BM chimeric mice activated in vitro with α-CD3 displayed a 2- to 6-fold increase in the GFP and luciferase activity in both T and B cells (data not shown), suggesting that there may be a lower number of cells localized at the tumor site than the number estimated above (22). [18F]FHBG imaging may be able to detect immune cell activation in addition to localization and expansion.
PET imaging provides tomographic scans with sufficient resolution to interpret with confidence at any site throughout the body. The use of FDG as a radiotracer has become a standard procedure for the assessment of cancer and metastasis (5). Interestingly, the increased glucose metabolism indicative of malignant tumor cells has also been reported to be seen in macrophages (23) and lymphocytes (24) upon activation in vitro. CD28 costimulation of anti-CD3-activated human peripheral blood T cells provides a signal to increase their glucose uptake through the glucose transporter, Glut1, by >5-fold (24). Our study using CB-17SCID/SCID mice (Fig. 2C) was able to demonstrate that lymphocytes were primarily responsible for the [18F]FDG accumulation in the tumor DLN. Similarly, other in vivo studies have suggested a role for inflammatory cells in [18F]FDG sequestration at sites of malignancy (25, 26). Two studies in humans have detected activated lymph nodes during early and asymptomatic HIV-1 infection (27). [18F]FDG imaging can potentially be used as a diagnostic tool of global immune cell activation.
Our approach of imaging a primary immune response by using BM chimeric mice is a promising tool but has limitations. Specific immune cell types may be seen if the expression of the tri-reporter gene is driven by more specific promoters. New PET-imaging probes that could selectively identify immune cells in vivo may provide added specificity. Others have reported novel PET approaches to monitor immune cells. The [18F]-galacto-RGD probe was used to image αvβ3 expression in the cutaneous delayed-type hypersensitivity reaction, monitoring not only macrophages, lymphocytes, and granulocytes, but also angiogenesis (28). [123I]IL-2 imaging of activated T lymphocytes in patients with inflammatory bowel diseases, autoimmune thyroid diseases, insulin-dependent diabetes and melanoma restricted users to monitoring both CD4+ and CD8+ T cells in an activated state (29). One study has demonstrated the ability to follow a specific immune cell subset by using the [11C]peripheral benzodiazepine ligand. The PET probe binds to benzodiazepine receptors, which are abundant on macrophages during the development of HIV encephalitis and expressed at low levels in the noninfected brain (30). Future developments in this area could offer the ability to evaluate immunotherapies in the clinic by monitoring a subset of immune cells in a spatio-temporal manner.
Supplementary Material
Acknowledgments
We are grateful to Dr. Waldemar Ladno and Judy Edwards for micro-PET imaging, the chemists and cyclotron crew for production of radioisotopes, and David Stout for technical advice. We thank David Baltimore (California Institute of Technology, Pasadena) for the gift of Δ8.9 and pVSVG vectors. We thank Shirley Quan, James Johnson, LaKeisha Perkins, Donghui Cheng, and Mireille Riedinger for outstanding technical assistance. We thank Barbara Anderson for excellent preparation of the manuscript. We are grateful to Dr. Michael Phelps for his inspiration and support. This work was supported by National Institutes of Health Research Training in Pharmacological Sciences Training Grant PHS T32 CM008652 (to C.J.S.) and by a Howard Hughes Medical Institute Research Training Fellowship for Medical Students (to A.N.). We also acknowledge support of the National Cancer Institute (NCI) Small Animal Imaging Resource Program (to S.S.G.) and NCI Grant ICMIC P50 (to S.S.G.). C.G.R. was supported by a Cancer Research Institute fellowship. O.N.W. is an Investigator of the Howard Hughes Medical Institute. This research was supported in part by the Office of Science (Office of Biological and Environmental Research), U.S. Department of Energy, Cooperative Agreement No. DE-FC02-ER63420.
Conflict of interest statement: No conflicts declared.
Abbreviations: PET, positron emission tomography; BLI, bioluminescence imaging; [18F]FDG, 2-[18F]fluoro-2-deoxy-d-glucose; [18F]FHBG, 9-[4-[18F]fluoro-3-(hydroxymethyl) butyl]guanine; M-MSV/M-MuLV, Moloney murine sarcoma virus/Moloney murine leukemia virus; LN, lymph node; DLN, draining LN; DEX, dexamethasone; HSC, hematopoietic stem cell; ROI, region of interest; %ID/g, percent injected dose per gram of tissue.
References
- 1.Butcher, E. C., Williams, M., Youngman, K., Rott, L. & Briskin, M. (1999) Adv. Immunol. 72, 209-253. [DOI] [PubMed] [Google Scholar]
- 2.Therasse, P., Arbuck, S. G., Eisenhauer, E. A., Wanders, J., Kaplan, R. S., Rubinstein, L., Verweij, J., Van Glabbeke, M., van Oosterom, A. T., Christian, M. C. & Gwyther, S. G. (2000) J. Natl. Cancer Inst. 92, 205-216. [DOI] [PubMed] [Google Scholar]
- 3.Iparraguirre, A. & Weninger, W. (2003) Int. Arch. Allergy Immunol. 132, 277-293. [DOI] [PubMed] [Google Scholar]
- 4.Shah, K., Jacobs, A., Breakefield, X. O. & Weissleder, R. (2004) Gene Ther. 11, 1175-1187. [DOI] [PubMed] [Google Scholar]
- 5.Gambhir, S. S. (2002) Nat. Rev. Cancer 2, 683-693. [DOI] [PubMed] [Google Scholar]
- 6.Blasberg, R. G. (2003) Mol. Cancer Ther. 2, 335-343. [PubMed] [Google Scholar]
- 7.Blasberg, R. G. & Tjuvajev, J. G. (2003) J. Clin. Invest. 111, 1620-1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hildebrandt, I. J. & Gambhir, S. S. (2004) Clin. Immunol. 111, 210-224. [DOI] [PubMed] [Google Scholar]
- 9.Dubey, P., Su, H., Adonai, N., Du, S., Rosato, A., Braun, J., Gambhir, S. S. & Witte, O. N. (2003) Proc. Natl. Acad. Sci. USA 100, 1232-1237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Koehne, G., Doubrovin, M., Doubrovina, E., Zanzonico, P., Gallardo, H. F., Ivanova, A., Balatoni, J., Teruya-Feldstein, J., Heller, G., May, C., et al. (2003) Nat. Biotechnol. 21, 405-413. [DOI] [PubMed] [Google Scholar]
- 11.Ray, P., De, A., Min, J. J., Tsien, R. Y. & Gambhir, S. S. (2004) Cancer Res. 64, 1323-1330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fefer, A., McCoy, J. L. & Glynn, J. P. (1967) Cancer Res. 27, 1626-1631. [PubMed] [Google Scholar]
- 13.Gorczynski, R. M. & Knight, R. A. (1975) Eur. J. Immunol. 5, 148-155. [DOI] [PubMed] [Google Scholar]
- 14.Kim, Y. J., Dubey, P., Ray, P., Gambhir, S. S. & Witte, O. N. (2004) Mol. Imaging Biol. 6, 331-340. [DOI] [PubMed] [Google Scholar]
- 15.Lois, C., Hong, E. J., Pease, S., Brown, E. J. & Baltimore, D. (2002) Science 295, 868-872. [DOI] [PubMed] [Google Scholar]
- 16.Bhaumik, S. & Gambhir, S. S. (2002) Proc. Natl. Acad. Sci. USA 99, 377-382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Uchida, N., Jerabek, L. & Weissman, I. L. (1996) Exp. Hematol. 24, 649-659. [PubMed] [Google Scholar]
- 18.Gambhir, S. S., Bauer, E., Black, M. E., Liang, Q., Kokoris, M. S., Barrio, J. R., Iyer, M., Namavari, M., Phelps, M. E. & Herschman, H. R. (2000) Proc. Natl. Acad. Sci. USA 97, 2785-2790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Qi, J., Leahy, R. M., Cherry, S. R., Chatziioannou, A. & Farquhar, T. H. (1998) Phys. Med. Biol. 43, 1001-1013. [DOI] [PubMed] [Google Scholar]
- 20.Schepers, K., Toebes, M., Sotthewes, G., Vyth-Dreese, F. A., Dellemijn, T. A., Melief, C. J., Ossendorp, F. & Schumacher, T. N. (2002) J. Immunol. 169, 3191-3199. [DOI] [PubMed] [Google Scholar]
- 21.McKay, L. I. & Cidlowski, J. A. (1999) Endocr. Rev. 20, 435-459. [DOI] [PubMed] [Google Scholar]
- 22.Su, H., Forbes, A., Gambhir, S. S. & Braun, J. (2004) Mol. Imaging Biol. 6, 139-148. [DOI] [PubMed] [Google Scholar]
- 23.Ahmed, N., Kansara, M. & Berridge, M. V. (1997) Biochem. J. 327, 369-375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frauwirth, K. A., Riley, J. L., Harris, M. H., Parry, R. V., Rathmell, J. C., Plas, D. R., Elstrom, R. L., June, C. H. & Thompson, C. B. (2002) Immunity 16, 769-777. [DOI] [PubMed] [Google Scholar]
- 25.Mamede, M., Saga, T., Ishimori, T., Nakamoto, Y., Sato, N., Higashi, T., Mukai, T., Kobayashi, H. & Konishi, J. (2003) Neoplasia 5, 179-183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kubota, R., Yamada, S., Kubota, K., Ishiwata, K., Tamahashi, N. & Ido, T. (1992) J. Nucl. Med. 33, 1972-1980. [PubMed] [Google Scholar]
- 27.Iyengar, S., Chin, B., Margolick, J. B., Sabundayo, B. P. & Schwartz, D. H. (2003) Lancet 362, 945-950. [DOI] [PubMed] [Google Scholar]
- 28.Pichler, B. J., Kneilling, M., Haubner, R., Braumuller, H., Schwaiger, M., Rocken, M. & Weber, W. A. (2005) J. Nucl. Med. 46, 184-189. [PubMed] [Google Scholar]
- 29.Signore, A., Procaccini, E., Annovazzi, A., Chianelli, M., van der Laken, C. & Mire-Sluis, A. (2000) Cytokine 12, 1445-1454. [DOI] [PubMed] [Google Scholar]
- 30.Venneti, S., Lopresti, B. J., Wang, G., Bissel, S. J., Mathis, C. A., Meltzer, C. C., Boada, F., Capuano, S., 3rd, Kress, G. J., Davis, D. K., et al. (2004) J. Clin. Invest. 113, 981-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.