Abstract
Coronary artery disease (CAD) and its most important complication, myocardial infarction (MI), are the leading cause of premature death in the Western world. CAD has a substantial genetic basis, especially when it occurs early. We investigated the genetic determinants of premature CAD by performing a genomewide linkage analysis of 4,175 affected subjects from 1,933 families recruited throughout the United Kingdom. Each family had at least two available siblings with CAD, with validated onset before age 66 years. Linkage analysis was performed using 416 microsatellite markers. We observed suggestive linkage, for both CAD and MI, to a region on chromosome 2. For CAD, a LOD score of 1.86 was observed at marker D2S2271, which, in an ordered subset analysis, increased to 2.70 in families (n=1,698) with a minimum age at diagnosis of 56 years or younger. For MI, an overlapping peak with a LOD score of 1.15 was observed at marker D2S2216, which increased to 2.1 in families (n=801) with a minimum age at diagnosis of 59 years or younger. Exclusion mapping showed that 100% of the autosomal genome could be excluded for locus-specific sibling relative risks of 1.5 and 1.6 for CAD and MI, respectively. The region identified on chromosome 2 overlaps linked regions observed in two other smaller genome scans for CAD. Together, these findings strongly suggest that there is a locus on chromosome 2 that influences coronary atherosclerosis risk. The exclusion of a common locus that increases risk of CAD to siblings by >50% has important implications for strategies for further defining the genetic basis of CAD.
Introduction
Atherosclerotic coronary artery disease (CAD) and its most important complication, acute myocardial infarction (MI), are the most common cause of premature death and morbidity in the Western world. In addition to well-recognized lifestyle risk factors, genetic factors play a key role in the pathogenesis of CAD, particularly early-onset CAD. First-degree relatives of patients who have had an acute MI prior to age 55 years have 2–7 times the risk that their peers have (Lusis et al. 2004). Furthermore, twin studies indicate an eightfold increase in risk of death from an MI if a first twin dies of an MI before age 55 years (Marenberg et al. 1994).
With use of a case-control approach, variants in many candidate genes have been investigated for their effects on risks of CAD and MI. Polymorphisms in genes encoding proteins involved in lipid metabolism, thrombosis, and vascular biology, among others, have been associated with increased risk of CAD and/or MI in certain populations (Topol et al. 2001; Ozaki et al. 2002, 2004; Yamada et al. 2002; Lusis et al. 2004). However, many of the findings made with this approach require independent verification and identification of the functional basis of the association. At the same time, a number of genomewide linkage scans—mainly of affected sib pairs—to map CAD and/or MI loci have also been attempted (Pajukanta et al. 2000; Francke et al. 2001; Broeckel et al. 2002; Harrap et al. 2002; Hauser et al. 2004a; Helgadottir et al. 2004; Wang et al. 2004). These have suggested the presence of such loci on chromosomes 1, 2, 3, 12, 13, 14, 16, and X. However, in only one case (chromosome 13) has the putative underlying gene yet been identified (Helgadottir et al. 2004).
Taken together, the findings to date provide grounds for belief that further analysis of the genetic basis of CAD may provide novel insights into its pathogenesis, improve risk stratification of individuals, identify novel targets for drug development, and influence treatment strategies. The BHF Family Heart Study was conceived primarily to provide a major resource of affected sib pairs for mapping genes involved in the pathogenesis of premature CAD through a linkage-based approach. The 1,933 families analyzed represent one of the largest collections yet assembled for this purpose. Because of the possibility that some loci may confer risk only for MI rather than for CAD overall (Lusis et al. 2004; Wang et al. 2004) and that some may manifest their effect only at a younger age, we prospectively specified additional analyses in relation to these groups. Here, we report the BHF Family Heart Study results for these main analyses of the initial genome scan.
Subjects and Methods
Recruitment
The objective of the BHF Family Heart Study was to establish a substantial resource of families with at least two siblings affected with premature CAD, defined as “the occurrence of MI or angina (verified by exercise stress test or angiography) or the need for coronary artery bypass surgery (CABG) or percutaneous transluminal coronary angioplasty (PTCA) before the 66th birthday.” Recruitment was performed on a national basis in the United Kingdom (UK) through (1) a direct appeal to the public via the media and (2) mailing all general practices (family physicians) in the UK with information about the study. In an initial pilot phase, potential participants were also identified and approached through local CAD databases at the two lead centers (Leeds and Leicester). Interested subjects could register in the study either through a Freephone or Freepost basis. An attempt was made to include all available affected siblings of participating families. Unaffected siblings were also recruited, if available, but are not included in the present analysis. In total, we collected 6,285 subjects. After validation, 4,216 individuals from 1,958 families were genotyped, and the final analysis included 4,175 affected subjects from 1,933 families (see the “Results” section). The study was approved by the Yorkshire Multicentre Research Ethics Committee and by 206 local research ethics committees across the UK. Subjects provided written informed consent, and the study was performed under the principles of the Declaration of Helsinki and complied with UK legislation for data protection.
Data Collection, Validation, and Handling
After a preliminary screen to establish suitability, participating members of families that met the criteria for the study were interviewed by telephone by a trained nurse using a standardized questionnaire. Information obtained included personal and family history of CAD, including the nature and time of first and any subsequent events, and information about risk-factor profile. Arrangements were made, usually via the general practitioner (GP), to obtain a blood sample for DNA. For each subject, independent verification of history of CAD was obtained either via direct examination of hospital records or via the GP; only subjects for whom such verification could be obtained were included in the study. All clinical data were stored on a Microsoft SQL server relational database specifically designed for the study. Each subject was given a unique identity number and only anonymized clinical data were merged with the genotype data for analysis.
DNA Extraction and Genotyping
Leukocyte DNA was extracted from 18 ml of blood collected in EDTA with use of the PURGENE_DNA Extraction Kits (Gentra Systems) and was quantified by spectrophotometry. In the first phase of the genome scan, 398 of 400 microsatellite markers from the ABI-Prism Linkage Mapping set v2.5-MD10 (Applied Biosystems) spaced at ∼10 cM were typed. For technical reasons, two markers from the set, D14S68 and D6S434, were not typed. PCR was performed, using 10 ng of DNA per reaction, under conditions described by the manufacturers, and the products were pooled in panels and were analyzed on ABI-Prism 3700 DNA sequencers. Genotypes from each marker were examined by the GeneMapper software v 2.0 (Applied Biosystems). In the second phase, a further 18 markers (D1S2850, D3S3609, D3S3706, D5S1960, D6S282, D6S1671, D8S543, D8S1705, D11S4190, D12S313, D12S1725, D14S1044, D17SS927, D18S465, D18S1127, DXS1055, DXS1061, and DXS1062) were genotyped, to close any gaps >19 cM. These markers were selected from ABI-Prism Linkage Mapping set v2.5-HD5. Finally, an additional eight markers (D2S139, D2S2264, D2S293, D2S2329, D2S2271, D2S2215, D2S1326, and D2S2241) were typed, to investigate and refine the linkage region observed on chromosome 2.
Quality Control of Genotype Data
All genotypes, including those that passed GeneMapper’s internal quality control, were manually read by at least one individual, and those genotypes that did not fully pass the program’s quality control were read independently by two individuals. Genotypes were rejected and the samples rerun if there was not complete agreement on allele calls. The Graphical Representation of Relationship (Abecasis et al. 2001) and RELATIVE (Goring and Ott 1997) programs were used to confirm family relationships. Genotypes inconsistent with Mendelian laws were detected using the PedCheck program (O’Connell and Weeks 1998). Those genotypes were reexamined, were corrected if obviously incorrect, or were deleted. Absence of parental data meant that such errors were detectable only in larger sibships, so unlikely genotypes were also identified using Merlin (Abecasis et al. 2002). When these “unlikely” genotypes were reexamined, clear errors were corrected and uncertain genotypes were deleted, but genotypes that appeared correct on reexamination were left unchanged. The mean proportion of alleles per marker taken to analysis was 96.0% (a range of 90.2%–99.4%), and the mean information content across all loci assessed in the genome scan was 64% (38%–84%).
Linkage Analysis
Recombination fractions between markers were estimated from CEPH (Dib et al. 1996) and deCode (Kong et al. 2002) genotype data by use of the CRI-MAP program (Lander and Green 1987) and were implemented in MAP-O-MAT (Kong et al. 2004; Kong and Matise 2005), under licensed agreement with deCODE. Maximum-likelihood estimates of marker-allele frequencies were obtained using MENDEL v5.5 (Lange et al. 2001). This method uses genotype data from all individuals and takes their genetic relationships into account. Genomewide nonparametric multipoint linkage analysis was performed using the Allegro program (Gudbjartsson et al. 2000). LOD scores were calculated on the basis of the exponential model of Kong and Cox (1997). This LOD score was chosen because, in cases for which the inheritance information is incomplete, it does not have the conservative properties of the nonparametric linkage (NPL) score implemented in Genehunter (Kruglyak et al. 1996). The SALL statistic (Whittemore and Halpern 1994), which captures the identity-by-descent–sharing information in a family, was used to calculate the LOD score. SALL considers the family as a whole and gives extra weight if the same allele is shared between different pairs of relatives.
For the CAD phenotype, all families were included in the analysis. For the MI phenotype, analysis was restricted to those families with at least two individuals with a confirmed MI.
Reported P values are estimates of pointwise significance that are based on the theoretical distribution of the statistic, provided by Allegro (Kong et al. 2004). In addition, genomewide significance was assessed by simulating data under the null hypothesis of no linkage with the observed pedigree structure and marker characteristics (including the pattern of missing genotype data). One thousand such data sets were simulated using the program Merlin (Abecasis et al. 2002) and then were analyzed using the Kong and Cox (1997) LOD score. Genomewide significance was estimated by the proportion of times that a particular LOD score was reached anywhere in the genome.
Analysis for Age Effect
Ordered subset linkage analysis (OSA) (Hauser et al. 2004b) was employed for consideration of age at diagnosis. The OSA groups families into subsets on the basis of the value of a covariate—in this case, the minimum age at diagnosis in each family. In each subset, all families in which the minimum age is below a certain value are included. Therefore, as the age increases, the size of the subset also increases. Linkage analysis was performed for each subset, and the maximum LOD score on each chromosome across all subsets was reported. OSA has been used elsewhere as a test for the effect of a covariate (such as age at onset) on linkage, conditional on overall linkage (Hauser et al. 2004b). Our aim was to use this method differently, to test for linkage, possibly confined to a subset defined by young age at onset, against the null hypothesis of no linkage. For this reason, we assessed the chromosomewide significance of our findings by conducting simulations in which we examined how often observed LOD scores were exceeded anywhere on the same chromosome in any subset defined by ordering on the covariate. In each simulation, the pedigree structures and ages at onset were fixed, but genotypes were assigned according to the null hypothesis of no linkage (so there was no conditioning on the observed linkage signal).
Exclusion Mapping
In addition to the linkage analysis, exclusion mapping was performed for a number of different values of λs (the sibling relative risk) and an exclusion criterion of LOD<-2, to exclude genomic regions unlikely to harbor loci contributing to various levels of CAD or MI risk. This was performed for all 22 autosome pairs, with use of GENEHUNTER (Kruglyak et al. 1996), with use of one sibling pair from each family.
Results
Subjects
In total, we collected 6,285 subjects. After validation, 4,216 individuals from 1,958 families were genotyped. We identified 100 half-sibling pairs, 16 identical pairs (MZ twins or sample duplications), 11 individuals with incorrectly specified sex, and 16 families with unrelated pairs of individuals, presumably because of sample swaps or adoption. Half siblings in 93 sibships were included in the analysis after correct specification of the relationship. Other relationships were corrected when possible; otherwise, the individuals were removed from subsequent analyses. In the final analysis, we included 4,175 affected subjects from 1,933 families: 1,675 affected sibling pairs, 220 affected trios, and 38 sibships or extended families with more than three affected individuals.
The demographics of the families and individuals analyzed are shown in table 1. There was a 3:1 ratio of males:females. The mean age (±SD) at the first validated evidence of CAD was 51.7 (7.7) years for men and 54.1 (7.3) years for women (P<.001). The mean age at time of the study was 61.0 (7.0) years. Of the subjects, 2,556 (61.2%) had suffered an MI. A majority (60.5%) had also had a revascularization procedure. As expected, the major coronary risk factors were commonly present, and the majority of subjects had multiple risk factors (table 1). A majority (60%) of the subjects had a family history (one or both parents) of premature CAD. On the basis of self-reported ethnicity, 98.5% of subjects were of European origin.
Table 1.
BHF Family Heart Study Population Characteristics
|
Findings for Subjects with |
||
| Characteristic | CAD Phenotype | MI Subphenotype |
| No. of study families | 1,933 | 847 |
| No. of affected individuals | 4,175 | 1,776 |
| Percentage of affected males/females | 73.8/26.2 | 77.2/22.8 |
| Mean age, in years (SD), at onset for affected males | 51.7 (7.7) | 50.7 (7.9) |
| Mean age, in years (SD), at onset for affected females | 54.1 (7.4) | 53.4 (7.8) |
| Mean age, in years (SD), at exam for affected individuals | 61.0 (7.4) | 61.6 (7.8) |
| Percentages of subjects, by ethnicity (European/Asian/other) | 98.5/1.4 /.1 | 98.6/1.2/.1 |
| No. (%) of validated and analyzed families, by number of affected siblings: | ||
| Two | 1,675 (86.5) | 772 (91.1) |
| Three | 220 (11.4) | 69 (8.1) |
| More than three | 38 (2.0) | 6 (.8) |
| No. (%) of subjects affected at age <66 years: | ||
| With MI with revascularization (percutaneous coronary intervention/CABG) | 1,365 (32.7) | 937 (52.8) |
| With MI without revascularization | 1,191 (28.5) | 839 (47.2) |
| No. (%) of subjects affected by CAD but not MI at age <66 years: | ||
| CAD with revascularization (percutaneous coronary intervention/CABG) | 1,161 (27.8) | … |
| CAD (angiography or exercise stress test) without revascularization | 458 (11.0) | … |
| No. (%) of subjects with parental history of CAD at age <66 years: | ||
| Neither parent affected | 774 (40.1) | 343 (40.4) |
| Father affected | 617 (31.9) | 260 (30.7) |
| Mother affected | 320 (16.6) | 146 (17.2) |
| Both affected | 221 (11.4) | 98 (11.6) |
| No. (%) of subjects with cardiovascular risk factors: | ||
| Smoking history | 3,051 (73.1) | 1,355 (76.3) |
| Diabetes mellitus (receiving treatment) | 490 (11.7) | 151 (11.8) |
| Hypertension (receiving treatment) | 1,924 (46.1) | 789 (44.4) |
| Hyperlipidemia (>5.0 mmol/liter or receiving treatment) | 3,384 (81.1) | 1,415 (79.7) |
| Obesity (BMI >25 kg/m2) | 3,018 (72.3) | 1,283 (72.2) |
Results of Genome Scans for CAD and MI
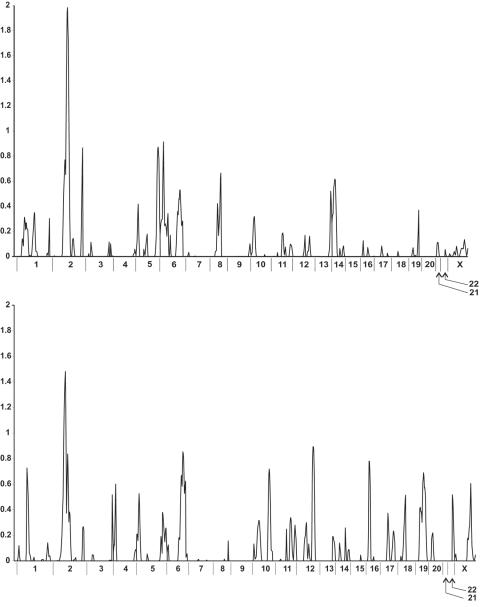
The results of the multipoint 10-cM genome scan for the whole population with CAD (n=1,933 families) and for the families with only the MI phenotype (n=847) are summarized in figure 1. For each phenotype, we found a single LOD score >1.0—on chromosome 2 (see below). Other loci with LOD scores of 0.5–1.0 were observed for the CAD phenotype on chromosomes 2, 5, 6, 8, and 14 and for the MI phenotype on chromosomes 1, 2, 4, 5, 6, 10, 12, 16, 18, 19, and X (fig. 1 and table 2). None of the results were statistically significant at the genomewide level.
Figure 1.
Summary of results of linkage analysis for CAD (upper panel) and MI (lower panel). The peaks and corresponding LOD scores obtained for each chromosome from the multipoint linkage analysis of the initial genome scan (n=398 markers) are shown.
Table 2.
Summary of the LOD Scores >.5 for CAD and MI[Note]
| Phenotype Chromosome,and Position(cM) | Nearest Marker(s) | LOD | Pointwise P |
| CADa: | |||
| 2: | |||
| 149 | D2S347–D2S112 | 1.9819 | .001449 |
| 274.5 | D2S338–D2S125 | .7275 | .03886 |
| 5: | |||
| 207.6 | D5S400–D5S1960 | .8709 | .0278 |
| 6: | |||
| 22.2 | D6S289 | .9125 | .025396 |
| 182.8 | D6S1581–D6S264 | .5304 | .0636 |
| 8: | |||
| 124.8 | D8S1784 | .6639 | .0455 |
| 14: | |||
| 70.0 | D14S63–D14S258 | .6159 | .05125 |
| 36.5 | D14S70 | .5194 | .0655 |
| MI: | |||
| 1: | |||
| 118.8 | D1S2841 | .7266 | .033095 |
| 2: | |||
| 119.3 | D2S2216 | 1.4814 | .0046 |
| 4: | |||
| 0 | D4S412 | .5167 | .0602 |
| 40.6 | D4S419 | .6002 | .046941 |
| 5: | |||
| 27.4 | D5S630 | .5254 | .058574 |
| 6: | |||
| 182.8 | D6S1581–D6S264 | .8529 | .023688 |
| 10: | |||
| 174.7 | D10S587–D10S217 | .7187 | .0338 |
| 12: | |||
| 157.0 | D12S86 | .8932 | .02136 |
| 16: | |||
| 40.3 | D16S3068 | .7794 | .0287 |
| 18: | |||
| 93.8 | D18S64 | .514 | .0606 |
| 19: | |||
| 89.6 | D19S571–D19S418 | .6889 | .0366 |
| X: | |||
| 0 | DXS106 | .5171 | .0613 |
| 155.3 | DXS1062 | .6049 | .0475 |
Note.— For each region, the position of the maximum LOD score is listed. The position is the distance from the most telomeric marker genotyped (set to 0 cM). The pointwise P values are derived from the theoretical distribution of the LOD score under the null hypothesis.
Includes MI.
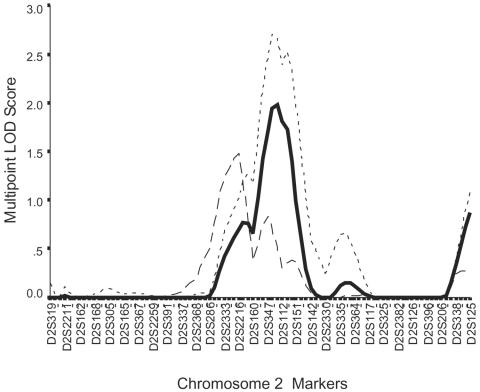
Chromosome 2
Figure 2 shows in detail the results for the main peaks on chromosome 2. The peak LOD score for the CAD phenotype was between neighboring markers D2S347 and D2S112, with a LOD score of 1.98 (pointwise P=.0014). In simulation studies, a maximum LOD score of ⩾1.98 was found in 310/1,000 analyses of data simulated under the null hypothesis of no linkage, for a genomewide P value of .31. With inclusion of the extra markers, the LOD fell slightly to 1.86 (pointwise P=.0021) at marker D2S2271, which lies between the two previous flanking markers. For MI, the peak multipoint LOD score was at marker D2S2216, with a LOD score of 1.48 (pointwise P=.0046) (fig. 2), falling to 1.15 (pointwise P=.011) with the addition of the extra markers.
Figure 2.
More-detailed representation of the linkages observed on chromosome 2 from the initial genome scan. The solid line represents the main CAD phenotype; the long-dashed line represents the MI subphenotype; the short-dashed line represent the CAD phenotype for families with minimum age at onset of <56 years.
Effect of Age at Onset
The OSA for CAD had a LOD score of 2.70 on chromosome 2 at the same location as in the overall analysis of families with a minimum age at diagnosis of ⩽56 years (fig. 2). There were 1,698 families in this subgroup. For MI, a peak LOD score of 2.1 was found, again at marker D2S2216, in 801 families with a minimum age at diagnosis of ⩽59 years.
Other loci identified from OSA for CAD included a LOD of 3.42 between flanking markers D9S175 and D9S167 on chromosome 9. However, the subset was small, with only 16 families with at least one individual with an age at diagnosis of ⩽30 years. A LOD score of 3.24 was also seen between D17S787 and D17S944 on chromosome 17. Again, this was in a small subset of 26 families with at least one individual with an age at diagnosis of ⩽31 years. OSA also identified (1) a locus for MI, with a LOD score of 2.4 between markers D5S630 and D5S416 on chromosome 5, in a subset of 205 families with at least one individual with an age at diagnosis of ⩽42 years and (2) a locus for MI on the X chromosome with a LOD score of 2.4 between markers DXS991 and DXS986 in a subset of 232 families with at least one individual with an age at diagnosis of ⩽43 years. Chromosomewide P values were estimated to be .08 for chromosome 2, .006 for chromosome 9, and .01 for chromosome 17. The results for chromosomes 9 and 17 are thus significant at the chromosomewide but not genomewide level.
Exclusion Mapping
The results of exclusion mapping for CAD and MI (the full length of the map used was 3,747.4 cM) for this population showed that, for CAD, 100% of the autosomal genome could be excluded for a locus-specific λs of 1.5 and, for MI, because of the smaller sample size, 100% of the autosomal genome could be excluded only for a λs of 1.6. For CAD, the entire genome, apart from a region surrounding the peak on chromosome 2, could be excluded, for a λs of 1.4; this represents 99.6% of the autosomal genome. For a λs of 1.3, 98.8% of the genome was excluded, dropping to 95.6% for a λs of 1.25. Although 86.1% of the genome is excluded for a λs of 1.2, there remain regions on most chromosomes that could harbor disease genes with this size of effect.
Discussion
Identification of genetic factors that predispose to CAD and understanding their interactions with environmental risk factors has the potential to identify of novel therapeutic targets, to improve risk stratification of individuals, and to allow better tailoring of preventive and treatment options. In this study, we report the largest genome scan to date, investigating the genetic basis of CAD and MI in a cohort of subjects with a risk-factor profile typical of patients with premature CAD (Yusuf et al. 2004). Our results are consistent with the complex genetic etiology of CAD and suggest that there is either no single locus or a small number of major loci that explain the familial risk of these diseases.
Our most interesting finding was on chromosome 2, where we observed suggestive linkage for both CAD and MI. Several observations suggest that these observed linkages are informative, despite LOD scores that do not reach genomewide significance. Both CAD and MI mapped to the same site, and the peaks persisted, with slightly reduced LOD scores, when additional markers in the region were genotyped. The fact that the peak locations of the linkages for CAD and MI were slightly displaced from each other should not be considered an indication of the presence of separate loci for the two conditions. Simulation studies have shown that there is considerable imprecision in the location of peaks in linkage studies, and it is more likely that this explains the lack of coincidence (Roberts et al. 1999). Furthermore, the peaks for both CAD and MI (fig. 2) increased when analysis was restricted to families with earlier onset of disease, consistent with the observation that the genetic contribution to CAD and MI is likely to be more important in early disease. Importantly, there is concordance of our findings on chromosome 2 with two other genome scans related to atherosclerotic coronary disease. Wang et al. (2004), in a study of 928 families, reported a LOD score of 3.82 at the same locus in a U.S. cohort characterized by familial premature CAD and MI, although that was not their major peak. More strikingly, the linkage observed with CAD to 2q21.1-22 in a genetically isolated Finnish population of 156 families (Pajukanta et al. 2000) is very close to our region. Interestingly, a genomewide linkage study conducted by Fox et al. (2004) to localize QTLs influencing carotid intima-media thickness, a marker of atherosclerosis, revealed a LOD score of 1.57 at marker D2S1790 (103 cM). These findings, taken together, strongly suggest that there is a locus influencing coronary atherosclerosis risk in this region of chromosome 2. However, it should be noted that our main locus of interest does not overlap with that of another genome scan that reported suggestive linkage to chromosome 2 for acute coronary syndrome (Harrap et al. 2002).
Genome scans of the type undertaken here have also reported significant linkages to CAD and/or MI for chromosomes 1p34-36 (Wang et al. 2004), 1q25 (Hauser et al. 2004a), 3q (Hauser et al. 2004a), 14q (Broeckel et al. 2002), and 16p13-pter (Francke et al. 2001). Despite the much larger size of our study, we have not replicated those results. There are several potential explanations. First, it is notable that the different scans published elsewhere also vary in the loci identified, with little replication between them. Although the linkages described reach genomewide significance, this should be interpreted with caution, since some could still be false-positive results. However, there could also be genuine differences between populations. Although the majority of our subjects were of European origin, it is conceivable that they represent a more heterogeneous group than did subjects of some of the other studies. Differences in classification of endpoints or genotyping approaches seem unlikely sources of discrepancy. Our definitions of CAD and MI were standard and were verified in all subjects by direct assessment of patient records. Genotyping was performed to a high degree of completeness, and several checks were employed (see the “Subjects and Methods” section) to ensure accuracy. There are some variations in the precise statistical tests used. We used the extension to the NPL statistic that was proposed by Kong and Cox (1997), which tests for excess sharing of alleles among the set of affected relatives within a family. This test has been shown to compare well with other methods, which can be biased toward the null hypothesis (Cordell 2004).
It was somewhat surprising that we found neither distinct nor stronger linkages to MI, given the more precise definition of this phenotype (Samani 1998) and the findings of some previous studies, in which linkages were observed only for MI and not for CAD (Wang et al. 2004). This is despite the fact that even with the reduced number of families for the MI phenotype, our study is still one of the largest that has investigated this phenotype. Although the presence of coronary atherosclerosis provides the main substrate for the development of MI, other factors, including thrombotic potential, influence the risk of MI, and it is not entirely unexpected that specific loci could influence the risk of each phenotype. That said, it is interesting to note that both CAD and MI mapped to the same locus in our study, which suggests that, if a common gene is involved, it affects pathways that influence the risk of both phenotypes.
The genetic influence on CAD is more apparent the earlier the disease onset, although the relationship is not necessarily linear and may vary from locus to locus. Several approaches can be used to look for genetic loci that influence the risk of more-premature disease. In this study, we used the OSA method recently described by Hauser et al. (2004b). The approach is valuable in that it does not define any arbitrary “age cutoff” to look for an effect; instead, it maximizes the use of data and allows modeling of age effect for each locus. Our analysis strengthened the findings on chromosome 2 and suggests that this locus especially predisposes to disease in subjects aged <56 years. However, it should be noted that the peak LOD score of 2.70 in this analysis also did not reach genomewide significance in simulation studies. OSA also identified a number of other potential loci. Of particular interest is the observation of a locus on chromosome X linked to MI where another study has also reported linkage (Pajukanta et al. 2000). The findings on chromosomes 9 and 17, for which the results relate to only a small number of families, need to be treated with appropriate caution. However, the recent report (Wang et al. 2003) of a possibly monogenic form of early-onset CAD does indicate that some families may harbor mutations of this type. Although an initial examination of these families does not identify any obvious distinguishing clinical features, a more detailed evaluation of the linkages in these families would seem worthwhile.
Our findings raise several issues about the genomewide-linkage approach to common complex diseases such as CAD. The presence of genetic heterogeneity is clearly an important limitation and the most likely explanation for our failure to find significant linkages despite the comparatively large size of our study. Our analyses of subjects with MI and the use of age as a criterion were attempts to address this issue. With use of even more narrowly defined phenotypes, less genetically heterogeneous groups could be identified, but any choice should be based on strong biological plausibility. For CAD, other risk factors and variables, such as smoking history, dyslipidemia, and BMI, could be used to try to define more-homogeneous groups and will be the subject of further analysis of our data.
An immediate challenge posed by our findings is determination of the best approach for further interrogation of the locus on chromosome 2. The locus, encompassing both the MI and CAD peaks, spans ∼80 Mb and contains 446 putative genes, many of which are poorly characterized. However, there are several attractive potential candidate genes with cardiovascular effects. These include, for example, the interleukins 1A and 1B clusters and protein C. Interleukin 1B polymorphisms have recently been found to associate with risk of premature MI (Iacoviello et al. 2005). Apart from analysis of obvious candidates, the systematic typing of SNPs at regular intervals could narrow the region of interest. This approach led to the identification of the 5′-lipoxygenase activating protein (FLAP) as the likely gene on chromosome 13 that confers risk of MI and stroke (Helgadottir et al. 2004). Of note is that the peak LOD score of 2.48 observed in the initial genome scan in the study by Helgadottir et al. (2004) was also only suggestive of linkage.
Genome scans with polymorphic markers in affected families have the inherent advantage of allowing a nonbiased search for disease-causing loci. However, they have lower power than case-control association studies. Until recently, genome scans provided a complementary approach to candidate-gene association studies. The increasing identification of SNPs and availability of high-throughput SNP-typing platforms has opened the possibility of undertaking genomewide association studies (Hinds et al. 2005), which makes the previous advantage of genomewide linkage scans less certain. However, even for such association studies, the key issues of accurate phenotype and genetic risk remain. In this regard, the BHF Family Heart Study has significant advantages, not only because of its size but also because of its selection of subjects enriched for genetic risk of CAD.
In summary, we describe the assembly of a large resource of families with premature CAD for identification of genetic factors that predispose to this major disease. We report the results of our initial genome scan and mapping of a locus for both CAD and MI, with suggestive linkage to chromosome 2p12-2q23.3.
Acknowledgments
The Family Heart Study was supported by British Heart Foundation program grant RG2000010. We are grateful to Sainsbury UK Ltd. for sponsoring a bus to advertise the study, to the staff in hospitals and general practices throughout the UK for helping to collect DNA samples and provide records to validate the history of participants, and to the BHF regional volunteers for supporting the project. Finally, and most importantly, we thank all the families and subjects who participated in the study.
Members of the BHF Family Heart Study Research Group are as follows: Writing Group—Leicester: Nilesh J. Samani, Paul Burton, and Massimo Mangino; Leeds: Stephen G. Ball, Anthony J. Balmforth, Jenny Barrett, Timothy Bishop, and Alistair Hall. Collection Group—Leicester: N. J. Samani (lead clinician), Julian Stribling, and Pat de Souza; coordinators Ravi Singh, Jenny Ogleby, Cathy Ridge, Elaine Logtens, Laura Hopwood, and Julie Faulkes; Leeds: A. S. Hall (lead clinician), Christine Morrell, and Beryl M. Jackson; coordinators Lynne Barthorpe, Natalie Burtonwood, Micha Dorsch, Nigel Durham, Claire Forest, Natasha Kelly, Vera Hall, Richard Lawrance, Julia Oldham, Elizabeth Rennie, Adrian Smith, and Samantha Thompson. Genotyping Group—A. J. Balmforth (coordinator); Leicester: M. Mangino and Sue Adams (laboratory leads), Peter Braund, Jenny-Rebecca Clemitson, Claire Bodycote, Andrea Koekemoer, and Stuart Raleigh; Leeds: Azhar Maqbool (laboratory lead), Nadira Yuldasheva, Stacey Ellis, Samantha Mason, Lynne Midgley, Natalie Pleasants, and Richard Cuthbert. Data Handling Group—Peter F. Tooze (lead data manager), P. Braund, M. Mangino, Mark Platts, Joanne Fox, and Rick Dixon. Statistics Group—J. Barrett (lead statistician); Leicester: P. Burton, M. Mangino, Nuala Sheehan, and Katrina Scurrah; Leeds: T. Bishop, Sarah Pickett, Kevin Walters, and Jeremie Nsengimana. DNA/Data Guardians—A. S. Hall (coordinator), N. J. Samani, A. J. Balmforth, P. Burton, T. Bishop, and S. G. Ball.
Web Resources
The URL for data presented herein is as follows:
- Applied Biosystems, http://www.appliedbiosystems.com/
References
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR (2001) GRR: graphical representation of relationship errors. Bioinformatics 17:742–743 10.1093/bioinformatics/17.8.742 [DOI] [PubMed] [Google Scholar]
- ——— (2002) Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet 30:97–101 10.1038/ng786 [DOI] [PubMed] [Google Scholar]
- Broeckel U, Hengstenberg C, Mayer B, Holmer S, Martin LJ, Comuzzie AG, Blangero J, Nurnberg P, Reis A, Riegger GA, Jacob HJ, Schunkert H (2002) A comprehensive linkage analysis for myocardial infarction and its related risk factors. Nat Genet 30:210–214 10.1038/ng827 [DOI] [PubMed] [Google Scholar]
- Cordell HJ (2004) Bias toward the null hypothesis in model-free linkage analysis is highly dependent on the test statistic used. Am J Hum Genet 74:1294–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, Marc S, Hazan J, Seboun E, Lathrop M, Gyapay G, Morissette J, Weissenbach J (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 10.1038/380152a0 [DOI] [PubMed] [Google Scholar]
- Fox CS, Cupples LA, Chazaro I, Polak JF, Wolf PA, D’Agostino RB, Ordovas JM, O’Donnell CJ (2004) Genomewide linkage analysis for internal carotid artery intimal medial thickness: evidence for linkage to chromosome 12. Am J Hum Genet 74:253–261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francke S, Manraj M, Lacquemant C, Lecoeur C, Lepretre F, Passa P, Hebe A, Corset L, Yan SL, Lahmidi S, Jankee S, Gunness TK, Ramjuttun US, Balgobin V, Dina C, Froguel P (2001) A genome-wide scan for coronary heart disease suggests in Indo-Mauritians a susceptibility locus on chromosome 16p13 and replicates linkage with the metabolic syndrome on 3q27. Hum Mol Genet 10:2751–2765 10.1093/hmg/10.24.2751 [DOI] [PubMed] [Google Scholar]
- Goring HH, Ott J (1997) Relationship estimation in affected sib pair analysis of late-onset diseases. Eur J Hum Genet 5:69–77 [PubMed] [Google Scholar]
- Gudbjartsson DF, Jonasson K, Frigge ML, Kong A (2000) Allegro, a new computer program for multipoint linkage analysis. Nat Genet 25:12–13 10.1038/75514 [DOI] [PubMed] [Google Scholar]
- Harrap SB, Zammit KS, Wong ZY, Williams FM, Bahlo M, Tonkin AM, Anderson ST (2002) Genome-wide linkage analysis of the acute coronary syndrome suggests a locus on chromosome 2. Arterioscler Thromb Vasc Biol 22:874–878 10.1161/01.ATV.0000016258.40568.F1 [DOI] [PubMed] [Google Scholar]
- Hauser ER, Crossman DC, Granger CB, Haines JL, Jones CJ, Mooser V, McAdam B, et al (2004a) A genomewide scan for early-onset coronary artery disease in 438 families: the GENECARD Study. Am J Hum Genet 75:436–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hauser ER, Watanabe RM, Duren WL, Bass MP, Langefeld CD, Boehnke M (2004b) Ordered subset analysis in genetic linkage mapping of complex traits. Genet Epidemiol 27:53–63 10.1002/gepi.20000 [DOI] [PubMed] [Google Scholar]
- Helgadottir A, Manolescu A, Thorleifsson G, Gretarsdottir S, Jonsdottir H, Thorsteinsdottir U, Samani NJ, et al (2004) The gene encoding 5-lipoxygenase activating protein confers risk of myocardial infarction and stroke. Nat Genet 36:233–239 10.1038/ng1311 [DOI] [PubMed] [Google Scholar]
- Hinds DA, Stuve LL, Nilsen GB, Halperin E, Eskin E, Ballinger DG, Frazer KA, Cox DR (2005) Whole-genome patterns of common DNA variation in three human populations. Science 307:1072–1079 10.1126/science.1105436 [DOI] [PubMed] [Google Scholar]
- Iacoviello L, Di Castelnuovo A, Gattone M, Pezzini A, Assanelli D, Lorenzet R, Del Zotto E, Colombo M, Napoleone E, Amore C, D’Orazio A, Padovani A, de Gaetano G, Giannuzzi P, Donati MB (2005) Polymorphisms of the interleukin-1beta gene affect the risk of myocardial infarction and ischemic stroke at young age and the response of mononuclear cells to stimulation in vitro. Arterioscler Thromb Vasc Biol 25:222–227 [DOI] [PubMed] [Google Scholar]
- Kong A, Cox NJ (1997) Allele-sharing models: LOD scores and accurate linkage tests. Am J Hum Genet 61:1179–1188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Gudbjartsson DF, Sainz J, Jonsdottir GM, Gudjonsson SA, Richardsson B, Sigurdardottir S, Barnard J, Hallbeck B, Masson G, Shlien A, Palsson ST, Frigge ML, Thorgeirsson TE, Gulcher JR, Stefansson K (2002) A high-resolution recombination map of the human genome. Nat Genet 31:241–247 [DOI] [PubMed] [Google Scholar]
- Kong X, Matise TC (2005) MAP-O-MAT: internet-based linkage mapping. Bioinformatics 21:557–559 10.1093/bioinformatics/bti024 [DOI] [PubMed] [Google Scholar]
- Kong X, Murphy K, Raj T, He C, White PS, Matise TC (2004) A combined linkage-physical map of the human genome. Am J Hum Genet 75:1143–1148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruglyak L, Daly MJ, Reeve-Daly MP, Lander ES (1996) Parametric and nonparametric linkage analysis: a unified multipoint approach. Am J Hum Genet 58:1347–1363 [PMC free article] [PubMed] [Google Scholar]
- Lander ES, Green P (1987) Construction of multilocus genetic linkage maps in humans. Proc Natl Acad Sci USA 84:2363–2367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange K, Cantor R, Horvath S, Perola M, Sabatti C, Sinsheimer J, Sobel E (2001) Mendel version 4.0: a complete package for the exact genetic analysis of discrete traits in pedigree and population data sets. Am J Hum Genet Suppl 69:A1886 [Google Scholar]
- Lusis AJ, Mar R, Pajukanta P (2004) Genetics of atherosclerosis. Annu Rev Genomics Hum Genet 5:189–218 10.1146/annurev.genom.5.061903.175930 [DOI] [PubMed] [Google Scholar]
- Marenberg ME, Risch N, Berkman LF, Floderus B, de Faire U (1994) Genetic susceptibility to death from coronary heart disease in a study of twins. N Engl J Med 330:1041–1046 10.1056/NEJM199404143301503 [DOI] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ozaki K, Inoue K, Sato H, Iida A, Ohnishi Y, Sekine A, Odashiro K, Nobuyoshi M, Hori M, Nakamura Y, Tanaka T (2004) Functional variation in LGALS2 confers risk of myocardial infarction and regulates lymphotoxin-α secretion in vitro. Nature 429:72–75 10.1038/nature02502 [DOI] [PubMed] [Google Scholar]
- Ozaki K, Ohnishi Y, Iida A, Sekine A, Yamada R, Tsunoda T, Sato H, Hori M, Nakamura Y, Tanaka T (2002) Functional SNPs in the lymphotoxin-α gene that are associated with susceptibility to myocardial infarction. Nat Genet 32:650–654 10.1038/ng1047 [DOI] [PubMed] [Google Scholar]
- Pajukanta P, Cargill M, Viitanen L, Nuotio I, Kareinen A, Perola M, Terwilliger JD, Kempas E, Daly M, Lilja H, Rioux JD, Brettin T, Viikari JS, Rönnemaa T, Laakso M, Lander ES, Peltonen L (2000) Two loci on chromosomes 2 and X for premature coronary heart disease identified in early- and late-settlement populations of Finland. Am J Hum Genet 67:1481–1493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SB, MacLean CJ, Neale MC, Eaves LJ, Kendler KS (1999) Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet 65:876–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani NJ (1998) Molecular genetics of coronary artery disease: measuring the phenotype. Clin Sci (Lond) 95:645–646 [PubMed] [Google Scholar]
- Topol EJ, McCarthy J, Gabriel S, Moliterno DJ, Rogers WJ, Newby LK, Freedman M, Metivier J, Cannata R, O’Donnell CJ, Kottke-Marchant K, Murugesan G, Plow EF, Stenina O, Daley GQ (2001) Single nucleotide polymorphisms in multiple novel thrombospondin genes may be associated with familial premature myocardial infarction. Circulation 104:2641–2644 [DOI] [PubMed] [Google Scholar]
- Wang L, Fan C, Topol SE, Topol EJ, Wang Q (2003) Mutation of MEF2A in an inherited disorder with features of coronary artery disease. Science 302:1578–1581 10.1126/science.1088477 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Q, Rao S, Shen G-Q, Li L, Moliterno DJ, Newby LK, Rogers WJ, Cannata R, Zirzow E, Elston RC, Topol EJ (2004) Premature myocardial infarction novel susceptibility locus on chromosome 1P34–36 identified by genomewide linkage analysis. Am J Hum Genet 74:262–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whittemore AS, Halpern J (1994) A class of tests for linkage using affected pedigree members. Biometrics 50:118–127 [PubMed] [Google Scholar]
- Yamada Y, Izawa H, Ichihara S, Takatsu F, Ishihara H, Hirayama H, Sone T, Tanaka M, Yokota M (2002) Prediction of the risk of myocardial infarction from polymorphisms in candidate genes. N Engl J Med 347:1916–1923 10.1056/NEJMoa021445 [DOI] [PubMed] [Google Scholar]
- Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, McQueen M, Budaj A, Pais M, Varigos J, Lisheng L for the INTERHEART Study Investigators (2004) Effect of potentially modificable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet 364:937–952 10.1016/S0140-6736(04)17018-9 [DOI] [PubMed] [Google Scholar]