Abstract
Bardet-Biedl syndrome (BBS) is an autosomal recessive, genetically heterogeneous, pleiotropic human disorder characterized by obesity, retinopathy, polydactyly, renal and cardiac malformations, learning disabilities, and hypogenitalism. Eight BBS genes representing all known mapped loci have been identified. Mutation analysis of the known BBS genes in BBS patients indicate that additional BBS genes exist and/or that unidentified mutations exist in the known genes. To identify new BBS genes, we performed homozygosity mapping of small, consanguineous BBS pedigrees, using moderately dense SNP arrays. A bioinformatics approach combining comparative genomic analysis and gene expression studies of a BBS-knockout mouse model was used to prioritize BBS candidate genes within the newly identified loci for mutation screening. By use of this strategy, parathyroid hormone-responsive gene B1 (B1) was found to be a novel BBS gene (BBS9), supported by the identification of homozygous mutations in BBS patients. The identification of BBS9 illustrates the power of using a combination of comparative genomic analysis, gene expression studies, and homozygosity mapping with SNP arrays in small, consanguineous families for the identification of rare autosomal recessive disorders. We also demonstrate that small, consanguineous families are useful in identifying intragenic deletions. This type of mutation is likely to be underreported because of the difficulty of deletion detection in the heterozygous state by the mutation screening methods that are used in many studies.
Introduction
Bardet-Biedl syndrome (BBS [MIM 209900]) is a pleiotropic, autosomal recessive disorder characterized by obesity, pigmentary retinopathy, polydactyly, renal abnormalities, learning disabilities, and hypogenitalism (Bardet 1920; Biedl 1922; Green et al. 1989). The disorder is also associated with an increased susceptibility to diabetes mellitus, hypertension, and congenital heart disease (Harnett et al. 1988; Green et al. 1989; Elbedour et al. 1994). BBS shows variable expressivity within and between families, a finding that suggests genetic complexity. The disorder also displays extensive genetic heterogeneity, and the involvement of two mutations at one locus and a third mutation at a second locus has been suggested (Katsanis et al. 2001). To date, eight BBS loci have been mapped, and the causative gene at each of these loci has been identified (Kwitek-Black et al. 1993; Sheffield et al. 1994; Carmi et al. 1995; Young et al. 1999; Katsanis et al. 2000; Slavotinek et al. 2000; Nishimura et al. 2001; Mykytyn et al. 2001, 2002; Ansley et al. 2003; Badano et al. 2003; Chiang et al. 2004; Fan et al. 2004; Li et al. 2004). Mutation screening of the eight known genes has resulted in the identification of approximately half of the causative mutations, indicating that additional BBS genes and mutations have yet to be identified (Katsanis 2004; Hichri et al. 2005; V.C.S. and E.M.S., unpublished data; L.G.B., unpublished data).
The initial identification of BBS genes relied heavily on positional cloning, a process that begins with the localization of genetic intervals, with the use of genetic linkage studies of large multiplex families (Katsanis et al. 2000; Slavotinek et al. 2000; Mykytyn et al. 2001; Nishimura et al. 2001). Subsequently, bioinformatic comparisons of protein sequences aided in the identification of additional BBS genes. For example, the positional cloning of the BBS1 gene was aided by the finding of limited sequence homology to BBS2 (Mykytyn et al. 2002). BBS7 and BBS8 were identified by performing database searches for proteins with partial homology to BBS2 and BBS4, respectively (Ansley et al. 2003; Badano et al. 2003). The BBS3 and BBS5 genes were recently identified using a combination of comparative genomics and positional cloning (Chiang et al. 2004; Fan et al. 2004; Li et al. 2004). The identification of additional BBS genes is hindered by the extensive genetic heterogeneity and by the paucity of additional large multiplex families for genetic mapping. To overcome these obstacles in this study, we performed homozygosity mapping by the use of genomewide SNP analysis of small (one or two affected individuals), consanguineous BBS families, to identify potential new BBS candidate regions, and we applied a bioinformatics approach that combines comparative genome analysis with gene expression analysis of a Bbs4-null mouse model to identify and prioritize BBS candidate genes. The identification of mutations in a gene found within a candidate positional interval validates the use of small families for the identification of new BBS genes. Small, consanguineous pedigrees also proved useful for the identification of patients with intragenic gene deletions in three previously known BBS genes.
Material and Methods
Human Subjects
Signed informed consents approved by the institutional review boards at the University of Iowa and collaborating institutions were obtained from all study participants at the time of their entry into the study. The diagnosis of BBS in individuals was based on the presence of at least three of the cardinal features of BBS (obesity, polydactyly, renal anomalies, retinopathy, hypogonadism, and learning disabilities). The features most consistently observed in the patients recruited for the study were obesity, polydactyly, and retinal degeneration. The 110 control samples (220 chromosomes) used for allele frequency estimation in the general population included a subset of the DNA Polymorphism Discovery Resource (Coriell Cell Repositories) and additional samples matched to the BBS patient population. The panel was designed to reflect the diversity present in the human population (Collins et al. 1998).
SNP Homozygosity Mapping
Genomic DNA from affected individuals of consanguineous BBS families was used as the template to generate probes for use on Affymetrix 10K 2.0 SNP microarrays (Affymetrix). Sample processing and labeling were performed in accordance with the manufacturer's specifications (Affymetrix). The arrays were hybridized, washed, and scanned at the University of Iowa DNA Facility. Chip images were processed by the GeneChip DNA Analysis Software (GDAS) to generate SNP allele calls. For the homozygosity analysis, a sliding window of 30 SNPs was used to identify regions of homozygosity. To allow for potential SNP miscalls, we incorporated the use of quality control data to identify potential miscalls. To be conservative, we ignored SNPs that could not be scored by GDAS for this analysis. For the samples examined, an average call rate of >95% was obtained. When more than one sample was available from a family, a further constraint was placed on the analysis, in that all the samples from a family were required to be homozygous for the same allele at each SNP, consistent with identity by descent.
STRP Genotyping
PCR amplification for the analysis of STRPs was performed using 40 ng genomic DNA in 8.4 μl reactions containing 1.25 μl 10× PCR buffer (100 mM Tris-HCl [pH 8.8], 500 mM KCl, 15 mM MgCl2, 0.01% gelatin [w/v]); 200 μM each of dATP, dCTP, dGTP, and dTTP; 2.5 pmol of each primer; and 0.2 units of Taq polymerase. Samples were subjected to 35 cycles of 94°C for 30 s (50°C, 52°C, 55°C, or 57°C, as required, for 30 s) and 72°C for 30 s. Amplification products were separated on 6% polyacrylamide gels containing 7.7 M urea at 60 W, for ∼2 h. The bands were visualized by silver staining. Oligonucleotide primers for the STRPs were custom designed and synthesized commercially (IDT).
Comparative Genomic Analysis
Identification and prioritization of BBS candidate genes in this study used a computational comparative genomics technique that is similar to previous methods (Avidor-Reiss et al. 2004; Chiang et al. 2004; Fan et al. 2004; Li et al. 2004). A basic principle of the comparative genomics approach is that a specific biological feature is evolutionarily conserved among a group of organisms. A second group of organisms that explicitly lack this same characteristic is then identified. These two groups of organisms serve as “positive” and “negative” reference sets, respectively, for compiling a list of candidate genes. The organisms for our analysis were selected with the goal of minimizing genome complexity. In addition, ciliated organisms were included, since evidence indicates that BBS genes are involved in cilia function and/or maintenance (Avidor-Reiss et al. 2004; Blacque et al. 2004; Li et al. 2004; Mykytyn et al. 2004). In a modification of the comparative genomics approach used to identify genes involved in cilia function (Chiang et al. 2004; Fan et al. 2004; Li et al. 2004), Giardia lamblia was included in the negative set despite the presence of flagella, since no apparent BBS orthologues are found in the Giardia genome. The inclusion of Giardia allows for a more specific screen for BBS genes because other cilia-related genes in the Giardia genome are removed from the BBS candidate gene set. The computational sequence similarity tool BLAST (Altschul et al. 1990) was used to determine the orthologs of human proteins from Ensembl release 28.35a.1 to those of the model organisms Trypanasoma cruzi, Leishmania major, G. lamblia, and Saccharomyces cerevisiae. The positive and negative reference sets were compared using a threshold of e-37 to identify BBS gene candidates. This value was selected empirically on the basis of observed levels of similarity among the known BBS proteins in this group of organisms. Protein sequences for the model organisms were obtained from The Institute for Genomic Research (T. cruzi), the Sanger Institute (L. major), GiardiaDB (G. lamblia), and the Saccharomyces Genome Database (SGD) (S. cerevisiae).
Bbs4 Microarray Analysis
The Bbs4−/− mouse model has been described elsewhere (Mykytyn et al. 2004). Whole eyes were dissected from euthanized Bbs4−/− and wild-type mice at 8 mo of age. One eye from each animal was processed for histological analysis to verify retinal degeneration, while the second eye was used for the isolation of total RNA. Total RNA from pooled Bbs4−/− and wild-type eyes (three in each group) was extracted using TRIzol reagent (Invitrogen) and was purified through RNeasy columns (Qiagen). Integrity of the RNA was assessed on an Agilent 2100 bioanalyzer (Agilent Technologies). Pooling of eyes was necessary because of the low RNA yield from individual eyes and also to minimize the effects of biological variability among samples.
The two RNA pools were hybridized to Affymetrix mouse 430 2.0 arrays. Five micrograms of total RNA was used for the synthesis of double-stranded cDNA by the Superscript Choice system (Invitrogen) and T7-(dT)24 primer (Sigma-Genosys). In vitro transcription was performed in the presence of biotinylated UTP, and the cRNA was fragmented and hybridized for 16 h to the arrays by use of the Affymetrix GeneChip Instrumention System at the DNA Core Facility of the University of Iowa. After stringent washes, the arrays were stained with streptavidin-phycoerythrin (Molecular Probes) and then scanned. Data were acquired using the Affymetrix data collection and analysis programs GeneChip Operating Software (GCOS) and GDAS. Probe sets that demonstrated a greater than twofold decrease in expression in the knockout pool, as compared with the wild-type pool, were identified. Such probe sets were further constrained, in that the signal for at least one of the pools was required to be scored as present by GDAS.
DNA Sequencing and Mutation Screening
PCR products for sequencing were amplified in a 20-μl reaction volume and were separated on a 2.0% agarose gel, as described elsewhere (Nishimura et al. 1998). The corresponding bands were excised and purified using the QIAquick gel extraction kit (Qiagen). As the template for sequencing reactions using dye-terminator chemistry (Applied Biosystems), 4.5 μl of purified PCR product was used. PCR product sequencing reactions were precipitated in the presence of glycogen and isopropanol. The reactions were analyzed on an ABI 3730XL DNA Sequencer. All sequence variants were verified by direct DNA sequencing and/or by restriction enzyme digestion. Primer sequences used to screen the entire B1 gene are available upon request.
In some cases, the coding sequence of candidate genes was also screened by SSCP analysis. Amplicons for SSCP analysis were designed to be ∼200 bp in size. For SSCP, PCR products were separated on SSCP gels (7 ml 50% glycerol, 3.5 ml 5× TBE, 8.8 ml 37.5:1 acrylamide/bis, and 50.7 ml ddH20) for 3–4 h in 0.5× TBE at room temperature, with the temperature controlled by a cooling fan. The gels were silver stained to visualize DNA bands. Abnormal variants were sequenced and compared with a control sample (CEPH sample 1331–01) for the detection of any changes from that of the normal sequence.
B1 Gene Expression Studies
Northern blots containing 2 μg of poly (A) RNA isolated from a broad array of human and mouse tissues were purchased from Clontech. Following hybridization, the blots were stripped of radioactivity and rehybridized with a cDNA probe for β-actin for the verification of equal loading of RNA (Swiderski et al. 1999). RT-PCR was performed with the SuperScript II Reverse Transcriptase kit (Invitrogen) in accordance with the manufacturer’s instructions. Products were separated on 2.0% agarose gels. Bands were excised from the agarose gels and sequenced, to confirm their identities or to discern their sequences, following the protocols described above.
Results
Comparative Genomic Analysis
The existence of eight known BBS genes provided an opportunity to use bioinformatic methods to identify additional BBS genes. We hypothesized that the genomes of model organisms that retained orthologues of the known BBS genes were likely to contain yet-to-be-identified BBS genes, and the genomes of organisms that did not contain orthologues of known BBS genes were unlikely to contain orthologues of additional BBS genes. We examined a number of model organisms for potential inclusion in our comparative genomics analysis. Model organisms such as T. cruzi, L. major, G. lamblia, and S. cerevisiae were attractive for two reasons: These organisms have relatively small genome sizes, and the paucity of intervening intronic sequence facilitates the de novo identification of novel protein coding sequences. Table 1 illustrates the similarity of human BBS proteins to their putative orthologues in these four organisms. With the exception of the MKKS protein sequence, the known BBS protein sequences have high levels of similarity to sequences in T. cruzi and L. major. From these observations, a threshold of ⩽e-37 was used for considering a gene found in a model organism to be an orthologue of the human BBS gene. Based on this cutoff, BBS orthologues were not found in the genomes of G. lamblia and S. cerevisiae.
Table 1.
Characteristics of BBS Proteins
| ProteinSymbol | Locus | UniGene | Location | T. cruzia | L. majora | G. lambliaa | Yeasta | FCb | Changec |
| BBS1 | BBS1 | Hs.502915 | 11q13.2 | 1e-62 | 2e-72 | 1.9 | 9.4 | 3.0 | D |
| BBS2 | BBS2 | Hs.333738 | 16q12.2 | 3e-87 | 1e-76 | 0.2 | 0.068 | 2.6 | D |
| ARL6 | BBS3 | Hs.373801 | 3q11.2 | 3e-41 | 9e-45 | 5e-32 | 3e-36 | 4.9 | D |
| BBS4 | BBS4 | Hs.208681 | 15q24.1 | 3e-72 | 6e-80 | 5e-07 | 1e-08 | 9.9 | D |
| BBS5 | BBS5 | Hs.233398 | 2q31.1 | 6e-69 | 2e-66 | 3e-17 | 1.5 | 5.3 | D |
| MKKS | BBS6 | Hs.472119 | 20p12.2 | 2e-11 | 5e-07 | 1e-11 | 5e-13 | 1.2 | NC |
| BBS7 | BBS7 | Hs.58974 | 4q27 | 8e-41 | 1e-39 | 1.7 | 0.44 | 2.3 | D |
| TTC8 | BBS8 | Hs.303055 | 14q32.11 | 1e-79 | 3e-64 | 7e-20 | 5e-04 | 2.3 | D |
Expected value of best match.
Fold change in Bbs4−/− eyes versus wild-type control eyes.
Change in expression in Bbs4−/− eyes versus wild-type control eyes. D = decrease and NC = no change.
We next performed BLAST analysis of the set of 35,838 human proteins from Ensembl release 28.35a.1 against the proteomes of T. cruzi, L. major, G. lamblia, and S. cerevisiae. For the positive BBS candidate set, we included all human proteins that exceeded the threshold of e-37 to orthologues in T. cruzi and L. major. We then removed from the BBS candidate set those human proteins that did not meet the e-37 threshold in either G. lamblia or S. cerevisiae, two organisms that do not contain orthologues to the known BBS genes. The analysis of G. lamblia orthologous proteins was especially helpful for the comparative genomic analysis, in that this organism has four pairs of flagella yet lacks identifiable BBS orthologues. Several lines of evidence, including evaluation of BBS mouse models and protein localization, indicate that BBS genes are involved in cilia synthesis, function, and/or maintenance (Avidor-Reiss et al. 2004; Blacque et al. 2004; Li et al. 2004; Mykytyn et al. 2004). The use of the Giardia sequence allows removal of cilia-related genes that may not be involved in BBS. The removal of G. lamblia and S. cerevisiae proteins resulted in a set of 366 human proteins, 239 of which are unique. The genes corresponding to these protein sequences will be referred to as the “comparative genomics” set of BBS gene candidates.
Refinement of the BBS Candidate Gene Set with Gene Expression Data
We have previously described elsewhere a knockout mouse model for BBS4 (Mykytyn et al. 2004). Homozygous knockout animals exhibit an early onset form of retinal degeneration, with loss of the photoreceptor layer by 8 mo of age. Gene expression studies were performed by microarray analysis of whole eye RNA between Bbs4−/− versus age- and sex-matched controls. Photoreceptor loss in the Bbs4−/− animals was confirmed by histology. As shown in table 1, seven of the eight known BBS genes demonstrate a twofold or greater decrease in expression in eyes from Bbs4 knockout mice aged 8 mo, as compared with control eyes. Only the Bbs6 gene, Mkks, failed to show a significant decrease in gene expression in this model system. Of the 45,000+ probe sets represented on the Mouse Genome 430 2.0 Array, 667 unique probe sets demonstrated a twofold or greater decrease in gene expression in the knockout eyes, as compared with the controls. This group of genes is referred to as the “retinal expression candidate gene set.”
We next hypothesized that, by considering the intersection of genes between the comparative genomics BBS candidate gene set and the retinal expression candidate gene set, we would be able to identify a refined set of genes that was highly enriched for potential new BBS genes. There are a total of 19 genes that are found in common in the bioinformatics and expression candidate gene sets, including seven of the eight known BBS genes. Because this set of 19 genes (table 2) contains known BBS genes, it was hypothesized that this refined set of genes also included undiscovered BBS genes.
Table 2.
BBS Gene Candidates Identified by Bioinformatics Analysis
| GeneSymbol | UniGene | BBS | Location | T. cruzia | L. majora | G. lambliaa | Yeasta | FDb |
| PLCL4 | Hs.170156 | … | 1p36.32 | 2e-40 | 8e-43 | 0.033 | 2e-35 | 1.9 |
| BBS5 | Hs.233398 | BBS5 | 2q31.1 | 6e-69 | 2e-66 | 3e-17 | 1.5 | 5.3 |
| ARL6 | Hs.373801 | BBS3 | 3q11.2 | 3e-41 | 9e-45 | 5e-32 | 3e-36 | 4.9 |
| PDE6B | Hs.59872 | … | 4p16.3 | 6e-54 | 3e-54 | 2e-10 | 1e-13 | 84.5 |
| BBS7 | Hs.58974 | BBS7 | 4q27 | 8e-41 | 1e-39 | 1.7 | 0.44 | 2.3 |
| WDR17 | Hs.532056 | … | 4q34.2 | 2e-58 | 3e-69 | 5e-19 | 3e-18 | 34.3 |
| PDE8B | Hs.78106 | … | 5q13.3 | 2e-42 | 1e-41 | 2e-21 | 4e-16 | 2.3 |
| PDE6A | Hs.151710 | … | 5q33.1 | 5e-48 | 1e-52 | 3e-07 | 3e-11 | 32.0 |
| B1 | Hs.372360 | BBS9 | 7p14.3 | 3e-83 | 2e-67 | 0.43 | 1.6 | 2.6 |
| CCDC2 | Hs.145402 | … | 9p21.2 | 5e-56 | 7e-48 | 5e-21 | 2e-19 | 1.7 |
| PDE6C | Hs.93173 | … | 10q23.33 | 8e-53 | 1e-51 | 1e-11 | 9e-12 | 3.3 |
| DPCD | Hs.549129 | … | 10q24.32 | 4e-38 | 1e-40 | 6e-25 | 0.92 | 1.7 |
| BBS1 | Hs.502915 | BBS1 | 11q13.2 | 1e-62 | 2e-72 | 1.9 | 9.4 | 3.0 |
| TTC8 | Hs.303055 | BBS8 | 14q32.11 | 1e-79 | 3e-64 | 7e-20 | 5e-04 | 2.3 |
| BBS4 | Hs.208681 | BBS4 | 15q24.1 | 3e-72 | 6e-80 | 5e-07 | 1e-08 | 9.9 |
| PDE8A | Hs.9333 | … | 15q25.3 | 3e-44 | 9e-43 | 2e-25 | 2e-17 | 2.3 |
| BBS2 | Hs.333738 | BBS2 | 16q12.2 | 3e-87 | 1e-76 | 0.2 | 0.068 | 2.6 |
| UNC119 | Hs.410455 | … | 17q11.2 | 1e-38 | 1e-41 | 9e-07 | 0.91 | 6.5 |
| MGC26694 | Hs.303669 | … | 19p13.11 | 1e-49 | 5e-49 | None | 8e-33 | 2.0 |
Expected value of best match.
Fold decrease in Bbs4−/− eyes versus wild-type control eyes.
Homozygosity Mapping with Small, Consanguineous BBS Pedigrees
Most of the previously identified BBS genes were identified using positional information obtained from genetic mapping studies using multiplex families. We do not have access to additional large families suitable for positional cloning studies. However, small, consanguineous BBS families with one or two affected individuals can be used to identify loci that are homozygous in the affected individual(s). Loci that are homozygous by descent in affected individuals from small, consanguineous families are candidates for disease loci. Mapping data from a small family typically identifies a number of large candidate intervals, only one of which includes the gene that is mutated in that family. The intersection of putative candidate intervals identified by homozygosity mapping with BBS candidate gene sets can lead to the identification of the highest priority candidate genes for mutation screening.
To identify putative BBS loci, we performed genomewide SNP genotyping at 10,000 SNP density in two groups of consanguineous BBS families. The first group contained seven families with two affected individuals (consanguineous, affected sibling pairs [CSP]), while the second group contained families with only one affected individual. Homozygosity mapping studies in the first group of seven families revealed 20 candidate regions (table 3). Only a single candidate region at 9q22 was identified in more than one CSP family; all other candidate regions were identified in only a single CSP family. The second group consisted of nine consanguineous BBS families, each with one affected individual (consanguineous affected proband [CAP]). Genotyping of the nine CAP samples revealed 79 regions of homozygosity, 19 of which overlapped (at least partially) with regions identified in the CSP families (see table 3). The data from the homozygosity mapping studies were then compared with the candidate gene set and were used to prioritize candidate genes for mutation screening in selected BBS patients. Three genes were initially selected for candidate gene screening—DPCD (Hs.549129, 10q24.32), UNC119 (Hs.410455, 17q11.2), and B1 (Hs.372360, 7p14.3)—on the basis that these genes were those most strongly supported from the homozygosity mapping data (each mapped to an overlapping region of homozygosity in one CSP family and in one CAP family, or in two CAP families).
Table 3.
BBS Candidate Regions (CSP Families)
| Candidate Region | Markersa | Size (Mb) | CSPFamily | CAPsb |
| 2p25.3-p24.1 | 87 | 21.5 | 7 | … |
| 4q24-q32.2 | 203 | 57.9 | 5 | 5, 6, 7 |
| 5q14.3-q23.1 | 124 | 32.1 | 7 | 1, 7 |
| 5q31.3-q32 | 37 | 2.7 | 6 | 3 |
| 7p22.2-p14.3 | 130 | 30.8 | 4 | 3, 8 |
| 7q32.3-q34 | 37 | 8.6 | 7 | … |
| 8p21.3-p12 | 45 | 9.7 | 7 | … |
| 8p12-q11.23 | 41 | 18.5 | 3 | … |
| 9q21.2-q22.1 | 265 | 11.4 | 2 | 4, 7 |
| 9q21.32-q31.2 | 76 | 23.6 | 7 | 1 |
| 9q33.1 | 83 | 2.4 | 1 | … |
| 10q22.1-q22.2 | 33 | 6.9 | 6 | 7 |
| 11p15.5-p15.3 | 45 | 9.6 | 5 | … |
| 12q15-q22 | 99 | 26.2 | 4 | 2, 4 |
| 14q11.2-q12 | 35 | 7.3 | 4 | … |
| 14q23.1-q24.2 | 51 | 12.7 | 3 | 3, 7 |
| 15q21.3-q22.2 | 61 | 4.8 | 6 | 3, 4 |
| 15q26.1-q26.3 | 158 | 9.9 | 2 | … |
| 16q22.1-q23.1 | 36 | 12.7 | 5 | … |
| 18q21.33-q22.1 | 43 | 2.7 | 6 | 3 |
No. of contiguous homozygous SNPs defining the candidate region.
CAPs with a region of homozygosity that overlaps the CSP-defined candidate region.
Identification of B1 as the BBS9 Gene
The coding sequences of three candidate genes were determined in DNA samples from those affected BBS individuals who had a region of homozygosity that overlapped with the specific candidate gene. The DPCD and UNC119 genes were sequenced without the finding of BBS sequence variants. When the B1 gene was sequenced in a consanguineous Arab BBS family (CAP-9) displaying homozygosity within the region of chromosome 7 containing the B1 gene, we detected a homozygous G→A transversion that affects the splice donor site of intron 17 (IVS17+1G→A). This change was not found in 110 control samples (220 control chromosomes). The splice donor mutation is predicted to prevent splicing at the donor site, thus leading to the incorporation of intron 17 into the mature mRNA and resulting in premature termination within intron 17. It was not possible to experimentally confirm the effects of this mutation, since cells or tissue were not available from this patient or family members.
We next used SSCP to screen the B1 gene in 95 unrelated BBS probands with no known consanguinity (unrelated affected probands [UAP]). We detected five probands, each with a different mutation in the homozygous state (see fig. 1). These mutations include a homozygous G→A transversion in exon 4 (421G→A) predicting G141R, a homozygous C→T transversion in exon 9 (1063C→T) predicting Q355X, a homozygous C→T transversion predicting R598X, a homozygous 1-bp insertion (2046insC) predicting K683fsX687, and a homozygous 4-bp deletion (1887_1880delAACA) predicting K626fsX647. The 1877_1880delAACA mutation was also found in an affected proband (UAP-74) in the heterozygous state. The UAP-74 family was also found to harbor a splice donor mutation (IVS4+1G→C) in intron 4 in the heterozygous state. In total, 6 of the 95 probands (6.3%) had either homozygous (5) or compound heterozygous (1) mutations. In each case, the mutation segregated with the disease in the family (fig. 1). None of these variants was detected in 110 control DNA samples (table 4). All mutations refer to GenBank reference sequence NM_198428.
Figure 1.
Pedigrees of BBS families with B1 mutations. Specific mutations detected in each family are indicated below the family identifier. All mutations were found in the homozygous state, with the exception of that in UAP-74. mt = Mutant allele corresponding to the mutation in each family. wt = Wild-type (normal) allele. In UAP-74, mt1 represents the K626fsX647 and mt2 represents the IVS4+1G→C mutation.
Table 4.
BBS Mutations Detected in the B1 Gene
| Gene Mutation | Location | Protein | BBSa | Controlsb |
| 421G→A | Exon 5 | G141R | 2 | 0 |
| IVS5+1G→C | Intron 5 | Splice site | 1 | 0 |
| 1063C→T | Exon 10 | Q355X | 2 | 0 |
| IVS17+1G→A | Intron 17 | Splice site | 2 | 0 |
| 1792C→T | Exon 18 | R598X | 2 | 0 |
| 1877_1880delAACA | Exon 18 | K626fsX647 | 3 | 0 |
| 2046insC | Exon 19 | K683fsX687 | 2 | 0 |
No. of mutant alleles detected in 95 BBS samples.
No. of alleles containing each variant detected in 110 unrelated controls.
B1 Gene Structure and Expression
Previous studies have shown that the B1 gene encodes multiple splice isoforms (Adams et al. 1999). A 1.8-kb isoform appears to be specific to bone or bone-derived cells. Two different isoforms of the B1 gene are listed in UniGene as RefSeq ID numbers NM_014451 and NM_198428. These two isoforms differ, in that exons 15 and 16 are present in NM_198428 but are absent in NM_014451. Furthermore, two additional downstream exons that are not present in either of the UniGene sequences have also been observed (Vernon et al. 2003). In an attempt to clarify the gene structure and expression pattern of the B1 gene, we performed bioinformatics analysis of B1 EST sequences and performed both northern blot and RT-PCR studies on human tissues. These studies indicate that the B1 gene consists of 25 exons that span >700 kb of genomic DNA. With the exception of the first exon, the remaining 24 exons contribute to the various B1 protein isoforms. The NM_198428 isoform (presence of exons 15 and 16) appeared to be the major isoform in all tissues examined, with little to no expression of the NM_014451 isoform. We found evidence for three splice isoforms at the 3′ end of the B1 gene, as depicted in figure 2. These three isoforms involve differential utilization of exons 22, 23, 24, and 25, along with the use of alternative 3′ UTR sequences.
Figure 2.
Genomic structure of the B1 gene, with exons depicted as boxes. Filled boxes, Coding regions. Open boxes, UTRs. Exon sizes are shown below the exons in the genomic structure. The three isoforms of B1 presented in this study are depicted below the genomic structure. The location of the B1 northern blot probe (hatched box) is shown. Locations of the BBS mutations detected in this study are indicated at the bottom of the figure.
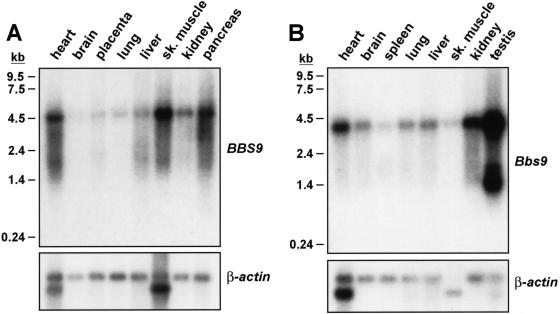
Multiple B1 transcripts have been previously reported elsewhere in a broad range of human tissues (Adams et al. 1999). To study B1 gene expression patterns in tissues related to the BBS phenotype in mouse and human, we prepared northern blot hybridization probes directed at exons 18–20, as depicted in figure 1. We observed expression of a 4.5-kb transcript in all tissues studied, as well as several smaller transcripts ranging in size from ∼2 kb to 4 kb, as shown in figure 3A. Similarly, in mouse tissues, expression of an ∼4.5-kb mRNA was noted (fig. 3B). Two additional smaller transcripts were detected solely in the testis. B1 gene expression was also noted in the eye and in many regions of the mouse brain, including the hypothalamus (data not shown).
Figure 3.
Northern blot analysis of B1 gene expression by northern blot analysis. A, Human B1 probe hybridized to human RNA. B, Mouse B1 probe, corresponding to the same location as the human B1 probe, hybridized to mouse RNA samples.
Alu-Mediated Deletions as a BBS Mutation Mechanism
In addition to the identification of a novel BBS gene, homozygosity mapping of individual patients from consanguineous matings was useful for facilitating the identification of previously undetected mutations in known BBS genes. Examination of the SNP data from nine unrelated BBS patients revealed that six were homozygous for at least one locus containing known BBS genes (table 5). We sequenced the known BBS gene at the homozygous locus in each of these individuals. No point mutations were detected within the coding regions or the flanking intron regions of the BBS genes that were examined. However, it was noticed that some of the amplicons consistently failed to amplify from the DNA samples of three affected patients. Since the samples used for this analysis were obtained from consanguineous BBS families, the presence of deletions in the homozygous state was considered as the cause of the failed amplification. In each case, deletions were confirmed and characterized by the examination of regions that flanked the exons that failed to amplify in the BBS-affected individuals. The regions of the BBS genes containing these deletions are shown in figure 4.
Table 5.
Screening of Known BBS Genes in CAP Patients
| Gene | CAP Family | Mutationa | Location | Markersb | Candidate Region Size(Mb) |
| BBS3 | 4 | NC | 3p13-q12.3 | 68 | 31.7 |
| BBS4 | 1 | DEL | 15q22.31-q26.1 | 99 | 29.6 |
| BBS4 | 4 | NC | 15q14-q26.1 | 197 | 54.5 |
| BBS5 | 2 | DEL | 2q12.3-q31.1 | 226 | 67.8 |
| BBS6 | 1 | NC | 20p13-q13.13 | 152 | 47.2 |
| BBS6 | 7 | NC | 20p12.3-p12.1 | 187 | 6.9 |
| BBS7 | 5 | NC | 4q25-q28.1 | 37 | 13.1 |
| BBS7 | 7 | NC | 4q26-q31.1 | 567 | 26.1 |
| BBS7 | 8 | DEL | 4q26-q26.3 | 372 | 16.6 |
Results of screening the coding region of a known BBS gene. NC = no change and DEL = large intragenic deletion.
No. of contiguous homozygous SNPs defining the candidate region.
Figure 4.
Intragenic deletions found in BBS4, BBS5, and BBS7. The extent of each deletion is depicted within the partial genomic structure for each gene. The locations of the exons are shown below the line representing the genomic sequence (AC016402, AC093899, and AC079341). Within the genomic sequence, repetitive element family members are indicated as follows: blue, AluS; red, AluJ; green, AluY; yellow, FLAM; and pink, SVA. Translation of the genomic region in the three forward reading frames are shown below each gene figure with start codons (green) and stop codons (red).
Exons 9, 10, 11, and 12 failed to amplify from a proband (CAP-2) that demonstrated homozygosity for a ∼68-Mb region on chromosome 2 that contains the BBS5 gene. Characterization of the deletion confirmed that the breakpoints were located within intron 8 and a region beyond the BBS5 3′ UTR. The breakpoint in intron 8 was found to have occurred within an Alu element, while the downstream breakpoint occurred within an SVA repeat element. There was no apparent sequence similarity at the site of the breakpoints other than a short region that flanked the breakpoints. No other family members were available for study.
Exons 1, 2, 3, and 4 failed to amplify from a proband (CAP-8) that demonstrated homozygosity for a 17-Mb region on chromosome 4 that contained the BBS7 gene. Characterization of this region of homozygosity confirmed a four-exon deletion with breakpoints that were located upstream of exon 1 and within intron 4. The breakpoint upstream of exon 1 was found to have occurred immediately 3′ of an Alu element, whereas the breakpoint within intron 4 appeared to occur within a nonrepetitive sequence. There was no apparent sequence similarity at the breakpoint regions. There were four other family members available for study, none of which had BBS. The BBS7 deletion was not found in the homozygous state in any of these family members.
Evaluation of the BBS4 gene in a proband (CAP-1) with a 30-Mb region of homozygosity on chromosome 15 revealed the amplification failure of exons 3 and 4. Further analysis confirmed a two-exon deletion with breakpoints within introns 2 and 4. This mutation was identical to that reported in two other families with a deletion in BBS4 (Mykytyn et al. 2001). The small sizes of these deletions (10 kb–20 kb), in comparison with the average 300-kb spacing of SNPs on the array used for this study, precluded their direct detection by SNP genotyping.
We examined the regions containing the breakpoints in each of the three deletion events. Three of the six breakpoints occurred within Alu-repetitive elements, whereas a fourth was found to occur just outside an Alu element. The two breakpoints involved in the BBS4 deletion appear to have occurred within the Alu element in regions that share sequence similarity (fig. 5). It has been previously noted elsewhere that this sequence shows a striking resemblance to the prokaryotic χ-sequence, which is thought be involved in stimulating recombination in E. coli (Rudiger et al. 1995; Kolomietz et al. 2002). The close proximity of Alu elements to at least one of the regions involved in each of the deletion events suggests that such elements play some role in their occurrence.
Figure 5.
Sequence flanking the breakpoints for the BBS4 deletion. The sequences of the 5′ and 3′ Alu elements are shown as well as the sequence of the junction fragment of the BBS4 deletion event in family CAP-1. Box, The region in which the breakpoint has occurred within each Alu element. The 26-bp core sequence within the Alu element identified by Rudiger et al. (1995) is shown at the bottom of the figure, as well as a comparison to the prokaryotic χ-sequence.
Discussion
Although BBS is uncommon in the general population, some clinical features of the disorder, such as hypertension, obesity, and diabetes, are prevalent. Several BBS genes have been identified to date, yet our knowledge of BBS gene function remains limited. The identification of additional BBS genes remains a worthy goal for a number of reasons, including that (1) accurate molecular diagnosis of this disorder is not currently possible for many patients, (2) additional clues to a common molecular mechanism involved in BBS pathogenesis may be elucidated by the identification of additional genes, (3) a clearer understanding of the genetic complexity of BBS will be aided by the identification of additional genes, and (4) identification of additional BBS genes is likely to give insights into the pathways involved in individual components of the BBS phenotype.
Before the current study, eight BBS loci were known, and the disease-causing gene at each of these loci was identified using positional cloning and candidate gene analysis. On the basis of mutation analysis of coding sequence from the eight known genes, the disease-causing mutation has been identified in approximately half of the patients. Possible explanations for such a result include the existence of as-yet-unidentified BBS genes, the existence of undetected mutations in one or more of the known BBS genes, and/or misdiagnosis of BBS patients. The current study was based on the hypothesis that additional BBS genes are yet to be discovered.
The genetic attributes of BBS presents obstacles that impede mapping efforts, including the genetic heterogeneity of the disorder, the relative rarity of the disorder, and the paucity of multiplex families. To overcome these obstacles, we generated and utilized three types of data. First, we performed homozygosity mapping in small, consanguineous BBS pedigrees with one or two affected individuals. Second, we utilized comparative genomics data to generate a set of BBS candidate genes that are conserved in the genomes of model organisms in a pattern similar to the currently known BBS genes. Finally, we used comparative expression data generated by microarray analysis of Bbs4−/− and control mouse eyes. The combination of these three types of data allowed us to select high-priority genes for mutation screening in selected BBS patients.
To take maximum advantage of small pedigrees to identify new genetic loci involved in BBS, we selected pedigrees in which the parents of the patients were known to be related. The use of consanguineous pedigrees allowed homozygosity mapping to be performed. The size of the pedigrees did not provide enough power to unambiguously map new loci. However, modest density SNP genotyping of the small, consanguineous pedigrees did identify candidate BBS loci on the basis of the identification of regions with apparent homozygosity by descent. SNP genotyping of individual patients revealed numerous homozygous SNPs. In most cases, individual SNPs are homozygous by state. However, large stretches of consecutive homozygous SNPs are more likely to be homozygous by descent in the offspring of consanguineous matings than they are to be homozygous by state. In this study, we used a sliding window of 30 SNPs to stringently identify regions of homozygosity, since even relatively uninformative SNP markers are unlikely to be homozygous by state for 30 consecutive markers. The requirement of 30 consecutive homozygous SNPs limited the number of false positive regions yet allowed for the identification of large regions of homozygosity. Because any individual is likely to have multiple homozygous intervals, genetic intervals identified in this manner can be considered only as putative disease loci. However, such loci are useful for prioritizing the selection of candidate genes for mutation screening, and this application of homozygosity data was used successfully in this study. In future studies, the use of higher-density SNP genotyping and the use of genetic and physical map position of the SNPs may allow for the detection of smaller disease candidate regions that are homozygous by descent in other families.
Comparative genomics has been utilized to facilitate the identification of the two most recently identified BBS genes, BBS3 and BBS5 (Chiang et al. 2004; Fan et al. 2004; Li et al. 2004). In the current study, we have refined this approach by selecting two core sets of genomes of model organisms for comparative genomic analysis. An important improvement was the use of the genome of G. lamblia, a bi-nucleated organism, with four pairs of flagella, and one of the most primitive known eukaryotic organisms. It contains no orthologues of the known BBS genes, despite the presence of flagella. This observation suggests that BBS genes are not required for biogenesis or maintenance of the flagella in Giardia. Because Giardia has flagella, the Giardia genome should contain the genes that are required for generating and maintaining flagella. It is the subtraction of this particular set of genes during the comparative genomics analysis that made the genome sequence of Giardia a key resource in defining BBS candidate genes.
A third component to our approach to identifying novel BBS genes was the use of gene expression analysis of a BBS mouse knockout model. We have previously characterized the Bbs4−/− mouse model and found that the homozygous mutant mice underwent an early onset retinal degeneration, with loss of the photoreceptor layer by 8 mo of age (Mykytyn et al. 2004). The loss of the photoreceptor layer provided an opportunity to use this animal model to identify genes that are expressed in the photoreceptor layer by using microarray analysis to compare knockout and control animals. In addition to the Bbs4 gene, six of the seven known mouse BBS genes showed significantly decreased expression in the Bbs4-knockout eye. This suggests that the BBS genes are expressed predominantly in the photoreceptor layer, though they may also be expressed at lower levels in other regions of the eye. Importantly, we would expect that other, as-yet-unidentified BBS genes would demonstrate similar expression in the photoreceptor layer, which would, in turn, make their identification by microarray analysis possible.
Seven of the eight known BBS genes are found in both the comparative genomics and gene expression candidate gene sets. Each set individually has hundreds of potential candidate genes. By identifying genes that were found in both candidate gene sets, it was possible to define a set of 19 gene candidates that had a very high probability of being mutated in patients with BBS. Only the BBS6 gene, MKKS, is not contained in this list of BBS gene candidates. In fact, the MKKS gene does not satisfy criteria for inclusion into either the comparative genomics or gene expression candidate gene sets.
We conducted homozygosity mapping studies in 16 consanguineous BBS families, each containing only one or two affected individuals. The information gained from the mapping studies was used to select those BBS patients that were most likely to harbor mutations in specific candidate genes. This strategy resulted in the identification of the B1 gene on chromosome 7p as a novel BBS gene.
Multiple lines of evidence support the conclusion that the B1 gene is BBS9. A total of seven mutations were detected in the B1 gene in DNA samples from BBS patients. None of these mutations was detected in controls. Of note, all but one of the seven probands in which mutations were found were homozygous for their specific mutation (one patient was a compound heterozygote), despite the fact that the majority of these families were not known to be consanguineous. This finding was predicted for disease-causing loci on the basis of the observation of Sir Archibald Garrod (1902) that patients with rare autosomal recessive disorders were more likely than not to result from consanguineous unions. Furthermore, the tissue expression pattern was similar to that of other BBS genes. The B1 gene has also been shown to be expressed in ciliated cells in C. elegans, a finding that has been observed for other BBS genes (Blacque et al. 2004). The B1 protein has no similarity to the other known BBS proteins and the specific function of the B1 gene is unknown.
In addition to identifying a novel BBS gene, we also used homozygosity mapping to identify three families with homozygous deletions in three different BBS genes (BBS4, BBS5, and BBS7). These deletions are predicted to result in null alleles. They were discovered fortuitously in the consganguineous samples, because of the inability to amplify certain exons from samples with the deletions. However, in the heterozygous state, such deletions would not be detected by current screening methods, since the remaining copy of the BBS gene would provide an adequate template for PCR amplification. Thus, it is likely that some samples for which only a single BBS mutation is detected may actually harbor an undetected deletion or duplication. The existence of patients with BBS who are heterozygous for a single BBS mutation has been used as an argument for the triallelic hypothesis of BBS inheritance (Beales et al. 2003). Large deletions (or duplications) may be the second mutation in some samples for which only a single mutation has been detected. Other types of mutations that are not detected by standard mutation screens include promoter and other regulatory mutations, some of which may be located at a large distance from the first exon of the gene. Together, these classes of mutations might explain cases in which only a single causative BBS mutation has been identified.
In addition to the identification of regions of homozygosity, it is possible to use SNP genotyping for the identification of deleted regions of the genome. In the current study, we did not identify novel BBS candidate regions on the basis of the identification of microdeletions with the use of SNP genotyping. However, the use of higher-density SNP arrays could facilitate the identification of relatively small deletions, either by detecting regions of apparent homozygosity or, in the case of homozygous deletions, by the failure of contiguous SNPs to give a detectable signal on the array.
In summary, we have used a multifaceted approach that combines comparative genomics analysis with gene expression information from a BBS-knockout model, to identify a focused set of BBS candidate genes. Small, consanguineous BBS families were used to identify candidate regions for new BBS loci. By use of this information, the B1 gene was identified as a new BBS gene, BBS9. The identification of BBS9 demonstrates that small, consanguineous BBS families can be used successfully to identify new BBS genes. The mapping information was also used to identify three intragenic deletions within the BBS4, BBS5, and BBS7 genes. Since this type of mutation is not detected by the commonly used mutation detection strategies, the frequency of such mutations in the BBS population is largely unknown and may represent a class of mutation that is underestimated. The approaches used in this study would be applicable to numerous other autosomal recessive disorders.
Acknowledgments
We are grateful to the patients and their families for participating in this study. We thank M. Andrews, G. Beck, K. Bugge, T. Kucaba, R. Mullins, A. Nalley, and M. Olvera, for technical assistance, and D. Aguiar-Crouch, for administrative assistance. This work was supported by the following grants and organizations: National Institutes of Health grants P50-HL-55006 (to V.C.S.) and R01-EY-11298 (to V.C.S. and E.M.S.); Carver Endowment for Molecular Ophthalmology (to E.M.S. and V.C.S.); Research to Prevent Blindness (to the Department of Ophthalmology, University of Iowa); the University of Iowa (to D.Y.N.); and the intramural program of the National Human Genome Research Institute (to L.G.B.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/BLAST/
- Ensembl Genome Browser, http://www.ensembl.org/
- GenBank, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=Nucleotide/ (for NM_198428)
- GiardiaDB, http://gmod.mbl.edu/perl/site/giardia?page-intro/ (for G. lamblia)
- OMIM, http://www.ncbi.nih.gov/entrez/Omim/ (for BBS)
- Sanger Institute, http://www.sanger.ac.uk/ (for L. major)
- SGD, http://www.yeastgenome.org/(for S. cerevisiae)
- The Institute for Genomic Research, http://www.tigr.org/ (for T. cruzi)
- UniGene, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unigene/ (for RefSeq ID numbers NM_014451 and NM_198428)
References
- Adams AE, Rosenblatt M, Suva LJ (1999) Identification of a novel parathyroid hormone-responsive gene in human osteoblastic cells. Bone 24:305–313 10.1016/S8756-3282(98)00188-4 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- Ansley SJ, Badano JL, Blacque OE, Hill J, Hoskins BE, Leitch CC, Kim JC, Ross AJ, Eichers ER, Teslovich TM, Mah AK, Johnsen RC, Cavender JC, Lewis RA, Leroux MR, Beales PL, Katsanis N (2003) Basal body dysfunction is a likely cause of pleiotropic Bardet-Biedl syndrome. Nature 425:628–633 10.1038/nature02030 [DOI] [PubMed] [Google Scholar]
- Avidor-Reiss T, Maer AM, Koundakjian E, Polyanovsky A, Keil T, Subramaniam S, Zuker CS (2004) Decoding cilia function: defining specialized genes required for compartmentalized cilia biogenesis. Cell 117:527–539 10.1016/S0092-8674(04)00412-X [DOI] [PubMed] [Google Scholar]
- Badano JL, Ansley SJ, Leitch CC, Lewis RA, Lupski JR, Katsanis N (2003) Identification of a novel Bardet-Biedl syndrome protein, BBS7, that shares structural features with BBS1 and BBS2. Am J Hum Genet 72:650–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bardet G (1920) Sur un syndrome d’obesite infantile avec polydactylie et retinite pigmentaire (contribution a l’etude des formes cliniques de l’obesite hypophysaire). Thesis, Paris [Google Scholar]
- Beales PL, Badano JL, Ross AJ, Ansley SJ, Hoskins BE, Kirsten B, Mein CA, Froguel P, Scambler PJ, Lewis RA, Lupski JR, Katsanis N (2003) Genetic interaction of BBS1 mutations with alleles at other BBS loci can result in non-Mendelian Bardet-Biedl syndrome. Am J Hum Genet 72:1187–1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedl A (1922) Ein Geschwisterpaar mit adipose-genitaler Dystrophie. Dtsch Med Wochenschr 48:1630 [Google Scholar]
- Blacque OE, Reardon MJ, Li C, McCarthy J, Mahjoub MR, Ansley SJ, Badano JL, Mah AK, Beales PL, Davidson WS, Johnsen RC, Audeh M, Plasterk RH, Baillie DL, Katsanis N, Quarmby LM, Wicks SR, Leroux MR (2004) Loss of C. elegans BBS-7 and BBS-8 protein function results in cilia defects and compromised intraflagellar transport. Genes Dev 18:1630–1642 10.1101/gad.1194004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmi R, Rokhlina T, Kwitek-Black AE, Elbedour K, Nishimura D, Stone EM, Sheffield VC (1995) Use of a DNA pooling strategy to identify a human obesity syndrome locus on chromosome 15. Hum Mol Genet 4:9–13 [DOI] [PubMed] [Google Scholar]
- Chiang AP, Nishimura D, Searby C, Elbedour K, Carmi R, Ferguson AL, Secrist J, Braun T, Casavant T, Stone EM, Sheffield VC (2004) Comparative genomic analysis identifies an ADP-ribosylation factor-like gene as the cause of Bardet-Biedl syndrome (BBS3). Am J Hum Genet 75:475–484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins FS, Brooks LD, Chakravarti A (1998) A DNA polymorphism discovery resource for research on human genetic variation. Genome Res 8:1229–1231 [DOI] [PubMed] [Google Scholar]
- Elbedour K, Zucker N, Zalzstein E, Barki Y, Carmi R (1994) Cardiac abnormalities in the Bardet-Biedl syndrome: echocardiographic studies of 22 patients. Am J Med Genet 52:164–169 10.1002/ajmg.1320520208 [DOI] [PubMed] [Google Scholar]
- Fan Y, Esmail MA, Ansley SJ, Blacque OE, Boroevich K, Ross AJ, Moore SJ, Badano JL, May-Simera H, Compton DS, Green JS, Lewis RA, van Haelst MM, Parfrey PS, Baillie DL, Beales PL, Katsanis N, Davidson WS, Leroux MR (2004) Mutations in a member of the Ras superfamily of small GTP-binding proteins causes Bardet-Biedl syndrome. Nat Genet 36:989–993 10.1038/ng1414 [DOI] [PubMed] [Google Scholar]
- Garrod AE (1902) The incidence of alkaptonuria: a study in chemical individuality. Lancet ii:1616–1620 [Google Scholar]
- Green JS, Parfrey PS, Harnett JD, Farid NR, Cramer BC, Johnson G, Heath O, McManamon PJ, O’Leary E, Pryse-Phillips W (1989) The cardinal manifestations of Bardet-Biedl syndrome, a form of Laurence-Moon-Biedl syndrome. N Engl J Med 321:1002–1009 [DOI] [PubMed] [Google Scholar]
- Harnett JD, Green JS, Cramer BC, Johnson G, Chafe L, McManamon P, Farid NR, Pryse-Phillips W, Parfrey PS (1988) The spectrum of renal disease in Laurence-Moon-Biedl syndrome. N Engl J Med 319:615–618 [DOI] [PubMed] [Google Scholar]
- Hichri H, Stoetzel C, Laurier V, Caron S, Sigaudy S, Sarda P, Hamel C, Martin-Coignard D, Gilles M, Leheup B, Holder M, Kaplan J, Bitoun P, Lacombe D, Verloes A, Bonneau D, Perrin-Schmitt F, Brandt C, Besancon AF, Mandel JL, Cossee M, Dollfus H (2005) Testing for triallelism: analysis of six BBS genes in a Bardet-Biedl syndrome family cohort. Eur J Hum Genet 13:607–616 10.1038/sj.ejhg.5201372 [DOI] [PubMed] [Google Scholar]
- Katsanis N (2004) The oligogenic properties of Bardet-Biedl syndrome. Hum Mol Genet 13 Spec No 1:R65–71 10.1093/hmg/ddh092 [DOI] [PubMed] [Google Scholar]
- Katsanis N, Ansley SJ, Badano JL, Eichers ER, Lewis RA, Hoskins BE, Scambler PJ, Davidson WS, Beales PL, Lupski JR (2001) Triallelic inheritance in Bardet-Biedl syndrome, a Mendelian recessive disorder. Science 293:2256–2259 10.1126/science.1063525 [DOI] [PubMed] [Google Scholar]
- Katsanis N, Beales PL, Woods MO, Lewis RA, Green JS, Parfrey PS, Ansley SJ, Davidson WS, Lupski JR (2000) Mutations in MKKS cause obesity, retinal dystrophy and renal malformations associated with Bardet-Biedl syndrome. Nat Genet 26:67–70 10.1038/79201 [DOI] [PubMed] [Google Scholar]
- Kolomietz E, Meyn MS, Pandita A, Squire JA (2002) The role of Alu repeat clusters as mediators of recurrent chromosomal aberrations in tumors. Genes Chromosomes Cancer 35:97–112 10.1002/gcc.10111 [DOI] [PubMed] [Google Scholar]
- Kwitek-Black AE, Carmi R, Duyk GM, Buetow KH, Elbedour K, Parvari R, Yandava CN, Stone EM, Sheffield VC (1993) Linkage of Bardet-Biedl syndrome to chromosome 16q and evidence for non-allelic genetic heterogeneity. Nat Genet 5:392–396 10.1038/ng1293-392 [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N, Dutcher SK (2004) Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117:541–552 10.1016/S0092-8674(04)00450-7 [DOI] [PubMed] [Google Scholar]
- Mykytyn K, Braun T, Carmi R, Haider NB, Searby CC, Shastri M, Beck G, Wright AF, Iannaccone A, Elbedour K, Riise R, Baldi A, Raas-Rothschild A, Gorman SW, Duhl DM, Jacobson SG, Casavant T, Stone EM, Sheffield VC (2001) Identification of the gene that, when mutated, causes the human obesity syndrome BBS4. Nat Genet 28:188–191 10.1038/88925 [DOI] [PubMed] [Google Scholar]
- Mykytyn K, Mullins RF, Andrews M, Chiang AP, Swiderski RE, Yang B, Braun T, Casavant T, Stone EM, Sheffield VC (2004) Bardet-Biedl syndrome type 4 (BBS4)-null mice implicate Bbs4 in flagella formation but not global cilia assembly. Proc Natl Acad Sci USA 101:8664–8669 10.1073/pnas.0402354101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mykytyn K, Nishimura DY, Searby CC, Shastri M, Yen HJ, Beck JS, Braun T, Streb LM, Cornier AS, Cox GF, Fulton AB, Carmi R, Luleci G, Chandrasekharappa SC, Collins FS, Jacobson SG, Heckenlively JR, Weleber RG, Stone EM, Sheffield VC (2002) Identification of the gene (BBS1) most commonly involved in Bardet-Biedl syndrome, a complex human obesity syndrome. Nat Genet 31:435–438 [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Searby CC, Carmi R, Elbedour K, Van Maldergem L, Fulton AB, Lam BL, Powell BR, Swiderski RE, Bugge KE, Haider NB, Kwitek-Black AE, Ying L, Duhl DM, Gorman SW, Heon E, Iannaccone A, Bonneau D, Biesecker LG, Jacobson SG, Stone EM, Sheffield VC (2001) Positional cloning of a novel gene on chromosome 16q causing Bardet-Biedl syndrome (BBS2). Hum Mol Genet 10:865–874 10.1093/hmg/10.8.865 [DOI] [PubMed] [Google Scholar]
- Nishimura DY, Swiderski RE, Alward WL, Searby CC, Patil SR, Bennet SR, Kanis AB, Gastier JM, Stone EM, Sheffield VC (1998) The forkhead transcription factor gene FKHL7 is responsible for glaucoma phenotypes which map to 6p25. Nat Genet 19:140–147 10.1038/493 [DOI] [PubMed] [Google Scholar]
- Rudiger NS, Gregersen N, Kielland-Brandt MC (1995) One short well conserved region of Alu-sequences is involved in human gene rearrangements and has homology with prokaryotic chi. Nucl Acids Res 23:256–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheffield VC, Carmi R, Kwitek-Black A, Rokhlina T, Nishimura D, Duyk GM, Elbedour K, Sunden SL, Stone EM (1994) Identification of a Bardet-Biedl syndrome locus on chromosome 3 and evaluation of an efficient approach to homozygosity mapping. Hum Mol Genet 3:1331–1335 [DOI] [PubMed] [Google Scholar]
- Slavotinek AM, Stone EM, Mykytyn K, Heckenlively JR, Green JS, Heon E, Musarella MA, Parfrey PS, Sheffield VC, Biesecker LG (2000) Mutations in MKKS cause Bardet-Biedl syndrome. Nat Genet 26:15–16 10.1038/79116 [DOI] [PubMed] [Google Scholar]
- Swiderski RE, Ying L, Cassell MD, Alward WL, Stone EM, Sheffield VC (1999) Expression pattern and in situ localization of the mouse homologue of the human MYOC (GLC1A) gene in adult brain. Brain Res Mol Brain Res 68:64–72 10.1016/S0169-328X(99)00085-6 [DOI] [PubMed] [Google Scholar]
- Vernon EG, Malik K, Reynolds P, Powlesland R, Dallosso AR, Jackson S, Henthorn K, Green ED, Brown KW (2003) The parathyroid hormone-responsive B1 gene is interrupted by a t(1;7)(q42;p15) breakpoint associated with Wilms’ tumour. Oncogene 22:1371–1380 10.1038/sj.onc.1206332 [DOI] [PubMed] [Google Scholar]
- Young TL, Penney L, Woods MO, Parfrey PS, Green JS, Hefferton D, Davidson WS (1999) A fifth locus for Bardet-Biedl syndrome maps to chromosome 2q31. Am J Hum Genet 64:900–904 [DOI] [PMC free article] [PubMed] [Google Scholar]