Abstract
Mitochondrial DNA (mtDNA) mutations are a major cause of human disease. A large number of different molecular defects ultimately compromise oxidative phosphorylation, but it is not clear why the same biochemical defect can cause diverse clinical phenotypes. There is emerging evidence that nuclear genes modulate the phenotype of primary mtDNA disorders. Here, we define an X-chromosomal haplotype that interacts with specific MTND mutations to cause visual failure in the most common mtDNA disease, Leber hereditary optic neuropathy. This effect is independent of the mtDNA genetic background and explains the variable penetrance and sex bias that characterizes this disorder.
Leber hereditary optic neuropathy (LHON [MIM 535000]) typically presents in young adult life with sequential bilateral visual failure due to focal degeneration of retinal ganglion cells within the optic nerve (Nikoskelainen et al. 1996; Howell 1997). In 95% of cases worldwide, LHON is due to one of three point mutations of mtDNA in genes that code for complex I of the respiratory chain: 3460G→A in MTND1, 11778G→A in MTND4, and 14484T→C in MTND6 (Mackey et al. 1996). Only ∼50% of male and ∼10% of female mutation carriers develop symptoms, which indicates that additional genetic or environmental factors are required for the phenotypic expression of LHON (Newman 2002). Epidemiological studies have failed to substantiate anecdotal reports of a link with excess alcohol and tobacco (Kerrison et al. 2000), but segregation analyses are consistent with a nuclear-encoded X-linked susceptibility allele in some pedigrees (Bu and Rotter 1991; Nakamura et al. 1993). Early attempts to map the locus were inconclusive (Chen et al. 1989; Vilkki et al. 1991; Carvalho et al. 1992), but these studies used widely spaced, often noninformative markers and failed to account for mtDNA heteroplasmy, which can influence the expression of LHON. Later studies were more comprehensive, but exclusion mapping was based on an X-linked recessive model, which cannot explain the segregation pattern in all pedigrees (Mackey 1993), and study size limited the power to exclude a substantial portion of the X chromosome (Juvonen et al. 1993; Chalmers et al. 1996).
LHON mutations are found in ∼1 in 8,500 individuals in the general population and are inherited through the maternal line (Man et al. 2003). Large multigenerational pedigrees with LHON are well recognized, with affected individuals present in ⩾10 generations. It is therefore unlikely that a nuclear modifier locus would be strictly coinherited with the primary mtDNA mutation throughout the whole pedigree. Given the relative rarity of the primary mtDNA mutation, it is far more likely that the nuclear modifier is common in the general population and moves in and out of the maternal pedigree through random mating between mothers who transmit the LHON mtDNA mutation (who largely remain unaffected) and unrelated male partners not harboring the LHON mtDNA mutation. The nuclear modifier is therefore likely to be one or more ancient genetic variants that may be present at a high frequency in the population. We therefore used a nonparametric complex-disease-mapping strategy to identify the locus.
A total of 389 DNA samples were collected from affected and unaffected individuals from 100 families with LHON (6 families from Finland, 23 from the United Kingdom, 54 from Italy, 15 from Hungary, 1 from Spain, and 1 from Slovenia). The clinical phenotype was determined by local ophthalmologists. Affected individuals had the characteristic phenotype of LHON, presenting with acute or subacute painless sequential or bilateral visual failure in the presence of an established pathogenic mtDNA mutation, with no other inflammatory, metabolic, or structural cause identified. Unaffected individuals had no visual symptoms and were older than the median age at onset for LHON (24 years). The diagnosis in affected individuals was confirmed by direct sequencing of the MTND genes or by PCR-RFLP analysis. The percentage with heteroplasmy was determined by fluorescent primer-extension assay (Man et al. 2003). Only samples with >70% mutated mtDNA were included in the linkage study, because this level of mutated mtDNA is associated with the same penetrance as homoplasmic mutated mtDNA (Chinnery et al. 2001a). Subjects were homoplasmic for the following primary LHON mtDNA mutations: 11778G→A (n=290 subjects; 51% male), 14484T→C (n=18; 61% male), 3460A→G (n=75; 49% male), and 14495A→G (n=6; 50% male).
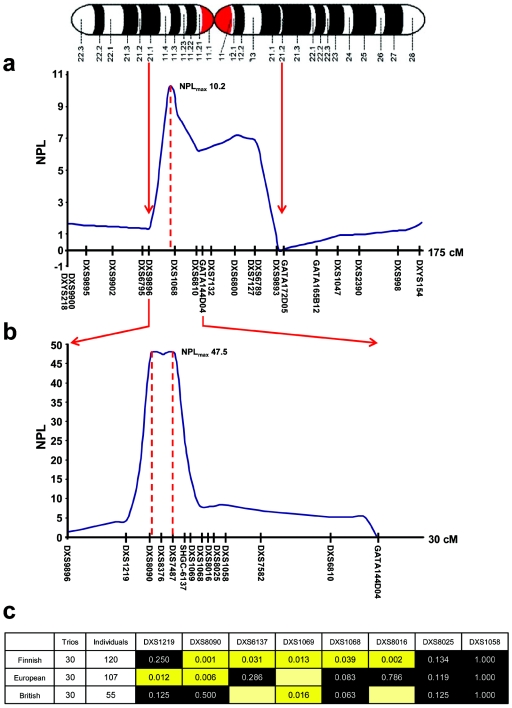
Linkage disequilibrium (LD) blocks in young, geographically isolated populations are generally larger than average, which facilitates low-density linkage mapping of complex traits (Wright et al. 1999). We therefore initially performed nonparametric linkage (NPL) analysis in six Finnish families. X-chromosome microsatellite analysis was performed using part of the 9-cM Weber human genome screening set (version 10aRG [Invitrogen]) in accordance with the manufacturer’s conditions, and markers were sized using a Beckman Coulter CEQ 8000 fluorescent DNA analyzer. NPL analysis was performed using Genehunter, version 1.3. Markers between DXS9896 and DXS9893 had multipoint NPL scores >2, identifying a region with a maximal NPL score of 10.2 (DXS1068 in fig. 1a). The region was mapped further with 12 additional markers identified from the UniSTS database, synthesized with a forward fluorescent tag (Proligo), and sized using the same method. Fine mapping confirmed our findings, with an NPL score >45 spanning 1 cM for markers DXS8090, DXS8376, and DXS7487 (fig. 1b).
Figure 1.
Mapping of the nuclear modifier locus in Finnish pedigrees transmitting homoplasmic LHON mtDNA mutations. a, Human X chromosome with the corresponding NPL scores for the initial linkage screen (maximum NPL score = 10.2). b, Fine mapping of the linked locus, confirming earlier findings and revealing a maximum NPL score of 47.5. c, XRC-TDT for informative markers in the Finnish, mainland European, and British cohorts. As expected, the Finnish families show a large block of linkage and association corresponding to the linked region. There is significant transmission distortion for specific markers at the same locus for the British and mainland European cohorts, which confirms our initial findings and demonstrates the smaller block of LD present outside Finland. A black background indicates that the association is nonsignificant; a yellow background indicates that it is significant at the .05 level; and a yellow box without a value inside it indicates a marker that is uninformative when the number of alleles is <4.
The X-linked reconstruction-combination transmission/disequilibrium test (XRC-TDT) (Horvath et al. 2000) with the use of informative markers confirmed that 2.4 Mb of the X chromosome flanked by DXS8090 and DXS8016 was being preferentially transmitted to affected male offspring in the Finnish families (fig. 1c). We then turned to two independent European cohorts with LHON. In 30 trios from the European mainland, two markers (DXS1219 and DXS8090) showed significant transmission disequilibrium in the XRC-TDT, and, in 30 British trios, DXS1069 showed significant transmission disequilibrium in the XRC-TDT, confirming linkage and association with the same region of the X chromosome (fig. 1c). Differences in the strength of the statistical association between adjacent markers in the different populations is likely a reflection of different marker allele frequencies in the different subgroups.
By comparing microsatellite allele frequencies in 146 European men, we identified the major high-risk haplotype defined by DXS8090 and DXS1068. Of the 89 men affected with LHON, 37 (42%) had the DXS8090(166)-DXS1068(258) haplotype, compared with only 3 (5%) of 57 unaffected men (Fisher’s exact P = .0001; odds ratio [OR] 12.81; 95% CI 3.72–44.11).
The 3460A→G mutation is considered to be the most severe molecular defect, with a consistently reduced complex I activity (Howell et al. 1991), a less marked sex bias (Newman et al. 1991; Johns et al. 1993), and no association with the background mtDNA haplotype (Man et al. 2004). We therefore thought that a nuclear-mitochondrial interaction would be more prominent in non-3460A→G pedigrees. In keeping with this, there was no significant increase in the risk of visual impairment in 3460A→G males with the DXS8090(166)-DXS1068(258) haplotype, but non-3460A→G males were >35-fold more likely to develop visual failure if they had the DXS8090(166)-DXS1068(258) haplotype (table 1). In contrast to 3460A→G, both 11778G→A and 14484T→C are strongly associated with mtDNA haplogroup J (Man et al. 2004). We therefore used logistic regression to determine whether the DXS8090(166)-DXS1068(258) haplotype exerted its effect independent of haplogroup J status, modeling the effect of the nuclear or mtDNA haplotypes alone and through additive, synergistic, and antagonistic interactions. An additive interaction between DXS8090(166)-DXS1068(258) and mtDNA haplogroup was the most compelling model (tables 2 and 3). Non-3460A→G males who did not harbor DXS8090(166)-DXS1068(258) and who belonged to mtDNA haplogroup J had a 35.1% risk of visual failure. The presence of DXS8090(166)-DXS1068(258) or not belonging to mtDNA haplogroup J was associated with a 58.9% risk of visual failure, but 100% of individuals who were both non-mtDNA haplogroup J and had DXS8090(166)-DXS1068(258) were visually impaired (P<.0001). The effect of the nuclear haplotype (OR 33.84; 95% CI 4.38–261.53) was >10-fold greater than that of the mtDNA haplotype (OR 2.45; 95% CI 1.03–5.82).
Table 1.
Frequency of the DXS8090(166)-DXS1068(258) Haplotype in European Males with Different Primary LHON mtDNA Mutations (British and Mainland European Data Combined)[Note]
|
No. of Males |
||||
| LHON mtDNA Mutationand X-Chromosomal Haplotype | Unaffected | Affected | Pa | OR (95% CI)b |
| 3460A→G: | ||||
| Non DXS8090(166)-DXS1068(258) | 7 | 8 | ||
| DXS8090(166)-DXS1068(258) | 2 | 4 | .66 | 1.75 (.24–12.64) |
| Non 3460A→G: | ||||
| Non DXS8090(166)-DXS1068(258) | 47 | 44 | ||
| DXS8090(166)-DXS1068(258) | 1 | 33 | <.0001 | 35.25 (4.62–268.80) |
Note.— 14495A→G has been described in two families with LHON, and, like 14484T→C, it affects the MTND6 gene and is associated with a good prognosis (Chinnery et al. 2001b). The single 14495A→G family was pooled with the other non-3460A→G LHON pedigrees, because of the limited number of 14484T→C and 14495A→G pedigrees. Although it is possible that limited power explains the lack of association between DXS8090(166)-DXS1068(258) and visual failure in 3460A→G males, the increased penetrance of LHON in 3460A→G families (in this study, 62% of males and 29% of females are affected) suggests that a nuclear modifier locus is less influential in this subgroup.
Fisher’s exact P.
Corresponding to the risk of visual failure.
Table 2.
Logistic Regression Model of the Additive Effects of mtDNA Haplotype and the DXS8090(166)-DXS1068(258) Haplotype in Males Homoplasmic for 11778A→G, 14484T→C, and 14495A→G[Note]
| Variables in the Equation | P | OR (95% CI) |
| Non mtDNA haplogroup J | .043 | 2.45 (1.03–5.82) |
| DXS8090(166)-DXS1068(258) | .001 | 33.84 (4.38–261.53) |
| Constant | <.01 | .02 |
Note.— As expected, the frequency of non-3460A→G haplogroup J pedigrees (23%) was greater than the European average (found to be 9%–14% in the European populations studied here). The effect of each genetic variable was approximately the same when modeled independently as when both variables were included in the same regression model. Modeling did not identify a significant direct interaction between mtDNA haplotype and DXS8090(166)-DXS1068(258). χ2 was greatest for the additive model (χ2=35.68; P<.0001). Together, these findings indicate that each genetic factor that exerts its effect is independent and additive. Logistic regression showed an increased risk of visual failure in non–haplogroup J pedigrees in this cohort. A formal study of the penetrance of LHON in haplogroup J pedigrees versus non-J pedigrees had not been performed previously.
Table 3.
Logistic Regression Models of the Effects of mtDNA Background Haplotype and the DXS8090(166)-DXS1068(258) Nuclear Haplotype in Males Homoplasmic for 11778A→G, 14484T→C, and 14495A→G[Note]
| A. Independent Effect of the Nuclear Haplotype DXS8090(166)-DXS1068(258), with χ2 = 31.42, P < .001 | ||
| Variables in the Equation | P | OR (95% CI) |
| DXS8090(166)-DXS1068(258) | .001 | 35.25 (4.62–268.73) |
| Constant | .001 | .03 |
| B. Independent Effect of the mtDNA Haplotype, with χ2 = 7.53, P = .006 | ||
| Variables in the Equation | P | OR (95% CI) |
| Non mtDNA haplotype J | .007 | 2.89 (1.34–6.19) |
| Constant | .001 | .43 |
| C. Synergistic Interaction between DXS8090(166)-DXS1068(258) and Non-J mtDNA Background, with χ2 = 27.38, P < .001 | ||
| Variables in the Equation | P | OR (95% CI) |
| (Non mtDNA haplogroup J) × DXS8090(166)-DXS1068(258) | .65 | 9,340.46 (0–1.91 × 1021) |
| Constant | .65 | 0 |
| D. Antagonistic Interaction between DXS8090(166)-DXS1068(258) and Non-J mtDNA Background, with χ2 = 2.86, P = .09 | ||
| Variables in the Equation | P | OR (95% CI) |
| (mtDNA haplogroup J) × DXS8090(166)-DXS1068(258) | .152 | 4.73 (.56–39.61) |
| Constant | .031 | .467 |
Note.— The additive model is shown in the text and has the greatest χ2 of 35.68 (P < .0001).
To confirm our observations in males, we turned our attention to non-3460A→G female subjects. Of 107 females, 20 (19.05%) were heterozygous for DXS8090(166)-DXS1068(258). Given that LHON was 97% penetrant in non-3460A→G males with LHON who were hemizygous for DXS8090(166)-DXS1068(258), we predicted that approximately four females would be homozygous for DXS8090(166)-DXS1068(258) and that most of them would also be visually impaired. Five women were homozygous, four of whom were also visually impaired, which supports a recessive haploinsufficiency model. Molecular testing of unaffected maternal relatives in known homoplasmic pedigrees with LHON has been of limited clinical use to date, confirming the expected inheritance of the mutation but not altering well-established recurrence risks. Here, we show that identification of DXS8090(166)-DXS1068(258) in hemizygous males or homozygous females greatly increases the chance of visual failure occuring during life. This may be of use in prenatal diagnosis and predictive testing.
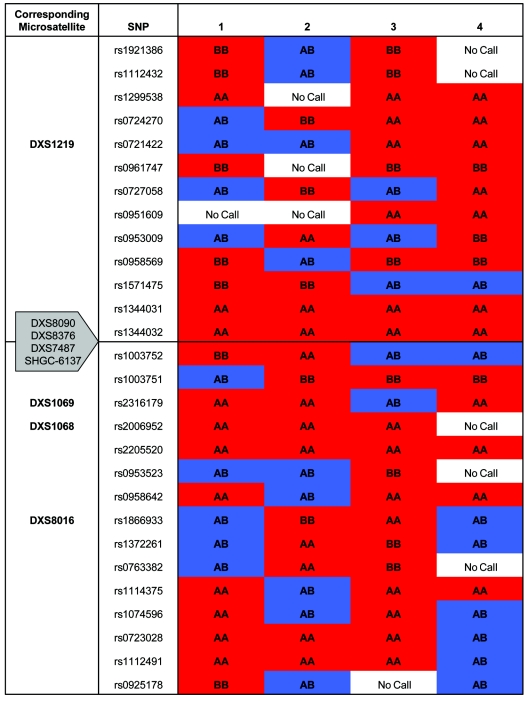
We also identified a second, less-frequent predisposing haplotype in Europeans, DXS1068(258)-DXS8016(218), present in 14.7% of males in this study. All males harboring this haplotype were affected (Fisher’s exact P<.0001), and the majority (74%) of these individuals also had DXS8090(166), indicating that this is a subgroup of the common disease haplotype. It is possible that diallelic mutation of microsatellites and recombination have generated other, less-frequent high-risk marker haplotypes that remain undetected in this study, contributing to marker heterogeneity and confounding attempts to map the locus. We therefore performed homozygosity mapping with the 10K SNP microarray (Affymetrix) for four unrelated women with visual failure due to the 11778A→G mutation who had more than three male offspring, all of whom were clinically affected. All four women were homozygous for SNPs adjacent to the critical region, which pointed toward a small, common shared haplotype block (fig. 2). On the other hand, the striking difference between the Finns and the other European subjects points to allelic heterogeneity at the nuclear visual-loss susceptibility locus, and not just at the flanking markers. Of the affected Finnish men, 63% had a high-risk haplotype defined by DXS8090(168)-DXS1069(262) (P=.0259; OR 7.71; 95% CI 1.28–46.37), found in only 3.7% of mainland Europeans in this study. Different nuclear alleles may behave in subtly different ways that are specific to particular LHON mtDNA mutations or geographic/ethnic regions.
Figure 2.
Homozygosity mapping with the 10K SNP Affymetrix microarray for four unrelated women with visual failure due to a homoplasmic LHON mutation. Subjects 1, 2, and 3 are European. Subject 4 is Finnish. All four individuals had more than three affected sons and no unaffected sons. All four show homozygosity for the two SNPs adjacent to the principal linked markers that define the high-risk disease haplotype.
Epigenetic mechanisms provide an alternative explanation for the reduced penetrance and sex bias in LHON. Although two studies failed to find skewed X inactivation in the blood (Pegoraro et al. 1996) and ocular tissue (Pegoraro et al. 2003) from women affected with LHON, many imprinted disease genes do not show X skewing (Renieri et al. 2003). In families with no known disease, the failure to fully correct a maternal imprint leads to the preferential transmission of grandpaternal chromosomes to male offspring (Naumova et al. 1998). The inheritance of a partially silenced grandpaternal gene could explain the characteristic penetrance of primary LHON mutations in males and could also explain the further reduced penetrance in women when it is superimposed on the normal female random inactivation pattern. X-chromosomal imprinting has been implicated in a number of common neurodevelopmental diseases (Falls et al. 1999), resulting in “parent of origin” effects. We therefore manually reconstructed the 3-generation X-chromosomal haplotypes in 52 males with homoplasmic LHON mtDNA mutations. Overall, we observed the same distortion of transmission of grandpaternal alleles that is well described in control populations (proportion of grandpaternal alleles = 0.60; 95% CI 0.45–0.73; P=.87, for comparison with control data of Naumova et al. [1998]). There was no difference in the frequency of grandpaternal alleles between affected and unaffected male offspring (table 4) nor between affected and unaffected female offspring, which suggests that imprinting is not the principal mechanism underpinning the phenotypic variability seen in LHON.
Table 4.
Grandparental Origin of X-Chromosomal Markers in Males from 3-Generation Pedigrees with Homoplasmic LHON mtDNA Mutations
|
Unaffected Males(n=24) |
Affected Males(n=28) |
||||
| Marker | Grandpaternal | Grandmaternal | Grandpaternal | Grandmaternal | Pa |
| DXS1219 | 14 | 10 | 15 | 13 | .7848 |
| DXS8090 | 14 | 10 | 17 | 11 | 1 |
| DXS8376 | 14 | 10 | 17 | 11 | 1 |
| DXS7487 | 14 | 10 | 17 | 11 | 1 |
| SHGC-6137 | 14 | 10 | 17 | 11 | 1 |
| DXS1069 | 14 | 10 | 17 | 11 | 1 |
| DXS1068 | 14 | 10 | 17 | 11 | 1 |
| DXS8016 | 14 | 10 | 17 | 11 | 1 |
| DXS8025 | 14 | 10 | 17 | 11 | 1 |
| DXS1058 | 16 | 10 | 15 | 13 | .5929 |
| DXS7582 | 16 | 10 | 15 | 13 | .5929 |
| DXS6810 | 14 | 10 | 15 | 13 | .7848 |
Fisher’s exact P comparing the frequency of origin of grandpaternal and grandmaternal alleles in affected and unaffected individuals.
In conclusion, we have identified a region of the X chromosome containing a high-risk haplotype that explains the reduced penetrance and sex bias that characterizes LHON in the majority of pedigrees. Clarification of the underlying sequence variation is likely to have broader implications for our understanding of nuclear-mitochondrial interactions in health and disease, opening new avenues for therapeutic intervention.
Acknowledgments
P.F.C. is a Wellcome Trust Senior Fellow in Clinical Science. P.F.C. also receives funding from Ataxia (UK), the Alzheimer’s Research Trust, the Association Française contre les Myopathies, the United Mitochondrial Diseases Foundation, and the European Union Research Framework Programme (EU FP) EUmitocombat. P.F.C., H.J.M.S., and I.F.M.d.C. are partners in the EU FP6 program MITOCIRCLE. R.H. and V.K. share German-Hungarian Collaborative research grant DFG/436UNG113/153. We are grateful to Mike Gerards, Michael Knapp, Robert Vlietink, Andrew Carothers, and Alan Wright, for their advice, and to the clinicians who sent DNA samples and clinical data, particularly Karin Writzl and John Tolmie.
Web Resources
The URLs for data presented herein are as follows:
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for LHON) [PubMed]
- UniSTS Database, http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?db=unists
References
- Bu X, Rotter JI (1991) X chromosome-linked and mitochondrial gene control of Leber hereditary optic neuropathy: evidence from segregation analysis for dependence on X chromosome inactivation. Proc Natl Acad Sci USA 88:8198–8202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho MR, Muller B, Rotzer E, Berninger T, Kommerell G, Blankenagel A, Savontaus ML, Meitinger T, Lorenz B (1992) Leber’s hereditary optic neuroretinopathy and the X-chromosomal susceptibility factor: no linkage to DXS7. Hum Hered 42:316–320 [DOI] [PubMed] [Google Scholar]
- Chalmers RM, Davis MB, Sweeney MG, Wood NW, Harding AE (1996) Evidence against an X-linked visual loss susceptibility locus in Leber hereditary optic neuropathy. Am J Hum Genet 59:103–108 [PMC free article] [PubMed] [Google Scholar]
- Chen JD, Cox I, Denton MJ (1989) Preliminary exclusion of an X-linked gene in Leber optic atrophy by linkage analysis. Hum Genet 82:203–207 10.1007/BF00291154 [DOI] [PubMed] [Google Scholar]
- Chinnery PF, Andrews RM, Turnbull DM, Howell N (2001a) Leber’s hereditary optic neuropathy: does heteroplasmy influence the inheritance and expression of the G11778A mitochondrial DNA mutation? Am J Med Genet 98:235–243 [DOI] [PubMed] [Google Scholar]
- Chinnery PF, Brown DT, Andrews RM, Singh-Kler R, Riordan-Eva P, Lindley J, Applegarth D, Turnbull DM, Howell N (2001b) The mitochondrial ND6 gene is a hotspot for mutations that cause Leber’s hereditary optic neuropathy. Brain 124:209–218 10.1093/brain/124.1.209 [DOI] [PubMed] [Google Scholar]
- Falls JG, Pulford DJ, Wylie AA, Jirtle RL (1999) Genomic imprinting: implications for human disease. Am J Pathol 154:635–647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath S, Laird NM, Knapp M (2000) The transmission/disequilibrium test and parental-genotype reconstruction for X-chromosomal markers. Am J Hum Genet 66:1161–1167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell N (1997) Leber hereditary optic neuropathy: how do mitochondrial DNA mutations cause degeneration of the optic nerve? J Bioenerg Biomembr 29:165–173 10.1023/A:1022690030664 [DOI] [PubMed] [Google Scholar]
- Howell N, Bindoff LA, McCullough DA, Kubacka I, Poulton J, Mackey D, Taylor L, Turnbull DM (1991) Leber hereditary optic neuropathy: identification of the same mitochondrial ND1 mutation in six pedigrees. Am J Hum Genet 49:939–950 [PMC free article] [PubMed] [Google Scholar]
- Johns DR, Heher KL, Miller NR, Smith KH (1993) Leber’s hereditary optic neuropathy: clinical manifestations of the 14484 mutation. Arch Ophthalmol 111:495–498 [DOI] [PubMed] [Google Scholar]
- Juvonen V, Vilkki J, Aula P, Nikoskelainen E, Savontaus ML (1993) Reevaluation of the linkage of an optic atrophy susceptibility gene to X-chromosomal markers in Finnish families with Leber hereditary optic neuroretinopathy (LHON) [letter]. Am J Hum Genet 53:289–292 [PMC free article] [PubMed] [Google Scholar]
- Kerrison JB, Miller NR, Hsu F, Beaty TH, Maumenee IH, Smith KH, Savino PJ, Stone EM, Newman NJ (2000) A case-control study of tobacco and alcohol consumption in Leber hereditary optic neuropathy. Am J Ophthalmol 130:803–812 10.1016/S0002-9394(00)00603-6 [DOI] [PubMed] [Google Scholar]
- Mackey D (1993) Blindness in offspring of women blinded by Leber’s hereditary optic neuropathy [letter]. Lancet 341:1020–1021 10.1016/0140-6736(93)91105-U [DOI] [PubMed] [Google Scholar]
- Mackey DA, Oostra RJ, Rosenberg T, Nikoskelainen E, Bronte-Stewart J, Poulton J, Harding AE, Govan G, Bolhuis PA, Norby S (1996) Primary pathogenic mtDNA mutations in multigeneration pedigrees with Leber hereditary optic neuropathy. Am J Hum Genet 59:481–485 [PMC free article] [PubMed] [Google Scholar]
- Man PY, Griffiths PG, Brown DT, Howell N, Turnbull DM, Chinnery PF (2003) The epidemiology of Leber hereditary optic neuropathy in the north east of England. Am J Hum Genet 72:333–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Man PY, Howell N, Mackey DA, Norby S, Rosenberg T, Turnbull DM, Chinnery PF (2004) Mitochondrial DNA haplogroup distribution within Leber hereditary optic neuropathy pedigrees. J Med Genet 41:e41 10.1136/jmg.2003.011247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura M, Fujiwara Y, Yamamoto M (1993) The two locus control of Leber hereditary optic neuropathy and a high penetrance in Japanese pedigrees. Hum Genet 91:339–341 [DOI] [PubMed] [Google Scholar]
- Naumova AK, Leppert M, Barker DF, Morgan K, Sapienza C (1998) Parental origin–dependent, male offspring–specific transmission-ratio distortion at loci on the human X chromosome. Am J Hum Genet 62:1493–1499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman NJ (2002) From genotype to phenotype in Leber hereditary optic neuropathy: still more questions than answers. J Neuroophthalmol 22:257–261 [DOI] [PubMed] [Google Scholar]
- Newman NJ, Lott MT, Wallace DC (1991) The clinical characteristics of pedigrees of Leber’s hereditary optic neuropathy with the 11778 mutation. Am J Ophthalmol 111:750–762 [DOI] [PubMed] [Google Scholar]
- Nikoskelainen EK, Huoponen K, Juvonen V, Lamminen T, Nummelin K, Savontaus ML (1996) Ophthalmologic findings in Leber hereditary optic neuropathy, with special reference to mtDNA mutations. Ophthalmology 103:504–514 [DOI] [PubMed] [Google Scholar]
- Pegoraro E, Carelli V, Zevianni M, Cortelli P, Montagna P, Barboni P, Angelini C, Hoffman EP (1996) X-inactivation patterns in female Leber’s hereditary optic neuropathy patients do not support a strong X-linked determinant. Am J Med Genet 61:356–362 [DOI] [PubMed] [Google Scholar]
- Pegoraro E, Vettori A, Valentino ML, Molon A, Mostacciuolo ML, Howell N, Carelli V (2003) X-inactivation pattern in multiple tissues from two Leber’s hereditary optic neuropathy (LHON) patients. Am J Med Genet A 119:37–40 10.1002/ajmg.a.10211 [DOI] [PubMed] [Google Scholar]
- Renieri A, Meloni I, Longo I, Ariani F, Mari F, Pescucci C, Cambi F (2003) Rett syndrome: the complex nature of a monogenic disease. J Mol Med 81:346–354 [DOI] [PubMed] [Google Scholar]
- Vilkki J, Ott J, Savontaus ML, Aula P, Nikoskelainen EK (1991) Optic atrophy in Leber hereditary optic neuroretinopathy is probably determined by an X-chromosomal gene closely linked to DXS7. Am J Hum Genet 48:486–491 [PMC free article] [PubMed] [Google Scholar]
- Wright AF, Carothers AD, Pirastu M (1999) Population choice in mapping genes for complex diseases. Nat Genet 23:397–404 10.1038/70501 [DOI] [PubMed] [Google Scholar]