Abstract
Neurofibromatosis type 1 (NF1) demonstrates phenotypic overlap with Noonan syndrome (NS) in some patients, which results in the so-called neurofibromatosis-Noonan syndrome (NFNS). From a genetic point of view, NFNS is a poorly understood condition, and controversy remains as to whether it represents a variable manifestation of either NF1 or NS or is a distinct clinical entity. To answer this question, we screened a cohort with clinically well-characterized NFNS for mutations in the entire coding sequence of the NF1 and PTPN11 genes. Heterozygous NF1 defects were identified in 16 of the 17 unrelated subjects included in the study, which provides evidence that mutations in NF1 represent the major molecular event underlying this condition. Lesions included nonsense mutations, out-of-frame deletions, missense changes, small inframe deletions, and one large multiexon deletion. Remarkably, a high prevalence of inframe defects affecting exons 24 and 25, which encode a portion of the GAP-related domain of the protein, was observed. On the other hand, no defect in PTPN11 was observed, and no lesion affecting exons 11–27 of the NF1 gene was identified in 100 PTPN11 mutation-negative subjects with NS, which provides further evidence that NFNS and NS are genetically distinct disorders. These results support the view that NFNS represents a variant of NF1 and is caused by mutations of the NF1 gene, some of which have been demonstrated to cause classic NF1 in other individuals.
The so-called neurofibromatosis-Noonan syndrome (NFNS [MIM 601321]) is a peculiar clinical association, first noted in 1985 by Allanson and colleagues (1985), who described subjects with features of both neurofibromatosis type 1 (NF1 [MIM 162200]) and Noonan syndrome (NS [MIM 163950]) (Opitz and Weaver 1985). Since that report, a number of NFNS cases, including a few families transmitting the trait, have been documented (Kaplan and Rosenblatt 1985; Mendez 1985; Saul 1985; Meinecke 1987; Quattrin et al. 1987; Shuper et al. 1987; Abuelo and Meryash 1988; Stern et al. 1992; Colley et al. 1996).
It has been long speculated whether NFNS is a variant of either NF1 (Riccardi 1992) or NS (Allanson 1987), there is a chance association, or they are distinct disorders (Opitz and Weaver 1985; Colley et al. 1996; Carey 1998). Colley et al. (1996) and Bahuau et al. (1996, 1998) documented independent segregation of NF1 and NS traits in two families. Stern et al. (1992) and Colley et al. (1996) reported a few families in which only a fraction of affected members with NF1 exhibited some NS features. Carey et al. (1997) first reported a two-generation family in which the NFNS trait cosegregated with a mutation within the NF1 gene, the gene responsible for all cases of NF1 (Xu et al. 1990). Subsequently, Baralle et al. (2003) examined the NF1 gene in six subjects with NFNS and found mutations in two cases. Very recently, Bertola et al. (2005) reported a patient with NF1 and NS features who carried a heterozygous mutation in both NF1 and PTPN11 (MIM 176876), the latter of which is responsible for half of the cases of NS (Tartaglia et al. 2001), providing the molecular evidence for concurrence of both disorders in one individual. However, from a genetic point of view, NFNS still remains a poorly understood disorder.
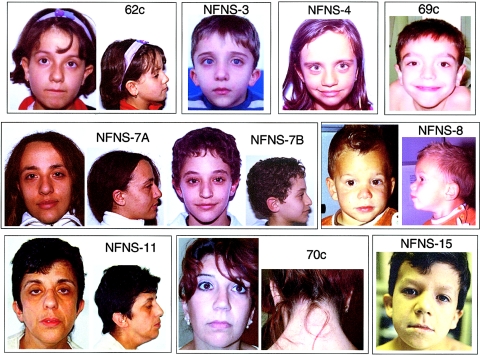
To delineate the genetic cause of NFNS, we screened a well-characterized NFNS cohort for mutations affecting the NF1 and PTPN11 genes. Included in the study were 14 unrelated Italians with sporadic NFNS and three families transmitting the trait. All subjects were evaluated by clinical dysmorphologists experienced with both NF1 and NS (G.Z., A.B., C.D., and B.D.). For each patient, clinical assessment included family history; physical, anthropometric, neurological, and cardiac evaluation (including chest x-ray, electrocardiogram, and 2-dimensional and color Doppler echocardiography); renal ultrasonography; and radiological and magnetic resonance imaging (MRI) studies. The phenotype was evaluated through accurate clinical examination for NS facial and other dysmorphisms, such as hypertelorism (interpupillary distance >2 SD), ptosis (abnormally low lid position), downslanting palpebral fissures, low-set and posteriorly rotated ears, short neck, low posterior hair line, and thoracic and other skeletal anomalies. NF1 was diagnosed on the presence of features fitting the NF1 diagnostic criteria (Stumpf et al. 1988; Gutmann et al. 1997), whereas, for diagnosis of NS, the criteria introduced by van der Burgt et al. (1994) and discussed by Jongmans et al. (2005) were used. The clinical features of the study population are summarized in table 1 and are shown in figure 1. Café-au-lait spots (CLSs) and low-set posteriorly rotated ears were observed in all subjects. CLSs were numerous and stochastically dispersed but variable in number (from 15 to 59) and size (from pointlike to 8×2.5 cm) among different-aged patients. Among the NF1 features, a variable number (1–30) of neurofibromas and Crowe sign (freckling of axillary and inguinal regions) were present in 41% and 73% of subjects, respectively. Lisch nodules were detected in 60% of patients, whereas optic gliomas or other MRI findings, such as unidentified bright objects (UBOs), were found in 28% and 69%, respectively. A single plexiform neurofibroma was observed in one patient. Among the NS features, a variable combination of facial dysmorphisms was observed in all subjects. Short stature and congenital heart defect (CHD) were present in 45% and 32% of the subjects, respectively. In the latter, pulmonary valve stenosis (PVS) was the prevailing defect (50% of cases). Short and/or webbed neck and thoracic abnormalities were also common, present in 64% and 50% of subjects, respectively. Among the features common to both conditions, macrocephaly and scoliosis were observed in 64% and 41% of patients, respectively, whereas mental retardation or learning difficulties were documented in half of all patients. All patients fulfilled the National Institutes of Health Consensus Criteria for the diagnosis of NF1, except for the four youngest patients (individuals NFNS-3, NFNS-4, NFNS-8, and NFNS-10), who presented with only one NF1 criteria, almost certainly because of their young age. In particular, patient NFNS-3, aged 6 years, showed >60 CLSs and mild thickening of the optic nerves on MRI investigation; patient NFNS-4, aged 2.2 years, presented with >20 CLSs and UBOs; whereas patients NFNS-8 and NFNS-10 presented with >15 CLSs at age 2.2 and 4 years, respectively.
Table 1.
Clinical Features of 22 Subjects with NFNS[Note]
|
Findings for |
|||||||||||||||||||||||
| Family 6 |
Family 7 |
Family 15 |
|||||||||||||||||||||
| Characteristics | NFNS1 | 62c | NFNS3 | NFNS4 | 69c | NFNS6a | NFNS6b | NFNS6c | NFNS7a | NFNS7b | NFNS7c | NFNS8 | NFNS9 | NFNS10 | NFNS11 | NFNS12 | 70c | 128 | NFNS15a | NFNS15b | NFNS16 | NFNS17 | Total |
| Age (in years) at observation | 9 | 6.5 | 6 | 2.2 | 8 | 7 | 11 | 41 | 18 | 13 | 47 | 2.2 | 6 | 4 | 40 | 9.5 | 38 | 10 | 17.7 | 45 | 7 | 12 | |
| Sex | M | F | M | F | M | F | M | F | F | M | M | M | F | F | F | F | F | M | M | M | M | M | 12M, 10F |
| NF1 features: | |||||||||||||||||||||||
| CLS | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 22/22 |
| Neurofibromas | + | + (1) | − | − | − | − | − | + | + | − | + | − | − | − | + | − | + | − | + | + | − | − | 9/22 |
| Plexiform neurofibromas | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | − | + (1) | − | − | − | − | − | 1/22 |
| Axillary freckling | + | + | − | − | + | + | + | + | + | − | + | − | − | − | + | + | + | + | + | + | + | + | 16/22 |
| Lisch nodules | − | − | − | − | NE | + | + | + | + | + | + | − | + | − | + | − | NE | + | − | + | + | − | 12/20 |
| Optic gliomas | BIL | MON | * | − | − | − | − | NE | − | − | NE | NE | NE | NE | + | NE | NE | − | − | NE | + | − | 4/14 |
| Other MRI findings | UBOs | UBOs | * | UBOs | NE | − | − | NE | UBOs, SM | UBOs | NE | NE | NE | NE | UBOs | NE | NE | UBOs | − | NE | UBOs | UBOs | 9/13 |
| Mental retardation | − | − | − | − | Mild | − | + | − | − | − | − | − | − | − | − | − | − | − | + | − | + | − | 4/22 |
| Learning difficulties | − | + | + | NV | + | + | + | − | − | + | − | NV | − | − | + | − | − | − | + | + | + | + | 11/20 |
| Scoliosis | − | − | − | − | + | + | + | + | + | − | − | − | − | − | + | + | − | − | + | + | − | + | 9/22 |
| Other tumors | − | − | − | − | − | − | NE | NBL | − | − | − | − | − | − | − | − | − | − | − | − | − | − | 1/21 |
| NS features: | |||||||||||||||||||||||
| Short stature | − | − | + | − | + | Mild | + | + | − | − | − | − | + | − | + | + | + | − | − | − | − | + | 10/22 |
| Macrocephaly | + | + | − | + | + | − | + | − | + | + | + | + | − | + | − | + | − | + | + | + | − | − | 14/22 |
| Cardiac defect | − | − | − | PVS | − | PVS | − | − | MVP | − | − | PVS | − | ASD | − | PVSa | MVT | A | − | − | − | − | 7/22 |
| Hypertelorism | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | − | − | + | + | + | + | 20/22 |
| Downslanting palpebral fissures | − | + | + | + | + | + | + | + | + | + | − | + | + | − | + | + | + | − | + | + | + | + | 18/22 |
| Ptosis | + | + | − | + | − | − | − | + | + | − | − | + | + | + | + | + | − | + | + | + | + | + | 15/22 |
| Malar hypoplasia | − | − | + | − | + | − | − | + | − | − | − | − | + | − | + | − | − | − | − | − | − | − | 5/22 |
| Epicanthic folds | − | + | − | + | − | + | + | − | + | − | − | + | + | + | + | − | + | − | + | − | − | + | 12/22 |
| Other facial dysmorphisms | TL | TL, FNB | − | TL | HP, MS, LE, MO | − | − | − | TL | − | − | TL | − | TL | − | − | HP | TL | TL | TL | SLE | − | |
| Low posterior hairline | + | − | + | + | + | + | + | + | + | + | + | + | + | − | + | + | + | − | + | − | − | + | 17/22 |
| Short/broad/webbed neck | − | − | + | + | + | + | + | + | + | − | − | + | + | − | + | + | + | − | + | − | − | + | 14/22 |
| Low-set posteriorly rotated ears | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | + | 22/22 |
| Pectus/thoracic abnormality | − | + | − | − | + | − | − | + | − | − | − | − | + | − | + | + | + | + | + | − | + | + | 11/22 |
| Cubitus valgus | − | − | − | − | + | + | + | + | − | − | − | − | − | − | + | − | − | − | − | − | + | + | 7/22 |
| Cryptorchidism | − | − | − | + | − | − | − | − | − | − | − | − | 1/12 | ||||||||||
| Urinary system abnormality | − | − | − | − | − | − | − | − | − | − | − | − | − | − | RK | − | − | RC | − | − | − | − | 2/22 |
| Additional features | HN | − | HYP, HJ | − | − | Myopia | − | − | − | − | − | H | E | SD | − | KP, PPD, | − | − | SE, BIL | − | − | − | |
| SD, RBA | CD | ||||||||||||||||||||||
| Family history | S | S | S | S | S | FA | FA | FA | FA | FA | FA | S | S | S | S | S | S | S | FA | FA | S | S | 13 S; 3 FA |
Note.— S=sporadic; FA=familial; a plus sign (+)=present; a minus sign (−)=absent; NE=not evaluated; NV=not evaluable; BIL=bilateral; MON=monolateral. Craniofacial features: HP=high arched palate; MO=malocclusion; TL=thick lips; FNB=flat nasal bridge; LE=large ears. Ectodermal features: KP=keratosis pilaris (face); HN=hairy nevus; E=eczema; SLE=sparse lateral eyebrows. Cardiac defects: ASD=atrial septal defect (ostium secundum); A=arrhythmia (right-branch block); MVT=mitral valve thickening; MVP=mitral valve prolapse. Other features: an asterisk (*)=mild thickening of the optic nerves; SM=thoracic syringomyelia; NBL=neuroblastoma; SD=sensorineural deafness; CD=conductive deafness, SE=seizures; RBA=retarded bone age; HYP=hypotonia; HJ=hyperextensible joints; H=hyperactivity; PPD=postaxial polydactyly of hands; MS=macrostomia; RC=renal cyst; RK=Rokitansky.
With dysplastic leaflets.
Figure 1.
Facial characteristics and other clinical features of study individuals with NFNS
Genomic DNA was isolated from peripheral-blood lymphocytes by phenolchloroform extraction and ethanol precipitation. The entire coding sequence and flanking intronic portions of the NF1 and PTPN11 genes were screened by denaturing high-performance liquid chromatography (DHPLC) analysis, by use of a 3100 or 3500HT WAVE DNA fragment analysis system (Transgenomic). PCR settings, amplicons length, and resolution temperatures for DHPLC analysis were reported elsewhere (Tartaglia et al. 2002, 2004a; De Luca et al. 2003, 2004). Bidirectional sequencing of purified PCR products (Qiagen) was performed using the ABI BigDye Terminator Sequencing Kit v.1.1 (Applied Biosystems) and an ABI 3700 Capillary Array Sequencer or ABI Prism 3100 Genetic Analyzer (Applied Biosystems).
No sequence variation affecting the coding sequence of PTPN11 (exons 1–15 and their flanking intronic stretches) was observed. On the contrary, 15 different NF1 defects were identified in 16 of the 17 unrelated subjects (table 2). Mutations cosegregated with the disease in the three families transmitting NFNS. Consistently, none of the three lesions was observed in >200 Italian control individuals. For sporadic cases with mutations, parental DNA was available for patients NFNS-1, NFNS-9, NFNS-10, and 128; no parent was identified as carrying the mutation. A large deletion involving a portion of the NF1 gene was observed in one subject (NFNS-16). The deletion was suspected because of lack of heterozygosity along the entire NF1 gene. Patient NFNS-16 and both his parents were genotyped using a battery of STR markers, either intragenic to NF1 or flanking the gene (see table 3), and loss of heterozygosity due to loss of the maternal allele of marker 3′-NF1-1 (López Correa et al. 1999), which is located 200 kb downstream of the gene, was documented. STR results were compatible with a partial deletion of ⩽1.7 Mb of the maternal chromosome, between markers D17S1849 (telomeric to exon 23-1) and D17S798 (telomeric to NF1). The deletion was confirmed by quantitative real-time PCR performed on exon 25 of the gene (data not shown; primers, probe, and experimental procedure are available on request). In the remaining patient (NFNS-17), no intragenic defect was identified. This individual exhibited a heterozygous condition for five exonic and intronic polymorphisms (IVS3, 288+41G→A; Ex5, 702A→G; IVS10b, 1528-29insA; IVS29, 5547+19T→A; and IVS41, 7395-29G→A), which would not support occurrence of a deletion involving the entire NF1 gene. On the whole, NF1 gene defects accounted for 16 (94.1%) of the 17 NFNS cases included in the study.
Table 2.
NF1 Gene Mutations in NFNS[Note]
| Subject | Exon | NucleotideMutation | Predicted Amino Acid Change | Type/Effect | Reference |
| NFNS-1 | 4b | c.581T→G | L194R | Missense | Novel |
| 62c | 11 | 1721+3A→G | Splicing/truncating | Purandare et al. 1994 | |
| NFNS-3 | 12a | 1756delACTA | Deletion/truncating | Park and Pivnick 1998 | |
| NFNS-4 | 12b | 1862delC | Deletion/truncating | Novel | |
| 69c | 13 | 2153delA | Deletion/truncating | Novel | |
| NFNS-6a | 17 | 2970delAAT | 991delMb | Inframe deletion | Carey et al. 1997 |
| NFNS-7a | 24 | 4243G→T | E1415X | Nonsense/truncating | Fahsold et al. 2000 |
| NFNS-8 | 24 | 4267A→G | K1423Eb | Missense | Li et al. 1992 |
| NFNS-9 | 24 | 4267A→G | K1423Eb | Missense | Li et al. 1992 |
| NFNS-10 | 25 | 4289A→C | N1430T | Missense | Novel |
| NFNS-11 | 25 | 4294G→C | V1432L | Missense | Novel |
| NFNS-12 | 25 | 4312delGAA | 1438delEb | Inframe deletion | Baralle et al. 2003 |
| 70c | 29 | 5339T→G | L1780X | Nonsense/truncating | Fahsold et al. 2000 |
| 128 | 35 | 6641+1G→A | Splicing/truncating | Novel | |
| NFNS-15c | 45 | 7877delG | Deletion/truncating | Novel | |
| NFNS-16 | Partial NF1 gene deletiond | Novel |
Note.— Mutations affecting residues located within the GAP-related domain are shown in bold italics.
From a family with three affected members.
Mutations found to recur in NFNS.
From a family with two affected members.
Centromeric breakpoint maps between marker D17S1849 (intron 23-1) and exon 25, and telomeric breakpoint localizes between markers 3′-NF1 (200 kb downstream of the NF1 gene) and D17S798 (1.6 Mb downstream of the NF1 gene).
Table 3.
Markers on Chromosome 17q11.2
|
Microsatellite Marker |
|||
| Markers | Position Relative to the NF1 Gene | Genotype in NFNS-16 | Microsatellite Positiona (bp) |
| D17S1873 | Centromeric | Homozygote | 24481471 |
| D17S841 | Centromeric | Homozygote | 24566579 |
| D17S1863 | Centromeric | Heterozygote | 25954520 |
| D17S635 | Centromeric | Homozygote | 26281121 |
| NF1 start point | 26443243 | ||
| D17S1849 | IVS-23-1 | Heterozygote | 26594230 |
| D17S1166 | IVS-27b | Homo/hemizygote | 26673142 |
| SNP-IVS-29 | IVS-29 | Homo/hemizygote | 26679002 |
| IVS-38 | IVS-38 | Homo/hemizygote | 26692648 |
| NF1 stop point | 26725590 | ||
| 3′ NF1-1 | Telomeric | Hemizygote | 26943296 |
| D17S1800 | Telomeric | Homo/hemizygote | 26960896 |
| 3′ NF1-2 | Telomeric | Homo/hemizygote | 26963296 |
| D17S798 | Telomeric | Heterozygote | 28313925 |
| D17S250 | Telomeric | Heterozygote | 34405617 |
Positions of STR markers are based on the UCSC Genome Browser human reference sequence.
NF1 intragenic lesions included nonsense and frameshift mutations as well as missense mutations and small inframe deletions. According to the Human Gene Mutation Database, 8 of the 15 defects were novel, whereas the remaining 7 had been documented in patients with NF1, either with or without NS features. Eight defects, including four small out-of-frame deletions and two splice-site and two nonsense mutations, were predicted to result in a truncated protein. Five mutations of this group—1721+3A→G, 1756delACTA, 1862delC, 2153delA, and 4243G→T—would result in a protein missing a portion of or the entire GAP-related domain, which has a major role in controlling RAS function by promoting conversion of active guanosine triphosphate (GTP)–binding RAS to inactive guanosine diphosphate (GDP)–binding RAS (Martin et al. 1990). Of note, four of these mutations had been reported elsewhere in patients with NF1, none of whom exhibited features of the NFNS condition. Six different missense mutations or small inframe deletions were identified in seven probands. Remarkably, these lesions were not randomly distributed; the majority involved portions of the gene coding for two functional domains of the protein. Specifically, five defects affected the GAP-related domain (exons 21–27a), the majority clustering in a short amino acid stretch (residues 1423–1438) of this domain, whereas the single-residue deletion affecting codon 991 was located within the putative cysteine/serine–rich domain (exons 11–17). Comparison with NF1 orthologs demonstrated that all the affected residues were highly conserved among vertebrates. Three of these mutations—991delM, K1423E, and 1438delE—were reported elsewhere (Li et al. 1992; Carey et al. 1997; Baralle et al. 2003). Among them, 991delM and 1438delE had been documented in patients with NFNS (Carey et al. 1997; Baralle et al. 2003), which indicates a genotype-phenotype correlation.
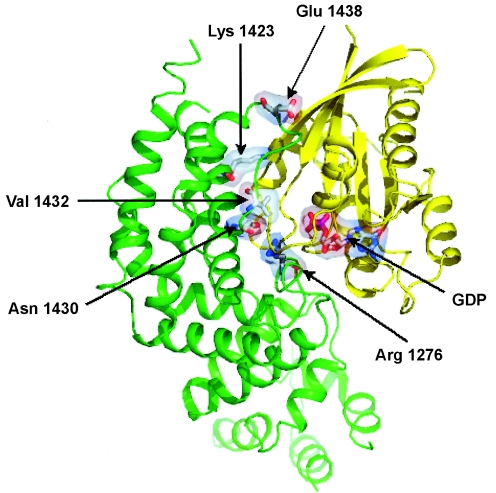
Because of relevant clustering of inframe mutations within a small portion of the GAP-related domain, the predicted spatial location and function of four affected residues—K1423, N1430, V1432, and E1438—were analyzed. To model the structure of the neurofibromin/RAS complex, RAS was docked to the structure of the neurofibromin GAP-related domain (Protein Data Bank [PDB] code 1NF1), according to the experimental topology of binding between the homologous p120GAP and HRAS (PDB code 1WQ1). This model is in agreement with the model of the neurofibromin GRD/RAS complex described by Scheffzek et al. (1998). As shown in figure 2, all the affected residues lie at the interface of interaction between the GAP-related domain of neurofibromin and RAS, close to the catalytic site of the former. Such impressive clustering suggests that mutations affecting these residues might impair the ability of the neurofibromin GAP-related domain in stimulation of the GTPase activity of RAS, affecting either stability of the neurofibromin/RAS complex or neurofibromin catalytic activity. Structural data indicate that K1423 is directly involved in the intermolecular interaction, since it forms a salt bridge with residue D38 of RAS. Such interaction is lost in the K1423E mutant, since the positive charge of the lysine is replaced with the negative charge of a glutamic acid residue. Because of the electrostatic repulsion between the two negatively charged residues E1423 and D38, this mutation causes further destabilization of the neurofibromin/RAS complex. It has been consistently demonstrated that the K1423E mutation results in a dramatic reduction of GAP activity (Poullet et al. 1994). The structural reorganization caused by the N1430T mutation might affect the local environment in proximity to residue R1276, which is known as the arginine finger of the GAP-related domain and therefore might perturb the catalytic activity of the protein. Disruptive effects on both substrate binding and catalysis can also be expected for the substitution of V1432, which lies on the surface of the GAP-related domain in proximity to the arginine finger. In regard to the 1438delE mutation, E1438 is located in a region of the domain with local negative electrostatic potential. Loss of this ionizable negative group is predicted to cause decreased repulsion to the negative surface of RAS, which promotes an increased stability of the neurofibromin/RAS complex. Other mutations (R1413G/K1436R, N1430H, R1491K, and R1276G) with a similar perturbing role on neurofibromin binding to RAS were described elsewhere (Morcos et al. 1996). Such an abnormally increased stability of the complex would result in the saturation of the GAP active site by lower RAS concentrations and, as a consequence, in decreased availability of the protein for stimulation of the GTPase activity of further incoming RAS molecules.
Figure 2.
Three-dimensional model showing location of residues affected by mutations in the neurofibromin GAP-related domain/RAS complex. Ribbon-like structures of neurofibromin GAP-related domain and RAS are shown in green and yellow, respectively. The mutated residues (Lys1423, Asn1430, Val1432, and Glu1438), the catalytic arginine finger (Arg1276), and GDP are represented by sticks and transparent surfaces. Molecular alignment and representation were made with the programs SwissPdb Viewer v. 3.7 (Guex and Peitsch 1997) and PyMOL (DeLano 2002), respectively.
Because of the wide clinical spectrum associated with different NF1 mutations, we investigated possible genotype-phenotype relationships. Comparison with the data reported by Fahsold et al. (2000) demonstrated a significantly higher prevalence of missense mutations and inframe deletions among patients with NFNS than among individuals with NF1 (7/16 vs. 28/278; χ2=13.31; P=.0003). Within the NFNS cohort, comparison of the clinical features between patients carrying missense mutations or inframe deletions and those with truncating mutations showed a preferential but not significant association, with a slightly increased risk of CHD (odds ratio [OR]=2.40; 95% CI 0.27–23.27; Fisher's exact test one-tailed P=.32). Among the families transmitting the trait, intrafamilial phenotypic variability was evident. Patient NFNS-17, who apparently did not carry any NF1 gene mutation or deletion, fulfilled the NF1 diagnostic criteria and presented with CLSs, axillary freckling, learning difficulties, and the typical NS-associated facial dysmorphisms, short stature, webbed neck, and thoracic abnormality.
Because of the clinical overlap between NFNS and NS, as well as the antagonistic modulatory function of neurofibromin and SHP-2 in RAS signaling, we also investigated the possible contribution of NF1 gene lesions to NS. One hundred unrelated patients with NS but no symptoms of NF1 were screened for those exons (exons 11–27) and flanking intronic sequences that represent the major hotspot regions in patients with NFNS. All subjects had been documented elsewhere to be negative for mutations affecting the PTPN11 coding sequence (Sarkozy et al. 2003; M.T., unpublished data). No NF1 pathogenetic mutation was detected.
As discussed elsewhere by Carey (1998), despite the amount of work done and number of cases documented, including a few families transmitting the trait, debate still continues about the genetic cause and nosologic entity of NFNS. It has been suggested that the NFNS phenotype might represent the result of a chance association of two common autosomal dominant disorders—NF1 and NS—or that certain NF1 features in subjects with NFNS might occur as a component of NS. In NS, a similar association was documented for the occurrence of bony and soft-tissue giant cell lesions or CLSs, as observed, respectively, in Noonan-like/multiple giant cell lesion syndrome (NL/MGCLS [MIM 163955]) and LEOPARD syndrome (LS [MIM 151100]). Indeed, both NL/MGCLS and LS are caused by missense mutations in the PTPN11 gene (Digilio et al. 2002; Legius et al. 2002; Tartaglia et al. 2002; Sarkozy et al. 2004; Lee et al. 2005), which is mutated in a large percentage of subjects with NS (Tartaglia and Gelb 2005). These conditions can now be viewed as either a part of the NS phenotypic spectrum (NL/MGCLS) or an allelic variant of NS (LS).
In contrast, with one exception, no mutation affecting PTPN11 has been identified in NFNS thus far (Baralle et al. 2003; Bertola et al. 2005; present study), which strongly supports the hypothesis that PTPN11 is not a major disease gene contributing to or causing NFNS and that NFNS and NS are distinct genetic disorders. Accordingly, large clinical studies of NS make no reference to patients with neurofibromas, which suggests that NF1 features do not occur frequently in classic NS (Sharland et al. 1992). Coexistence of NS and NF1 features has also been explained by consideration of certain NS signs in subjects with NFNS as part of the phenotypic variability of NF1 or as a distinct and well-delineated condition. Both these possibilities imply mutations in NF1 as a common molecular event underlying the condition. However, in the former, one would expect both a number of mutations shared with NF1 and a similar distribution. In the latter, specific mutations not occurring (or rare) in patients with NF1 would be expected. The identification of NF1 lesions in 16 of 17 subjects in the present cohort provides evidence of a major role of NF1 in NFNS. With combination of the present and previous data, 18 distinct NF1 gene mutations have been described in 22 unrelated patients with NFNS (Carey et al. 1997; Baralle et al. 2003; Castle et al. 2003; Bertola et al. 2005; present study). These lesions include nonsense mutations, out-of-frame deletions, insertions, or splicing mutations, as well as missense mutations and small inframe deletions. Among them, a statistically significant incidence (42%) of missense mutations and small inframe deletions is observed. Remarkably, inframe defects account for a considerably lower percentage (10%) of NF1 lesions among patients with NF1 than among those with NFNS (10/24 patients with NFNS studied [Carey et al. 1997; Baralle et al. 2003; Castle et al. 2003; Bertola et al. 2005; present study] vs. 28/278 patients with NF1 [Fahsold et al. 2000]; χ2=17.28; P=.00003). In NFNS, most (60%) of these inframe mutations cluster to the GAP-related domain, which does not represent the major (25%) mutational hotspot region for missense mutations in patients with NF1 (6/10 patients with NFNS [Carey et al. 1997; Baralle et al. 2003; Castle et al. 2003; Bertola et al. 2005; present study] vs. 7/28 patients with NF1 [Fahsold et al. 2000]; Fisher's exact test one-tailed P=.055).
A nonsignificant trend of association between inframe mutations and CHD was also observed. Of note, two of the mutations (991delM and 1438delE) we documented in subjects with CHD had been associated with CHD elsewhere (Carey et al. 1997; Castle et al. 2003). In particular, 991delM had been associated with PVS in patients with NFNS (Carey et al. 1997; present study) and in patients with Watson syndrome, a condition characterized by PVS, CLSs, and mental retardation and caused by NF1 gene mutations (Tassabehji et al. 1993; Castle et al. 2003), thus suggesting an independent relationship between 991delM and PVS by the associated phenotype
Remarkably, the clinical phenotype of patients reported here is characterized by a peculiar presentation of NF1 and NS features. Specifically, whereas plexiform neurofibroma was found only in a single patient, neither pseudoarthrosis of the tibia nor NF1 gene mutation–related tumors—other than neurofibromas, optic gliomas, and one neuroblastoma—occurred, either in pediatric or in adult patients. These findings partly overlap with the conclusions of Carey (1998), who reported the absence of Lisch nodules, the small number of dermal neurofibromas, and the lack of internal tumors as distinct features of NFNS. Among NS features, typical dysmorphisms were hypertelorism, ptosis, and low-set ears, whereas short neck and stature and thoracic anomalies were present in only half of the patients. The identification of specific NF1 alleles recurring in NFNS, the evidence that these alleles cosegregate with the condition in families, and the observation of a peculiar mutational spectrum strongly suggest that the term “NFNS” does characterize a phenotypic variant of NF1, which manifests with a low incidence of plexiform neurofibromas, skeletal anomalies, and internal tumors, in association with hypertelorism, ptosis, low-set ears, and CHDs. However, it should be noted that some of the mutations identified in patients with NFNS have also been reported in NF1 without any feature suggestive of NS. From a molecular point of view, the clinical overlap between NFNS and NS is not surprising. Increasing evidence supports the hypothesis that NF1 and PTPN11 gene products—neurofibromin and SHP-2 (a cytoplasmic protein tyrosine phosphatase functioning as a transducer)—elicit their modulatory role through a common pathway. Indeed, whereas neurofibromin stimulates the intrinsic GTP hydrolysis of RAS proteins required for their functional silencing, SHP-2 promotes their sustained activation. The antagonistic function of neurofibromin and SHP-2 on RAS-mediated transduction cascades modulates cell response to several growth-factor and cytokine receptors, which control a number of developmental processes (Dasgupta and Gutmann 2003; Neel et al. 2003; Arun and Gutmann 2004; Tartaglia et al. 2004b). Consistent with the crucial role of neurofibromin and SHP-2 in modulating cell proliferation, children with NF1 or NS are predisposed to distinct but overlapping spectra of hematologic malignancies (Shannon et al. 1994; Side et al. 1998; Tartaglia et al. 2003, 2004a, 2005). For both NF1 and NS, deregulation of RAS signaling appears to occur in a ligand-dependent manner, which suggests that the differential contribution of these proteins to modulation of transduction pathways elicited by distinct signals might account for the phenotypic differences observed in NF1/NFNS and NS.
On the whole, the present study provides the first molecular evidence of a major role of NF1 mutations in NFNS, emphasizing the extreme phenotypic variability associated with lesions in the NF1 gene. We hypothesize that mutations affecting regulatory portions of the gene might also have a pathogenetic role in NFNS. Although mutations affecting the NF1 promoter have not been reported to date (Horan et al. 2000), lesions in other noncoding portions are to be expected. Gross rearrangements consistently comprise up to 5% of all NF1 mutations documented in NF1 (Korf 1998; Upadhyaya and Cooper 1998; Kluwe et al. 2004). Even though the present data do not allow exclusion of the possibility that NFNS is genetically heterogeneous, they definitely exclude defects in the coding sequence of the PTPN11 gene as a recurrent molecular event underlying NFNS. Similarly, the recently reported chance occurrence of mutations in both NF1 and PTPN11 represent a rare event in NFNS, probably accounting for a minority of these cases (Bertola et al. 2005). Finally, mutations in NF1 are unlikely to play an important role in NS, which further supports the view that NFNS is genetically distinct from NS.
Acknowledgments
We thank the patients and families who participated in this study and the physicians who referred the subjects. This work was supported in part by grants from the Italian Ministry of Health (RC 2004 and RC 2005) and the Italian Ministry of Education, University and Research (to B.D.) and Telethon-Italy (GGP04172) and Programma di Collaborazione Italia-USA/malattie rare (to M.T.).
Web Resources
The URLs for data presented herein are as follows:
- Human Gene Mutation Database, http://archive.uwcm.ac.uk/uwcm/mg/hgmd0.html
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for NFNS, NF1, NS, PTPN11, NL/MGCLS, and LS)
- PDB, http://www.rcsb.org/pdb/
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- Abuelo DN, Meryash DL (1988) Neurofibromatosis with fully expressed Noonan syndrome. Am J Med Genet 29:937–941 10.1002/ajmg.1320290426 [DOI] [PubMed] [Google Scholar]
- Allanson JE (1987) Noonan syndrome. J Med Genet 24:9–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allanson JE, Hall JG, Van Allen MI (1985) Noonan phenotype associated with neurofibromatosis. Am J Med Genet 21:457–462 10.1002/ajmg.1320210307 [DOI] [PubMed] [Google Scholar]
- Arun D, Gutmann DH (2004) Recent advances in neurofibromatosis type 1. Curr Opin Neurol 17:101–105 10.1097/00019052-200404000-00004 [DOI] [PubMed] [Google Scholar]
- Bahuau M, Flintoff W, Assouline B, Lyonnet S, Le Merrer M, Prieur M, Guilloud-Bataille M, Feingold N, Munnich A, Vidaud M, Vidaud D (1996) Exclusion of allelism of Noonan syndrome and neurofibromatosis-type 1 in a large family with Noonan syndrome-neurofibromatosis association. Am J Med Genet 66:347–355 [DOI] [PubMed] [Google Scholar]
- Bahuau M, Houdayer C, Assouline B, Blanchet-Bardon C, Le Merrer M, Lyonnet S, Giraud S, Recan D, Lakhdar H, Vidaud M, Vidaud D (1998) Novel recurrent nonsense mutation causing neurofibromatosis type 1 (NF1) in a family segregating both NF1 and Noonan syndrome. Am J Med Genet 75:265–272 [DOI] [PubMed] [Google Scholar]
- Baralle D, Mattocks C, Kalidas K, Elmslie F, Whittaker J, Lees M, Ragge N, Patton MA, Winter R, ffrench-Constant C (2003) Different mutations in the NF1 gene are associated with neurofibromatosis-Noonan syndrome (NFNS). Am J Med Genet 119:1–8 10.1002/ajmg.a.20023 [DOI] [PubMed] [Google Scholar]
- Bertola DR, Pereira AC, Passetti F, de Oliveira PS, Messiaen L, Gelb BD, Kim CA, Krieger JE (2005) Neurofibromatosis-Noonan syndrome: molecular evidence of the concurrence of both disorders in a patient. Am J Med Genet A 136:242–245 [DOI] [PubMed] [Google Scholar]
- Carey JC (1998) Neurofibromatosis-Noonan syndrome. Am J Med Genet 75:263–264 [DOI] [PubMed] [Google Scholar]
- Carey JC, Stevenson DA, Ota M, Neil S, Viskochil DH (1997) Is there an Noonan syndrome: part 2: documentation of the clinical and molecular aspects of an important family. Proc Greenwood Genet Center 17:52–53 [Google Scholar]
- Castle B, Baser ME, Huson SM, Cooper DN, Upadhyaya M (2003) Evaluation of genotype-phenotype correlations in neurofibromatosis type 1. J Med Genet 40:109 10.1136/jmg.40.10.e109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colley A, Donnai D, Evans DGR (1996) Neurofibromatosis/Noonan phenotype: a variable feature of type 1 neurofibromatosis. Clin Gen 49:59–64 [DOI] [PubMed] [Google Scholar]
- Dasgupta B, Gutmann DH (2003) Neurofibromatosis 1: closing the GAP between mice and men. Curr Opin Genet Dev 13:20–27 10.1016/S0959-437X(02)00015-1 [DOI] [PubMed] [Google Scholar]
- De Luca A, Buccino A, Gianni D, Mangino M, Giustini S, Richetta A, Divona L, Calvieri S, Mingarelli R, Dallapiccola B (2003) NF1 gene analysis based on DHPLC. Hum Mutat 21:171–172 [DOI] [PubMed] [Google Scholar]
- De Luca A, Schirinzi A, Buccino A, Bottillo I, Sinibaldi L, Torrente I, Ciavarella A, Dottorini T, Porciello R, Giustini S, Calvieri S, Dallapiccola B (2004) Novel and recurrent mutations in the NF1 gene in Italian patients with neurofibromatosis type 1. Hum Mutat 23:629 10.1002/humu.9245 [DOI] [PubMed] [Google Scholar]
- DeLano WL (2002) Unraveling hot spots in binding interfaces: progress and challenges. Curr Opin Struct Biol 12:14–20 10.1016/S0959-440X(02)00283-X [DOI] [PubMed] [Google Scholar]
- Digilio MC, Conti E, Sarkozy A, Mingarelli R, Dottorini T, Marino B, Pizzuti A, Dallapiccola B (2002) Grouping of multiple-lentigines/LEOPARD and Noonan syndromes on the PTPN11 gene. Am J Hum Genet 71:389–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fahsold R, Hoffmeyer S, Mischung C, Gille C, Ehlers C, Kücükceylan N, Abdel-Nour M, Gewies A, Peters H, Kaufmann D, Buske A, Tinschert S, Nürnberg P (2000) Minor lesion mutational spectrum of the entire NF1 gene does not explain its high mutability but points to a functional domain upstream of the GAP-related domain. Am J Hum Genet 66:790–818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guex N, Peitsch (1997) MC SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723 10.1002/elps.1150181505 [DOI] [PubMed] [Google Scholar]
- Gutmann DH, Aylsworth A, Carey JC, Korf B, Marks J, Pyeritz RE, Rubenstein A, Viskochil D (1997) The diagnostic evaluation and multidisciplinary management of neurofibromatosis 1 and neurofibromatosis 2. JAMA 278:51–57 10.1001/jama.278.1.51 [DOI] [PubMed] [Google Scholar]
- Horan MP, Cooper DN, Upadhyaya M (2000) Hypermethylation of the neurofibromatosis type 1 (NF1) gene promoter is not a common event in the inactivation of the NF1 gene in NF1-specific tumours. Hum Genet 107:33–39 10.1007/s004390050007 [DOI] [PubMed] [Google Scholar]
- Jongmans M, Sistermans EA, Rikken A, Nillesen WM, Tamminga R, Patton M, Maier EM, Tartaglia M, Noordam K, van der Burgt I (2005) Genotypic and phenotypic characterization of Noonan syndrome: new data and review of the literature. Am J Med Genet A 134:165–170 [DOI] [PubMed] [Google Scholar]
- Kaplan P, Rosenblatt B (1985) A distinctive facial appearance in neurofibromatosis von Recklinghausen. Am J Med Genet 21:463–470 10.1002/ajmg.1320210308 [DOI] [PubMed] [Google Scholar]
- Kluwe L, Siebert R, Gesk S, Friedrich RE, Tinschert S, Kehrer-Sawatzki H, Mautner VF (2004) Screening 500 unselected neurofibromatosis 1 patients for deletions of the NF1 gene. Hum Mutat 23:111–116 10.1002/humu.10299 [DOI] [PubMed] [Google Scholar]
- Korf BR (1998) The NF1 Genetic Analysis Consortium. In: Upadhyaya M, Cooper DN (eds) Neurofibromatosis type 1: from genotype to phenotype. BIOS Scientific, Oxford, United Kingdom, pp 57–62 [Google Scholar]
- Lee JS, Tartaglia M, Gelb BD, Fridrich K, Sachs S, Stratakis CA, Muenke M, Robey PG, Collins MT, Slavotinek A (2005) Phenotypic and genotypic characterisation of Noonan-like/multiple giant cell lesion syndrome. J Med Genet 42:e11 10.1136/jmg.2004.024091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legius E, Schrander-Stumpel C, Schollen E, Pulles-Heintzberger C, Gewillig M, Fryns JP (2002) PTPN11 mutations in LEOPARD syndrome. J Med Genet 39:571–574 10.1136/jmg.39.8.571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Bollag G, Clark R, Stevens J, Conroy L, Fults D, Ward K, Freidman E, Samowits W, Robertson M, Bradley P, McCormick F, White R, Cawthon R (1992) Somatic mutations in the neurofibromatosis type 1 gene in human tumours. Cell 69:275–281 10.1016/0092-8674(92)90408-5 [DOI] [PubMed] [Google Scholar]
- López Correa C, Brems H, Lázaro C, Estivill X, Clementi M, Mason S, Rutkowski JL, Marynen P, Legius E (1999) Molecular studies in 20 submicroscopic neurofibromatosis type 1 gene deletions. Hum Mutat 14:387–393 [DOI] [PubMed] [Google Scholar]
- Martin GA, Viskochil D, Bollag G, McCabe PC, Crosier WJ, Haubruck H, Conroy L, Clark R, O'Connell P, Cawthon RM, Innis MA, McCormick F (1990) The GAP-related domain of the neurofibromatosis type 1 gene product interacts with ras p21. Cell 63:843–849 10.1016/0092-8674(90)90150-D [DOI] [PubMed] [Google Scholar]
- Meinecke P (1987) Evidence that the “neurofibromatosis-Noonan syndrome” is a variant of von Recklinghausen neurofibromatosis. Am J Med Genet 26:741–745 10.1002/ajmg.1320260331 [DOI] [PubMed] [Google Scholar]
- Mendez HM (1985) The neurofibromatosis-Noonan syndrome. Am J Med Genet 21:471–476 10.1002/ajmg.1320210309 [DOI] [PubMed] [Google Scholar]
- Morcos P, Thapar N, Tusneem N, Stacey D, Tamanoi F (1996) Identification of neurofibromin mutants that exhibit allele specificity or increased Ras affinity resulting in suppression of activated ras alleles. Mol Cell Biol 16:2496–2503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neel BG, Gu H, Pao L (2003) The ‘Shp’ing news: SH2 domain-containing tyrosine phosphatases in cell signaling. Trends Biochem Sci 28:284–293 10.1016/S0968-0004(03)00091-4 [DOI] [PubMed] [Google Scholar]
- Opitz JM, Weaver DD (1985) The neurofibromatosis-Noonan syndrome. Am J Med Genet 21:477–490 10.1002/ajmg.1320210310 [DOI] [PubMed] [Google Scholar]
- Park VM, Pivnick EK (1998) Neurofibromatosis type 1 (NF1): a protein truncation assay yielding identification of mutations in 73% of patients. J Med Genet 35:813–820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poullet P, Lin B, Esson K, Tamanoi F (1994) Functional significance of lysine 1423 of neurofibromin and characterization of a second site suppressor which rescues mutations at this residue and suppresses RAS2Val-19-activated phenotypes. Mol Cell Biol 14:815–821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purandare SM, Lanyon WG, Connor JM (1994) Characterisation of inherited and sporadic mutations in neurofibromatosis type-1. Hum Mol Genet 3:1109–1115 [DOI] [PubMed] [Google Scholar]
- Quattrin T, McPherson E, Putnam T (1987) Vertical transmission of the neurofibromatosis/Noonan syndrome. Am J Med Genet 26:645–649 10.1002/ajmg.1320260320 [DOI] [PubMed] [Google Scholar]
- Riccardi VM (1992) Neurofibromatosis: phenotype, natural history, and pathogenesis, 2nd ed. John Hopkins University, Baltimore [Google Scholar]
- Sarkozy A, Conti E, Digilio MC, Marino B, Morini E, Pacileo G, Wilson M, Calabro R, Pizzuti A, Dallapiccola B (2004) Clinical and molecular analysis of 30 patients with multiple lentigines LEOPARD syndrome. J Med Genet 41:e68 10.1136/jmg.2003.013466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkozy A, Conti E, Seripa D, Digilio MC, Grifone N, Tandoi C, Fazio VM, Di Ciommo V, Marino B, Pizzuti A, Dallapiccola B (2003) Correlation between PTPN11 gene mutations and congenital heart defects in Noonan and LEOPARD syndromes. J Med Genet 40:704–708 10.1136/jmg.40.9.704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saul RA (1985) Noonan syndrome in a patient with hyperplasia of the myenteric plexuses and neurofibromatosis. Am J Med Genet 21:491–492 10.1002/ajmg.1320210311 [DOI] [PubMed] [Google Scholar]
- Scheffzek K, Ahmadian MR, Wiesmuller L, Kabsch W, Stege P, Schmitz F, Wittinghofer A (1998) Structural analysis of the GAP-related domain from neurofibromin and its implications. EMBO J 17:4313–4327 10.1093/emboj/17.15.4313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shannon KM, O’Connell P, Martin GA, Paderanga D, Olson K, Dinndorf P, McCormick F (1994) Loss of the normal NF1 allele from the bone marrow of children with type 1 neurofibromatosis and malignant myeloid disorders. N Engl J Med 330:597–601 10.1056/NEJM199403033300903 [DOI] [PubMed] [Google Scholar]
- Sharland M, Burch M, McKenna WM, Paton MA (1992) A clinical study of Noonan syndrome. Arch Dis Child 67:178–183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shuper A, Mukamel M, Mimouni M, Steinherz R (1987) Noonan’s syndrome and neurofibromatosis. Arch Dis Child 62:196–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Side LE, Emanuel PD, Taylor B, Franklin J, Thompson P, Castleberry RP, Shannon KM (1998) Mutations of the NF1 gene in children with juvenile myelomonocytic leukemia without clinical evidence of neurofibromatosis, type 1. Blood 92:267–272 [PubMed] [Google Scholar]
- Stern HJ, Saal HM, Lee JS, Fain PR, Goldgar DE, Rosenbaum KN, Barker DF (1992) Clinical variability of type 1 neurofibromatosis: is there a neurofibromatosis-Noonan syndrome? J Med Genet 29:184–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stumpf DA, Alksne JF, Annegers JF, Brown SS, Conneally PM, Housman D, Leppert MF, Miller JP, Moss ML, Pileggi AJ, Rapin I, Strohman RC, Swanson LW, Zimmerman A (1988) Neurofibromatosis: conference statement. Arch Neurol 45:575–5783128965 [Google Scholar]
- Tartaglia M, Gelb BD (2005) Noonan syndrome and related disorders: genetics and pathogenesis. Annu Rev Genomics Hum Genet 6:45–68 10.1146/annurev.genom.6.080604.162305 [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Kalidas K, Shaw A, Song X, Musat DL, van der Burgt I, Brunner HG, Bertola DR, Crosby A, Ion A, Kucherlapati RS, Jeffery S, Patton MA, Gelb BD (2002) PTPN11 mutations in Noonan syndrome: molecular spectrum, genotype-phenotype correlation, and phenotypic heterogeneity. Am J Hum Genet 70:1555–1563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartaglia M, Martinelli S, Cazzaniga G, Cordeddu V, Iavarone I, Spinelli M, Palmi C, Carta C, Pession A, Arico M, Masera G, Basso G, Sorcini M, Gelb BD, Biondi A (2004a) Genetic evidence for lineage-related and differentiation stage-related contribution of somatic PTPN11 mutations to leukemogenesis in childhood acute leukemia. Blood 104:307–313 10.1182/blood-2003-11-3876 [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Martinelli S, Iavarone I, Cazzaniga G, Spinelli M, Giarin E, Petrangeli V, Carta C, Masetti R, Arico M, Locatelli F, Basso G, Sorcini M, Pession A, Biondi A (2005) Somatic PTPN11 mutations in childhood acute myeloid leukaemia. Br J Haematol 129:333–339 10.1111/j.1365-2141.2005.05457.x [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Mehler EL, Goldberg R, Zampino G, Brunner HG, Kremer H, van der Burgt I, Crosby AH, Ion A, Jeffery S, Kalidas K, Patton MA, Kucherlapati RS, Gelb BD (2001) Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet 29:465–468 10.1038/ng772 [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Niemeyer CM, Fragale A, Song X, Buechner J, Jung A, Hahlen K, Hasle H, Licht JD, Gelb BD (2003) Somatic mutations in PTPN11 in juvenile myelomonocytic leukemia, myelodysplastic syndromes and acute myeloid leukemia. Nat Genet 34:148–150 10.1038/ng1156 [DOI] [PubMed] [Google Scholar]
- Tartaglia M, Niemeyer CM, Shannon KM, Loh ML (2004b) SHP-2 and myeloid malignancies. Curr Opin Hematol 11:44–50 10.1097/00062752-200401000-00007 [DOI] [PubMed] [Google Scholar]
- Tassabehji M, Strachan T, Sharland M, Colley A, Donnai D, Harris R, Thakker N (1993) Tandem duplication within a neurofibromatosis type 1 (NF1) gene exon in a family with features of Watson syndrome and Noonan syndrome. Am J Hum Genet 53:90–95 [PMC free article] [PubMed] [Google Scholar]
- Upadhyaya M, Cooper DN (1998) The mutational spectrum in neurofibromatosis type 1 and its underlying mechanisms. In: Upadhyaya M, Cooper DN (eds) Neurofibromatosis type 1: from genotype to phenotype. BIOS Scientific, Oxford, United Kingdom, pp 65–82 [Google Scholar]
- van der Burgt I, Berends E, Lommen E, van Beersum S, Hamel B, Mariman E (1994) Clinical and molecular studies in a large Dutch family with Noonan syndrome. Am J Med Genet 53:187–191 10.1002/ajmg.1320530213 [DOI] [PubMed] [Google Scholar]
- Xu G, O’Connell P, Viskochil D, Cawthorn R, Robertson M, Culver M, Dunn D, Stevens J, Gesteland R, White R, Weiss R (1990) The neurofibromatosis type 1 gene encodes a protein related to GAP. Cell 62:599–608 10.1016/0092-8674(90)90024-9 [DOI] [PubMed] [Google Scholar]