Abstract
Peeling skin syndrome is an autosomal recessive genodermatosis characterized by the shedding of the outer epidermis. In the acral form, the dorsa of the hands and feet are predominantly affected. Ultrastructural analysis has revealed tissue separation at the junction between the granular cells and the stratum corneum in the outer epidermis. Genomewide linkage analysis in a consanguineous Dutch kindred mapped the gene to 15q15.2 in the interval between markers D15S1040 and D15S1016. Two homozygous missense mutations, T109M and G113C, were found in TGM5, which encodes transglutaminase 5 (TG5), in all affected persons in two unrelated families. The mutation was present on the same haplotype in both kindreds, indicating a probable ancestral mutation. TG5 is strongly expressed in the epidermal granular cells, where it cross-links a variety of structural proteins in the terminal differentiation of the epidermis to form the cornified cell envelope. An established, in vitro, biochemical cross-linking assay revealed that, although T109M is not pathogenic, G113C completely abolishes TG5 activity. Three-dimensional modeling of TG5 showed that G113C lies close to the catalytic domain, and, furthermore, that this glycine residue is conserved in all known transglutaminases, which is consistent with pathogenicity. Other families with more-widespread peeling skin phenotypes lacked TGM5 mutations. This study identifies the first causative gene in this heterogeneous group of skin disorders and demonstrates that the protein cross-linking function performed by TG5 is vital for maintaining cell-cell adhesion between the outermost layers of the epidermis.
Introduction
Peeling skin syndrome (PSS [MIM 270300]) is an autosomal recessive disorder characterized by the continuous shedding of the outer layers of the epidermis from birth and throughout life (Kurban and Azar 1969; Levy and Goldsmith 1982; Abdel-Hafez et al. 1983; Silverman et al. 1986; Mevorah et al. 1987; Judge et al. 2004). Histological and ultrastructural analyses in a number of cases have shown that the level of blistering in PSS is high in the epidermis, at the junction of the stratum granulosum (the last living layer) and the stratum corneum (the terminally differentiated and continuously shed, cornified squamous layers). Thus, the disorder is histologically distinguishable from the various forms of epidermolysis bullosa, in which blistering occurs in the basal keratinocytes of the epidermis (for simplex forms, see Lane and McLean 2004), within or close to the dermal-epidermal basement membrane (for junctional variants, see Uitto and Richard 2004), or in the upper papillary dermis (for dystrophic forms, see Uitto and Richard 2004). In some cases of PSS, skin peeling is accompanied by erythema, vesicular lesions, or, in rare cases, other ectodermal features, like fragile hair and nail abnormalities. Two main subtypes, noninflammatory type A and inflammatory type B, have been suggested (Traupe 1989; Judge et al. 2004); however, it is clear from the dermatology literature that there are additional subtypes. In some families, an acral form of PSS (APSS) has been reported, in which skin peeling is strictly limited to the dorsa of the hands and feet, and, again, ultrastructural and histological analysis shows a level of blistering high in the epidermis at the stratum granulosum–stratum corneum junction (Shwayder et al. 1997; Hashimoto et al. 2000).
Transglutaminases (TGs) are involved in protein cross-linking by catalyzing the formation of gamma-glutamyl-lysine isodipeptide bonds between adjacent polypeptides (Candi et al. 2005; Eckert et al. 2005). This process is particularly important in the terminal differentiation of the epidermis, where TGs heavily cross-link keratins and a range of differentiation-specific structural proteins, such as involucrin, loricrin, filaggrin, and small proline-rich proteins, in the formation of the cornified cell envelope in the biogenesis of the stratum corneum, the outermost, “dead” layer of the epidermis (Kalinin et al. 2002). This continuously shed outer layer of the epidermis performs the main barrier function of the skin. Recessive loss-of-function mutations in TGM1 have been shown to cause lamellar ichthyosis, a disease characterized by excessive scaling and shedding of the outer epidermis (Huber et al. 1995; Parmentier et al. 1995; Russell et al. 1995). No genetic disease associations have been reported so far for the other major epidermal TGs, TG3 and TG5.
Here, we describe how we mapped a gene for APSS to 15q15.2 and identified the same loss-of-function mutations in the TGM5 gene in two unrelated families.
Material and Methods
Genetic Linkage
A genomewide screen was performed as described elsewhere (van Steensel et al. 1999; Hamada et al. 2002) with the use of the ABI Linkage Mapping Set version 2, consisting of 400 fluorescent microsatellite markers placed at ∼10-cM intervals. PCRs for microsatellite analysis were performed in a total volume of 7.5 μl containing 100 ng of template DNA, 1× GeneAmp PCR buffer II (Applied Biosystems), 330 nM of each primer, 250 μM dinucleotide triphosphates (dNTPs), 2.5 mM MgCl2, and 0.30 units of Amplitaq Gold DNA polymerase (Applied Biosystems). Samples were amplified in a Touchdown ThermoCycler (Hybaid) in accordance with the following conditions: After an initial denaturation at 94°C for 2 min, samples were subjected to 35 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 30 s, followed by a final incubation of 72°C for 5 min. PCR fragments were sized on an ABI 3100 DNA sequencer with the use of Genescan and Genotyper software (Applied Biosystems). Two-point LOD scores were calculated with the MLINK algorithm of LINKAGE version 5.1, with the assumption that the mutant allele frequency was 0.0001 with 100% penetrance. Markers in the homozygous linked interval gave a maximum LOD score of 5.6 at θ=0. The following primers were used to amplify “homemade” microsatellites in the locus: TGM5-MS1 (5′-GAATTGCTTGAACCTCGGAG-3′ and 5′-GAGTCAAGCTTTCAACCTGG-3′; ∼265 bp); TGM5-MS2 (5′-GCTAGGGGTAGTGCTGAAGG-3′ and 5′-GGCTGGAAGCATAGTTCAGG-3′; ∼311 bp); and TGM5-MS3 (5′-CAGGGCAAATTGGTGAACAG-3′ and 5′-CCTCCAATTCCAAATGCTTA-3′; ∼401 bp).
TGM5 Mutation Analysis
All 13 exons, including splice sites and branch points, were PCR amplified from genomic DNA and were directly sequenced with both forward and reverse primers listed in table 1. For PCR, 200 ng of patient or control genomic DNA was added to a premix containing 1× PCR buffer (Promega), 10 nmol of each dNTP, 20 pmol of each primer, and 1 unit of Taq polymerase (Promega), in a total volume of 50 μl. After an initial denaturation at 94°C for 2 min, samples were subjected to 35 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 60 s, followed by a final incubation of 72°C for 5 min. The PCR products were examined by agarose gel electrophoresis, were purified using spin columns (Qiagen), and were directly sequenced using BigDye Terminators on an ABI 3100 genetic analyzer (Applied Biosystems).
Table 1.
Primers for TGM5 Amplification
| Primer | Sequence 5′→3′ | PCR Size (bp) |
| TGM5E1L | CGCTTCTCCCTCCTGACTTC | … |
| TGME1R | CTGTCTCCTGGTTCTCAAGC | 537 |
| TGM5E2L | CTATTTTGTCTAACCCTGGC | … |
| TGM5E2R | CCACGAAGATGATGTTGTCC | 205 |
| TGM5E3L | GGACAACATCATCTTCGTGG | … |
| TGM5E3R | CTTGAGCCTGTCTCTCTGGC | 447 |
| TGM5E4L | GGTGACTTTTCTGCTTCGTG | … |
| TGM5E4R | CGTTGGTCAGCAGTTCTAGG | 258 |
| TGM5E5L | CATACTCCCACGTGACAAGC | … |
| TGM5E5R | CACATTCTGGCGTCAGTCTC | 321 |
| TGM5E6L | GGAAGTCCACAGTAAAATGC | … |
| TGM5E6R | GATGGGAATGGTTCAGAAGG | 389 |
| TGM5E7L | CCAAAGTCCAGGAATATACG | … |
| TGM5E7R | GGGCTTGGCTCTGATGTGTG | 262 |
| TGM5E8L | GTTCATTTGCTCATCCCTCC | … |
| TGM5E9R | GATGCTCTCTGGCTCCCTGG | 609 |
| TGM5E10L | GCCTGAAGTTGTTGGTGGAG | … |
| TGM5E10R | CTTGACCCCACTGACATTGC | 534 |
| TGM5E11L | CTACTCTCACCCCATCCTTG | … |
| TGM5E11R | GAGCACCAGAAGAGGCAAAG | 327 |
| TGM5E12L | GTGCCCTTCGTAAATATCTG | … |
| TGM5E13R | CTCTGTTGTGGTGGGGAATG | 661 |
Construction of TG5 Expression Vectors
Full-length, wild-type human TG5 complementary DNA (cDNA) (Candi et al. 2001) cloned in a mammalian expression vector pCDNA3.1 was used as the template for the two mutant forms, T109M and G113C. Mutations were inserted by PCR amplification of the wild-type plasmid (Platinum HiFi Taq-Polymerase [Invitrogen]) with the use of primers carrying the selected mutations. The PCR product was digested (DpnI [Promega]), was ligated (DNA Ligation Kit v.2.0 [Takara]), and was transformed into a DH5-α bacterial strain (Invitrogen). Positive clones were selected by direct sequencing of mutagenesis sites. The following primers were used for the introduction of mutations: G113C +5′-ATCCACATCGACTCCTTCCAGGGG-3′ and −5′-TTTCAAGAGGTACCGACACACGGC-3′; and T109M +5′-CGGTACCTCTTGAAAATCCACATC-3′ and −5′-ACCCACGGCCGCCATGGGAG-3′. The following PCR program was used for both mutants: a denaturation step of 4 min at 95°C, followed by 16 cycles of 94°C for 40 s, 61°C for 45 s, and 70°C for 2 min and 30 s, and followed by a final step of 10 min at 72°C.
Cell Culture and Transfection
Cryopreserved NHEK cells obtained from BioWhittaker were grown in calf skin collagen–coated dishes (Sigma Chemical) in serum-free keratinocyte medium (BioWhittaker) at 0.05 mM Ca2+ and supplemented with Single-Quots (BioWhittaker) containing 7.5 μg/ml bovine pituitary extract, 0.5 mg/ml insulin, 0.5 mg/ml hydrocortisone, and 0.1 μg/ml hEGF. Third-passage cells were used for transfection experiments. HEK-293 cells were grown in Dulbecco’s modified Eagle medium, supplemented with 10% (v/v) fetal bovine serum. Cells were transiently transfected using Effectene (Qiagen) reagents in accordance with the manufacturer's instructions, with the use of 1.5 μg of pCDNA3.1-TG5-myc and mutant expression vectors. Aliquots of the extracted enzymes were used for the TG assay.
TG Activity Assay
TG activity was determined by measuring the incorporation of [3H]putrescine (Amersham) into N,N′-dimethyl-casein (Sigma Chemicals), as described elsewhere (Candi et al. 2001). The reaction mixtures contained 100 mM Tris-HCl at pH 8.5, 100 mM NaCl, 5 mM DTT, 10 mM CaCl2, 25 μl of casein (12.5 mg/ml), and 0.2 mM putrescine containing 1 μCi [3H]putrescine. Different samples were incubated with the reaction mixture in a final volume of 100 μl at 37°C. After 10 min of incubation, the reaction was stopped by spotting 25-μl aliquots on Whatman 3MM filter paper. Unbound [3H]putrescine was removed by washing with large volumes of 15%, 10%, and 5% trichloroacetic acid and absolute ethanol. Filters were then air dried, and the radioactivity was measured by liquid scintillation counting. The TG assay was performed in triplicate. The activities measured in different samples were normalized by protein content and by transfection efficiency to compare cells transfected with different expression vectors.
TG5 Three-Dimensional Modeling
A three-dimensional model of human TG5 was generated using MODELLER (Sali and Blundell 1993). The crystal structure of human TG2 (Protein Data Bank identification number 1KV3) was used as a starting point. Human TG2 and TG5 have a 41% sequence identity, and their sequences were aligned using LALIGN v.2.0u6. The alignment was input into MODELLER to model TG5 by satisfying spatial restraints. The model was optimized using the molecular probability density function by conjugate gradient and simulated annealing. Relative to TG2, TG5 has a 29–amino-acid insertion between the β-barrel 1 and the catalytic core, which could not be modeled.
Results
APSS Phenotype and Linkage Mapping
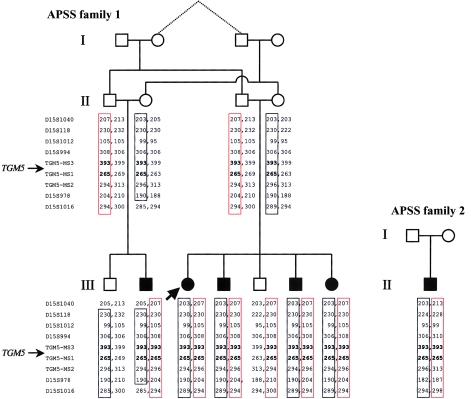
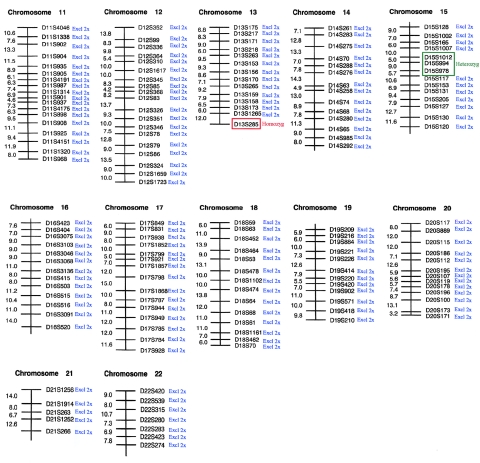
Here, we studied two families with noninflammatory APSS, where peeling was accompanied by erythema and was limited to the acral regions of the skin, predominantly over the hands and feet (fig. 1). Affected patients experienced periodic, painless, and superficial skin peeling. Although the skin peeling occurred spontaneously, the manual removal of strips of skin was easy. The abnormality was exacerbated by elevated ambient temperature and humidity. The peeled areas showed some residual erythema for a few days but healed spontaneously and without scarring. Similar families have been described elsewhere (Shwayder et al. 1997; Hashimoto et al. 2000), and so APSS represents a clinically recognizable entity. Family 1 was of Dutch origin and included five affected individuals in two interbred sibships (fig. 2). Genealogical analysis was able to establish distant consanguinity dating back to the 1780s (data not shown). Family 2 was of Scottish origin (fig. 2) and presented with clinical features essentially identical to those observed in family 1 (data not shown). In this case, consanguinity was unknown. A genomewide screen was performed on family 1 with the use of 400 microsatellite markers spaced at ∼10-cM intervals. Because of the initial uncertainty over consanguinity, the regions where all five affected individuals showed homozygosity or were heterozygous for the same two marker alleles were considered to be potentially linked loci. The genome scan data are summarized in figures 3 and 4. In the end, only two loci showed homozygosity, D1S2797 and D13S285, both of which were poorly informative, and these loci were subsequently excluded by nearby flanking markers (data not shown). One linked locus was identified on 15q15.2 between markers D15S1012 and D15S978, in which all five affected individuals were heterozygous for the same alleles and their unaffected sibs had differing genotypes (fig. 2). High-density microsatellite analysis in this region confirmed compound heterozygous linkage over a 10.2-Mb region, with visible recombination seen with markers D15S1040 and D15S1016 (fig. 2). However, “homemade” microsatellites (TGM5-MS1 and TGM5-MS3) in the central ∼270-kb region within this interval showed homozygosity-by-descent in all affected individuals and heterozygosity in the unaffected parents and siblings, which was consistent with consanguinity in the pedigree and was predictive of a homozygous mutation. These markers were chosen because they flank a strong candidate gene, TGM5, as described below. Linkage analysis gave a statistically significant maximum two-point LOD score of 5.6 at θ=0 for informative markers within the area of homozygosity-by-descent in family 1. Surprisingly, the proband in family 2 was found to also be homozygous for the same alleles of markers TGM5-MS1 and TGM5-MS3 within the linked area, predicting the same genetic lesion in both families.
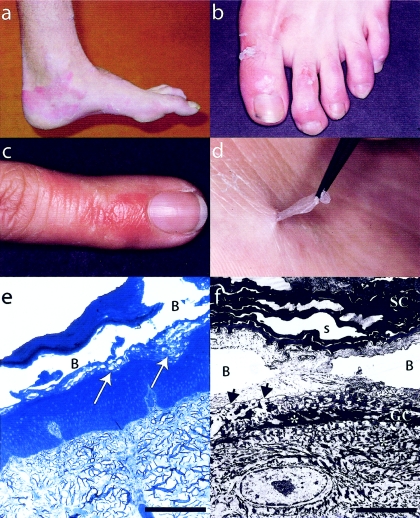
Figure 1.
Clinicopathological features of APSS. a–d, Superficial peeling of the skin, leaving residual, painless erythema (best seen in a and d). d, Painless, manual skin removal is also possible, demonstrating that the peeling is limited to the stratum corneum. e, Light microscopy of a representative skin lesion on the dorsum of the foot (proband of family 2). The arrows point to the level of dehiscence, where cytolysis can be seen. No other abnormalities are evident. f, Electron microscopy of a similar lesion (proband of family 1, 20,000× magnification), where B denotes a blister. The level of separation is evidently between the uppermost granular layer cells (GC) and the acellular stratum corneum (SC). The asterisks mark tonofilaments that appear intact. Widened spaces (s) were observed in the stratum corneum, but these can be seen in skin from normal individuals (not shown) and are probably artefactual.
Figure 2.
Pedigrees of APSS families 1 and 2 showing haplotype information in the vicinity of the TGM5 locus on 15q15. In family 1, of Dutch origin, two brothers married two sisters in generation II, and both resulting sibships produced affected children with APSS. At the outset, consanguinity could not be formally established, so compound heterozygous linkage was sought. This was identified for markers in the interval between D15S1040 (recombinant) and D15S1016 (recombinant), where all affected persons inherited the same two haplotypes (red and black boxes), whereas the unaffected individuals III-1 and III-5 inherited only one or the other of these haplotypes. Within the interval, homozygosity-by-descent was identified with markers flanking the TGM5 gene (bold type). Subsequently, a genealogical connection between the grandparents in generation I (dotted lines) was traced back to the 1780s, confirming the distant consanguinity implied by the genotype data. Interestingly, the proband in family 2 was homozygous for the same haplotype close to the TGM5 gene. This family was nonconsanguineous and of Scottish origin, with no history of Dutch ancestry.
Figure 3.
Summary of genome scan data obtained for APSS family 1, chromosomes 1–10, with microsatellite markers from Applied Biosystems Linkage Mapping Set version 2. Blue, locus excluded for both homozygous and compound heterozygous linkage. Red, locus showing homozygosity for all affected persons tested. Homozygosity with D1S2797 was found to be due to uninformativeness in the family. Region excluded by closely flanked markers D1S2713 (north) and D1S2652 (south).
Figure 4.
Summary of genome scan data obtained for APSS family 1, chromosomes 11–22, with microsatellite markers from Applied Biosystems Linkage Mapping Set version 2. Blue, locus excluded for both homozygous and compound heterozygous linkage. Red, locus showing homozygosity for all affected persons tested. Green, locus showing compound heterozygous linkage. Homozygosity with D13S285 was found to be due to uninformativeness in the family. Region excluded by closely flanking markers D13S1315 (north) and D13S293 (south). Heterozygous linkage in the region D15S1012–D15S978 was confirmed with high-density markers in the region. An area of homozygosity was identified within this region, as detailed in figure 2 in this article.
APSS Maps to a TG Gene Cluster
The critical interval of shared haplotypes contained nine genes (UCSC Genome Browser, May 2004 build). Located centrally in the locus was a small cluster of TG genes, TGM5, TGM7, and EPB42 (Grenard et al. 2001). These TG genes represented good candidates for APSS. Of the three genes in this cluster, TGM7 and EPB42 had expression patterns inconsistent with an epidermal phenotype (Grenard et al. 2001). Furthermore, loss-of-expression mutations in EPB42 are known to cause recessive spherocytic elliptocytosis (MIM 177070) (Rybicki et al. 1988; Bouhassira et al. 1992). TGM5 is widely expressed but, significantly, it has been shown to be strongly expressed in the epidermis, with maximal expression in the vicinity of the junction between the stratum granulosum and the stratum corneum (Candi et al. 2002), which is precisely where intraepidermal splitting was seen to occur in our APSS families (fig. 1e and 1f). Furthermore, TGM5 is also known to cross-link the structural proteins loricrin, involucrin, and small proline-rich proteins in vitro, all of which localize to this region of the epidermis (Candi et al. 2001). Thus, we considered TGM5 to be a prime candidate gene for APSS, in terms of its function and expression pattern.
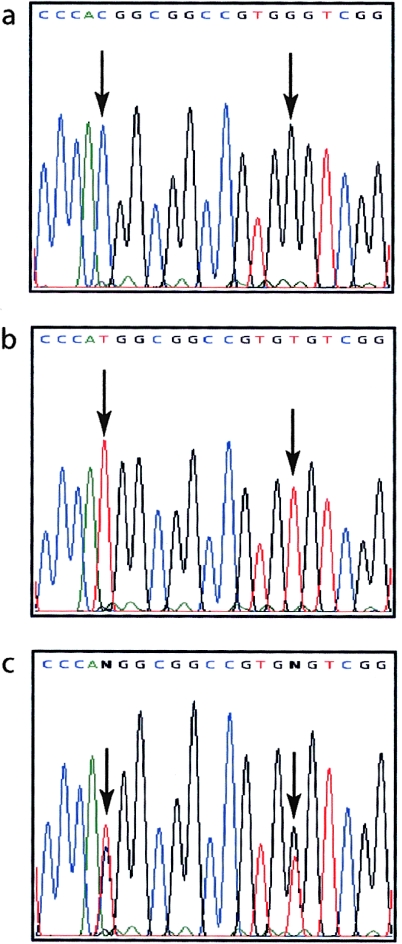
TGM5 consists of 13 exons spanning 33.7 kb of genomic DNA and encoding two major splice variants (Grenard et al. 2001). Mutation analysis of all exons and splice sites of TGM5 revealed two homozygous missense mutations, T109M and G113C, in all six affected members of families 1 and 2 (fig. 5). The clinically unaffected parents and siblings available for study were heterozygous for these changes, as consistent with the linkage data (fig. 2). Screening of 100 Dutch and 50 British controls by sequencing revealed one Dutch individual who was heterozygous for both mutations and had no history of skin peeling. On the basis of these data, we would predict an incidence of ∼1 in 90,000 affected persons in the population, a ratio that is compatible with the rarity of APSS when compared with other epidermal blistering conditions such as epidermolysis bullosa simplex, which has an incidence of ∼1 in 20,000 in the Scottish population (Horn et al. 1997).
Figure 5.
Identification of homozygous missense mutations in TGM5 in persons affected by APSS. a, Normal coding-strand DNA sequence derived from exon 3 of the TGM5 gene, showing codons 108–114 inclusive. b, The same region of TGM5, derived from the proband in family 1 (individual III-3). Two homozygous missense mutations are shown: 326C→T (left arrow) and 337G→T (right arrow), predicting the amino acid changes T109M and G113C, respectively. c, The same region of TGM5, derived from a heterozygous carrier in family 1 (individual III-5). All affected persons in families 1 and 2 were homozygous for both changes.
Functional Confirmation of Pathogenicity
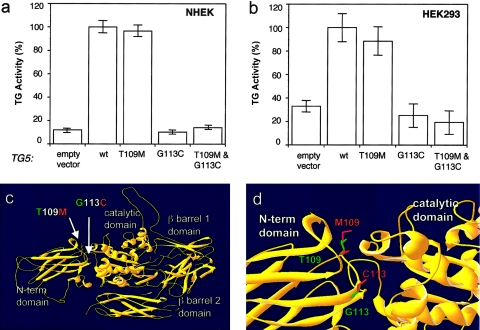
To further confirm that one or both of these missense mutations were, in fact, pathogenic, we made full-length expression constructs for TG5, into which we introduced the mutations, either singly or together. With the use of a well-established assay system (Candi et al. 2001), these constructs were used to measure TG5 enzyme activity following transfection into two different epithelial cell lines (fig. 6). In both cell lines, the missense change T109M did not significantly reduce TG5 activity; however, G113C completely abolished cross-linking activity, as did the double mutation (fig. 6). Thus, the G113C mutation is pathogenic, and T109M probably represents a rare polymorphism present on the same allele. Of the nine known human TGs, only TG5 has a threonine at this position; the other TGs have either asparagine (n=4), serine (n=2), lysine (n=1), or aspartic acid (n=1). Therefore, a variety of changes can apparently be tolerated in this position without affecting TG activity.
Figure 6.
Biochemical studies of TG5 mutant forms. NHEK (a) and HEK-293 (b) cells were transfected for 48 h with the use of wild-type TG5 and the three mutant forms indicated. Total TG5 activities were measured for insoluble and fractions and standardized for transfection efficiency (GFP). The control vector indicates the endogenous TG5 activity (cells transfected only with the GFP control vector). The data obtained are reported as percentages of TG activity, with 100% corresponding to the activity for cells transfected with the wild-type construct. The data presented are the averages of two independent experiments. c, View of the entire TG5 model with the four main structural domains. The mutations described here are located in the N-terminal domain at the interface between the N-terminal domain and the catalytic domain. d, Effects of the mutations in a local environment. The G113C mutation, close to the active site, would probably cause a local change in the main protein structure. The poorly conserved residue T109 is present on the surface and is, therefore, unlikely to be functionally critical. Green indicates the mutated residues, and red represents wild-type TG5 residues.
A computer model of human TG5 was generated from the published three-dimensional structure of human TG2 (fig. 6c and 6d). Not surprisingly, given the high degree of homology (41% identity), the overall predicted structure of TG5 closely matched that of TG2. This analysis showed that the mutation T109M is located at the surface of the enzyme and is, therefore, not likely to affect the structural integrity of the enzyme, which confirms the biochemical studies and possibly explains the poor evolutionary conservation of this residue. In contrast, Gly113 is conserved in all members of the TG family from many species and is located near the hinge region between the TG N-terminal domain and the catalytic domain. This new cysteine found in the mutant protein may form an unnatural disulfide bond that could disrupt the nearby active site. In addition, the mutation of the equivalent glycine residue in TG1 (G217S) has been described in a patient affected by autosomal recessive congenital ichthyosis (Laiho et al. 1997), showing that this highly conserved glycine is required for TG function.
Discussion
Here we have demonstrated that the abolition of TG5 activity in the skin, caused by homozygosity for the missense mutation G113C in exon 3 of TGM5, leads to an acral-dorsal site-restricted form of PSS. This not only explains the genetic defect underlying APSS but also demonstrates that the protein cross-linking function performed by TG5 (Candi et al. 2001) is vital for maintaining cell-cell adhesion between the granular layer of the epidermis and the overlying stratum corneum (fig. 1e and 1f). TG5 is able to utilize a variety of classical TG substrates in vitro, including loricrin, involucrin, and small proline-rich proteins (Candi et al. 2001). However, it is not yet clear which of these substrates is the predominant target of TG5 in vivo. TG5 is almost ubiquitously expressed (Grenard et al. 2001) and, in particular, is widely expressed in the epidermis (Candi et al. 2002). In view of this, it is surprising that APSS affects only the dorsal regions of the hands and feet (fig. 1). We speculate that there may be structurally important substrates expressed in these epidermal regions that are solely or preferentially cross-linked by TG5. Alternatively, the expression of TG1 and TG3 may be lower in these regions, so that TG5 activity predominates, although we have no experimental evidence to support this because of the ethical difficulties in obtaining normal biopsy material from these highly visible, cosmetically sensitive sites.
Our patients report that peeling is exacerbated by heat and humidity. Since an important function of the stratum corneum is to form a water barrier (Kalinin et al. 2002; Madison 2003), one might expect the hands and feet to be particularly prone to a barrier defect because of their greater environmental exposure to heat and water. Another possible explanation is that there are regional temperature differences in these areas of the skin that affect the activity of the various epidermal TGs. Sun exposure can probably be ruled out, since the feet, which are mostly protected from the sun in our patients, are affected about as much as the hands (fig. 1).
The mutation described here occurs in exon 3 of the TGM5 gene. This exon has been shown to be alternately spliced (Candi et al. 2001; Grenard et al. 2001). Grenard and colleagues reported isoform 1 (GenBank accession numbers NM_201631.1 and AF035961), which includes all 13 exons, and also isoform 2 (GenBank accession numbers NM_004245.1 and AF035960), which lacks exon 3 (Grenard et al. 2001). Candi and colleagues refer to isoform 2 as the “Δ3” form and also reported an isoform lacking exon 11, to which they refer as “Δ11” (Candi et al. 2001). There is no GenBank accession number for the Δ11 isoform. By means of the biochemical cross-linking assay for TG activity that was used here, it was shown that the Δ3 isoform lacks TG activity, at least that which was detectable by the putrescine incorporation assay (Candi et al. 2001). The fact that the lack of exon 3 leads to a loss of TG activity is further evidence that the protein domain where the G113C mutation is located is, in fact, a functionally critical part of the protein required for enzymatic activity. Nothing is currently known about the regional variation in the alternate splicing of TGM5 transcripts, which, again, might provide an explanation for the acral-limited phenotype observed here; that is, enzymatically active isoforms containing exon 3 might be preferentially expressed in the acral regions. Again, we have been unable to investigate this because of the lack of biopsy material, as discussed above.
Interestingly, we have excluded the TGM5 locus in a large, consanguineous kindred of Middle Eastern origin with more widespread peeling skin and, in addition, have also failed to detect TGM5 mutations in three small, outbred families with widespread peeling skin phenotypes (Andrew J. Cassidy and W. H. Irwin McLean, unpublished data). In all these families, the age of onset was in infancy and the phenotype was similar to the classical, generalized form of PSS described elsewhere (Kurban and Azar 1969; Levy and Goldsmith 1982; Abdel-Hafez et al. 1983; Silverman et al. 1986; Mevorah et al. 1987; Judge et al. 2004). One of the small outbred families was reported elsewhere in the literature (Levy and Goldsmith 1982). Thus, PSS is both clinically and genetically heterogeneous, as is suggested from diversity in clinical appearance reported in the literature (Kurban and Azar 1969; Levy and Goldsmith 1982; Abdel-Hafez et al. 1983; Silverman et al. 1986; Mevorah et al. 1987; Shwayder et al. 1997; Hashimoto et al. 2000; Judge et al. 2004). The causative genes in the cases presenting with generalized peeling skin may shed further light on the preferred cross-linking substrates for the epidermal TGs, particularly TG5, as well as the key protein-protein interactions that are necessary to maintain epidermal structural integrity and the important barrier function performed by the outermost layers of the epidermis.
Acknowledgments
We thank the patients and their families for their participation in this study. We also thank the Molecular Genetics Laboratory, Ninewells University Hospitals NHS Trust, for genomic DNA extraction. The Epithelial Genetics Group is funded by a Wellcome Trust Senior Research Fellowship, as well as by grants from the Dystrophic Epidermolysis Bullosa Research Association (DEBRA) UK and The Pachyonychia Congenita Project (to W.H.I.M.) M.A.M.v.S is supported by The Pachyonychia Congenita Project and Barrier Therapeutics NV. This work was partially supported by an Italian Telethon grant (to E.C.).
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for TGM5 isoform 1 [accession numbers NM_201631.1 and AF035961] and isoform 2 [accession numbers NM_004245.1 and AF035960]) [Google Scholar]
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PSS and spherocytic elliptocytosis)
- Protein Data Bank, http://www.rscb.org/pdb/ (for crystal structure of human TG2 [identification number 1KV3])
- UCSC Genome Browser, http://www.genome.ucsc.edu/ (for TGM5, TGM7, and EPB42)
References
- Abdel-Hafez K, Safer AM, Selim MM, Rehak A (1983) Familial continual skin peeling. Dermatologica 166:23–31 [DOI] [PubMed] [Google Scholar]
- Bouhassira EE, Schwartz RS, Yawata Y, Ata K, Kanzaki A, Qiu JJ, Nagel RL, Rybicki AC (1992) An alanine-to-threonine substitution in protein 4.2 cDNA is associated with a Japanese form of hereditary hemolytic anemia (protein 4.2NIPPON). Blood 79:1846–1854 [PubMed] [Google Scholar]
- Candi E, Oddi S, Paradisi A, Terrinoni A, Ranalli M, Teofoli P, Citro G, Scarpato S, Puddu P, Melino G (2002) Expression of transglutaminase 5 in normal and pathologic human epidermis. J Invest Dermatol 119:670–677 10.1046/j.1523-1747.2002.01853.x [DOI] [PubMed] [Google Scholar]
- Candi E, Oddi S, Terrinoni A, Paradisi A, Ranalli M, Finazzi-Agro A, Melino G (2001) Transglutaminase 5 cross-links loricrin, involucrin, and small proline-rich proteins in vitro. J Biol Chem 276:35014–35023 10.1074/jbc.M010157200 [DOI] [PubMed] [Google Scholar]
- Candi E, Schmidt R, Melino G (2005) The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol 6:328–340 10.1038/nrm1619 [DOI] [PubMed] [Google Scholar]
- Eckert RL, Sturniolo MT, Broome AM, Ruse M, Rorke EA (2005) Transglutaminase function in epidermis. J Invest Dermatol 124:481–492 10.1111/j.0022-202X.2005.23627.x [DOI] [PubMed] [Google Scholar]
- Grenard P, Bates MK, Aeschlimann D (2001) Evolution of transglutaminase genes: identification of a transglutaminase gene cluster on human chromosome 15q15: structure of the gene encoding transglutaminase X and a novel gene family member, transglutaminase Z. J Biol Chem 276:33066–33078 10.1074/jbc.M102553200 [DOI] [PubMed] [Google Scholar]
- Hamada T, McLean WHI, Ramsay M, Ashton GH, Nanda A, Jenkins T, Edelstein I, South AP, Bleck O, Wessagowit V, Mallipeddi R, Orchard GE, Wan H, Dopping-Hepenstal PJ, Mellerio JE, Whittock NV, Munro CS, van Steensel MA, Steijlen PM, Ni J, Zhang L, Hashimoto T, Eady RAJ, McGrath JA (2002) Lipoid proteinosis maps to 1q21 and is caused by mutations in the extracellular matrix protein 1 gene (ECM1). Hum Mol Genet 11:833–840 10.1093/hmg/11.7.833 [DOI] [PubMed] [Google Scholar]
- Hashimoto K, Hamzavi I, Tanaka K, Shwayder T (2000) Acral peeling skin syndrome. J Am Acad Dermatol 43:1112–1119 10.1067/mjd.2000.103645 [DOI] [PubMed] [Google Scholar]
- Horn HM, Priestly GC, Eady RAJ, Tidman MJ (1997) The prevalence of epidermolysis bullosa in Scotland. Br J Dermatol 136:560–564 10.1046/j.1365-2133.1997.d01-1235.x [DOI] [PubMed] [Google Scholar]
- Huber M, Rettler I, Bernasconi K, Frenk E, Lavrijsen SP, Ponec M, Bon A, Lautenschlager S, Schorderet DF, Hohl D (1995) Mutations of keratinocyte transglutaminase in lamellar ichthyosis. Science 267:525–528 [DOI] [PubMed] [Google Scholar]
- Judge MR, McLean WHI, Munro CS (2004) Disorders of keratinization. In: Burns T, Breathnach S, Cox C, Griffiths C (eds) Rook’s textbook of dermatology. Vol 2. Blackwell Scientific, Oxford, pp 34.54–34.56 [Google Scholar]
- Kalinin AE, Kajava AV, Steinert PM (2002) Epithelial barrier function: assembly and structural features of the cornified cell envelope. Bioessays 24:789–800 10.1002/bies.10144 [DOI] [PubMed] [Google Scholar]
- Kurban AK, Azar HA (1969) Familial continual skin peeling. Br J Dermatol 81:191–195 [DOI] [PubMed] [Google Scholar]
- Laiho E, Ignatius J, Mikkola H, Yee VC, Teller DC, Niemi KM, Saarialho-Kere U, Kere J, Palotie A (1997) Transglutaminase 1 mutations in autosomal recessive congenital ichthyosis: private and recurrent mutations in an isolated population. Am J Hum Genet 61:529–538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lane EB, McLean WHI (2004) Keratins and skin disorders. J Pathol 204:355–366 10.1002/path.1643 [DOI] [PubMed] [Google Scholar]
- Levy SB, Goldsmith LA (1982) The peeling skin syndrome. J Am Acad Dermatol 7:606–613 [DOI] [PubMed] [Google Scholar]
- Madison KC (2003) Barrier function of the skin: “la raison d’etre” of the epidermis. J Invest Dermatol 121:231–241 10.1046/j.1523-1747.2003.12359.x [DOI] [PubMed] [Google Scholar]
- Mevorah B, Frenk E, Saurat JH, Siegenthaler G (1987) Peeling skin syndrome: a clinical, ultrastructural and biochemical study. Br J Dermatol 116:117–125 [DOI] [PubMed] [Google Scholar]
- Parmentier L, Blanchet-Bardon C, Nguyen S, Prud’homme JF, Dubertret L, Weissenbach J (1995) Autosomal recessive lamellar ichthyosis: identification of a new mutation in transglutaminase 1 and evidence for genetic heterogeneity. Hum Mol Genet 4:1391–1395 [DOI] [PubMed] [Google Scholar]
- Russell LJ, DiGiovanna JJ, Rogers GR, Steinert PM, Hashem N, Compton JG, Bale SJ (1995) Mutations in the gene for transglutaminase 1 in autosomal recessive lamellar ichthyosis. Nat Genet 9:279–283 10.1038/ng0395-279 [DOI] [PubMed] [Google Scholar]
- Rybicki AC, Heath R, Wolf JL, Lubin B, Schwartz RS (1988) Deficiency of protein 4.2 in erythrocytes from a patient with a Coombs negative hemolytic anemia: evidence for a role of protein 4.2 in stabilizing ankyrin on the membrane. J Clin Invest 81:893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815 10.1006/jmbi.1993.1626 [DOI] [PubMed] [Google Scholar]
- Shwayder T, Conn S, Lowe L (1997) Acral peeling skin syndrome. Arch Dermatol 133:535–536 10.1001/archderm.133.4.535 [DOI] [PubMed] [Google Scholar]
- Silverman AK, Ellis CN, Beals TF, Woo TY (1986) Continual skin peeling syndrome: an electron microscopic study. Arch Dermatol 122:71–75 10.1001/archderm.122.1.71 [DOI] [PubMed] [Google Scholar]
- Traupe H (1989) The ichthyoses: a guide to clinical diagnosis, genetic counselling, and therapy. Springer-Verlag, Berlin [Google Scholar]
- Uitto J, Richard G (2004) Progress in epidermolysis bullosa: genetic classification and clinical implications. Am J Med Genet C Semin Med Genet 131C:61–74 [DOI] [PubMed] [Google Scholar]
- van Steensel MAM, Smith FJD, Steijlen PM, Kluijt I, Stevens HP, Messenger A, Kremer H, Dunnill MG, Kennedy C, Munro CS, Doherty VR, McGrath JA, Covello SP, Coleman CM, Uitto J, McLean WHI (1999) The gene for hypotrichosis of Marie Unna maps between D8S258 and D8S298: exclusion of the hr gene by cDNA and genomic sequencing. Am J Hum Genet 65:413–419 [DOI] [PMC free article] [PubMed] [Google Scholar]