Abstract
Bipolar, schizophrenia, and schizoaffective disorders are common, highly heritable psychiatric disorders, for which familial coaggregation, as well as epidemiological and genetic evidence, suggests overlapping etiologies. No definitive susceptibility genes have yet been identified for any of these disorders. Genetic heterogeneity, combined with phenotypic imprecision and poor marker coverage, has contributed to the difficulty in defining risk variants. We focused on families of Ashkenazi Jewish descent, to reduce genetic heterogeneity, and, as a precursor to genomewide association studies, we undertook a single-nucleotide polymorphism (SNP) genotyping screen of 64 candidate genes (440 SNPs) chosen on the basis of previous linkage or of association and/or biological relevance. We genotyped an average of 6.9 SNPs per gene, with an average density of 1 SNP per 11.9 kb in 323 bipolar I disorder and 274 schizophrenia or schizoaffective Ashkenazi case-parent trios. Using single-SNP and haplotype-based transmission/disequilibrium tests, we ranked genes on the basis of strength of association (P<.01). Six genes (DAO, GRM3, GRM4, GRIN2B, IL2RB, and TUBA8) met this criterion for bipolar I disorder; only DAO has been previously associated with bipolar disorder. Six genes (RGS4, SCA1, GRM4, DPYSL2, NOS1, and GRID1) met this criterion for schizophrenia or schizoaffective disorder; five replicate previous associations, and one, GRID1, shows a novel association with schizophrenia. In addition, six genes (DPYSL2, DTNBP1, G30/G72, GRID1, GRM4, and NOS1) showed overlapping suggestive evidence of association in both disorders. These results may help to prioritize candidate genes for future study from among the many suspected/proposed for schizophrenia and bipolar disorders. They provide further support for shared genetic susceptibility between these two disorders that involve glutamate-signaling pathways.
Introduction
Bipolar disorders (BP [MIM 125480]) and schizophrenia (SZ [MIM 181500]) are common and debilitating psychiatric disorders. Among the several forms of BP, bipolar 1 (BPI) is the most severe, has the highest interrater reliability, and is characterized by disabling bouts of mania and depression (American Psychiatric Association 1994). SZ is a disorder of thought and emotions, characterized by psychotic manifestations, including hallucinations and delusions, lack of motivation, and anhedonia. Both SZ and BPI show variable clinical expression. No symptom or behavior is pathognomic for either disorder. In addition, there can be overlap in clinical presentation (e.g., a majority of individuals with BPI experience hallucinations or delusions [Goodwin and Jamison 1990; Dunayevich and Keck 2000; Potash et al. 2001]). The schizoaffective disorder (SZA) diagnosis was developed to help categorize the group of patients who experienced both affective and SZ-like symptoms. The utility of this diagnosis remains controversial—that is, interrater reliability for the SZA disorder tends to be low, and diagnostic stability is worse than for SZ or BP (Kempf et al. 2005). In genetic studies, SZA cases are sometimes included with SZ cases and, other times, with BP cases (e.g., Lewis et al. 2003; Segurado et al. 2003). Multiple investigators have reported heritability estimates for SZ and BP, with ranges of 41%–87% for BP (Tsuang and Faraone 1990) and 70%–85% for SZ (Tsuang and Faraone 1990; McGue and Gottesman 1991; Tsuang et al. 1991; Cardno et al. 1999, 2001). Heritability estimates for SZA have been reported to be similar to those for SZ (Cardno et al. 1999).
Intensive efforts to localize or identify susceptibility genes for BPI have yielded neither consensus linkage regions nor consistent associations with candidate genes. The two meta-analyses of BPI linkage studies conducted thus far do not support overlapping linkage regions: Badner and Gershon (2002) identified regions on chromosomes 13q and 22q, whereas Segurado et al. (2003) identified regions on chromosomes 9p, 10q, and 14q. Recently, we identified suggestive linkage to BPI on chromosomes 1q23, 3p32, 11q12, and 18q22 among Ashkenazi Jewish (AJ) families (Fallin et al. 2004). Several candidate-gene associations for BP have been reported, but, in general, these have not been replicated, nor have causal variants been identified (Sklar et al. 2002; Maier et al. 2003; Hashimoto et al. 2005; Lasky-Su et al. 2005; Preisig et al. 2005).
By contrast, linkage evidence for an SZ/SZA locus has been identified across almost every human chromosome, with notable signals on chromosomes 1q, 6p, 6q, 8p, 13q, and 22q (for review, see Owen et al. 2004). Two meta-analyses of SZ genome scans do not find exactly overlapping regions, although one found evidence of linkage on chromosomes 8p, 13q, and 22q (Badner and Gershon 2002), and the other found the strongest evidence on chromosome 2q, with 15 other regions noted as important, including 8p and 22q (Lewis et al. 2003). We reported linkage evidence for SZ on chromosome 10q22 among 29 multiplex AJ families with SZ/SZA (Fallin et al. 2003). Other studies show suggestive evidence of linkage to SZ in this region, although the region is not usually among the highest signals (Lerer et al. 2003; Park et al. 2004) and was not identified in the meta-analyses. Interestingly, Hennah et al. (2004) found strong linkage to this 10q region for SZ in a Finnish sample, after conditioning on the disrupted-in-schizophrenia-1 gene (DISC1).
Several candidate genes have shown association with SZ/SZA, notably regulator of G-protein signaling 4 (RGS4) on chromosome 1q23, DISC1 on chromosome 1q42, dystrobrevin-binding protein 1 (DTNBP1) on chromosome 6p22, neuregulin 1 (NRG1) on chromosome 8p12, G30/G72 on chromosome 13q33, and catechol-O-methyltransferase (COMT) on chromosome 22q11 (Owen et al. 2004). Replication in at least one independent sample has been reported for each of these, along with multiple reports of failures to find association, and no sequence variant predicted to alter function has been established across populations. We have included SNPs for each of these genes in our study.
Several factors have likely contributed to the inconsistencies in linkage and association studies of BP and SZ/SZA, including locus and allelic heterogeneity, difficulties in diagnosis, and limitations in technology and marker availability that, until recently, have hindered adequate coverage of candidate genes and regions. To minimize these problems, we conducted a candidate-gene study of 440 SNPs in 64 candidate genes, using DNAs from case-parent trios of AJ descent, 337 with BPI, 217 with SZ, and 57 with SZA. We focused on families of AJ descent, to reduce allelic and locus heterogeneity, and we used precise and narrow disease definitions, such as restricting our BP trios to those with cases meeting criteria for BPI and considering a combined group of SZ/SZA as well as a restricted SZ-only set. We have taken advantage of more-affordable multiplex genotyping technology (Illumina Bead Array), as well as the emerging public SNP information, to achieve greater coverage of variation for a relatively large list of candidate genes, compared with previous candidate gene studies for BP or SZ, in the hope of increasing the potential to detect associations. Although the ultimate goal is to genotype the entire human genome to search for susceptibility, this candidate gene strategy has been advocated by others (Sklar et al. 2002; Botstein and Risch 2003) as a quick and efficient approach with the possibility of early success.
Methods
Recruitment
Over a 6-year period, we recruited individuals of AJ descent with BPI and SZ/SZA through advertisements in newspapers and Jewish newsletters, talks to community organizations, letters to leaders of the Jewish community, letters and talks to service providers, and a study Web site hosted by the Johns Hopkins Epidemiology-Genetics Program in Psychiatry. Case-parent trios were eligible for inclusion in these analyses if the proband met criteria for either a BPI diagnosis or an SZ or SZA diagnosis, according to the DSM-IV (American Psychiatric Association 1994), if both parents were available for DNA collection, and if four grandparents were known to be of AJ descent. All recruitment methods and protocols for the collection of blood samples and clinical data were approved by the Johns Hopkins institutional review board, and appropriate informed consent was obtained from all human subjects.
Diagnostic Instruments and Procedures
The data collection for this study was done in tandem for studies of BPI and SZ/SZA focusing on AJ participants. All diagnostic procedures for data collection were identical. Potentially affected individuals and their parents were examined in person by a doctoral-level clinical psychologist (clinical examiner) and were asked for a blood sample. Examiners were blind to the subject’s diagnosis; they did not know whether the family was being assessed for the study of BP or SZ/SZA. Most of the subjects were seen in their homes. Detailed clinical methods are available from prior publications (Fallin et al. 2003, 2004).
Demographic and Clinical Characteristics
The BPI analyses include 337 trios from 323 families. The BPI-affected subjects had an average age (±SD) at onset of 21.5±7.3 years, and 50.6% were female. Results reported for SZ/SZA are based on 274 trios from 263 families. Cases had an average age at onset of 19.5±4.5 years, and 25.8% were female. Of the SZ/SZA trios, 57 (21%) had probands with SZA, with an average age at onset of 19.3±5.4 years, and 33.3% were female. Included in our trio data are 14 of the 29 multiplex families with SZ/SZA and 8 of the 22 multiplex families with BPI that had been included in previous autosomal linkage scans for these disorders (Fallin et al. 2003, 2004). Five multiplex families contributed two trios each to the BPI analysis, and eight multiplex families contributed two trios each to the SZ/SZA analysis; no family contributed more than two trios to either the BPI analyses or the SZ/SZA analyses. Also, 14 families were “mixed”—that is, they contained both BPI- and SZ/SZA-affected subjects—but only the appropriate subject (proband with BPI for BPI analyses and proband with SZ/SZA for the SZ/SZA analyses) and his/her parents were included in either the SZ/SZA or BPI analyses. Of the BPI-affected cases, 66.7% had psychosis.
Ancestry questionnaires were completed for each proband to establish the country or region of origin of all four grandparents and to reduce the possibility of inclusion of non-AJ founders in our sample. Families were excluded from these analyses if any grandparent of an affected subject was known to be of non-AJ descent.
Gene Selection
We included candidate genes for BP and for SZ/SZA because of the emerging evidence for overlap in the genetic etiologies of these disorders. We chose candidate genes on the basis of two criteria: (1) evidence of association from previous studies of BP or SZ/SZA or location in candidate chromosome regions identified in previous linkage studies of BP or SZ/SZA, and (2) biological plausibility, including the presence of genes involved in neurodevelopment, in synaptic vesicle function, in antipsychotic drug metabolism, or in particular neurotransmitter systems (glutamatergic, serotonergic, dopaminergic, noradrenergic, GABAergic, or cholinergic). The genes we selected were located on 11 chromosomes (see table 1). Fourteen positional candidate genes in chromosome 22q11.21 (see table 1) were selected on the basis of their location in a candidate chromosomal region, as determined by linkage analysis, and because they are critically mapped within the boundaries of the typical ∼3-Mb deletion common in velocardiofacial syndrome (VCFS [MIM #192430]) (Driscoll et al. 1992; Lindsay et al. 1995; Edelmann et al. 1999), which has been associated with an increased risk of SZ (Shprintzen et al. 1992; Pulver et al. 1994b).
Table 1.
64 Candidate Genes for BPI and SZ and Suggestive SNP and Haplotype Results
| GeneSymbol | Region | Length(kB) | Exons | SNPsAttempted | SNPsTyped | SNPs/kB | MedianD′ | Medianr2 | LDBlocks | BlockCoveragea | HighlySuggestiveb | Suggestiveb |
| RGS4 | 1q23.3 | 8 | 5 | 4 | 3 | 1.9 | 1.00 | .36 | 1 | .95 | SZc | … |
| DISC1 | 1q42.1 | 414 | 13 | 22 | 19 | 20.7 | .16 | .00 | 10 | .46 | … | … |
| DRD3 | 3q13.3 | 50 | 7 | 5 | 5 | 8.4 | .88 | .30 | 1 | .55 | … | … |
| DRD1 | 5q35.1 | 4 | 3 | 5 | 2 | 1.3 | 1.00 | .99 | 1 | .36 | … | … |
| DTNBP1 | 6p22.3 | 140 | 10 | 16 | 16 | 8.3 | 1.00 | .05 | 4 | .59 | … | BP, SZ/SZA |
| SCA1 | 6p23 | 462 | 9 | 24 | 23 | 19.3 | .12 | .00 | 14 | .25 | SZ/SZA | … |
| KIF13A | 6p23 | 227 | 38 | 15 | 13 | 16.2 | .97 | .34 | 2 | .72 | … | BP |
| SynGAP1 | 6p21.32 | 37 | 19 | 3 | 1 | 18.7 | … | … | … | … | … | … |
| GRM4 | 6p21.3 | 112 | 10 | 13 | 6 | 16.0 | .61 | .08 | 3 | .24 | BP, SZ/SZA | … |
| FKBP5 | 6p21.31 | 163 | 12 | 7 | 5 | 27.2 | 1.00 | .58 | 1 | .34 | … | … |
| TFAP2B | 6p12.3 | 26 | 7 | 4 | 4 | 5.1 | 1.00 | .12 | 1 | .77 | … | … |
| GRM3 | 7q21 | 221 | 5 | 11 | 10 | 20.1 | .77 | .15 | 3 | .78 | BP | … |
| NAT1 | 8p22 | 53 | 17 | 5 | 2 | 17.7 | .04 | .00 | 2 | NA | … | … |
| PPP3CC | 8p21.3 | 100 | 13 | 10 | 9 | 10.0 | .97 | .88 | 2 | .86 | … | … |
| NEF3 | 8p21.2 | 7 | 3 | 4 | 1 | 3.5 | … | … | … | … | … | … |
| NEFL | 8p21.2 | 6 | 5 | 4 | 3 | 1.4 | 1.00 | .10 | 1 | 7.17 | … | … |
| GNRH1 | 8p21.2 | 7 | 3 | 5 | 3 | 1.8 | 1.00 | .17 | 1 | .34 | … | … |
| DPdYSL2 | 8p21.2 | 144 | 14 | 8 | 8 | 16.0 | .65 | .08 | 4 | .57 | SZ/SZA | BP |
| ADRA1A | 8p21.2 | 117 | 2 | 11 | 8 | 13.0 | .32 | .02 | 4 | .33 | … | SZ/SZA |
| CHRNA2 | 8p21.2 | 18 | 8 | 5 | 4 | 3.7 | .94 | .10 | 3 | .01 | … | … |
| PNOC | 8p21.1 | 26 | 4 | 7 | 7 | 3.3 | 1.00 | .09 | 1 | .67 | … | SZ/SZA |
| NRG1 | 8p12 | 1,128 | 12 | 15 | 12 | 86.8 | .26 | .01 | 6 | .04 | … | … |
| CHRNB3 | 8p11.21 | 40 | 6 | 5 | 4 | 8.0 | 1.00 | .23 | 1 | .88 | … | … |
| PPP3CB | 10q22.2 | 59 | 14 | 4 | 3 | 14.8 | 1.00 | .77 | 1 | .64 | … | … |
| MYOZ1 | 10q22.2 | 10 | 6 | 3 | 1 | 5.1 | … | … | … | … | … | … |
| NRG3 | 10q23.1 | 1,112 | 10 | 23 | 18 | 58.5 | .14 | .01 | 10 | .12 | … | SZ/SZA |
| GRID1 | 10q23.2 | 767 | 13 | 40 | 29 | 25.6 | .21 | .01 | 13 | .31 | SZc | BP, SZ/SZA |
| SNCG | 10q23.2 | 54 | 5 | 7 | 5 | 9.1 | .11 | .01 | 4 | .01 | … | … |
| HTR7 | 10q23.31 | 117 | 4 | 9 | 8 | 13.0 | 1.00 | .04 | 2 | .84 | … | … |
| LGI1 | 10q23.33 | 40 | 8 | 7 | 5 | 6.7 | .34 | .04 | 4 | .02 | … | SZ/SZA |
| INA | 10q24.33 | 13 | 3 | 5 | 3 | 3.3 | .97 | .88 | 1 | .81 | … | … |
| GFRA1 | 10q26 | 212 | 9 | 15 | 12 | 16.3 | .24 | .02 | 5 | .44 | … | SZ/SZA |
| DRD4 | 11p15.5 | 3 | 4 | 3 | 1 | 1.7 | … | … | … | … | … | … |
| DRD2 | 11q23 | 66 | 7 | 8 | 5 | 11.0 | .95 | .21 | 1 | .48 | … | … |
| GRIN2B | 12p12 | 419 | 13 | 34 | 31 | 13.1 | .22 | .02 | 10 | .51 | BP | … |
| DAO | 12q24 | 42 | 11 | 5 | 5 | 7.0 | .89 | .26 | 2 | .27 | BP | … |
| NOS1 | 12q24 | 244 | 29 | 10 | 9 | 24.4 | .49 | .07 | 4 | .20 | SZ/SZA | BP |
| HTR2A | 13q14.2 | 65 | 4 | 9 | 9 | 6.5 | .29 | .01 | 4 | .61 | … | SZ/SZA |
| SLC15A1 | 13q32.2 | 69 | 21 | 7 | 7 | 8.6 | .72 | .03 | 3 | .28 | … | SZ/SZA |
| ZIC2 | 13q32.3 | 5 | 3 | 4 | 3 | 1.2 | 1.00 | .18 | 1 | .64 | … | … |
| TPP2 | 13q33.1 | 82 | 29 | 6 | 6 | 11.8 | 1.00 | .25 | 1 | .73 | … | SZ/SZA |
| G30/G72 | 13q33.2 | 25 | 4 | 15 | 14 | 1.7 | .96 | .15 | 2 | 1.33 | … | BP, SZ/SZA |
| IL17R | 22q11.1 | 26 | 13 | 4 | 2 | 8.5 | 1.00 | .56 | 1 | .07 | … | … |
| TUBA8 | 22q11.21 | 69 | 5 | 5 | 2 | 22.9 | .53 | .03 | 2 | NA | BP | … |
| DGCR6 | 22q11.21d | 6 | 6 | 3 | 1 | 3.2 | … | … | … | … | … | … |
| PRODH | 22q11.21d | 24 | 15 | 11 | 6 | 3.4 | .66 | .03 | 4 | .07 | … | SZ/SZA |
| DGCR14 | 22q11.21d | 14 | 10 | 6 | 5 | 2.4 | .94 | .17 | 2 | .65 | … | … |
| GSCL | 22q11.21d | 1 | 3 | 3 | 1 | .6 | … | … | … | … | … | … |
| UFD1L | 22q11.21d | 30 | 12 | 5 | 5 | 4.9 | 1.00 | .12 | 1 | .73 | … | … |
| COMT | 22q11.21d | 28 | 6 | 10 | 7 | 3.5 | .87 | .36 | 3 | .73 | … | … |
| DGCR8 | 22q11.21d | 65 | 14 | 7 | 3 | 16.2 | .94 | .12 | 1 | .89 | … | BP |
| RTN4R | 22q11.21d | 27 | 1 | 6 | 4 | 5.4 | .69 | .42 | 2 | .07 | … | … |
| DGCR6L | 22q11.21d | 6 | 5 | 1 | 1 | 2.9 | … | … | … | … | … | … |
| ZNF74 | 22q11.21d | 21 | 6 | 4 | 3 | 5.2 | .96 | .46 | 1 | .51 | … | … |
| PCQAP | 22q11.21d | 80 | 18 | 9 | 6 | 11.4 | 1.00 | .20 | 1 | .68 | … | … |
| PIK4CA | 22q11.21d | 131 | 54 | 10 | 6 | 18.8 | 1.00 | .67 | 1 | .67 | … | … |
| SNAP29 | 22q11.21d | 32 | 5 | 5 | 4 | 6.4 | 1.00 | .98 | 1 | .98 | … | … |
| UBE2L3 | 22q11.21d | 56 | 1 | 5 | 2 | 18.8 | 1.00 | 1.00 | 1 | .30 | … | … |
| CABIN1 | 22q11.23 | 167 | 37 | 9 | 7 | 20.9 | .99 | .46 | 1 | .75 | … | BP |
| YWHAH | 22q12.3 | 13 | 2 | 9 | 8 | 1.5 | 1.00 | .35 | 2 | 1.23 | … | BP |
| IL2RB | 22q13.1 | 74 | 10 | 15 | 7 | 9.2 | .26 | .01 | 4 | .09 | BP | … |
| PRKCABP | 22q13.1 | 19 | 13 | 10 | 5 | 3.1 | 1.00 | .28 | 1 | .84 | … | … |
| BZRP | 22q13.2 | 12 | 4 | 8 | 6 | 1.7 | 1.00 | .26 | 1 | .99 | … | SZ/SZA |
| MLC1 | 22q13.33 | 27 | 12 | 10 | 7 | 3.3 | .94 | .09 | 2 | .93 | … | … |
| Total or Average | … | 7,849 | 10.5 | 580 | 440 | 11.6 | .75 | .25 | 2.91 | .66 | … | … |
Fraction of gene length covered by blocks defined by genotyped SNPs. Some fractions exceed 1 because the entire gene is within a larger block. NA indicates no blocks defined in that gene.
Genes were considered highly suggestive if any SNP or haplotype nominal P value was <.01 and suggestive if .01<P<.05.
Finding was for analyses restricted to only SZ diagnosis.
Located in the VCFS critical deletion region.
SNP Selection and Quality Control
We selected SNPs on the basis of public information available in August 2003 (dbSNP [genome build 33]). We initially selected 576 SNPs in and around our candidate genes, on the basis of the following criteria, in order of importance in our selection scheme: (1) known heterozygosity and minor allele frequency (MAF) > 0.05, (2) submission to dbSNP by more than one source, (3) validation reported, and (4) a minimum density of 1 SNP per 25 kb. Available nonsynonymous changes located in coding regions were also included when space on the array permitted. SNPs were preliminarily screened via a proprietary algorithm (Illumina) that predicts performance on the Illumina platform. The final set contained 22 coding SNPs, 12 nonsynonymous and 10 synonymous changes.
Genotyping
Genotyping was performed by Illumina, through use of their Integrated BeadArray System. We supplied Illumina with 22 96-well barcoded DNA microtiter plates containing 2,024 samples of DNA (4 μg) quantified with Pico Green to be at 100 ng/μl. Illumina delivered 1,165,824 genotypes, each with a quality score calculated by proprietary Illumina algorithms (GC_Score). GC_Score histograms, allele and genotype frequency estimation, and Hardy-Weinberg equilibrium (HWE) distribution testing were completed in SAS (v. 8.02) software scripts, with use of available parental data for all AJ probands with SZ/SZA or BPI (1,210 AJ parents).
Of 2,024 DNA samples, 59 failed to amplify, 11 failed a sex screen with the use of X- and Y-linked polymorphisms, and 3 pedigrees had Mendelian incompatibilities that could not be resolved; all were dropped from subsequent analyses. Ultimately, we included data from a total of 337 BPI trios and 274 SZ/SZA trios.
Of the 576 SNPs attempted, 531 (92%) returned genotype calls. Of these 531, 64 were discarded (22 were monomorphic, 4 had significant departures from HWE at the P<.01 level among parents, and 38 had MAF<5%). Across all SNP genotypes, the GC_Score ranged from 1.0×10-6 to 0.95, with a mean (±SD) of 0.80±0.11. From the distribution across all genotype calls, we chose a conservative threshold of 0.70 for inclusion of a genotype in our analyses, to minimize genotype error in our results. The resulting mean GC_Score was 0.84±0.06; minimum = 0.70, maximum = 0.95. Thirty-seven SNPs had a mean GC_Score below this threshold and were not included. Thus, a total of 435 (75.6%) of the original 531 Illumina-generated SNPs were useful for our final analyses. We also genotyped five SNPs on chromosome 22q11.21, by use of TaqMan (Applied Biosystems) assays in our own laboratories: two (rs2016118 and rs3788287) in PRODH, and three (rs165599, rs3788319, and rs4633) in COMT. For all 440 SNPs, we detected Mendelian inconsistencies by use of PedCheck (O’Connell and Weeks 1998). Genotypes for SNPs with incompatibility for any pedigree were deleted for that pedigree. Only 73 (0.0082%) of the 890,560 genotypes analyzed were omitted because of violation of Mendelian patterns of inheritance.
During the course of our analysis, we identified unrecognized microdeletions in the chromosome 22q11 VCFS critical region in one individual with SZ and in one individual with SZA, on the basis of their observed non-Mendelian incompatibilities. Both microdeletions were confirmed by FISH. Specific attention to this possibility is necessary when SZ genetic association in this region is considered. These families were not included in the chromosome 22q11 candidate-gene analyses for those SNPs showing this incompatibility. The families were included in all other candidate-gene analyses.
Graphical and Statistical Analyses
We measured pairwise linkage disequilibrium, using D′ and r2, and we used the program Haploview to generate graphical representations of the marker-to-marker disequilibrium and underlying haplotype block structure (Barrett et al. 2005). Both single-SNP and haplotype-based transmission/disequilibrium test (TDT) analyses were performed, since the value of SNPs versus haplotypes as markers for an underlying susceptibility allele depends on the nature of that allele in a particular setting and on the polymorphisms genotyped. In some situations, a single SNP will be the best marker of an association (e.g., r2=1 with the risk variant), whereas in other situations, a haplotype will provide the best information (e.g., r2=1 between the risk variant and a particular multi-SNP haplotype).
For single-SNP TDT analyses, including odds ratio (OR) estimation and calculation of global likelihood ratio test (LRT) P values, we used the genotypic TDT method, employing conditional logistic regression as implemented in the gtrr function of the GenAssoc STATA package made available by David Clayton (see David Clayton's Web site). We then performed 10,000 permutations of case-pseudocontrol status (with use of the permute function written for STATA by David Clayton), ran gtrr on each permuted data set, and calculated empirical P values for each SNP on the basis of 10,000 permutations.
Haplotype TDT analyses were performed using FBAT software for haplotype data (Horvath et al. 2001) Empirical global P values were obtained using 10,000 permutations, as implemented in FBAT. We delineated sets of SNPs for haplotype analysis in two ways: (1) creating two-, three-, and four-SNP windows tiled across each chromosome and (2) defining haplotype blocks of linkage disequilibrium (LD) according to the “solid spine of LD” approach, which requires all pairwise D′ values within the spine to be ⩾0.75. Currently, it is not clear which method best exploits the multimarker information in haplotypes (Schwartz et al. 2003). To maximize our information, we used both strategies. SNP haplotype windows were allowed to cross intergene gaps if the distance between genes was <1 Mb. This occurred for 24 intergene regions.
There are many ways to prioritize findings for interpretation and follow-up. Because of the varying prior evidence of association among these candidate genes and given a diverse correlation structure across SNPs, there is not a straightforward approach to correct for the complicated multiple testing in this study. We chose to rank results for each diagnosis separately by empirical P values at the single-SNP and haplotype levels. We then prioritized genes with haplotype or SNP empirical P values at the .05 and .01 levels as showing “suggestive” and “highly suggestive” evidence, respectively, for further consideration.
Results
Descriptive Analyses
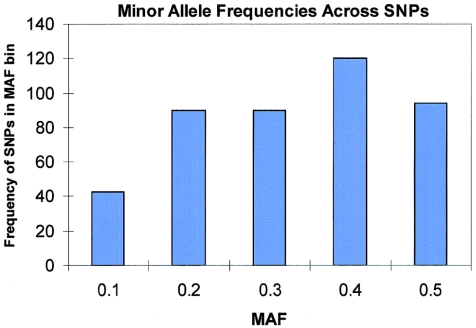
The candidate genes and their characteristics, SNP density, and LD information are provided in table 1. Gene sizes had a range from 1.3 kB to 1.13 Mb, and SNPs per gene had a range of 1–31 SNPs, with an average of 6.9 SNPs per gene and an average density of 1 SNP per 11.9 kB. The SNPs and their locations on dbSNP build 33, MAF, genotyped frequencies, and HWE P values are given in a tab-delimited ASCII file (online only) that can be imported into a spreadsheet. Of the 64 genes initially queried, 55 had at least 3 informative SNPs from the final set of 440 SNPs. The average heterozygosity across all 440 SNPs was 0.38, with an approximately uniform distribution of MAF (see fig. A1), which provides confidence that our SNP selection criteria yielded informative SNPs.
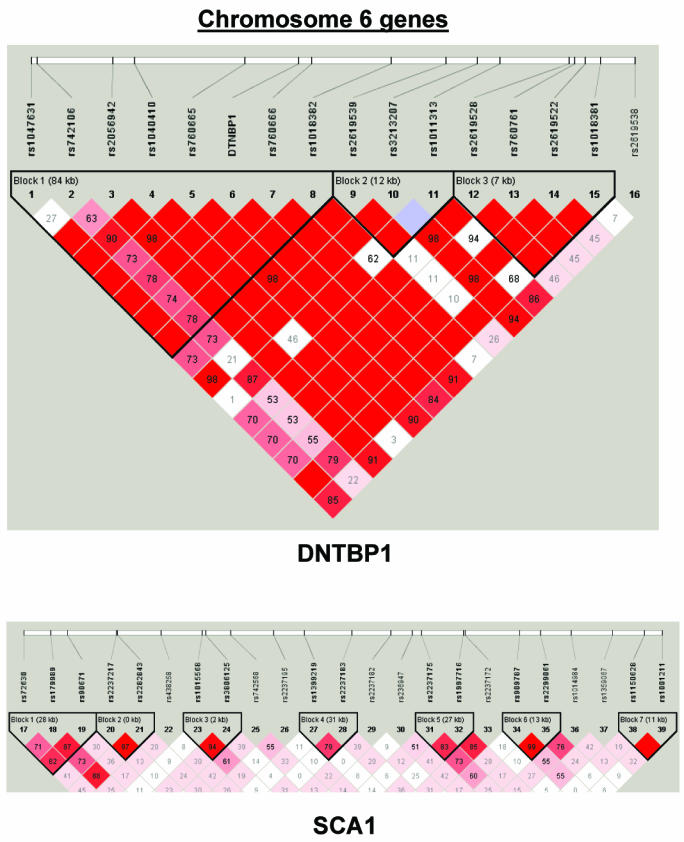
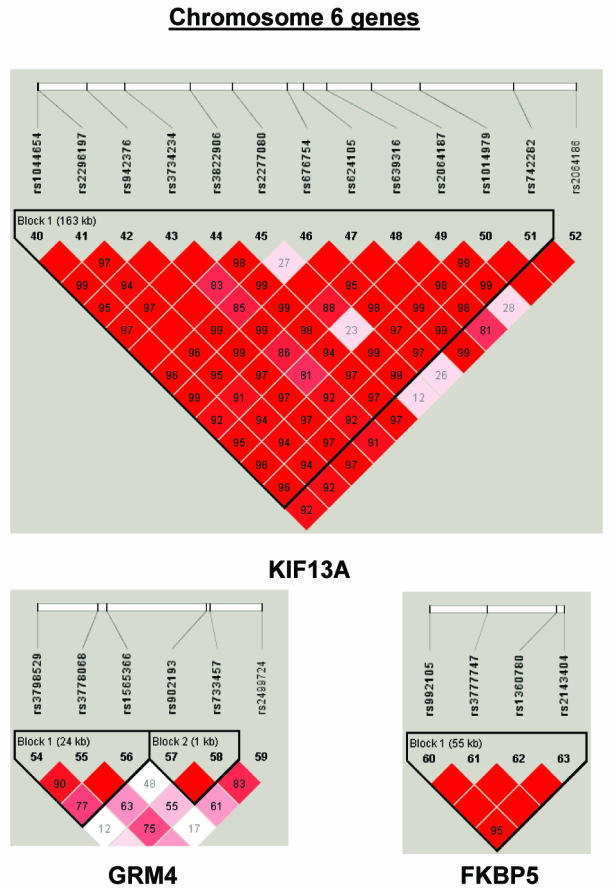
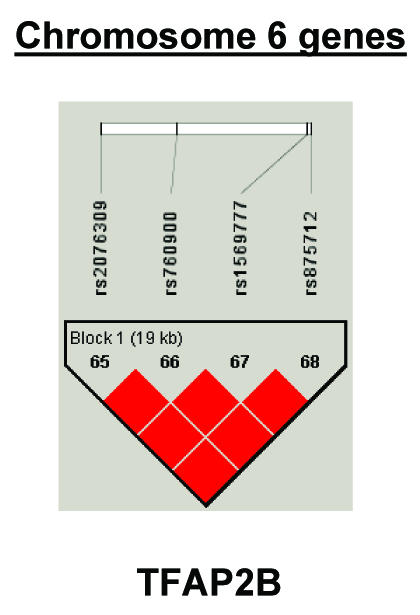
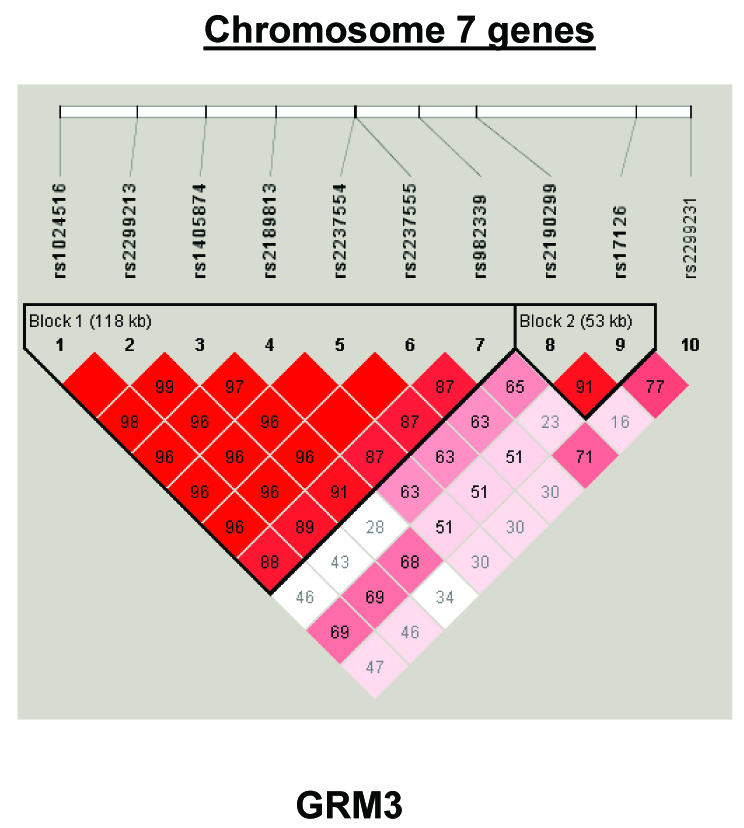
To summarize LD information within each gene, we calculated the median value for pairwise D′ and r2 between SNPs in each gene; the average across genes was D′=0.75, r2=0.25 (table 1). We identified a total of 171 LD blocks. We considered SNPs that were not part of an LD block with other SNPs in the data set to be a separate block of size 1 (69 of 171 were “orphan” SNPs). Of the 102 blocks with more than one SNP, the average length was 27.7 kB, with a range from 153 bp to 163.7 kB (table 1). The average number of blocks per gene was 2.9, with a range of 1–14 blocks per gene, according to the “solid spine” of D′>0.75 criterion. Graphs of pairwise LD values and haplotype block delineations for each gene are available in figure A2.
Association Analyses
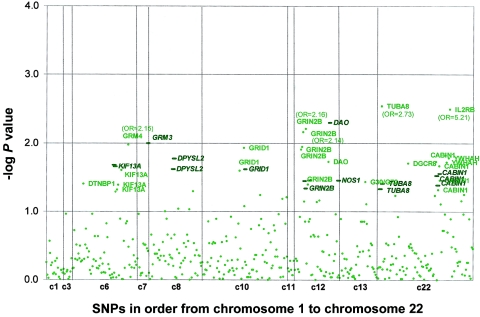
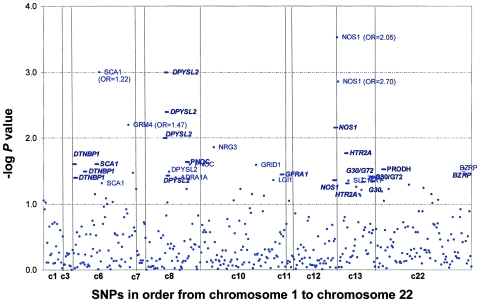
P values for the TDT and haplotype analyses for BPI and for SZ/SZA for all 440 SNPs are shown in figures 1 and 2. Of 387 single SNPs, 24 (6.2%) were significant at the .05 level (after removal of one of each SNP pair with r2=1 [53 SNPs]), and 8 (2.1%) were significant at the .01 level among the BPI trios, which suggests that there is not gross inflation of false-positive results from specified type I error levels. For the SZ/SZA sample, 23 (5.8%) of 399 SNPs were significant at the .05 level, and 5 (1.3%) were significant at the .01 level.
Figure 1.
SNP and haplotype results for BPI trios. Empirical P values from genotype TDT and HBAT analyses are plotted on the basis of 10,000 permutations. Circles, SNP results. Bold lines, 4-SNP haplotype windows. The corresponding genes are labeled for any result with a P value <.05, and OR estimates are provided for any SNP results with P value <.01. For ease of illustration, 4-SNP haplotype windows are plotted by reflecting only the best haplotype signal (of 4 possible windows) for each SNP.
Figure 2.
SNP and haplotype results for SZ/SZA trios. Empirical P values from genotype TDT and HBAT analyses are plotted on the basis of 10,000 permutations. Circles, SNP results. Bold lines, 4-SNP haplotype windows. The corresponding genes are labeled for any result with a P value <.05, and OR estimates are provided for any SNP results with P value <.01. For ease of illustration, 4-SNP haplotype windows are plotted by reflecting only the best haplotype signal (of 4 possible windows) for each SNP.
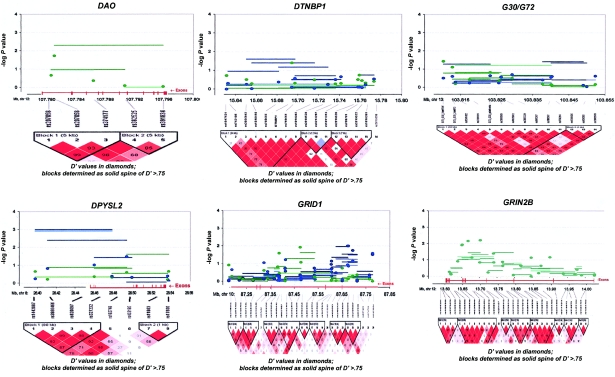
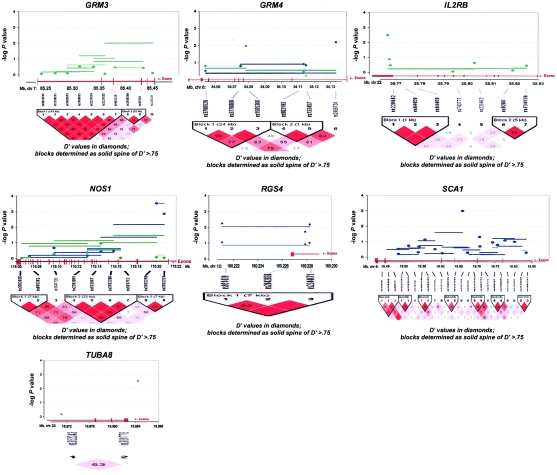
The genes with haplotype or SNP findings that met our “highly suggestive” criterion among BPI trios were DAO, IL2RB, GRM3, GRM4, GRIN2B, and TUBA8 (see table 1 and fig. 3). For the SZ/SZA analyses, the highly suggestive genes were SCA1, GRM4, DPYSL2, and NOS1. Because of concerns regarding heterogeneity in phenotype and genetic etiology between SZ and SZA, we performed analyses restricted to only trios with SZ subjects. There were no substantial differences from the combined SZ/SZA analysis, with the exception of RGS4 and GRID1, which both show highly suggestive associations only among SZ trios, although similar trends are seen for the combined group. P values for the three suggestive RGS4 SNPs among the SZ samples are P=.006, P=.019, and P=.007, versus P=.089, P=.120, and P=.096, respectively, for the SZ/SZA combined sample (fig. 3). For GRID1, two SNPs and one 4-SNP haplotype window achieved P=.01 among the SZ-only sample (fig. 3).
Figure 3.
Gene-specific results for the highest-ranked genes from each data set and for genes with overlapping evidence in BPI and SZ/SZA. P values for single-SNP genotype TDT (solid circles), HBAT with the use of 4-SNP haplotype windows (solid lines), and HBAT with the use of haplotypes according to LD structure (thatched lines) are plotted for each gene. Colors reflect the data set showing association, where blue indicates SZ/SZA, dark blue indicates SZ only, and green indicates BPI. The LD structure for the SNPs in each gene is also shown using the imaging software of Haploview.
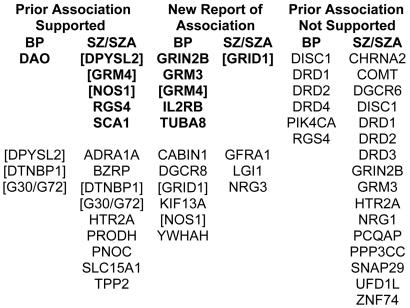
In addition, we compared association findings between BPI and SZ/SZA trios because aspects of the SZ/SZA and BPI phenotypes overlap. Also, there is emerging evidence of shared linkage and association signals for the two disorders (Maziade et al. 2005). Only one gene met our highly suggestive criterion for both disorders—GRM4, on chromosome 6, which had a highly suggestive SNP result for both BPI and SZ/SZA, although a different SNP was associated with each disorder and these are not in LD with each other (fig. 3). Five additional genes showed associations at the suggestive level for both disorders: DTNBP1, DPYSL2, G30/G72, GRID1, and NOS1. Candidate gene findings for BP and SZ/SZA, in the context of previous reports, are summarized in figure 4.
Figure 4.
Candidate gene findings for BP and SZ/SZA in the context of previous reports. Top, Genes shown in bold have P values <.01 for any SNP or haplotype. Bottom, Genes have P values <.05 for any SNP or haplotype. Genes in brackets have P values <.05 for both BPI and SZ/SZA. Because findings with P values in the .01–.10 range may reflect lack of power rather than lack of replication, we have tabulated only genes that did not provide evidence for association at even a liberal P<.10 level.
Known functions of the protein products of the highly suggestive and BPI-SZ/SZA overlapping genes are shown in table 2. Although genes were chosen for involvement in one of several neurotransmitter systems, the majority of genes among the highest ranks are involved in glutamatergic signaling pathways. This is consistent with previous implications of the glutamatergic signaling pathways in BPI and SZ (Harrison and Weinberger 2005; Owen et al. 2005).
Table 2.
Known Functions of 13 Candidate Genes Showing Highly Suggestive Association or Overlapping BPI/SZ Association
| GeneSymbol | Gene Name | Known Biological Systems |
| DTNBP1 | Dystrobrevin-binding protein 1 | Glutamate |
| GRID1 | Glutamate receptor, ionotropic, delta 1 | Glutamate |
| GRM3 | Glutamate receptor, metabotropic 3 | Glutamate |
| GRM4 | Glutamate receptor, metabotropic 4 | Glutamate |
| NOS1 | Nitric oxide synthase 1 (neuronal) | Glutamate, dopamine |
| DAO | D-amino acid oxidase | Glutamate, NMDA |
| G30/G72 | D-amino acid oxidase activator (DAOA) | Glutamate, NMDA |
| GRIN2B | Glutamate receptor, ionotropic, NMDA 2 | Glutamate, NMDA |
| RGS4 | Regulator of G-protein signaling proteins | Glutamate, signal transduction |
| DPYSL2 | Dihydropyrimidinase-like 2 | Neurodevelopmental |
| SCA1 | Spinocerebellar ataxia 1 (ATXN1) | Neurodegenerative repeat |
| IL2RB | Interleukin 2 receptor, beta | Immune response |
| TUBA8 | Alpha tubulin, peroxisome biogenesis factor 26 | Cell cytoskeleton |
Discussion
We have performed a screen of 64 region-specific or previously reported candidate genes for BPI and SZ/SZA disorders among AJ case-parent trios. We report six genes that meet our highly suggestive criterion for BPI association and four that meet this threshold for SZ/SZA. In addition, two genes meet the highly suggestive criterion only among SZ trios. Whereas only one gene met our highly suggestive criterion in both disorders, five others showed evidence at the suggestive level in both disorders, which may indicate that the genetic association is important for a subgroup of the affected individuals in each of the diagnostic categories. The subgroup may be associated with or defined by the presence of one or more of the overlapping clinical features (i.e., mania or psychosis).
This work has attempted to minimize several limitations often plaguing association studies in psychiatry. First, to minimize genetic heterogeneity, we have focused on cases of AJ descent. Although our sample may contain multiple susceptibility genes or alleles, we hypothesize that founder alleles will be a more common source of risk among AJ cases, thereby increasing our ability to detect gene effects. Founder effects have been described elsewhere for both monogenic and complex traits in the AJ population (Neuhausen et al. 1998; Stern et al. 2001; Rennert et al. 2002; Niell et al. 2003; Sugimura et al. 2003). Second, in an effort to minimize misclassification and phenotype heterogeneity, we have employed skilled diagnosticians and clinical consensus, have restricted the BP phenotype to the most severe and reliable diagnosis, and have considered SZ separately from SZA. Third, we have achieved greater gene coverage than previously reported for most of these candidate genes. We aimed to cover each gene as adequately as possible at the time of design, in accordance with available information, technology, and funding. Across these 64 genes, 58 had at least two SNPs genotyped and 55 had at least three SNPs, with an average of 6.9 SNPs per gene, an average density of 1 SNP/11.9 kB within genes, and 66% of each gene, on average, covered by LD blocks detectable with the SNPs genotyped. This is also one of the largest reported sets of SNP genotypes for AJ individuals, and these LD findings provide valuable information about LD in candidate genes for this population. With the use of an average SNP density of 11.9 kB, the mean block size was found to be 27.7 kB, which is slightly greater than that observed for more-outbred populations (Gabriel et al. 2002).
Prioritization
To analyze this large set of candidate gene SNPs, we used both single-SNP and haplotype-based methods. The comparable utility of these two methods is still debated and depends on whether the true risk allele is in higher LD with a single SNP or with a multi-SNP haplotype. Single-SNP analysis can be more statistically efficient if the former is true; however, there are many examples in which haplotypes provide more information than any single SNP genotyped. This will certainly depend on SNP density and LD in the region. Given the nature of this SNP study with varying densities and LD structures, it is important to pursue both methods. There are also many choices regarding the implementation of haplotype analyses. We chose a systematic approach of using sliding windows as well as haplotype sets defined by the LD structure in a gene. The former has the advantage of not relying heavily on a blocking algorithm, whereas the latter exploits LD specific to its structure. In our highest-ranked genes (see fig. 3), the sliding windows and block findings are similar.
This set of 440 SNPs introduces a challenge for multiple testing correction, as there is not a straightforward correction for these highly correlated tests, some of which are specific replications, and all of which are based on prior evidence of some kind. One suggested approach has been to identify the number of pseudoindependent signals within the data set through principal components analysis or the number of LD blocks (Nicodemus et al. 2004; Nyholt 2004). With the assumption that the 171 blocks represent independent information, an approximate correction may be α=.05/171=0.0003 for experimentwide significance at the α=0.05 level. Only one SNP (in TUBA8) and no haplotypes met this strict criterion for BPI, and only one SNP and one haplotype block reached this level for SZ/SZA (both in NOS1). Another suggestion would be to consider ranked P values and to estimate the false discovery rate for a determined threshold of P values. This method does not work well in the case of highly correlated tests, such as our candidate gene SNP approach (Efron 2004). To best interpret our results, we chose instead to rank our findings according to uncorrected empirical P values, with the realization that this method may include false positive results. However, most of these genes were included in this screen because of prior linkage or association reports and biological plausibility. The ultimate proof of association will be the consistent replication by several groups and in several populations and the identification of functional variants.
Genes Implicated in BPI
Genes achieving our highest ranks include some previously associated with this disorder as well as some new association findings (see fig. 4). DAO, which interacts with glutamate pathways, met our highly suggestive criterion among BPI trios and has been previously associated with both BPI and SZ risk (Chumakov et al. 2002; Schumacher et al. 2004). Our BPI-associated haplotypes overlap with the Schumacher et al. (2004) BP haplotype (rs3741775 in common) and the Chumakov et al. (2002) single-point SZ finding (rs3741775 and rs3918347 in common). The five additional genes meeting our highly suggestive criteria for BPI have not previously been reported to be associated with BP. These include two glutamate receptors (GRM3 and GRM4) and a glutamate receptor-related gene (GRIN2B), as well as two genes in the chromosome 22q11-13 SZ/SZA/BP region (IL2RB and TUBA8). Positive associations for GRIN2B have been reported for SZ (Ohtsuki et al. 2001a; Di Maria et al. 2004), but not for BP. GRM4, located in a SZ linkage region on chromosome 6p21, has also been associated with SZ (Ohtsuki et al. 2001b), but not with BP. TUBA8, located immediately proximal to the ∼3-Mb VCFS critical region on chromosome 22q11 (Stanchi et al. 2000), is a positional candidate, based on reported psychiatric disorders shown in VCFS-deleted patients. Finally, IL2RB, located more distally to the VCFS region, at chromosome 22q13, is involved in T-cell–mediated immune responses.
Genes Implicated in SZ/SZA
Among the six highly suggestive SZ/SZA or SZ-only genes, five confirm previous associations (RGS4, SCA1, GRM4, DPYSL2, and NOS1), and one can be considered a new association report (GRID1). Our strongest associations for SZ/SZA are for two SNPs (rs3782219 and rs3782221) 7.6 kb apart (D′=0.95) in NOS1 (P=.0003 and P=.0014, respectively). A SNP in NOS1 was previously shown elsewhere to be highly significant in a Japanese case-control study (Shinkai et al. 2002), although that SNP is 135 kb centromeric to our findings in this gene and was not significant in our analysis. It is, nonetheless, encouraging that two distinct populations show association for this gene, and it is not surprising that particular markers do not give similar results. Variants in RGS4 have been associated with both SZ/SZA and BP risk (Chowdari et al. 2002). A 4-SNP haplotype localized to a 10-kb region of the gene has been confirmed in an Irish sample (Morris et al. 2004; Williams et al. 2004). Our positive SZ association (omitting the SZA probands) includes one of those SNPs (rs951439) and another SNP ∼7 kb centromeric (rs2344671). SCA1 is of interest because of its role in spinocerebellar ataxia, type 1 (MIM 164400), a neurodegenerative disorder with ataxia, dysarthria, and progressive bulbar dysfunction associated in some patients with mild cognitive dysfunction (Zoghbi et al. 1991), and variants in SCA1 have been associated with SZ (Wang et al. 1996a). Previous association for DPYSL2 with schizophrenia used a Japanese case/control population and found support for the 2236T→C polymorphism and a 2-SNP haplotype (1506T-*2236C) (Nakata et al. 2003a). Although we did not genotype these SNPs, our associated haplotype spans the 1506T polymorphism, within 1.1 kb of the 2236C polymorphism.
Finally, GRID1 met our criterion for highly suggestive association with SZ, but it has not been previously reported as a risk factor for SZ or BP. SNPs and/or haplotypes in this gene were also suggestive of association with BPI and SZ/SZA combined. GRID1 is located 1.35 Mb from our peak chromosome 10q22-q23 SZ/SZA linkage signal among AJ families (D10S1774) (Fallin et al. 2003). This gene and its closest paralog, GRID2, encode two members of a subfamily of the glutamate ionotropic receptors; both were identified by sequence similarity with other glutamate receptors, but no ligand has yet been confirmed for either. In mice, mutations in GRID2 are responsible for cerebellar dysfunction (Selimi et al. 2003; Wang et al. 2003).
BPI and SZ
Several recent reports of linkage and association overlap between BP and SZ motivate closer consideration of possible similarities in associations between BPI and SZ/SZA among our samples. Whereas GRM4 is the only highly suggestive gene in common between the BPI and SZ/SZA studies, five additional genes have overlapping evidence of suggestive SNP or haplotype associations for both disorders. Only G30/G72 has previously been associated with both disorders (Hattori et al. 2003). The others are from various locations and have various functions, including glutamatergic, dopaminergic, and neurodevelopmental roles.
Relationship with Previous Findings
To summarize our results in the context of previous association studies, in figure 4 we show previous associations supported by our findings, as well as genes for which we found no support (P>.10) for previous associations. None of the dopamine receptor genes showed SNP or haplotype P values <.10 among the BPI trios, despite consistent previous associations (Spurlock et al. 1998; Jonsson et al. 2003). The same is true for DISC1, RGS4, and PIK4CA, although these genes have been less consistently replicated for association with BP in previous studies. Many of the previously associated SZ genes did not show association in our AJ families. This may indicate that these genes are less important in the genetic predisposition to SZ/SZA among individuals of AJ descent, or it could reflect a lack of power to detect subtle associations. Most notably, we observed no association with COMT, despite previous findings in AJ samples from Israel (Shifman et al. 2002) with several overlapping SNPs between the two studies (Shifman et al. 2002), including the functional val/met polymorphism (rs4680). This may be due, in part, to differences in the diagnostic exclusion criteria between the studies—that is, individuals diagnosed with SZA were excluded from the Shifman et al. (2002) study, but not from our study. Our analyses excluding SZA may not have had sufficient power to detect the association.
Genes were initially chosen for our study on the basis of biological plausibility, including their involvement in one of several neurotransmitter systems. Interestingly, the great majority of genes among the highest ranks are involved in the glutamatergic pathway. Three of the BPI-associated genes encode glutamate receptors, including both ionotropic (GRIN2B) and metabotropic receptors (GRM3 and GRM4) (Hollmann et al. 1994), whereas another (DAO) is involved in the regulation of glutamate receptor signaling. DAO is a major enzyme that controls the breakdown of D-amino acids such as D-serine, which has been shown to be a cotransmitter for the NMDA type of glutamate receptors (Mothet et al. 2000; Baranano et al. 2001), and, thus, DAO and G30/G72, which is a DAO activator, may regulate NMDA receptor function. For example, DAO-knockout mice have recently been reported to have defects in NMDA receptor signaling (Hashimoto et al. 2005). In addition, several genes associated with SZ/SZA are related to glutamatergic signaling. GRID1 is highly related to ionotropic glutamate receptor subunits (Hollmann et al. 1994), although the ligand for the GRID1 gene product has not been identified. Also, RGS4 regulates, and NOS1 is regulated by, glutamatergic signaling. The activation of several types of glutamate receptors has been shown to regulate nNOS activity (coded by NOS1) (Bredt and Snyder 1989; Snyder and Bredt 1991), whereas RGS4 has been shown to inhibit metabotropic glutamate receptor function (Saugstad et al. 1998). These results are consistent with recent findings implicating a potential disruption of excitatory synaptic function in SZ (Harrison and Weinberger 2005; Owen et al. 2005), and they suggest that genes involved in glutamatergic signaling pathways are promising candidates for an exhaustive study of genes associated with BPI and SZ/SZA.
Limitations
Despite the strengths of this study, several issues and limitations must be considered. First, although we have achieved adequate SNP coverage for many genes (see fig. A1 and fig. 4), gaps still exist, with the biggest gene having coverage as sparse as 1 SNP/87 kB (NRG1 [1.128 Mb]). This project was developed before the vast majority of HapMap data were available and, therefore, it relied on dbSNP, Celera, and Perlegen marker information at the time. These gaps can now be improved according to Caucasian marker and LD information for these genes in the HapMap project, assuming the Caucasian samples (families from Utah) used in HapMap are representative of the AJ population.
Second, we selected candidate genes in accordance with their prior association or location within a previously reported linkage region. Since there are many linkage regions, we prioritized linkage among AJ samples, such as our own SZ genome scan (Fallin et al. 2003) or linkage evidence from meta-analyses. Because of efficiency and budget constraints, some important regions were not considered, such as 2q, 6q, and 18q. These regions were of lower priority for this project because of the overlapping work of our collaborators and colleagues.
Third, although 337 and 274 case-parent trios are currently among the largest trio collections for BPI and SZ/SZA, respectively, small effect sizes, or marginal effects when interaction exists, may still be difficult to detect. For example, although our smaller data set (the SZ/SZA sample) provides 96% power to detect an OR=2.5 for a marker with 10% risk allele frequency in complete LD with the actual risk variant (assuming 0.0003 type I error rate), power to detect an OR of 1.5 is dramatically reduced (to 7%) under this same scenario (Gauderman 2002). Our inability to replicate some previous findings should be considered in this context.
Fourth, our focus on case-parent trios may result in a slightly different sample of cases than in other genetic studies that focused on either multiply affected families only or on unrelated cases and controls. We compared characteristics of the cases reported in this study (who had parental DNA available) with those of cases we have collected for other purposes, who did not have parents available for study (referred to as singletons). For both BPI and SZ/SZA cases, the cases in trios had a younger age at onset. In addition, among SZ cases in trios, there are more men (78% vs. 58%) and more cases with manic episodes (42% vs. 29%), compared with our singleton cases. This higher proportion of manic episodes may have contributed to the overlap in findings between BPI and SZ in our studies.
These prioritized genes deserve further evaluation to improve the understanding of their relative importance in both disorders in the AJ and other populations. They must be confirmed in other AJ samples, which we are in the process of collecting. In addition, the importance of these genes in risk for SZ and BP more generally must be evaluated in other, non-AJ populations. This genotype collection will also allow the evaluation of simultaneous effects in a way not previously possible, including conditional analyses to understand potential interactions and the relative importance of particular genes or clinical/environmental risk factors. Also, higher-order data-mining analyses to discover new hypotheses about interactions will be important. Further, the focus on parent/child trios will allow detection of parent-of-origin effects, which may become an important factor as epigenetic mechanisms are explored. These will be the foci of future work.
Supplementary Material
Acknowledgments
We thank the families for their participation in this research. We also thank Pamela Belmonte for assistance in the preparation of this manuscript. This project was funded by National Institutes of Mental Health (NIMH) grants R01MH057314 and R01MH58153.
Appendix A: Distribution of MAFs across 440 SNPs
Figure A1.
Distribution of MAFs across 440 SNPs
Appendix B: LD structure for each gene
Figure A2.
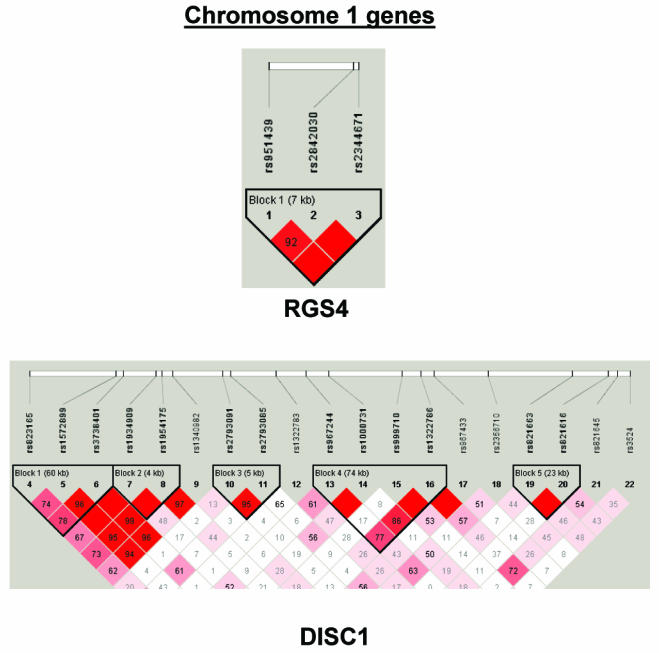
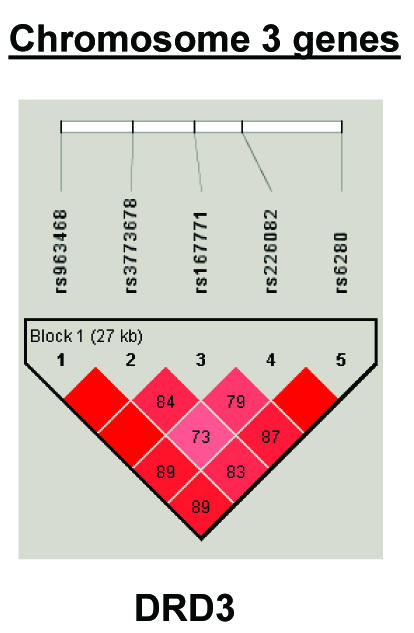
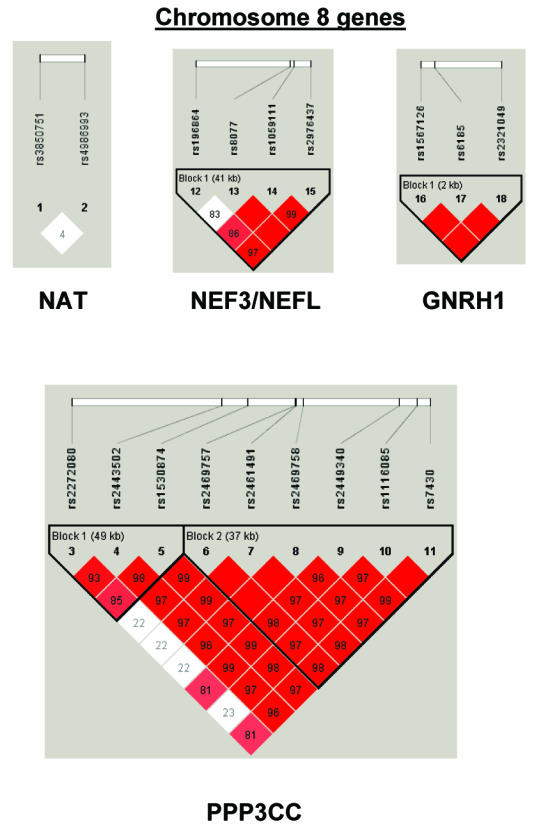
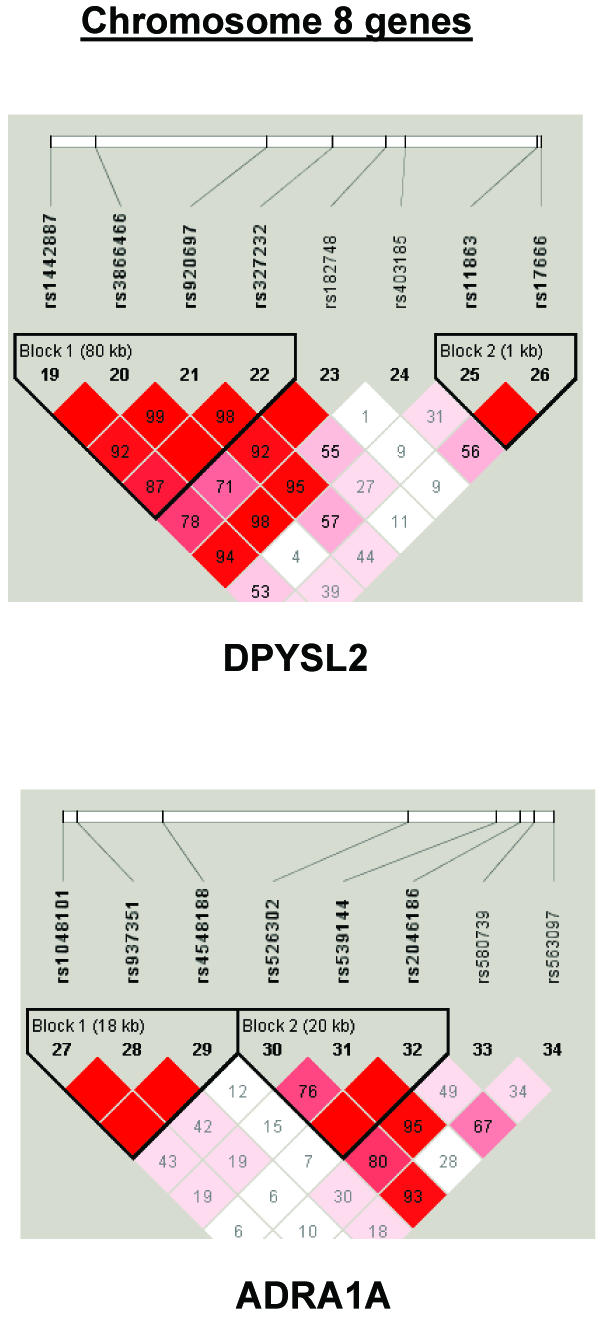
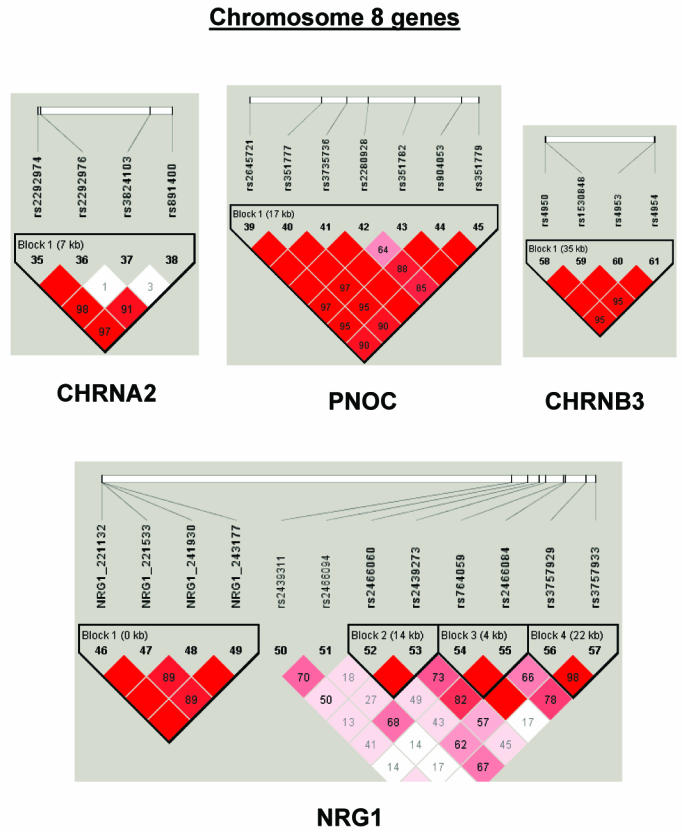
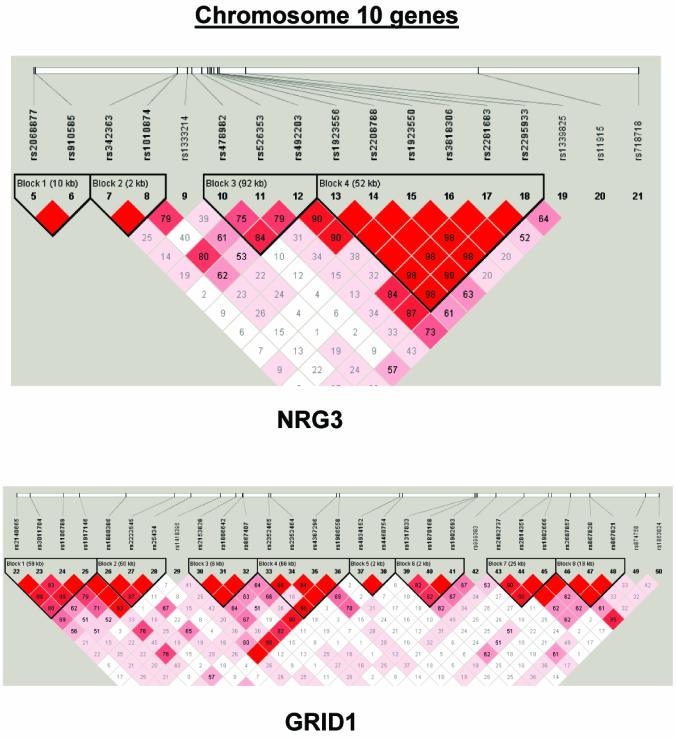
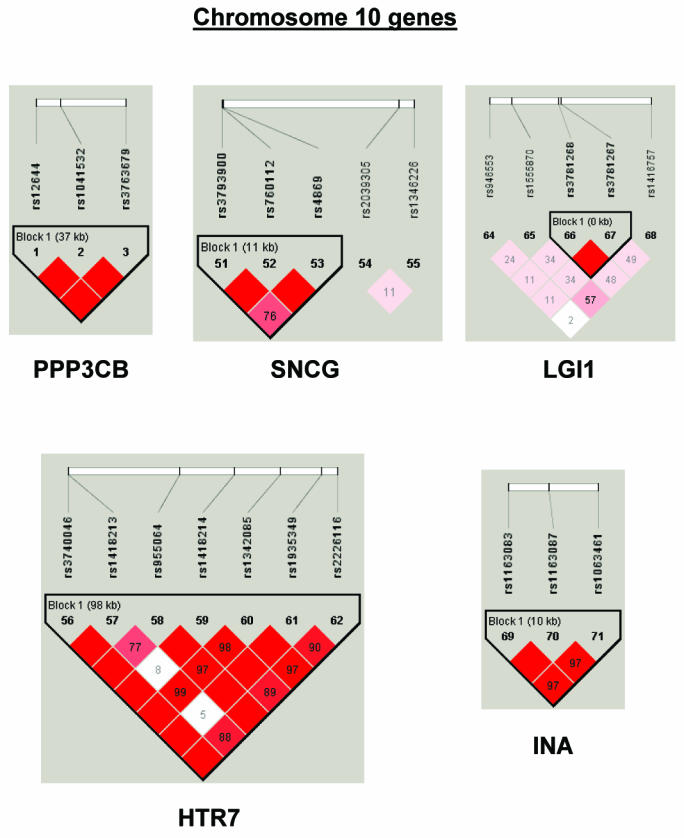
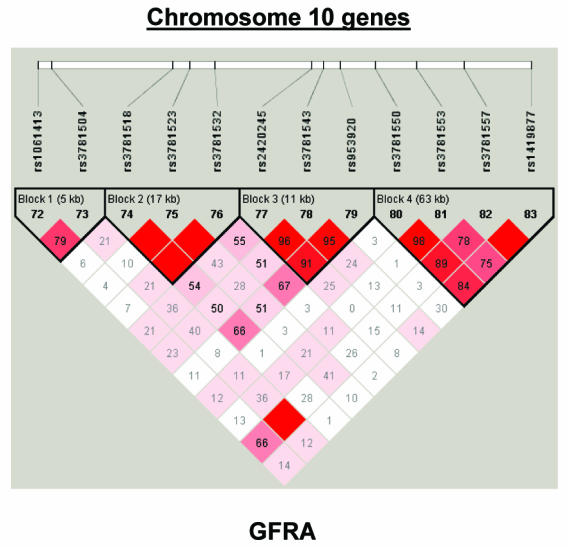
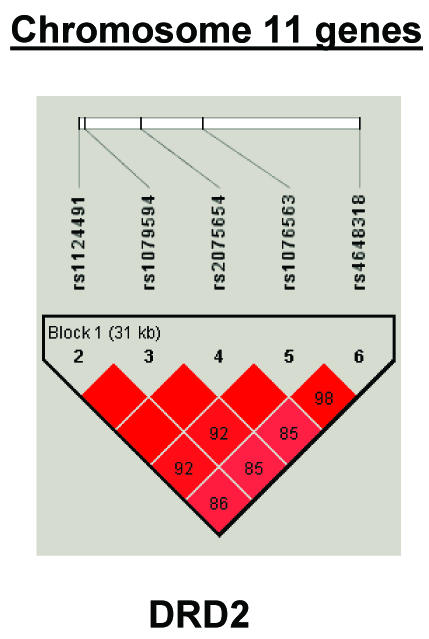
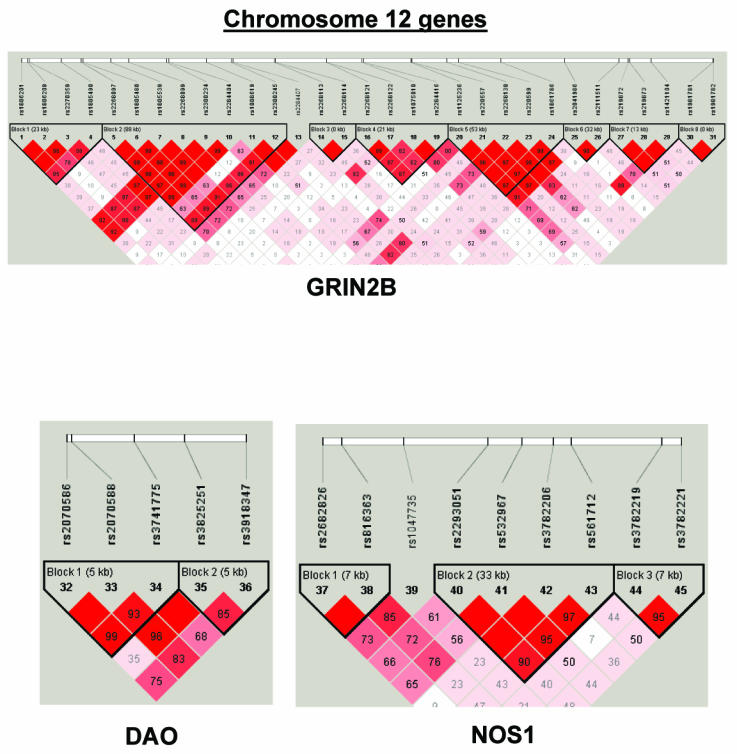
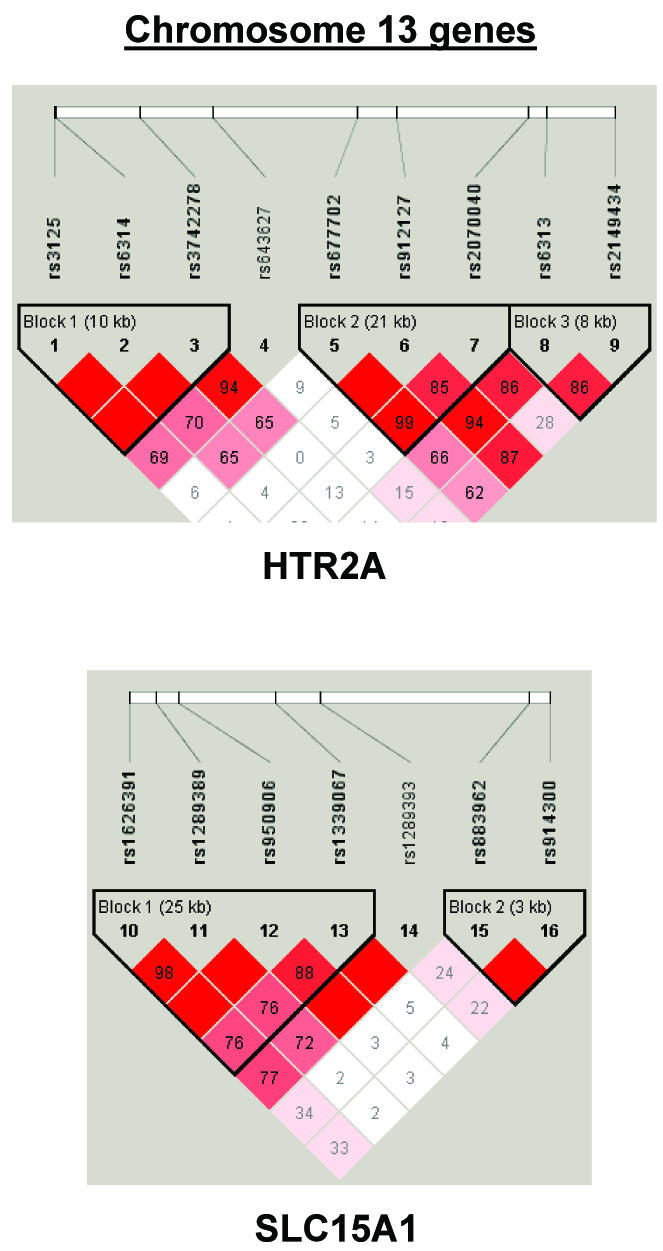
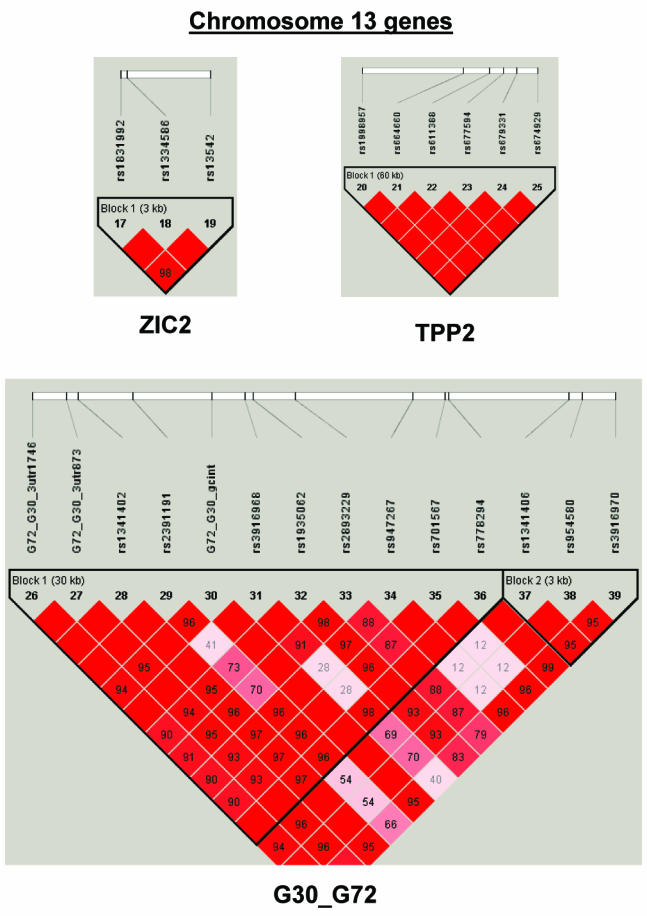
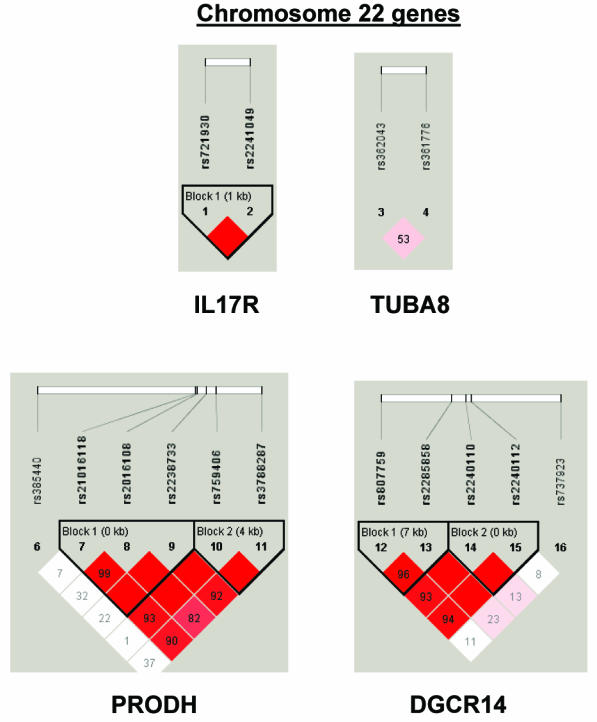
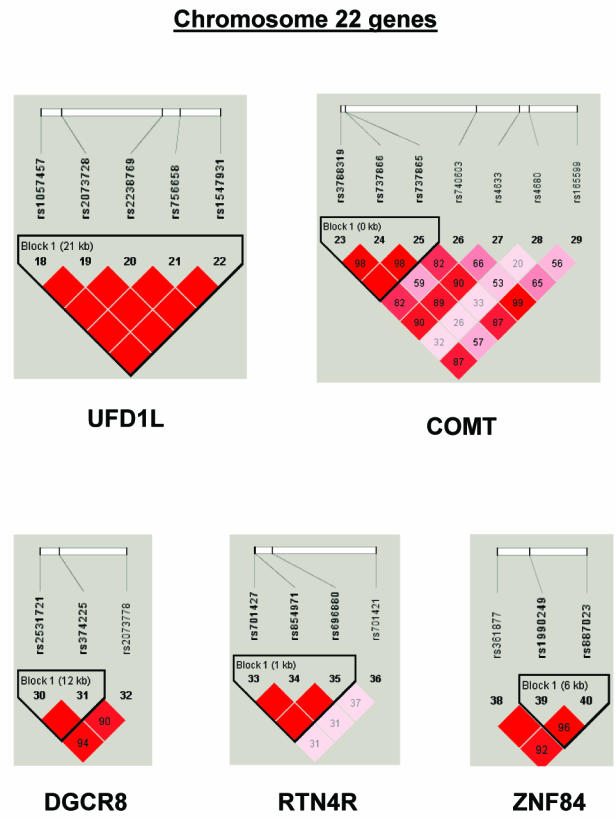
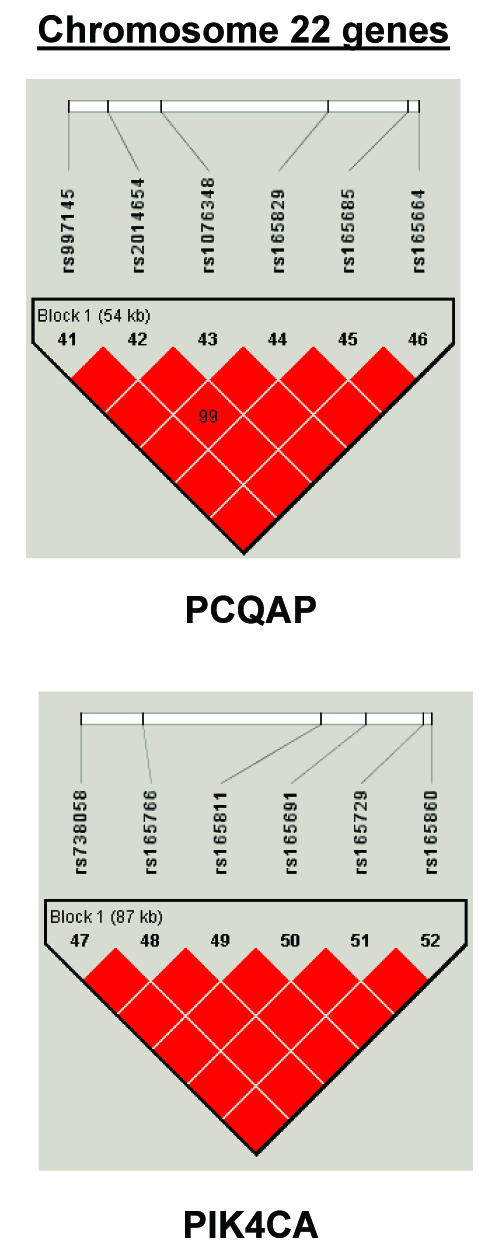
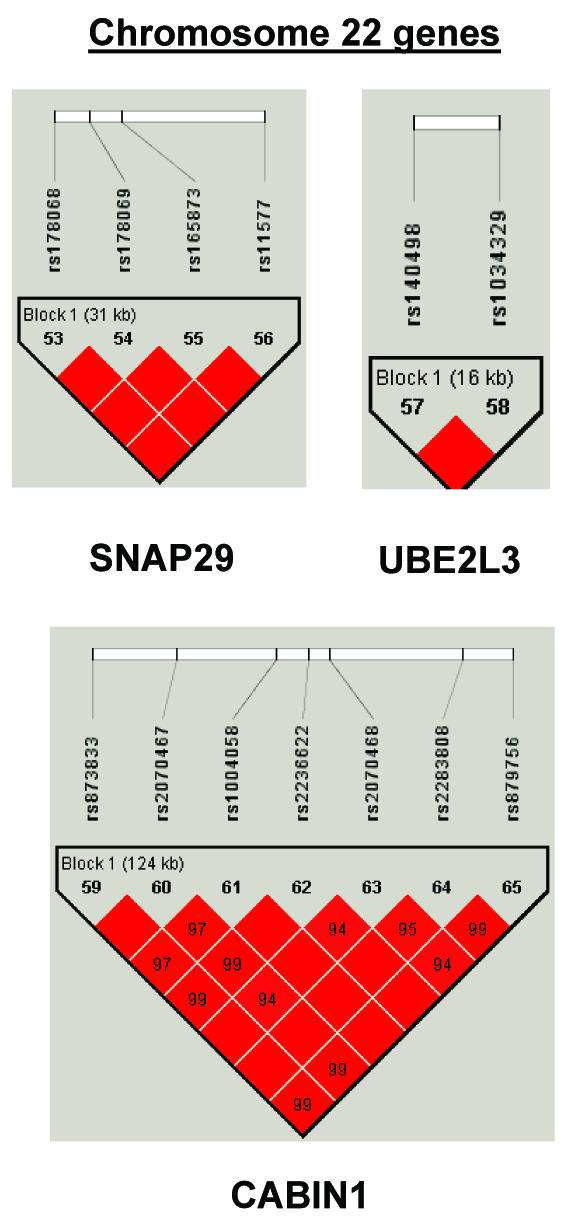
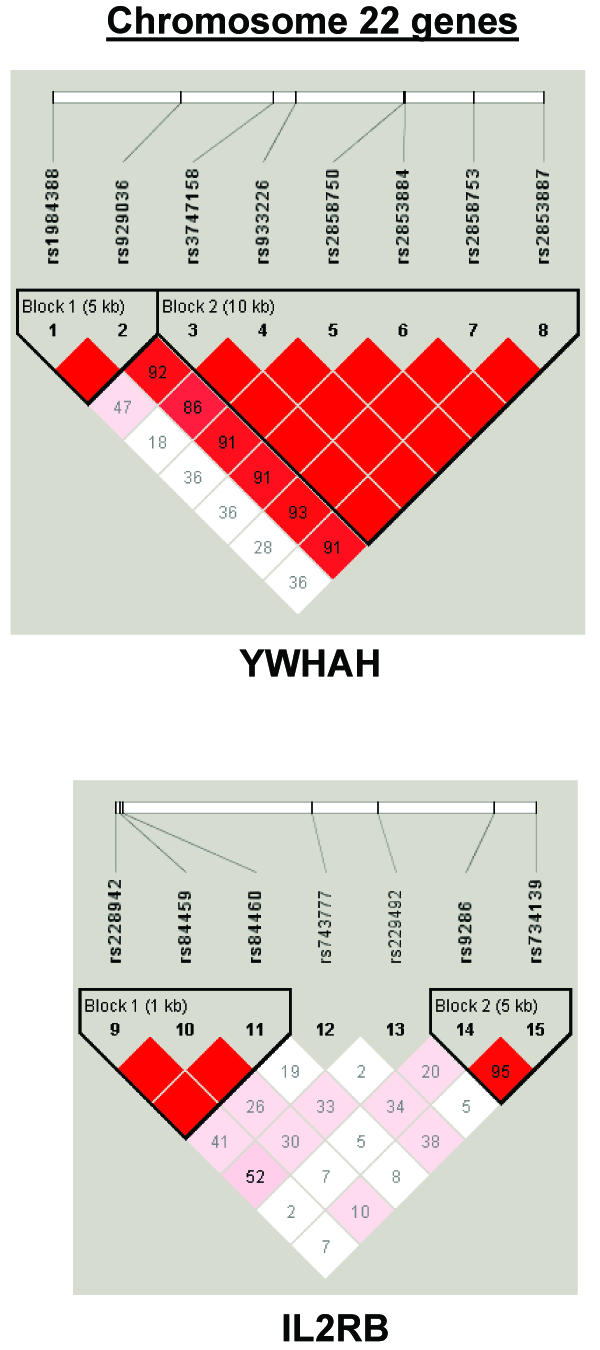
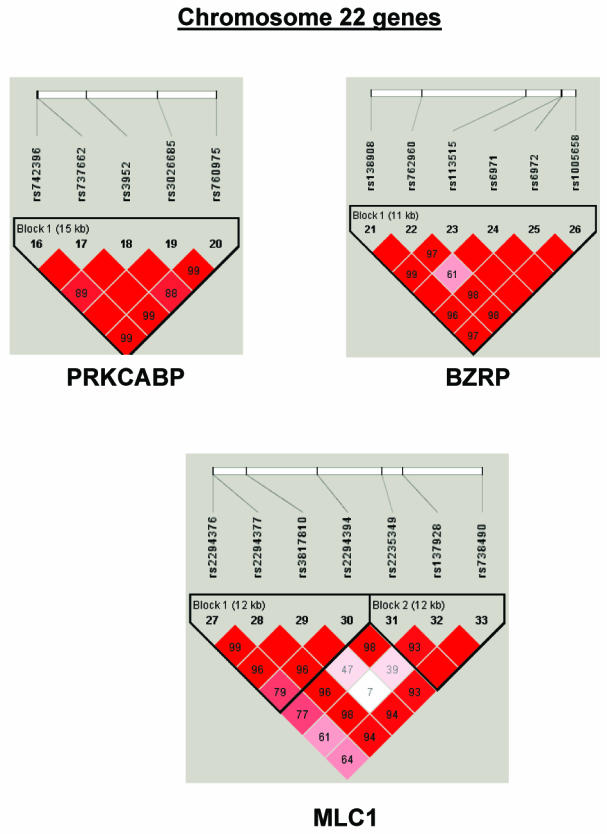
LD structure for each gene
Appendix C: 64 Genes: Reasons for Inclusion and Partial Reference List
Table A1.
64 Genes: Reasons for Inclusion and Partial Reference List
| Gene Symbol | Region | Reason for Inclusiona | Partial Reference Listb | |||
| RGS4 | 1q23.3 | SZ-L | BP-A | SZ-A | Mirnics et al. 2001; Brzustowicz et al. 2002; Cordeiro et al. 2005 | |
| DISC1 | 1q42.1 | BP-L | SZ-L | BP-A | SZ-A | Millar et al. 2000; Macgregor et al. 2004; Sachs et al. 2005; Thomson et al. 2005 |
| DRD3 | 3q13.3 | SZ-Ac | Ebstein et al. 1997; Jonsson et al. 2004 | |||
| DRD1 | 5q35.1 | BP-L | BP-A | SZ-A | Cichon et al. 1994; Cichon et al. 1996; Severino et al. 2005 | |
| DTNBP1 | 6p22.3 | BP-L | SZ-L | BP-A | SZ-A | Straub et al. 2002; Kirov et al. 2004; Raybould et al. 2005 |
| SCA1 | 6p23 | SZ-L | SZ-A | Wang et al. 1996b; Joo et al. 1999 | ||
| KIF13A | 6p23 | SZ-L | Jamain et al. 2001 | |||
| SynGAP1 | 6p21.32 | SZ-L | Kim et al. 1998; Porter et al. 2005 | |||
| GRM4 | 6p21.3 | SZ-L | Ohtsuki et al. 2001b | |||
| FKBP5 | 6p21.31 | SZ-L | Binder et al. 2004 | |||
| TFAP2B | 6p12.3 | SZ-L | 6p positional candidate | |||
| GRM3 | 7q21 | BP-L | SZ-A | Fujii et al. 2003; Bishop et al. 2005 | ||
| NAT1 | 8p22 | SZ-L | Blouin et al. 1998; Blaveri et al. 2001 | |||
| PPP3CC | 8p21.3 | BP-L | SZ-L | SZ-A | Gerber et al. 2003; Park et al. 2004; Eastwood et al. 2005 | |
| NEF3 | 8p21.2 | BP-L | SZ-L | 8p positional candidate; Blouin et al. 1998; Park et al. 2004 | ||
| NEFL | 8p21.2 | BP-L | SZ-L | Buckland et al. 2004 | ||
| GNRH1 | 8p21.2 | BP-L | SZ-L | 8p positional candidate; Blouin et al. 1998; Park et al. 2004 | ||
| DPYSL2 | 8p21.2 | BP-L | SZ-L | SZ-A | Nakata et al. 2003a; Nakata et al. 2003b | |
| ADRA1A | 8p21.2 | BP-L | SZ-L | SZ-A | Blouin et al. 1998; Clark et al. 2005 | |
| CHRNA2 | 8p21.2 | BP-L | SZ-L | SZ-A | Blaveri et al. 2001; Lohoff et al. 2005 | |
| PNOC | 8p21.1 | BP-L | SZ-L | Blaveri et al. 2001; Kawashima et al. 2002 | ||
| NRG1 | 8p12 | SZ-L | SZ-A | Stefansson et al. 2002; Stefansson et al. 2003; Tosato et al. 2005 | ||
| CHRNB3 | 8p11.21 | SZ-L | Koyama et al. 1994; Durner et al. 1999 | |||
| PPP3CB | 10q22.2 | BP-L | SZ-Lc | Wang et al. 1996b; Fallin et al. 2003 | ||
| MYOZ1 | 10q22.2 | BP-L | SZ-Lc | 10q positional candidate; Fallin et al. 2004 | ||
| NRG3 | 10q23.1 | BP-L | SZ-L | Zhang et al. 1997; Fallin et al. 2003; Falls 2003 | ||
| GRID1 | 10q23.2 | BP-L | SZ-L | Treadaway and Zuo 1998; Fallin et al. 2003 | ||
| SNCG | 10q23.2 | BP-L | SZ-L | Lavedan et al. 1998; Fallin et al. 2003 | ||
| HTR7 | 10q23.31 | BP-L | SZ-L | Vincent et al. 1999; Masellis et al. 2001; Fallin et al. 2003 | ||
| LGI1 | 10q23.33 | BP-L | SZ-L | Kalachikov et al. 2002; Fallin et al. 2003 | ||
| INA | 10q24.33 | BP-L | SZ-L | 10q positional candidate; Fallin et al. 2004 | ||
| GFRA1 | 10q26 | BP-L | SZ-L | Rossi et al. 2000 | ||
| DRD4 | 11p15.5 | BP-L | BP-A | SZ-A | Gelernter et al. 1992; Muglia et al. 2002; Jonsson et al. 2003 | |
| DRD2 | 11q23 | SZ-L | BP-A | SZ-A | Perez de Castro et al. 1995; Spurlock et al. 1998; Dubertret et al. 2004 | |
| GRIN2B | 12p12 | SZ-A | Ohtsuki et al. 2001a; Di Maria et al. 2004 | |||
| DAAO,DAO | 12q24 | BP-L | BP-A | SZ-A | Chumakov et al. 2002; Liu et al. 2004; Schumacher et al. 2004 | |
| NOS1 | 12q24 | BP-L | SZ-A | Shinkai et al. 2002; Buttenschon et al. 2004 | ||
| HTR2A | 13q14.2 | SZ-L | BP-A | SZ-A | Ranade et al. 2003; Abdolmaleky et al. 2004; Norton and Owen 2005 | |
| SLC15A1 | 13q32.2 | BP-L | SZ-L | Blouin et al. 1998; Maheshwari et al. 2002 | ||
| ZIC2 | 13q32.3 | BP-L | SZ-L | Blouin et al. 1998; Ogura et al. 2001 | ||
| TPP2 | 13q33.1 | BP-L | SZ-L | 13q positional candidate; Blouin et al. 1998 | ||
| G72/G30 | 13q33.2 | BP-L | SZ-L | BP-A | SZ-Ac | Chumakov et al. 2002; Hattori et al. 2003; Korostishevsky et al. 2005 |
| IL17R | 22q11.1 | BP-L | SZ-L | Kolls and Linden 2004 | ||
| TUBA8 | 22q11.21 | BP-L | SZ-L | Stanchi et al. 2000 | ||
| DGCR6 | 22q11.21 | BP-L | SZ-L | SZ-A | Chakravarti 2002; Liu et al. 2002 | |
| PRODH | 22q11.21 | BP-L | SZ-L | SZ-A | Gogos et al. 1999; Jacquet et al. 2002 | |
| DGCR14 | 22q11.21 | BP-L | SZ-L | Chieffo et al. 1997; Hoogendoorn et al. 2004 | ||
| GSCL | 22q11.21 | BP-L | SZ-L | Gottlieb et al. 1997; Williams et al. 2002 | ||
| UFD1L | 22q11.21 | BP-L | SZ-L | SZ-A | Pizzuti et al. 1997; De Luca et al. 2001 | |
| COMT | 22q11.21 | BP-L | SZ-L | SZ-Ac | Matthysse and Baldessarini 1972; Shifman et al. 2002; Munafo et al. 2005 | |
| DGCR8 | 22q11.21 | BP-L | SZ-L | Shiohama et al. 2003 | ||
| RTN4R | 22q11.21 | BP-L | SZ-L | Sinibaldi et al. 2004 | ||
| DGCR6L | 22q11.21 | BP-L | SZ-L | Edelmann et al. 2001 | ||
| ZNF74 | 22q11.21 | BP-L | SZ-L | SZ-A | Ravassard et al. 1999; Takase et al. 2001 | |
| PCQAP | 22q11.21 | BP-L | SZ-L | SZ-A | Berti et al. 2001; De Luca et al. 2003; Sandhu et al. 2004 | |
| PIK4CA | 22q11.21 | BP-L | SZ-L | BP-A | SZ-A | Saito et al. 2003 |
| SNAP29 | 22q11.21 | BP-L | SZ-L | SZ-A | Saito et al. 2001; Su et al. 2001; Wonodi et al. 2005 | |
| UBE2L3 | 22q11.21 | BP-L | SZ-L | 22q positional candidate; Karayiorgou et al. 1995 | ||
| CABIN1 | 22q11.23 | BP-L | SZ-L | Gerber et al. 2003 | ||
| YWHAH | 22q12.3 | BP-L | SZ-L | Duan et al. 2005 | ||
| IL2RB | 22q13.1 | BP-L | SZ-L | Pulver et al. 1994a; Schwab et al. 1995 | ||
| PRKCABP | 22q13.1 | BP-L | SZ-L | Perroy et al. 2002; Hong et al. 2004 | ||
| BZRP | 22q13.2 | BP-L | SZ-L | Kurumaji et al. 2000; Kurumaji et al. 2001 | ||
| MLC1 | 22q13.33 | BP-L | SZ-L | SZ-A | Meyer et al. 2001; Devaney et al. 2002; Leegwater et al. 2002 | |
BP-L is the region showing linkage to BP; SZ-L is the region showing linkage to SZ; BP-A indicates a previous association with BP; and SZ-A indicates a previous association with SZ.
Because of space limitations, a partial reference list was created. The references were chosen on the basis of the order of publication, with the attempt to choose one of the initial publications followed by a more recent publication and to give priority to a review paper or a meta-analysis, when available.
Previous association or linkage in the AJ population.
Web Resources
Accession numbers and URLs for data presented herein are as follows:
- David Clayton's Web site, http://www-gene.cimr.cam.ac.uk/clayton/software/stata/ (for GenAssoc STATA software)
- dbSNP, http://www.ncbi.nlm.nih.gov/SNP/
- Johns Hopkins Epidemiology-Genetics Program in Psychiatry, http://www.hopkinsmedicine.org/epigen
- FBAT, http://biosun1.harvard.edu/~fbat/fbat.htm
- Haploview, http://www.broad.mit.edu/mpg/haploview/index.php
- HapMap, http://www.hapmap.org
- Illumina, http://www.illumina.com/Products/prod_snp.ilmn
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for BP, SZ, VCFS, and spinocerebellar ataxia type 1)
References
- Abdolmaleky HM, Faraone SV, Glatt SJ, Tsuang MT (2004) Meta-analysis of association between the T102C polymorphism of the 5HT2a receptor gene and schizophrenia. Schizophr Res 67:53–62 10.1016/S0920-9964(03)00183-X [DOI] [PubMed] [Google Scholar]
- American Psychiatric Association (1994) Diagnostic and statistical manual of mental disorders. American Psychiatric Association, Washington, D.C. [Google Scholar]
- Badner JA, Gershon ES (2002) Meta-analysis of whole-genome linkage scans of bipolar disorder and schizophrenia. Mol Psychiatry 7:405–411 10.1038/sj.mp.4001012 [DOI] [PubMed] [Google Scholar]
- Baranano DE, Ferris CD, Snyder SH (2001) Atypical neural messengers. Trends Neurosci 24:99–106 10.1016/S0166-2236(00)01716-1 [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ (2005) Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics 21:263-265 10.1093/bioinformatics/bth457 [DOI] [PubMed] [Google Scholar]
- Berti L, Mittler G, Przemeck GK, Stelzer G, Gunzler B, Amati F, Conti E, Dallapiccola B, Hrabe de Angelis M, Novelli G, Meisterernst M (2001) Isolation and characterization of a novel gene from the DiGeorge chromosomal region that encodes for a mediator subunit. Genomics 74:320–332 10.1006/geno.2001.6566 [DOI] [PubMed] [Google Scholar]
- Binder EB, Salyakina D, Lichtner P, Wochnik GM, Ising M, Putz B, Papiol S, et al (2004) Polymorphisms in FKBP5 are associated with increased recurrence of depressive episodes and rapid response to antidepressant treatment. Nat Genet 36:1319–1325 10.1038/ng1479 [DOI] [PubMed] [Google Scholar]
- Bishop JR, Ellingrod VL, Moline J, Miller D (2005) Association between the polymorphic GRM3 gene and negative symptom improvement during olanzapine treatment. Schizophr Res 77:253–260 10.1016/j.schres.2005.04.001 [DOI] [PubMed] [Google Scholar]
- Blaveri E, Kalsi G, Lawrence J, Quested D, Moorey H, Lamb G, Kohen D, Shiwach R, Chowdhury U, Curtis D, McQuillin A, Gramoustianou ES, Gurling HM (2001) Genetic association studies of schizophrenia using the 8p21-22 genes: prepronociceptin (PNOC), neuronal nicotinic cholinergic receptor alpha polypeptide 2 (CHRNA2) and arylamine N-acetyltransferase 1 (NAT1). Eur J Hum Genet 9:469–472 10.1038/sj.ejhg.5200646 [DOI] [PubMed] [Google Scholar]
- Blouin JL, Dombroski BA, Nath SK, Lasseter VK, Wolyniec PS, Nestadt G, Thornquist M, et al (1998) Schizophrenia susceptibility loci on chromosomes 13q32 and 8p21. Nat Genet 20:70–73 10.1038/1734 [DOI] [PubMed] [Google Scholar]
- Botstein D, Risch N (2003) Discovering genotypes underlying human phenotypes: past successes for mendelian disease, future approaches for complex disease. Nat Genet 33 Suppl:228–237 [DOI] [PubMed] [Google Scholar]
- Bredt DS, Snyder SH (1989) Nitric oxide mediates glutamate-linked enhancement of cGMP levels in the cerebellum. Proc Natl Acad Sci USA 86:9030–9033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brzustowicz LM, Hayter JE, Hodgkinson KA, Chow EW, Bassett AS (2002) Fine mapping of the schizophrenia susceptibility locus on chromosome 1q22. Hum Hered 54:199–209 10.1159/000070665 [DOI] [PubMed] [Google Scholar]
- Buckland PR, Hoogendoorn B, Guy CA, Coleman SL, Smith SK, Buxbaum JD, Haroutunian V, O’Donovan MC (2004) A high proportion of polymorphisms in the promoters of brain expressed genes influences transcriptional activity. Biochim Biophys Acta 1690:238–249 [DOI] [PubMed] [Google Scholar]
- Buttenschon HN, Mors O, Ewald H, McQuillin A, Kalsi G, Lawrence J, Gurling H, Kruse TA (2004) No association between a neuronal nitric oxide synthase (NOS1) gene polymorphism on chromosome 12q24 and bipolar disorder. Am J Med Genet B Neuropsychiatr Genet 124:73–75 10.1002/ajmg.b.20040 [DOI] [PubMed] [Google Scholar]
- Cardno AG, Marshall EJ, Coid B, Macdonald AM, Ribchester TR, Davies NJ, Venturi P, Jones LA, Lewis SW, Sham PC, Gottesman, II, Farmer AE, McGuffin P, Reveley AM, Murray RM (1999) Heritability estimates for psychotic disorders: the Maudsley twin psychosis series. Arch Gen Psychiatry 56:162–168 10.1001/archpsyc.56.2.162 [DOI] [PubMed] [Google Scholar]
- Cardno AG, Sham PC, Murray RM, McGuffin P (2001) Twin study of symptom dimensions in psychoses. Br J Psychiatry 179:39–45 10.1192/bjp.179.1.39 [DOI] [PubMed] [Google Scholar]
- Chakravarti A (2002) A compelling genetic hypothesis for a complex disease: PRODH2/DGCR6 variation leads to schizophrenia susceptibility. Proc Natl Acad Sci USA 99:4755–4756 10.1073/pnas.092158299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chieffo C, Garvey N, Gong W, Roe B, Zhang G, Silver L, Emanuel BS, Budarf ML (1997) Isolation and characterization of a gene from the DiGeorge chromosomal region homologous to the mouse Tbx1 gene. Genomics 43:267–277 10.1006/geno.1997.4829 [DOI] [PubMed] [Google Scholar]
- Chowdari KV, Mirnics K, Semwal P, Wood J, Lawrence E, Bhatia T, Deshpande SN, B K T, Ferrell RE, Middleton FA, Devlin B, Levitt P, Lewis DA, Nimgaonkar VL (2002) Association and linkage analyses of RGS4 polymorphisms in schizophrenia. Hum Mol Genet 11:1373–1380 10.1093/hmg/11.12.1373 [DOI] [PubMed] [Google Scholar]
- Chumakov I, Blumenfeld M, Guerassimenko O, Cavarec L, Palicio M, Abderrahim H, Bougueleret L, et al (2002) Genetic and physiological data implicating the new human gene G72 and the gene for D-amino acid oxidase in schizophrenia. Proc Natl Acad Sci USA 99:13675–13680 10.1073/pnas.182412499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cichon S, Nothen MM, Rietschel M, Korner J, Propping P (1994) Single-strand conformation analysis (SSCA) of the dopamine D1 receptor gene (DRD1) reveals no significant mutation in patients with schizophrenia and manic depression. Biol Psychiatry 36:850–853 10.1016/0006-3223(94)90597-5 [DOI] [PubMed] [Google Scholar]
- Cichon S, Nothen MM, Stober G, Schroers R, Albus M, Maier W, Rietschel M, Korner J, Weigelt B, Franzek E, Wildenauer D, Fimmers R, Propping P (1996) Systematic screening for mutations in the 5′-regulatory region of the human dopamine D1 receptor (DRD1) gene in patients with schizophrenia and bipolar affective disorder. Am J Med Genet 67:424–428 [DOI] [PubMed] [Google Scholar]
- Clark DA, Arranz MJ, Mata I, Lopez-Ilundain J, Perez-Nievas F, Kerwin RW (2005) Polymorphisms in the promoter region of the alpha1A-adrenoceptor gene are associated with schizophrenia/schizoaffective disorder in a Spanish isolate population. Biol Psychiatry 58:435–439 10.1016/j.biopsych.2005.04.051 [DOI] [PubMed] [Google Scholar]
- Cordeiro Q, Talkowski ME, Chowdari KV, Wood J, Nimgaonkar V, Vallada H (2005) Association and linkage analysis of RGS4 polymorphisms with schizophrenia and bipolar disorder in Brazil. Genes Brain Behav 4:45–50 10.1111/j.1601-183x.2004.00096.x [DOI] [PubMed] [Google Scholar]
- De Luca A, Conti E, Grifone N, Amati F, Spalletta G, Caltagirone C, Bonaviri G, Pasini A, Gennarelli M, Stefano B, Berti L, Mittler G, Meisterernst M, Dallapiccola B, Novelli G (2003) Association study between CAG trinucleotide repeats in the PCQAP gene (PC2 glutamine/Q-rich-associated protein) and schizophrenia. Am J Med Genet B Neuropsychiatr Genet 116:32–35 10.1002/ajmg.b.10008 [DOI] [PubMed] [Google Scholar]
- De Luca A, Pasini A, Amati F, Botta A, Spalletta G, Alimenti S, Caccamo F, Conti E, Trakalo J, Macciardi F, Dallapiccola B, Novelli G (2001) Association study of a promoter polymorphism of UFD1L gene with schizophrenia. Am J Med Genet 105:529–533 10.1002/ajmg.1489 [DOI] [PubMed] [Google Scholar]
- Devaney JM, Donarum EA, Brown KM, Meyer J, Stober G, Lesch KP, Nestadt G, Stephan DA, Pulver AE (2002) No missense mutation of WKL1 in a subgroup of probands with schizophrenia. Mol Psychiatry 7:419–423 10.1038/sj.mp.4001022 [DOI] [PubMed] [Google Scholar]
- Di Maria E, Gulli R, Begni S, De Luca A, Bignotti S, Pasini A, Bellone E, Pizzuti A, Dallapiccola B, Novelli G, Ajmar F, Gennarelli M, Mandich P (2004) Variations in the NMDA receptor subunit 2B gene (GRIN2B) and schizophrenia: a case-control study. Am J Med Genet B Neuropsychiatr Genet 128:27–29 10.1002/ajmg.b.30028 [DOI] [PubMed] [Google Scholar]
- Driscoll DA, Spinner NB, Budarf ML, McDonald-McGinn DM, Zackai EH, Goldberg RB, Shprintzen RJ, Saal HM, Zonana J, Jones MC, et al (1992) Deletions and microdeletions of 22q11.2 in velo-cardio-facial syndrome. Am J Med Genet 44:261–268 10.1002/ajmg.1320440237 [DOI] [PubMed] [Google Scholar]
- Duan S, Gao R, Xing Q, Du J, Liu Z, Chen Q, Wang H, Feng G, He L (2005) A family-based association study of schizophrenia with polymorphisms at three candidate genes. Neurosci Lett 379:32–36 10.1016/j.neulet.2004.12.040 [DOI] [PubMed] [Google Scholar]
- Dubertret C, Gouya L, Hanoun N, Deybach JC, Ades J, Hamon M, Gorwood P (2004) The 3′ region of the DRD2 gene is involved in genetic susceptibility to schizophrenia. Schizophr Res 67:75–85 10.1016/S0920-9964(03)00220-2 [DOI] [PubMed] [Google Scholar]
- Dunayevich E, Keck PE Jr (2000) Prevalence and description of psychotic features in bipolar mania. Curr Psychiatry Rep 2:286–290 [DOI] [PubMed] [Google Scholar]
- Durner M, Zhou G, Fu D, Abreu P, Shinnar S, Resor SR, Moshe SL, Rosenbaum D, Cohen J, Harden C, Kang H, Wallace S, Luciano D, Ballaban-Gil K, Klotz I, Dicker E, Greenberg DA (1999) Evidence for linkage of adolescent-onset idiopathic generalized epilepsies to chromosome 8-and genetic heterogeneity. Am J Hum Genet 64:1411–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eastwood SL, Burnet PW, Harrison PJ (2005) Decreased hippocampal expression of the susceptibility gene PPP3CC and other calcineurin subunits in schizophrenia. Biol Psychiatry 57:702–710 10.1016/j.biopsych.2004.12.029 [DOI] [PubMed] [Google Scholar]
- Ebstein RP, Macciardi F, Heresco-Levi U, Serretti A, Blaine D, Verga M, Nebamov L, Gur E, Belmaker RH, Avnon M, Lerer B (1997) Evidence for an association between the dopamine D3 receptor gene DRD3 and schizophrenia. Hum Hered 47:6–16 [DOI] [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE (1999) A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet 8:1157–1167 10.1093/hmg/8.7.1157 [DOI] [PubMed] [Google Scholar]
- Edelmann L, Stankiewicz P, Spiteri E, Pandita RK, Shaffer L, Lupski JR, Morrow BE (2001) Two functional copies of the DGCR6 gene are present on human chromosome 22q11 due to a duplication of an ancestral locus. Genome Res 11:208–217 10.1101/gr.GR-1431R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Efron B (2004) Large-scale simultaneous hypothesis testing: the choice of a null hypothesis. JASA 99:96–104 [Google Scholar]
- Fallin MD, Lasseter VK, Wolyniec PS, McGrath JA, Nestadt G, Valle D, Liang KY, Pulver AE (2003) Genomewide linkage scan for schizophrenia susceptibility loci among Ashkenazi Jewish families shows evidence of linkage on chromosome 10q22. Am J Hum Genet 73:601–611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- ——— (2004) Genomewide linkage scan for bipolar-disorder susceptibility loci among Ashkenazi Jewish families. Am J Hum Genet 75:204–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falls DL (2003) Neuregulins: functions, forms, and signaling strategies. Exp Cell Res 284:14–30 10.1016/S0014-4827(02)00102-7 [DOI] [PubMed] [Google Scholar]
- Fujii Y, Shibata H, Kikuta R, Makino C, Tani A, Hirata N, Shibata A, Ninomiya H, Tashiro N, Fukumaki Y (2003) Positive associations of polymorphisms in the metabotropic glutamate receptor type 3 gene (GRM3) with schizophrenia. Psychiatr Genet 13:71–76 10.1097/00041444-200306000-00003 [DOI] [PubMed] [Google Scholar]
- Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229 10.1126/science.1069424 [DOI] [PubMed] [Google Scholar]
- Gauderman WJ (2002) Sample size requirements for association studies of gene-gene interaction. Am J Epidemiol 155:478–484 10.1093/aje/155.5.478 [DOI] [PubMed] [Google Scholar]
- Gelernter J, Kennedy JL, van Tol HH, Civelli O, Kidd KK (1992) The D4 dopamine receptor (DRD4) maps to distal 11p close to HRAS. Genomics 13:208–210 10.1016/0888-7543(92)90222-E [DOI] [PubMed] [Google Scholar]
- Gerber DJ, Hall D, Miyakawa T, Demars S, Gogos JA, Karayiorgou M, Tonegawa S (2003) Evidence for association of schizophrenia with genetic variation in the 8p21.3 gene, PPP3CC, encoding the calcineurin gamma subunit. Proc Natl Acad Sci USA 100:8993–8998 10.1073/pnas.1432927100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gogos JA, Santha M, Takacs Z, Beck KD, Luine V, Lucas LR, Nadler JV, Karayiorgou M (1999) The gene encoding proline dehydrogenase modulates sensorimotor gating in mice. Nat Genet 21:434–439 10.1038/7777 [DOI] [PubMed] [Google Scholar]
- Goodwin FK, Jamison KR (1990) Manic-depressive illness. Oxford University Press, New York [Google Scholar]
- Gottlieb S, Emanuel BS, Driscoll DA, Sellinger B, Wang Z, Roe B, Budarf ML (1997) The DiGeorge syndrome minimal critical region contains a goosecoid-like (GSCL) homeobox gene that is expressed early in human development. Am J Hum Genet 60:1194–1201 [PMC free article] [PubMed] [Google Scholar]
- Harrison PJ, Weinberger DR (2005) Schizophrenia genes, gene expression, and neuropathology: on the matter of their convergence. Mol Psychiatry 10:804 10.1038/sj.mp.4001686 [DOI] [PubMed] [Google Scholar]
- Hashimoto R, Suzuki T, Iwata N, Yamanouchi Y, Kitajima T, Kosuga A, Tatsumi M, Ozaki N, Kamijima K, Kunugi H (2005) Association study of the frizzled-3 (FZD3) gene with schizophrenia and mood disorders. J Neural Transm 112:303–307 10.1007/s00702-004-0264-2 [DOI] [PubMed] [Google Scholar]
- Hattori E, Liu C, Badner JA, Bonner TI, Christian SL, Maheshwari M, Detera-Wadleigh SD, Gibbs RA, Gershon ES (2003) Polymorphisms at the G72/G30 gene locus, on 13q33, are associated with bipolar disorder in two independent pedigree series. Am J Hum Genet 72:1131–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hennah W, Ekelund J, Hiekkalinna T, Paunio T, Kestilä M, Peltonen L (2004) Genome-wide scan conditioned on the presence of a DISC1 schizophrenia associating haplotype highlights 10 regions of interest. Paper presented at World Congress of Psychiatric Genetics. Dublin, October [Google Scholar]
- Hollmann M, Boulter J, Maron C, Heinemann S (1994) Molecular biology of glutamate receptors: potentiation of N-methyl-D-aspartate receptor splice variants by zinc. Ren Physiol Biochem 17:182–183 [PubMed] [Google Scholar]
- Hong CJ, Liao DL, Shih HL, Tsai SJ (2004) Association study of PICK1 rs3952 polymorphism and schizophrenia. Neuroreport 15:1965–1967 10.1097/00001756-200408260-00026 [DOI] [PubMed] [Google Scholar]
- Hoogendoorn B, Coleman SL, Guy CA, Smith SK, O’Donovan MC, Buckland PR (2004) Functional analysis of polymorphisms in the promoter regions of genes on 22q11. Hum Mutat 24:35–42 10.1002/humu.20061 [DOI] [PubMed] [Google Scholar]
- Horvath S, Xu X, Laird NM (2001) The family based association test method: strategies for studying general genotype—phenotype associations. Eur J Hum Genet 9:301–306 10.1038/sj.ejhg.5200625 [DOI] [PubMed] [Google Scholar]
- Jacquet H, Raux G, Thibaut F, Hecketsweiler B, Houy E, Demilly C, Haouzir S, Allio G, Fouldrin G, Drouin V, Bou J, Petit M, Campion D, Frebourg T (2002) PRODH mutations and hyperprolinemia in a subset of schizophrenic patients. Hum Mol Genet 11:2243–2249 10.1093/hmg/11.19.2243 [DOI] [PubMed] [Google Scholar]
- Jamain S, Quach H, Fellous M, Bourgeron T (2001) Identification of the human KIF13A gene homologous to Drosophila kinesin-73 and candidate for schizophrenia. Genomics 74:36–44 10.1006/geno.2001.6535 [DOI] [PubMed] [Google Scholar]
- Jonsson EG, Kaiser R, Brockmoller J, Nimgaonkar VL, Crocq MA (2004) Meta-analysis of the dopamine D3 receptor gene (DRD3) Ser9Gly variant and schizophrenia. Psychiatr Genet 14:9–12 10.1097/00041444-200403000-00002 [DOI] [PubMed] [Google Scholar]
- Jonsson EG, Sedvall GC, Nothen MM, Cichon S (2003) Dopamine D4 receptor gene (DRD4) variants and schizophrenia: meta-analyses. Schizophr Res 61:111–119 10.1016/S0920-9964(02)00287-6 [DOI] [PubMed] [Google Scholar]
- Joo EJ, Lee JH, Cannon TD, Price RA (1999) Possible association between schizophrenia and a CAG repeat polymorphism in the spinocerebellar ataxia type 1 (SCA1) gene on human chromosome 6p23. Psychiatr Genet 9:7–11 [DOI] [PubMed] [Google Scholar]
- Kalachikov S, Evgrafov O, Ross B, Winawer M, Barker-Cummings C, Martinelli Boneschi F, Choi C, Morozov P, Das K, Teplitskaya E, Yu A, Cayanis E, Penchaszadeh G, Kottmann AH, Pedley TA, Hauser WA, Ottman R, Gilliam TC (2002) Mutations in LGI1 cause autosomal-dominant partial epilepsy with auditory features. Nat Genet 30:335–341 10.1038/ng832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karayiorgou M, Morris MA, Morrow B, Shprintzen RJ, Goldberg R, Borrow J, Gos A, Nestadt G, Wolyniec PS, Lasseter VK, et al (1995) Schizophrenia susceptibility associated with interstitial deletions of chromosome 22q11. Proc Natl Acad Sci USA 92:7612–7616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima N, Fugate J, Kusnecov AW (2002) Immunological challenge modulates brain orphanin FQ/nociceptin and nociceptive behavior. Brain Res 949:71–78 10.1016/S0006-8993(02)02966-9 [DOI] [PubMed] [Google Scholar]
- Kempf L, Hussain N, Potash J (2005) Mood disorder with psychotic features, schizoaffective disorder and schizophrenia and mood features: trouble at the border. Int Rev Psychiatry 17:9–19 [DOI] [PubMed] [Google Scholar]
- Kim JH, Liao D, Lau LF, Huganir RL (1998) SynGAP: a synaptic RasGAP that associates with the PSD-95/SAP90 protein family. Neuron 20:683–691 10.1016/S0896-6273(00)81008-9 [DOI] [PubMed] [Google Scholar]
- Kirov G, Ivanov D, Williams NM, Preece A, Nikolov I, Milev R, Koleva S, Dimitrova A, Toncheva D, O’Donovan MC, Owen MJ (2004) Strong evidence for association between the dystrobrevin binding protein 1 gene (DTNBP1) and schizophrenia in 488 parent-offspring trios from Bulgaria. Biol Psychiatry 55:971–975 10.1016/j.biopsych.2004.01.025 [DOI] [PubMed] [Google Scholar]
- Kolls JK, Linden A (2004) Interleukin-17 family members and inflammation. Immunity 21:467–476 10.1016/j.immuni.2004.08.018 [DOI] [PubMed] [Google Scholar]
- Korostishevsky M, Kremer I, Kaganovich M, Cholostoy A, Murad I, Muhaheed M, Bannoura I, Rietschel M, Dobrusin M, Bening-Abu-Shach U, Belmaker RH, Maier W, Ebstein RP, Navon R (2005) Transmission disequilibrium and haplotype analyses of the G72/G30 locus: suggestive linkage to schizophrenia in Palestinian Arabs living in the North of Israel. Am J Med Genet B Neuropsychiatr Genet (http://www3.interscience.wiley.com/cgi-bin/abstract/110577213/ABSTRACT) (electronically published August 4, 2005) [DOI] [PubMed]
- Koyama K, Sudo K, Nakamura Y (1994) Mapping of the human nicotinic acetylcholine receptor beta 3 gene (CHRNB3) within chromosome 8p11.2. Genomics 21:460–461 10.1006/geno.1994.1300 [DOI] [PubMed] [Google Scholar]
- Kurumaji A, Nomoto H, Yamada K, Yoshikawa T, Toru M (2001) No association of two missense variations of the benzodiazepine receptor (peripheral) gene and mood disorders in a Japanese sample. Am J Med Genet 105:172–175 10.1002/ajmg.1194 [DOI] [PubMed] [Google Scholar]
- Kurumaji A, Nomoto H, Yoshikawa T, Okubo Y, Toru M (2000) An association study between two missense variations of the benzodiazepine receptor (peripheral) gene and schizophrenia in a Japanese sample. J Neural Transm 107:491–500 10.1007/s007020070090 [DOI] [PubMed] [Google Scholar]
- Lasky-Su JA, Faraone SV, Glatt SJ, Tsuang MT (2005) Meta-analysis of the association between two polymorphisms in the serotonin transporter gene and affective disorders. Am J Med Genet B Neuropsychiatr Genet 133:110–115 [DOI] [PubMed] [Google Scholar]
- Lavedan C, Leroy E, Dehejia A, Buchholtz S, Dutra A, Nussbaum RL, Polymeropoulos MH (1998) Identification, localization and characterization of the human gamma-synuclein gene. Hum Genet 103:106–112 10.1007/s004390050792 [DOI] [PubMed] [Google Scholar]
- Leegwater PA, Boor PK, Pronk JC, van der Knaap MS (2002) Association of WKL1/MLC1 with catatonic schizophrenia. Mol Psychiatry 7:1037–1038 10.1038/sj.mp.4001142 [DOI] [PubMed] [Google Scholar]
- Lerer B, Segman RH, Hamdan A, Kanyas K, Karni O, Kohn Y, Korner M, Lanktree M, Kaadan M, Turetsky N, Yakir A, Kerem B, Macciardi F (2003) Genome scan of Arab Israeli families maps a schizophrenia susceptibility gene to chromosome 6q23 and supports a locus at chromosome 10q24. Mol Psychiatry 8:488–498 10.1038/sj.mp.4001322 [DOI] [PubMed] [Google Scholar]
- Lewis CM, Levinson DF, Wise LH, DeLisi LE, Straub RE, Hovatta I, Williams NM, et al (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part II: schizophrenia. Am J Hum Genet 73:34–48 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindsay EA, Goldberg R, Jurecic V, Morrow B, Carlson C, Kucherlapati RS, Shprintzen RJ, Baldini A (1995) Velo-cardio-facial syndrome: frequency and extent of 22q11 deletions. Am J Med Genet 57:514–522 10.1002/ajmg.1320570339 [DOI] [PubMed] [Google Scholar]
- Liu X, He G, Wang X, Chen Q, Qian X, Lin W, Li D, Gu N, Feng G, He L (2004) Association of DAAO with schizophrenia in the Chinese population. Neurosci Lett 369:228–233 10.1016/j.neulet.2004.07.078 [DOI] [PubMed] [Google Scholar]
- Liu H, Heath SC, Sobin C, Roos JL, Galke BL, Blundell ML, Lenane M, Robertson B, Wijsman EM, Rapoport JL, Gogos JA, Karayiorgou M (2002) Genetic variation at the 22q11 PRODH2/DGCR6 locus presents an unusual pattern and increases susceptibility to schizophrenia. Proc Natl Acad Sci USA 99:3717–3722 10.1073/pnas.042700699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lohoff FW, Ferraro TN, McNabb L, Schwebel C, Dahl JP, Doyle GA, Buono RJ, Berrettini WH (2005) No association between common variations in the neuronal nicotinic acetylcholine receptor alpha2 subunit gene (CHRNA2) and bipolar I disorder. Psychiatry Res 135:171–177 10.1016/j.psychres.2005.04.004 [DOI] [PubMed] [Google Scholar]
- Macgregor S, Visscher PM, Knott SA, Thomson P, Porteous DJ, Millar JK, Devon RS, Blackwood D, Muir WJ (2004) A genome scan and follow-up study identify a bipolar disorder susceptibility locus on chromosome 1q42. Mol Psychiatry 9:1083–1090 10.1038/sj.mp.4001544 [DOI] [PubMed] [Google Scholar]
- Maheshwari M, Christian SL, Liu C, Badner JA, Detera-Wadleigh S, Gershon ES, Gibbs RA (2002) Mutation screening of two candidate genes from 13q32 in families affected with bipolar disorder: human peptide transporter (SLC15A1) and human glypican5 (GPC5). BMC Genomics 3:30 10.1186/1471-2164-3-30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier W, Zobel A, Rietschel M (2003) Genetics of schizophrenia and affective disorders. Pharmacopsychiatry 36 Suppl 3:S195–S202 [DOI] [PubMed] [Google Scholar]
- Masellis M, Basile VS, Meltzer HY, Lieberman JA, Sevy S, Goldman DA, Hamblin MW, Macciardi FM, Kennedy JL (2001) Lack of association between the T→C 267 serotonin 5-HT6 receptor gene (HTR6) polymorphism and prediction of response to clozapine in schizophrenia. Schizophr Res 47:49–58 10.1016/S0920-9964(00)00016-5 [DOI] [PubMed] [Google Scholar]
- Matthysse S, Baldessarini RJ (1972) S-adenosylmethionine and catechol-O-methyl-transferase in schizophrenia. Am J Psychiatry 128:1310–1312 [DOI] [PubMed] [Google Scholar]
- Maziade M, Roy MA, Chagnon YC, Cliche D, Fournier JP, Montgrain N, Dion C, Lavallee JC, Garneau Y, Gingras N, Nicole L, Pires A, Ponton AM, Potvin A, Wallot H, Merette C (2005) Shared and specific susceptibility loci for schizophrenia and bipolar disorder: a dense genome scan in Eastern Quebec families. Mol Psychiatry10:486-499 [DOI] [PubMed] [Google Scholar]
- McGue M, Gottesman II (1991) The genetic epidemiology of schizophrenia and the design of linkage studies. Eur Arch Psychiatry Clin Neurosci 240:174–181 10.1007/BF02190760 [DOI] [PubMed] [Google Scholar]
- Morris DW, Rodgers A, McGhee KA, Schwaiger S, Scully P, Quinn J, Meagher D, Waddington JL, Gill M, Corvin AP (2004) Confirming RGS4 as a susceptibility gene for schizophrenia. Am J Med Genet B Neuropsychiatr Genet 125:50–53 10.1002/ajmg.b.20109 [DOI] [PubMed] [Google Scholar]
- Meyer J, Huberth A, Ortega G, Syagailo YV, Jatzke S, Mossner R, Strom TM, Ulzheimer-Teuber I, Stober G, Schmitt A, Lesch KP (2001) A missense mutation in a novel gene encoding a putative cation channel is associated with catatonic schizophrenia in a large pedigree. Mol Psychiatry 6:302–306 10.1038/sj.mp.4000869 [DOI] [PubMed] [Google Scholar]
- Millar JK, Wilson-Annan JC, Anderson S, Christie S, Taylor MS, Semple CA, Devon RS, Clair DM, Muir WJ, Blackwood DH, Porteous DJ (2000) Disruption of two novel genes by a translocation co-segregating with schizophrenia. Hum Mol Genet 9:1415–1423 10.1093/hmg/9.9.1415 [DOI] [PubMed] [Google Scholar]
- Mirnics K, Middleton FA, Stanwood GD, Lewis DA, Levitt P (2001) Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol Psychiatry 6:293–301 10.1038/sj.mp.4000866 [DOI] [PubMed] [Google Scholar]
- Mothet JP, Parent AT, Wolosker H, Brady RO Jr, Linden DJ, Ferris CD, Rogawski MA, Snyder SH (2000) D-serine is an endogenous ligand for the glycine site of the N-methyl-D-aspartate receptor. Proc Natl Acad Sci USA 97:4926–4931 10.1073/pnas.97.9.4926 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muglia P, Petronis A, Mundo E, Lander S, Cate T, Kennedy JL (2002) Dopamine D4 receptor and tyrosine hydroxylase genes in bipolar disorder: evidence for a role of DRD4. Mol Psychiatry 7:860–866 10.1038/sj.mp.4001098 [DOI] [PubMed] [Google Scholar]
- Munafo MR, Bowes L, Clark TG, Flint J (2005) Lack of association of the COMT (Val(158/108) Met) gene and schizophrenia: a meta-analysis of case-control studies. Mol Psychiatry 10:765–770 10.1038/sj.mp.4001664 [DOI] [PubMed] [Google Scholar]
- Nakata K, Ujike H, Sakai A, Takaki M, Imamura T, Tanaka Y, Kuroda S (2003a) The human dihydropyrimidinase-related protein 2 gene on chromosome 8p21 is associated with paranoid-type schizophrenia. Biol Psychiatry 53:571–576 10.1016/S0006-3223(02)01729-8 [DOI] [PubMed] [Google Scholar]
- Nakata K, Ujike H, Tanaka Y, Takaki M, Sakai A, Nomura A, Katsu T, Uchida N, Imamura T, Fujiwara Y, Hamamura T, Kuroda S (2003b) No association between the dihydropyrimidinase-related protein 2 (DRP-2) gene and bipolar disorder in humans. Neurosci Lett 349:171–174 10.1016/S0304-3940(03)00824-3 [DOI] [PubMed] [Google Scholar]
- Neuhausen SL, Godwin AK, Gershoni-Baruch R, Schubert E, Garber J, Stoppa-Lyonnet D, Olah E, et al (1998) Haplotype and phenotype analysis of nine recurrent BRCA2 mutations in 111 families: results of an international study. Am J Hum Genet 62:1381–1388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicodemus KK, Liu W, Chase GA, Tsai YY, Fallin MD (2004) Comparison of type I error for multiple test corrections in large SNP studies using principal components versus haplotype blocking algorithms. Paper presented at Genetic Analysis Workshop 14, Noordwijkerhout, Holland, September 11–12 [Google Scholar]
- Niell BL, Long JC, Rennert G, Gruber SB (2003) Genetic anthropology of the colorectal cancer-susceptibility allele APC I1307K: evidence of genetic drift within the Ashkenazim. Am J Hum Genet 73:1250–1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norton N, Owen MJ (2005) HTR2A: association and expression studies in neuropsychiatric genetics. Ann Med 37:121–129 [DOI] [PubMed] [Google Scholar]
- Nyholt DR (2004) A simple correction for multiple testing for single-nucleotide polymorphisms in linkage disequilibrium with each other. Am J Hum Genet 74:765–769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Connell JR, Weeks DE (1998) PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet 63:259–266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura H, Aruga J, Mikoshiba K (2001) Behavioral abnormalities of Zic1 and Zic2 mutant mice: implications as models for human neurological disorders. Behav Genet 31:317–324 10.1023/A:1012235510600 [DOI] [PubMed] [Google Scholar]
- Ohtsuki T, Sakurai K, Dou H, Toru M, Yamakawa-Kobayashi K, Arinami T (2001a) Mutation analysis of the NMDAR2B (GRIN2B) gene in schizophrenia. Mol Psychiatry 6:211–216 10.1038/sj.mp.4000808 [DOI] [PubMed] [Google Scholar]
- Ohtsuki T, Toru M, Arinami T (2001b) Mutation screening of the metabotropic glutamate receptor mGluR4 (GRM4) gene in patients with schizophrenia. Psychiatr Genet 11:79–83 10.1097/00041444-200106000-00004 [DOI] [PubMed] [Google Scholar]
- Owen MJ, O’Donovan MC, Harrison PJ (2005) Schizophrenia: a genetic disorder of the synapse? Bmj 330:158–159 10.1136/bmj.330.7484.158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owen MJ, Williams NM, O’Donovan MC (2004) The molecular genetics of schizophrenia: new findings promise new insights. Mol Psychiatry 9:14–27 10.1038/sj.mp.4001444 [DOI] [PubMed] [Google Scholar]
- Park N, Juo SH, Cheng R, Liu J, Loth JE, Lilliston B, Nee J, Grunn A, Kanyas K, Lerer B, Endicott J, Gilliam TC, Baron M (2004) Linkage analysis of psychosis in bipolar pedigrees suggests novel putative loci for bipolar disorder and shared susceptibility with schizophrenia. Mol Psychiatry 9:1091–1099 10.1038/sj.mp.4001541 [DOI] [PubMed] [Google Scholar]
- Perez de Castro I, Santos J, Torres P, Visedo G, Saiz-Ruiz J, Llinares C, Fernandez-Piqueras J (1995) A weak association between TH and DRD2 genes and bipolar affective disorder in a Spanish sample. J Med Genet 32:131–134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perroy J, El Far O, Bertaso F, Pin JP, Betz H, Bockaert J, Fagni L (2002) PICK1 is required for the control of synaptic transmission by the metabotropic glutamate receptor 7. Embo J 21:2990–2999 10.1093/emboj/cdf313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzuti A, Novelli G, Ratti A, Amati F, Mari A, Calabrese G, Nicolis S, Silani V, Marino B, Scarlato G, Ottolenghi S, Dallapiccola B (1997) UFD1L, a developmentally expressed ubiquitination gene, is deleted in CATCH 22 syndrome. Hum Mol Genet 6:259–265 10.1093/hmg/6.2.259 [DOI] [PubMed] [Google Scholar]
- Porter K, Komiyama NH, Vitalis T, Kind PC, Grant SG (2005) Differential expression of two NMDA receptor interacting proteins, PSD-95 and SynGAP during mouse development. Eur J Neurosci 21:351–362 10.1111/j.1460-9568.2005.03874.x [DOI] [PubMed] [Google Scholar]
- Potash JB, Willour VL, Chiu YF, Simpson SG, MacKinnon DF, Pearlson GD, DePaulo JR Jr, McInnis MG (2001) The familial aggregation of psychotic symptoms in bipolar disorder pedigrees. Am J Psychiatry 158:1258–1264 10.1176/appi.ajp.158.8.1258 [DOI] [PubMed] [Google Scholar]
- Preisig M, Ferrero F, Malafosse A (2005) Monoamine oxidase a and tryptophan hydroxylase gene polymorphisms: are they associated with bipolar disorder? Am J Pharmacogenomics 5:45–52 [DOI] [PubMed] [Google Scholar]
- Pulver AE, Karayiorgou M, Lasseter VK, Wolyniec P, Kasch L, Antonarakis S, Housman D, Kazazian HH, Meyers D, Nestadt G, et al (1994a) Follow-up of a report of a potential linkage for schizophrenia on chromosome 22q12-q13.1: part 2. Am J Med Genet 54:44–50 10.1002/ajmg.1320540109 [DOI] [PubMed] [Google Scholar]
- Pulver AE, Karayiorgou M, Wolyniec PS, Lasseter VK, Kasch L, Nestadt G, Antonarakis S, et al (1994b) Sequential strategy to identify a susceptibility gene for schizophrenia: report of potential linkage on chromosome 22q12-q13.1: part 1. Am J Med Genet 54:36–43 10.1002/ajmg.1320540108 [DOI] [PubMed] [Google Scholar]
- Ranade SS, Mansour H, Wood J, Chowdari KV, Brar LK, Kupfer DJ, Nimgaonkar VL (2003) Linkage and association between serotonin 2A receptor gene polymorphisms and bipolar I disorder. Am J Med Genet B Neuropsychiatr Genet 121:28–34 10.1002/ajmg.b.20070 [DOI] [PubMed] [Google Scholar]
- Ravassard P, Cote F, Grondin B, Bazinet M, Mallet J, Aubry M (1999) ZNF74, a gene deleted in DiGeorge syndrome, is expressed in human neural crest-derived tissues and foregut endoderm epithelia. Genomics 62:82–85 10.1006/geno.1999.5982 [DOI] [PubMed] [Google Scholar]
- Raybould R, Green EK, MacGregor S, Gordon-Smith K, Heron J, Hyde S, Caesar S, Nikolov I, Williams N, Jones L, O’Donovan MC, Owen MJ, Jones I, Kirov G, Craddock N (2005) Bipolar disorder and polymorphisms in the dysbindin gene (DTNBP1). Biol Psychiatry 57:696–701 10.1016/j.biopsych.2005.01.018 [DOI] [PubMed] [Google Scholar]
- Rennert H, Bercovich D, Hubert A, Abeliovich D, Rozovsky U, Bar-Shira A, Soloviov S, Schreiber L, Matzkin H, Rennert G, Kadouri L, Peretz T, Yaron Y, Orr-Urtreger A (2002) A novel founder mutation in the RNASEL gene, 471delAAAG, is associated with prostate cancer in Ashkenazi Jews. Am J Hum Genet 71:981–984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossi J, Tomac A, Saarma M, Airaksinen MS (2000) Distinct roles for GFRalpha1 and GFRalpha2 signalling in different cranial parasympathetic ganglia in vivo. Eur J Neurosci 12:3944–3952 10.1046/j.1460-9568.2000.00292.x [DOI] [PubMed] [Google Scholar]
- Sachs NA, Sawa A, Holmes SE, Ross CA, Delisi LE, Margolis RL (2005) A frameshift mutation in Disrupted in Schizophrenia 1 in an American family with schizophrenia and schizoaffective disorder. Mol Psychiatry 10:758–764 10.1038/sj.mp.4001667 [DOI] [PubMed] [Google Scholar]
- Saito T, Guan F, Papolos DF, Rajouria N, Fann CS, Lachman HM (2001) Polymorphism in SNAP29 gene promoter region associated with schizophrenia. Mol Psychiatry 6:193–201 10.1038/sj.mp.4000825 [DOI] [PubMed] [Google Scholar]
- Saito T, Stopkova P, Diaz L, Papolos DF, Boussemart L, Lachman HM (2003) Polymorphism screening of PIK4CA: possible candidate gene for chromosome 22q11-linked psychiatric disorders. Am J Med Genet B Neuropsychiatr Genet 116:77–83 10.1002/ajmg.b.10042 [DOI] [PubMed] [Google Scholar]
- Sandhu HK, Hollenbeck N, Wassink TH, Philibert RA (2004) An association study of PCQAP polymorphisms and schizophrenia. Psychiatr Genet 14:169–172 10.1097/00041444-200409000-00010 [DOI] [PubMed] [Google Scholar]
- Saugstad JA, Marino MJ, Folk JA, Hepler JR, Conn PJ (1998) RGS4 inhibits signaling by group I metabotropic glutamate receptors. J Neurosci 18:905–913 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J, Jamra RA, Freudenberg J, Becker T, Ohlraun S, Otte AC, Tullius M, Kovalenko S, Bogaert AV, Maier W, Rietschel M, Propping P, Nothen MM, Cichon S (2004) Examination of G72 and D-amino-acid oxidase as genetic risk factors for schizophrenia and bipolar affective disorder. Mol Psychiatry 9:203–207 10.1038/sj.mp.4001421 [DOI] [PubMed] [Google Scholar]
- Schwab SG, Lerer B, Albus M, Maier W, Hallmayer J, Fimmers R, Lichtermann D, Minges J, Bondy B, Ackenheil M, et al (1995) Potential linkage for schizophrenia on chromosome 22q12-q13: a replication study. Am J Med Genet 60:436–443 10.1002/ajmg.1320600515 [DOI] [PubMed] [Google Scholar]
- Schwartz R, Halldorsson BV, Bafna V, Clark AG, Istrail S (2003) Robustness of inference of haplotype block structure. J Comput Biol 10:13–19 10.1089/106652703763255642 [DOI] [PubMed] [Google Scholar]
- Segurado R, Detera-Wadleigh SD, Levinson DF, Lewis CM, Gill M, Nurnberger JI Jr, Craddock N, et al (2003) Genome scan meta-analysis of schizophrenia and bipolar disorder, part III: bipolar disorder. Am J Hum Genet 73:49–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selimi F, Lohof AM, Heitz S, Lalouette A, Jarvis CI, Bailly Y, Mariani J (2003) Lurcher GRID2-induced death and depolarization can be dissociated in cerebellar Purkinje cells. Neuron 37:813–819 10.1016/S0896-6273(03)00093-X [DOI] [PubMed] [Google Scholar]
- Severino G, Congiu D, Serreli C, De Lisa R, Chillotti C, Del Zompo M, Piccardi MP (2005) A48G polymorphism in the D1 receptor genes associated with bipolar I disorder. Am J Med Genet B Neuropsychiatr Genet 134:37–38 [DOI] [PubMed] [Google Scholar]
- Shifman S, Bronstein M, Sternfeld M, Pisante-Shalom A, Lev-Lehman E, Weizman A, Reznik I, Spivak B, Grisaru N, Karp L, Schiffer R, Kotler M, Strous RD, Swartz-Vanetik M, Knobler HY, Shinar E, Beckmann JS, Yakir B, Risch N, Zak NB, Darvasi A (2002) A highly significant association between a COMT haplotype and schizophrenia. Am J Hum Genet 71:1296–1302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai T, Ohmori O, Hori H, Nakamura J (2002) Allelic association of the neuronal nitric oxide synthase (NOS1) gene with schizophrenia. Mol Psychiatry 7:560–563 10.1038/sj.mp.4001041 [DOI] [PubMed] [Google Scholar]
- Shiohama A, Sasaki T, Noda S, Minoshima S, Shimizu N (2003) Molecular cloning and expression analysis of a novel gene DGCR8 located in the DiGeorge syndrome chromosomal region. Biochem Biophys Res Commun 304:184–190 10.1016/S0006-291X(03)00554-0 [DOI] [PubMed] [Google Scholar]
- Shprintzen RJ, Goldberg R, Golding-Kushner KJ, Marion RW (1992) Late-onset psychosis in the velo-cardio-facial syndrome [letter]. Am J Med Genet 42:141–142 10.1002/ajmg.1320420131 [DOI] [PubMed] [Google Scholar]
- Sinibaldi L, De Luca A, Bellacchio E, Conti E, Pasini A, Paloscia C, Spalletta G, Caltagirone C, Pizzuti A, Dallapiccola B (2004) Mutations of the Nogo-66 receptor (RTN4R) gene in schizophrenia. Hum Mutat 24:534–535 10.1002/humu.9292 [DOI] [PubMed] [Google Scholar]
- Sklar P, Gabriel SB, McInnis MG, Bennett P, Lim YM, Tsan G, Schaffner S, Kirov G, Jones I, Owen M, Craddock N, DePaulo JR, Lander ES (2002) Family-based association study of 76 candidate genes in bipolar disorder: BDNF is a potential risk locus: brain-derived neutrophic factor. Mol Psychiatry 7:579–593 10.1038/sj.mp.4001058 [DOI] [PubMed] [Google Scholar]
- Snyder SH, Bredt DS (1991) Nitric oxide as a neuronal messenger. Trends Pharmacol Sci 12:125–128 10.1016/0165-6147(91)90526-X [DOI] [PubMed] [Google Scholar]
- Spurlock G, Williams J, McGuffin P, Aschauer HN, Lenzinger E, Fuchs K, Sieghart WC, Meszaros K, Fathi N, Laurent C, Mallet J, Macciardi F, Pedrini S, Gill M, Hawi Z, Gibson S, Jazin EE, Yang HT, Adolfsson R, Pato CN, Dourado AM, Owen MJ (1998) European Multicentre Association Study of Schizophrenia: a study of the DRD2 Ser311Cys and DRD3 Ser9Gly polymorphisms. Am J Med Genet 81:24–28 [DOI] [PubMed] [Google Scholar]
- Stanchi F, Corso V, Scannapieco P, Ievolella C, Negrisolo E, Tiso N, Lanfranchi G, Valle G (2000) TUBA8: a new tissue-specific isoform of alpha-tubulin that is highly conserved in human and mouse. Biochem Biophys Res Commun 270:1111–1118 10.1006/bbrc.2000.2571 [DOI] [PubMed] [Google Scholar]
- Stefansson H, Sarginson J, Kong A, Yates P, Steinthorsdottir V, Gudfinnsson E, Gunnarsdottir S, Walker N, Petursson H, Crombie C, Ingason A, Gulcher JR, Stefansson K, St Clair D (2003) Association of neuregulin 1 with schizophrenia confirmed in a Scottish population. Am J Hum Genet 72:83–87 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Sigurdsson E, Steinthorsdottir V, Bjornsdottir S, Sigmundsson T, Ghosh S, Brynjolfsson J, et al (2002) Neuregulin 1 and susceptibility to schizophrenia. Am J Hum Genet 71:877–892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern HS, Viertelhausen S, Hunter AG, O’Rourke K, Cappelli M, Perras H, Serfas K, Blumenthall A, Dewar D, Baumann E, Lagarde AE (2001) APC I1307K increases risk of transition from polyp to colorectal carcinoma in Ashkenazi Jews. Gastroenterology 120:392–400 10.1053/gast.2001.21170 [DOI] [PubMed] [Google Scholar]
- Straub RE, Jiang Y, MacLean CJ, Ma Y, Webb BT, Myakishev MV, Harris-Kerr C, Wormley B, Sadek H, Kadambi B, Cesare AJ, Gibberman A, Wang X, O’Neill FA, Walsh D, Kendler KS (2002) Genetic variation in the 6p22.3 gene DTNBP1, the human ortholog of the mouse dysbindin gene, is associated with schizophrenia. Am J Hum Genet 71:337–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Q, Mochida S, Tian JH, Mehta R, Sheng ZH (2001) SNAP-29: a general SNARE protein that inhibits SNARE disassembly and is implicated in synaptic transmission. Proc Natl Acad Sci USA 98:14038–14043 10.1073/pnas.251532398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugimura K, Taylor KD, Lin YC, Hang T, Wang D, Tang YM, Fischel-Ghodsian N, Targan SR, Rotter JI, Yang H (2003) A novel NOD2/CARD15 haplotype conferring risk for Crohn disease in Ashkenazi Jews. Am J Hum Genet 72:509–518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takase K, Ohtsuki T, Migita O, Toru M, Inada T, Yamakawa-Kobayashi K, Arinami T (2001) Association of ZNF74 gene genotypes with age-at-onset of schizophrenia. Schizophr Res 52:161–165 10.1016/S0920-9964(00)00191-2 [DOI] [PubMed] [Google Scholar]
- Thomson PA, Wray NR, Millar JK, Evans KL, Hellard SL, Condie A, Muir WJ, Blackwood DH, Porteous DJ (2005) Association between the TRAX/DISC locus and both bipolar disorder and schizophrenia in the Scottish population. Mol Psychiatry 10:616, 657–668 10.1038/sj.mp.4001699 [DOI] [PubMed] [Google Scholar]
- Tosato S, Dazzan P, Collier D (2005) Association between the neuregulin 1 gene and schizophrenia: a systematic review. Schizophr Bull 31:613-617 10.1093/schbul/sbi043 [DOI] [PubMed] [Google Scholar]
- Treadaway J, Zuo J (1998) Mapping of the mouse glutamate receptor delta1 subunit (Grid1) to chromosome 14. Genomics 54:359–360 10.1006/geno.1998.5599 [DOI] [PubMed] [Google Scholar]
- Tsuang MT, Faraone SV (1990) The genetics of mood disorders. Johns Hopkins Press, Baltimore [Google Scholar]
- Tsuang MT, Gilbertson MW, Faraone SV (1991) The genetics of schizophrenia: current knowledge and future directions. Schizophr Res 4:157–171 10.1016/0920-9964(91)90031-L [DOI] [PubMed] [Google Scholar]
- Vincent JB, Masellis M, Lawrence J, Choi V, Gurling HM, Parikh SV, Kennedy JL (1999) Genetic association analysis of serotonin system genes in bipolar affective disorder. Am J Psychiatry 156:136–138 [DOI] [PubMed] [Google Scholar]
- Wang S, Detera-Wadleigh SD, Coon H, Sun CE, Goldin LR, Duffy DL, Byerley WF, Gershon ES, Diehl SR (1996a) Evidence of linkage disequilibrium between schizophrenia and the SCa1 CAG repeat on chromosome 6p23 [letter]. Am J Hum Genet 59:731–736 [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Matsuda S, Drews V, Torashima T, Meisler MH, Yuzaki M (2003) A hot spot for hotfoot mutations in the gene encoding the delta2 glutamate receptor. Eur J Neurosci 17:1581–1590 10.1046/j.1460-9568.2003.02595.x [DOI] [PubMed] [Google Scholar]
- Wang MG, Yi H, Guerini D, Klee CB, McBride OW (1996b) Calcineurin A alpha (PPP3CA), calcineurin A beta (PPP3CB) and calcineurin B (PPP3R1) are located on human chromosomes 4, 10q21→q22 and 2p16→p15 respectively. Cytogenet Cell Genet 72:236–241 [DOI] [PubMed] [Google Scholar]
- Williams NM, Preece A, Spurlock G, Norton N, Williams HJ, McCreadie RG, Buckland P, Sharkey V, Chowdari KV, Zammit S, Nimgaonkar V, Kirov G, Owen MJ, O’Donovan MC (2004) Support for RGS4 as a susceptibility gene for schizophrenia. Biol Psychiatry 55:192–195 10.1016/j.biopsych.2003.11.002 [DOI] [PubMed] [Google Scholar]
- Williams NM, Spurlock G, Norton N, Williams HJ, Hamshere ML, Krawczak M, Kirov G, Nikolov I, Georgieva L, Jones S, Cardno AG, O’Donovan MC, Owen MJ (2002) Mutation screening and LD mapping in the VCFS deleted region of chromosome 22q11 in schizophrenia using a novel DNA pooling approach. Mol Psychiatry 7:1092–1100 10.1038/sj.mp.4001188 [DOI] [PubMed] [Google Scholar]
- Wonodi I, Hong LE, Avila MT, Buchanan RW, Carpenter WT Jr, Stine OC, Mitchell BD, Thaker GK (2005) Association between polymorphism of the SNAP29 gene promoter region and schizophrenia. Schizophr Res 78:339-341 10.1016/j.schres.2005.03.023 [DOI] [PubMed] [Google Scholar]
- Zhang D, Sliwkowski MX, Mark M, Frantz G, Akita R, Sun Y, Hillan K, Crowley C, Brush J, Godowski PJ (1997) Neuregulin-3 (NRG3): a novel neural tissue-enriched protein that binds and activates ErbB4. Proc Natl Acad Sci USA 94:9562–9567 10.1073/pnas.94.18.9562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zoghbi HY, Jodice C, Sandkuijl LA, Kwiatkowski TJ Jr, McCall AE, Huntoon SA, Lulli P, Spadaro M, Litt M, Cann HM, et al (1991) The gene for autosomal dominant spinocerebellar ataxia (SCA1) maps telomeric to the HLA complex and is closely linked to the D6S89 locus in three large kindreds. Am J Hum Genet 49:23–30 [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.