Abstract
We describe genomic structures of 59 X-chromosome segmental duplications that include the proteolipid protein 1 gene (PLP1) in patients with Pelizaeus-Merzbacher disease. We provide the first report of 13 junction sequences, which gives insight into underlying mechanisms. Although proximal breakpoints were highly variable, distal breakpoints tended to cluster around low-copy repeats (LCRs) (50% of distal breakpoints), and each duplication event appeared to be unique (100 kb to 4.6 Mb in size). Sequence analysis of the junctions revealed no large homologous regions between proximal and distal breakpoints. Most junctions had microhomology of 1–6 bases, and one had a 2-base insertion. Boundaries between single-copy and duplicated DNA were identical to the reference genomic sequence in all patients investigated. Taken together, these data suggest that the tandem duplications are formed by a coupled homologous and nonhomologous recombination mechanism. We suggest repair of a double-stranded break (DSB) by one-sided homologous strand invasion of a sister chromatid, followed by DNA synthesis and nonhomologous end joining with the other end of the break. This is in contrast to other genomic disorders that have recurrent rearrangements formed by nonallelic homologous recombination between LCRs. Interspersed repetitive elements (Alu elements, long interspersed nuclear elements, and long terminal repeats) were found at 18 of the 26 breakpoint sequences studied. No specific motif that may predispose to DSBs was revealed, but single or alternating tracts of purines and pyrimidines that may cause secondary structures were common. Analysis of the 2-Mb region susceptible to duplications identified proximal-specific repeats and distal LCRs in addition to the previously reported ones, suggesting that the unique genomic architecture may have a role in nonrecurrent rearrangements by promoting instability.
Introduction
Pelizaeus-Merzbacher disease (PMD [MIM 312080]) is caused predominantly by submicroscopic duplications of chromosome Xq22.2 that include the entire proteolipid protein 1 gene (PLP1). PLP1 is a dosage-sensitive gene, and either gain or loss of copy number can result in neurological disorders in both humans and animals (Raskind et al. 1991; Ellis and Malcolm 1994; Kagawa et al. 1994; Readhead et al. 1994). PMD is characterized by dysmyelination and demyelination within the CNS, and the clinical phenotype is variable—with signs including nystagmus, psychomotor developmental delay, ataxia, and spasticity—and onset is within the first year of life (Boulloche and Aicardi 1986; Hodes et al. 1993).
Variation in both duplication size and position of proximal and distal breakpoints has been shown by our work and that of others (Woodward et al. 1998, 2000; Inoue et al. 1999). Rearrangements are typically tandem head-to-tail duplications within Xq22.2, and a mechanism of unequal sister-chromatid exchange during spermatogenesis has been proposed (Woodward et al. 1998; Inoue et al. 1999; Mimault et al. 1999). Several atypical cases of apparent transposition events have also been described in which an additional copy of the PLP1 gene has integrated at noncontiguous sites on the X chromosome (Xp22 and Xq26) (Hodes et al. 2000; Woodward et al. 2003). The mechanism by which these atypical nontandem duplication events in PMD arise is currently unknown. We have also described triplication and quintuplication events that involve PLP1 (Wolf et al. 2005). The mechanism by which these are generated is also not known.
DNA rearrangements, such as duplications, deletions, and insertions, may occur in cells as a result of mechanisms that have evolved for the repair of DNA damage. Homologous recombination and nonhomologous end joining (NHEJ) are the two major types of repair mechanisms (Kanaar et al. 1998; Tsukamoto and Ikeda 1998). These pathways differ in their accuracy. Homologous recombination mechanisms ensure accurate repair by using homologous sequence as a template. Conversely, NHEJ is an error-prone mechanism that uses limited sequence homology or none at all (Roth et al. 1985; Roth and Wilson 1986). Junctions formed by NHEJ typically have microhomology, insertion, or deletion of a few base pairs of DNA (Lieber et al. 2003).
The nonrandom distribution of rearrangements in the genome suggests that DNA sequence and genomic architecture can predispose certain regions to chromosomal instability. Specific sequences and structures have been implicated, including low-copy repeats (LCRs) (usually 10–500 kb in size, with 95%–98% homology) (Stankiewicz and Lupski 2002), inverted repeats (Chuzhanova et al. 2003), palindromic AT-rich sequences (Edelmann et al. 2001; Kurahashi et al. 2003), and motifs involved in recombination (Abeysinghe et al. 2003). Charcot-Marie-Tooth disease type 1A (CMT1A) and other genomic diseases described to date mostly occur by nonallelic homologous recombination (NAHR) mediated by LCRs (Lupski et al. 1991; Chance et al. 1994; Lupski 1998; Perez Jurado et al. 1998; Christian et al. 1999; Edelmann et al. 1999; Inoue and Lupski 2002; Shaw and Lupski 2004).
In this study, we have determined the location and size of the Xq22 duplications in a cohort of 59 patients with PMD who have an increased dosage of the PLP1 gene. To identify structures and motifs causing the genomic instability and to determine whether the duplications occurred by homologous or nonhomologous mechanisms, we mapped the proximal and distal duplication breakpoints and sequenced the duplication junctions from 13 of these individuals with tandem duplications. Our results suggest that tandem head-to-tail Xq22 duplications in patients with PMD were formed by coupled homologous and nonhomologous recombination, rather than by NAHR like the common rearrangements associated with other genomic disorders.
Material and Methods
Patients
Peripheral blood samples, lymphoblastoid cell lines, or both were obtained from 59 patients with PMD from different families who were found to harbor PLP1 duplications by FISH or multiplex PCR (data not shown). This study was approved by the local ethics committees at the Institute of Child Health and the Alfred I. duPont Hospital for Children, and informed consent was obtained appropriately.
Mapping Strategy
Metaphase FISH was performed on a subset of the patients to confirm that the duplication was within Xq22. In the laboratory at the Institute of Child Health, the extent of duplication was mapped using several complementary methods. Most patients studied in this laboratory were examined by both interphase FISH and multiplex PCR (24/28 patients), and two of these were also characterized by fiber FISH (patients PMD9 and PMD24). The exceptions were three patients who were investigated only by PCR (PMD23, PMD30, and PMD42) and one patient who was screened only by FISH (PMD45). In the laboratory at the duPont Hospital, the extent of duplication was determined solely by multiplex PCR, but a different method was used. Reproducibility of the different mapping strategies was confirmed by three samples mapped by both laboratories independently. Further validation of mapping methods was obtained by testing 49 samples from both laboratories with the use of a third multiplex PCR approach.
FISH Analysis
FISH was performed as described elsewhere (Woodward et al. 2003). Cosmid, BAC, or PAC genomic clones were selected from the Ensembl Genome Browser Web site (Hubbard et al. 2002) and were obtained from the Wellcome Trust Sanger Institute (Hinxton, United Kingdom) (see The Wellcome Trust Sanger Institute: Human X Project Web site). Metaphase chromosomes and interphase nuclei from patient lymphoblastoid cell lines were cultured and harvested in accordance with standard protocols. DNA fibers were prepared from lymphoblastoid cell culture by use of a protocol available at The Wellcome Trust Sanger Institute: DNA Fibers Web site. DNA is not stretched uniformly using this method, and, therefore, inferences about signal size can be made only on the basis of comparison with other signals on the same fiber, since different fibers may be stretched to different degrees.
Multiplex PCR
Primers were designed using the human genome sequence (Ensembl and National Center for Biotechnology Information [NCBI] Web sites) and the Primer3 program, MacVector software (Accelrys), or inspection of the sequence. Primer sequences spanning ∼7.4 Mb from position 99.40 Mb to position 106.80 Mb on the X chromosome (Ensembl v29.35b; NCBI build 35), lying 3.4 Mb proximal and 4 Mb distal to PLP1, are shown in table 1. Several multiplex PCR approaches were used. The approach used in the laboratory at the Institute of Child Health was universal-primer quantitative fluorescent multiplex (UPQFM)–PCR (Heath et al. 2000), which involved two PCR reactions. The primary reaction amplified selected sequences by using unique primers with a universal tag. A typical reaction included three primers pairs around each of the breakpoints to be analyzed, a PLP1 exon 6 probe expected to be duplicated (primers PLP6F and PLP6R), and a control autosomal locus from the cystic fibrosis gene (CF) (CFF and CFR). The secondary reaction amplified the products from the first reaction by using universal primers with a fluorescent dye at the 5′ end (UNIVF and UNIVR). PCR products were separated on an ABI 377 DNA sequencer, and the relative amounts of the amplified peak areas were determined using Genotyper software (Applied Biosystems) and were compared with ratios obtained from normal controls and control duplication patients. For a male with a PLP1 duplication, the expected ratio value for a single-copy sequence, compared with PLP1, is 0.5, and the expected value for a duplicated sequence is 1. When the autosomal CF locus is used as a control, the male ratio expected for a nonduplicated sequence is 1, compared with a ratio of 2 for a duplicated sequence.
Table 1.
Primer Sequences Used in Mapping
| Primer Namea | Sequenceb(5′→3′) |
| UNIVF | TCCGTCTTAGCTGAGTGGCGTA |
| UNIVR | AGGCAGAATCGACTCACCGCTA |
| PLP6F | tccgtcttagctgagtggcgtaTGGAGCATATTACTGCTGTTGC |
| PLP6R | aggcagaatcgactcaccgctaAAGCTTCCCTCCAGCATTTC |
| CFF | tccgtcttagctgagtggcgtaCGAGGCTACAGCTTTGGAAC |
| CFR | aggcagaatcgactcaccgctaCATCACACTTGTGCCATTCC |
| dJ635g19F8797 | tccgtcttagctgagtggcgtaGTTGGGGCCTTCTGTGTTTA |
| dJ635g19R8962 | aggcagaatcgactcaccgctaACTCACCCTGGATGGTTCAG |
| dJ635g19F30902 | tccgtcttagctgagtggcgtaTCCTTCCTGCCTGACTCCTA |
| dJ635g19R31157 | aggcagaatcgactcaccgctaTCGCAGATGATGTCCCACTA |
| dJ635g19F48059 | tccgtcttagctgagtggcgtaGGTGGTCCCTCCCATTTAGT |
| dJ635g19R48309 | aggcagaatcgactcaccgctaATTGGTGTGCCAGATGTGAA |
| dJ635G19F60570 | tccgtcttagctgagtggcgtaAGAGTGGGTCTCTGGCTCAA |
| dJ635g16R60728 | aggcagaatcgactcaccgctaGGTGCCTCCAACTTTTGTGT |
| dJ635G19F68207 | tccgtcttagctgagtggcgtaAGGACAGCCAGCAAGAAAAA |
| dJ635G19R68454 | aggcagaatcgactcaccgctaCCAGTTTCCTCCCCCTTTAG |
| dJ635G19F69048 | tccgtcttagctgagtggcgtaCCACCCCCAATATCACTTTG |
| dJ635G19R69265 | aggcagaatcgactcaccgctaGCTTAGTTTCGCACCTGGAG |
| cU65A4F11590 | tccgtcttagctgagtggcgtaATTTTGATTGCGGTCATGGT |
| cU65A4R11756 | aggcagaatcgactcaccgctaCCAAAACAGTTTGCCACTTTC |
| cU65A4F9959 | tccgtcttagctgagtggcgtaGAAGCTTGGGGGAAAGAATC |
| cU65A4R9747 | aggcagaatcgactcaccgctaCAATCAGGGCATGAATGAGA |
| cU65A4F6833 | tccgtcttagctgagtggcgtaGGGCCATATGGAAGACAGAA |
| cU65A4R7109 | aggcagaatcgactcaccgctaGCCATCCAGTATTGGCACTT |
| cU65a4F4480 | tccgtcttagctgagtggcgtaGCTTGGAAGCTCCACTTGAC |
| cU65A4R4685 | aggcagaatcgactcaccgctaCATGTGGAGGAAGTGTGTGG |
| cU177E8F36491 | tccgtcttagctgagtggcgtaGTGAAACAGGGATGGGAAGAG |
| cU177E8R36712 | aggcagaatcgactcaccgctaCTAGTCCAGAGCATGGCACA |
| cU177E8F38464 | tccgtcttagctgagtggcgtaCTTCTCCCAACCCCAGTACA |
| cU177E8R38658 | aggcagaatcgactcaccgctaCAGCACATGGCAGTTCCTTA |
| cU177E8F20484 | tccgtcttagctgagtggcgtaTCTTGGTGCACAAAAACAGG |
| cU177E8R20744 | aggcagaatcgactcaccgctaAGGCCTTCACCTTCATCCTT |
| cU177E8F32435 | tccgtcttagctgagtggcgtaCTTTGCCAGCAATGAGAACA |
| cU177E8R32693 | aggcagaatcgactcaccgctaACCTCTGGCCCTTGTGTAAA |
| cU177E8F5951 | tccgtcttagctgagtggcgtaAGACTCTAGCCCACCAGCAA |
| cU177e8R5698 | aggcagaatcgactcaccgctaTGACGCTTGAGTCTG TGACC |
| dJ43H13F12256 | tccgtcttagctgagtggcgtaAGATGTGGATTTAGGGTCAACAGC |
| dJ43H13R12507 | aggcagaatcgactcaccgctaCAAGCTCCTAACCTCCATCTGTC |
| cV857G6F11085 | tccgtcttagctgagtggcgtaCTGTGCTAAACTGCGTGGAA |
| cV857G6R11289 | aggcagaatcgactcaccgctaTGCAAAGCCAAGCTACAAGA |
| cV857G6F41850 | tccgtcttagctgagtggcgtaCTTTCCTTGAGTCATTGAGGTTTT |
| cV857G6R42081 | aggcagaatcgactcaccgctaTTGTTGAGTGATTCTGATCTCCAT |
| cV857G6F32969 | tccgtcttagctgagtggcgtaCACAAATGTCCATCATTACAGGTT |
| cV857G6R33205 | aggcagaatcgactcaccgctaGCATCTCTAGAGGGCAATATCACT |
| cU246D9F8349 | tccgtcttagctgagtggcgtaGTTGGTATCAAATGGGCATATCAC |
| cU246D9R9530 | aggcagaatcgactcaccgctaGAACTCATGTGGCTCCTGAACTG |
| cU246D9F16048 | tccgtcttagctgagtggcgtaTGTCGATTGCATTATTCCTGTTA |
| cU246D9R16277 | aggcagaatcgactcaccgctaCTGCTCAGTGTTGACATCCATTT |
| dJ1055C14F1 | tccgtcttagctgagtggcgtaCATGCCCCACTGTTATAGGG |
| dJ1055C14R1 | aggcagaatcgactcaccgctaGCCCATTTTCTATTGGGTGA |
| dJ1055C14F3 | tccgtcttagctgagtggcgtaGTTAACACACGGGCCTCACT |
| dJ1055C14R3 | aggcagaatcgactcaccgctaTTGCATGCGCTATGATCTTC |
| dJ1055C14F4 | tccgtcttagctgagtggcgtaGCATTTTAGGGCAGCAAGAG |
| dJ1055C14R4 | aggcagaatcgactcaccgctaTGGTTAGGGATGAGGTGCTC |
| dJ1055C14F5 | tccgtcttagctgagtggcgtaAGAGCCAAGCTGGACAAAA |
| dJ1055C14R5 | aggcagaatcgactcaccgctaGTAGGGAACAGGTGGCAAAA |
| dJ1055C14F8 | tccgtcttagctgagtggcgtaGGAAGGTTCTGCAATCAGGA |
| dJ1055C14R8 | aggcagaatcgactcaccgctaGTGGCCACAAGAAAGGAAAA |
| dJ1055C14F10 | tccgtcttagctgagtggcgtaCCTCCAACCGCCATAGTAAA |
| dJ1055C14R10 | aggcagaatcgactcaccgctaAACGGAGGAAGAGGATTGGT |
| dJ1055C14F7 | tccgtcttagctgagtggcgtaGCTCTTACCTTTGCGTCCTG |
| dJ1055C14R7 | aggcagaatcgactcaccgctaACCAAACAGCCAATCCTTTG |
| dJ1055C14F6 | tccgtcttagctgagtggcgtaGGTTTGAAACTGGGGACAGA |
| dJ1055C14R6 | aggcagaatcgactcaccgctaAAGGGCCAGGACTTTTGTTT |
| dJ1055C14F2 | tccgtcttagctgagtggcgtaCGCAATAGGAAAAGGGATGA |
| dJ1055C14R2 | aggcagaatcgactcaccgctaTCCTGCTGCAGATTCAAATG |
| cU35G3F5631 | tccgtcttagctgagtggcgtaTCCATGGGGAAGTTCTTGAG |
| cU35G3R5805 | aggcagaatcgactcaccgctaAGCCAGGAGAGTTTGCGTTA |
| cV362H12F34770 | tccgtcttagctgagtggcgtaGTTGCATCTGGTAGCCATCC |
| cV362H12R34963 | aggcagaatcgactcaccgctaAACGTGGGGCAAAACTACTG |
| dJ839M11F37922 | tccgtcttagctgagtggcgtaGCTGTTCCTCAGAGGTCGTC |
| dJ839M11R38082 | aggcagaatcgactcaccgctaTCCAATCAGACGTGAAGCTG |
| cU240C4F29788 | tccgtcttagctgagtggcgtaGAGGTGCCAACAGGGAATAG |
| cU240C2R29983 | aggcagaatcgactcaccgctaGGTCACGTGCCCCTTACTTA |
| cU46H11F32229 | tccgtcttagctgagtggcgtaATTCCAGAGGCTCCATTCCT |
| cU46H11R32404 | aggcagaatcgactcaccgctaTCCTTGCCAGTTTTGACCTC |
| 35F15tag66750F | tccgtcttagctgagtggcgtaTAACCACCGTCCACTCACAA |
| 35F15tag66949R | aggcagaatcgactcaccgctaGAGCCTTGGTTCTGTGGAAG |
| 364B14F16714 | tccgtcttagctgagtggcgtaAGGATCTGGACCAACACAGG |
| 364B14R16959 | aggcagaatcgactcaccgctaTGAAGGCAGGGAACTCCTTA |
| 364B14F25512 | tccgtcttagctgagtggcgtaATGATCCAAGGGTTCCATGA |
| 364B14R25756 | aggcagaatcgactcaccgctaACTGGCAGGAATTGGCTAAC |
| 364B14F48225 | tccgtcttagctgagtggcgtaGATTCCCACCTACTGCTGGA |
| 364b14R48392 | aggcagaatcgactcaccgctaAATGCTGCAGGAGCCTAAGA |
| 364B14tag74126F | tccgtcttagctgagtggcgtaTTTCCAGTGGGCTTGGTTAG |
| 364B14tag74369R | aggcagaatcgactcaccgctaTAGAGGGCTCTGCGGAATTA |
| 595A18F26757 | tccgtcttagctgagtggcgtaTTTGTTTGCTGCTTCAGTGG |
| 595A18R27039 | aggcagaatcgactcaccgctaTTGGAAGGCGGATACAATTC |
| 595A18F54671 | tccgtcttagctgagtggcgtaAGGAAGGCCCATCAACTTC |
| 595A18R54825 | aggcagaatcgactcaccgctaGAGGCATCATAGGGGCAGTA |
| 595a18f101906 | tccgtcttagctgagtggcgtaGATGCCCCAGATTGTACCAC |
| 595a18r102140 | aggcagaatcgactcaccgctaTATTTGCCGAATTTCAACCA |
| 595a18f114741 | tccgtcttagctgagtggcgtaACTCTATGCGGTGGATGACC |
| 595a18r114935 | aggcagaatcgactcaccgctaGAAAGGTCTGAGCCAGTTGC |
| 291b3f26828 | tccgtcttagctgagtggcgtaTCCTACAAACGCGATTAGCC |
| 291b3r27044 | aggcagaatcgactcaccgctaACCCTGGTCCCGAGAGTAGT |
| 291B3F64796 | tccgtcttagctgagtggcgtaACTTGCCCCACTCTGTATGG |
| 291B3R65011 | aggcagaatcgactcaccgctaTGGGCTCATATGCATGTTGT |
| J127B14-F | TACAAGAGGTCAACGGAACATGGG |
| J127B14-R | GCCTAATTCATTCCAGGGACAGTG |
| U157D4-F | AACAGGCATGGTTCCTCTCACTTC |
| U157D4-R | TTGTTTCAGGACCCACCCACTATC |
| 237H1-F2 | GACAGAAACGCAGCTCTGATAATC |
| 237H1-R2 | TCATGGATGCAACTTTAGTTCCAG |
| 237H1-F3 | CAGAGGCTGCTCCTTTTGTGAGTG |
| 237H1-R3 | AGGTTGAACTGGCTCGGGAG |
| 237H1-F4 | TACAAACTGAATTCACCACGCCC |
| 237H1-R4 | TGTGTGAGAACTCTGTCCTGAGAG |
| 235H3-F3 | ATGACTGGGAAATCACCTGGCG |
| 235H3-R3 | GTGGCTGAAATGCTGGTTGC |
| 235H3-F2 | GATCCACTTCCTGGGTATTAAATG |
| 235H3-R2 | GCAAAATTGACCCTATCACTCTGC |
| 235H3-F | CTGTTGATAGGGAGATAACAAGGG |
| 235H3-R | GTTCTTTCCAGATCAGCAGCTAAC |
| U61F10-F | CATTCGGTCACTAAAAGTGCTGGG |
| U61F10-R | ATTCTGCTGTGAGTTTGGGAACCAC |
| 198P4-F | GCAAACCTCAGCAAGATTCAAAGC |
| 198P4-R | ACAGAATCAAGTGAGGAGAACTGC |
| 198P4-F2 | AGGCACACAGTATTCACCAGCATC |
| 198P4-R2 | GCGGAATCCTGAAAAATGAGTAGC |
| 198P4-F3 | GGTAGTCTAAAAACAAATCCTGGC |
| 198P4-R3 | TATTATGAGGCTTGTCCCTGTGTC |
| 221F2-F | CATGATCTTGTGTCAGGTCAAATC |
| 221F2-R | ACTACTTGCTGTGTTCCTTCTCTC |
| 221F2-F2 | CCCCAGAAGCATTTGCTTAATGAG |
| 221F2-R2 | GCAGACATGGTTTTGCATGATAAAG |
| 101D3-F6 | GTCCACTATGACTGAAGTGAAGAG |
| 101D3-R6 | CAAAGAATGGTGGTGATGTAATGG |
| 101D3-F5 | CCTCAGACGAGGAATTGCCAAAAG |
| 101D3-R5 | CGGAACGACACTCCATCTCAAAAC |
| 101D3-F4 | CTCTCTTGTTGTTGGAAATAGGAC |
| 101D3-R4 | GAACAAGAGCTGTGAATCTCTAAG |
| 101D3-F3 | ACAGCTTGACATACTCATAGCCTC |
| 101D3-R3 | GTCGTGGAAATAGAACCATTGTCC |
| 101D3-F | AACTTCATTAGGAGAAGCCAGAGG |
| 101D3-R | ACATAGCATGAGATGGTAGACACC |
| J635G19-F | GTGGGACATCATCTGCGAAAGTAG |
| J635G19-R | GGGAGTAGACACTGCAATGGTTTG |
| U177E8-F | CTCAAGCCTGATGGTGAGATACAG |
| U177E8-R | ACTAAGGTTTCACTTGGAGGCTTC |
| U177E8-F2 | GAACACATGTGAAGTCTACACCAC |
| U177E8-R2 | AGAAGTTAGGCTGAGTTTGCTGAG |
| 79P11-F2 | TATTATCAAGGAGGTGTTGTCTGG |
| 79P11-R2 | ACAAACATTGCTCAAGAGTCCCAC |
| 79P11-F3 | GCCTTAATGGATAGGACCACAGC |
| 79P11-R3 | ATAAAATGGGACCCCATGCACTC |
| 79P11-F5 | GATCTTTGTCCTTAACGTGCAGTG |
| 79P11-R5 | CAGATCAAGAGTGATAGCTGGAAG |
| 79P11-F4 | AACCAAGGCACACTCTCTAAGCTC |
| 79P11-R4 | GAGAGCAGAGCATAACATTCCCTC |
| 79P11-F | ACCATCAGGATTCAGGGCATAAGG |
| 79P11-R | TGCAGTATAGATGGGACATAATGC |
| 79P11-F8 | GTGACAGATTCAGCCATCAGAGTG |
| 79P11-R8 | CAGTCATCTTTGAGTGAGAGAATAG |
| U105G4-F8 | TCCCCTAATACTCCAAATGGCTTC |
| U105G4-R8 | TATACCTGGGCTTCTTAGCAGCTC |
| U105G4-F10 | GGATCTTCCCTATTAGTTCCTTAGG |
| U105G4-R10 | GAAAGATTAACTTACTGATAAGGCAAG |
| U105G4-F9 | GCAAAGTCTTCAACACCCGAATCC |
| U105G4-R9 | AACAGTAGAGCATCATTGTCACAG |
| U105G4-F2 | CAGGTTGGAGTGAAGGAACCACTG |
| U105G4-R2 | AATTGGAGAAAGATGAGGAGAAGG |
| U105G4-F3 | CACAAACATCTCCAGTTGAGAACG |
| U105G4-R3 | TCTACTTTATCCCACCTGACTTGC |
| U105G4-F5 | GGAATCTATTGGCACATTTCTCCC |
| U105G4-R5 | TAGCATAAGGACTTGAGAGGTCAC |
| U105G4-F6 | TATGCTTTGGGGTACATCTGTTGC |
| U105G4-R6 | TACTTTTGGAAGTTGCCACCTATG |
| U105G4-F7 | ACACTTCTGCTGACAAAGGATTTG |
| U105G4-R7 | GGGACCACCTCATTCATTATCACC |
| U105G4-F4 | ACAGCAACGTCACAGGTGAGTTG |
| U105G4R4 | GCTGCCTCTTCCTGATGATTAGAG |
| U105G4-F | ACGTAGATCTCATGTGACCAGGAG |
| U105G4-R | CTCCCACAAGTCTCTGCCTTAAAC |
| 349O20-F | TTTGCCTTATGCCTTGACTCCTGC |
| 349O20-R | GCTGACAGAAACTTACACTGAGTG |
| 349O20-F4 | GTTGGATGACTTCATGTTTTCACC |
| 349O20-R4 | GGGACATTAGCACATTGAGGTGTC |
| 349O20-F6 | CTGAATCCATGGTGTATGTTACAG |
| 349O20-R6 | CTGGCATTGAAGAAAATAGAACAG |
| 349O20-F7 | CTCTATACAAGTGTGTCATTGGGC |
| 349O20-R7 | GGTGGGCAAAGAATGTCTGTCTTC |
| 349O20-F5 | ACTGCTGGATCTGAAGAAAACCTG |
| 349O20-R5 | TTCAGATGTACTGATCCGCCTGTG |
| 349O20-F3 | AGTGTTATTCAGCCTACAGCACAG |
| 349O20-R3 | CCCTCACTCACTTGCTAGTATCTC |
| 349O20-F2 | TGATGTTCAGACTGGGGTCCTAATC |
| 349O20-R2 | TCTCCGTTTCTCTTCACTGTGTCAG |
| J823F3-F2 | GGGACATAGATTACTTGCCATGTG |
| J823F3-R2 | AGTCACTTCCTTGTGCAATCTGTC |
| 421I20-F | GTTGAAGTCTTGGTCCATGCTCAG |
| 421I20-R | TACTAGACCATCCAGACTTTGCAC |
| 421I20-F5 | ACTACCCAATTGTATCAGTGCCAC |
| 421I20-R5 | TGACCTTCAAATATGGAGTGGAAG |
| 421I20-F6 | GTGAGGTTGTACTTTTTTGTCCC |
| 421I20-R6 | GCTAGGAAATGAATAACACTGACG |
| 421I20-F2 | GGGGTATAGGCAAAACATCATCAC |
| 421I20-R2 | AGACTTTCATCGCTTCACTCTTGC |
| 421I20-F3 | TCCTTTCCACACTTCACTTTCTTG |
| 421I20-R3 | TAACTCCACGGGTTCAGTCATCTG |
| 421I20-F4 | GCCTTTCACTAGGTCATGTGTTTG |
| 421I20-R4 | GTCCTTTAGTTATTGTAATCTGGC |
| U250H12-F12 | ATCACCAGCCCTACCCCATAAGAC |
| U250H12-R12 | GTGGATTTCTGTGGATACTGACTGC |
| U250H12-F11 | GCTAAGGCAGGCATTGTGAAAAG |
| U250H12-R11 | ACTGGATTGGGGTCTACTCAGAGG |
| U250H12-F2 | ACTACCTTGATGAGTGTCAGTGTG |
| U250H12-R2 | AGGACTGGTTCTTTCAGGCTTCAC |
| U250H12-F* | ATTTCCCACAACATTCCTGGGAGG |
| U250H12-R* | GGTCAGCAGGAAAGCAAGATTTGC |
| U250H12-F5 | GCTGGAGCCAATCTCAGTTTCAC |
| U250H12-R5 | AGATAGCTGTGATGCCTCCTGATG |
| U250H12-F6 | ATAAGAATCCCATACGACAGCTGC |
| U250H12-R6 | ATGATTTCCCAAACTGTTCTCACC |
| U250H12-F3 | GCTTACAGGCATGATATTGAACAG |
| U250H12-R3 | TAAATGTTGGAACACAGCAGTAGG |
| U250H12-F10 | CATCGAACTCAAGGAATATCAGTC |
| U250H12-R10 | AAGACAGAGACACACAGGGAGAGC |
| U250H12-F9 | ACAGTGCCAGTGTTTGCTTCCTTC |
| U250H12-R9 | AGTGTAACTCACTACCATACCATC |
| U250H12-F4 | AAGGGTTGGATGAGTGCCTAATTC |
| U250H12-R4 | CTCTCGCTGGACAAAGATGAAGC |
| 43H13-F2 | ACCTTCTGACCAACTGTGTATTTC |
| 43H13-R2 | CATCAGCAGCTATGTGCTTTGAAG |
| 43H13-F | AGATGTGGATTTAGGGTCAACAGC |
| 43H13-R | CAAGCTCCTAACCTCCATCTGTC |
| 43H13-F4 | AATGTTCTCTACATTCTTGGGTGC |
| 43H13-R4 | TATACTGATGACTGTGGATGGGGC |
| 43H13-F5 | CTTAGCATCAGAATGAGGAACATC |
| 43H13-R5 | CATTGTCTGACAGCAAAATGTAAGG |
| 43H13-F3 | TTGAAAGTAGGCTCCTCTGTTTAG |
| 43H13-R3 | TGAGTGAGACTCTGACTCAACTTG |
| 246D9-F2 | GCCTACTAGATGCTACACATGTGG |
| 246D9-R2 | CCCTCAAGTTGACCTCAGTCC |
| 246D9-F3 | GATATGACACAAACAGCAACATCC |
| 246D9-R3 | TATGTATTACGCTATGACACAGGG |
| 246D9-F4 | ATGCTAAGCAGACATCTGTGGCAG |
| 246D9-R4 | CAACCTAAAGATAGGATTGTCAGG |
| 246D9-F | GTTGGTATCAAATGGGCATATCAC |
| 246D9-R | GAACTCATGTGGCTCCTGAACTG |
| 857G6-F | ATCCTGTGAGTTGTATGTGTGGTG |
| 857G6-R | AAGCTCTGAGATGTAGAGAGCAGG |
| U25D11-F | TGACCATACCCAGCTGTTTAGCTC |
| U25D11-R | GTGGTGAAAGGCAATTGAGAGGAC |
| 1055C14-F | AGAAAATGAAGAAGAGCCGCAGAG |
| 1055C14-R | CTCAAACAGGTCCTTCCTGGAAAG |
| 1055C14-F15 | AGATATTTCCATTCTGTTACCCTG |
| 1055C14-R15 | TGATATACAAAGGAAGTTCAAGGC |
| 1055C14-F14 | TCTTTAACTCTCCTCAAATCACCC |
| 1055C14-R14 | GCAAGTAATTTGTAGTGTGCCTTTC |
| 1055C14-F2 | ATAAACCTGGGACATTGGAGCATC |
| 1055C14-R2 | AGATGCAATTCTGGAGGAGTATGC |
| 1055C14-F11 | ACATGACACCACTGTCAGTGATTG |
| 1055C14-R11 | CTGTACATCTCACACACAATAGGC |
| 1055C14-F7 | GAACTGACAATCTTCGTTCACCTC |
| 1055C14-R7 | GATTTGAAGGCATACATGCTCACTC |
| 1055C14-F4 | GGAGGCTTCTTTCAAGTTCTCTAG |
| 1055C14-R4 | TTGCTTGGAGATCAGGACGCAAAG |
| 1055C14-F10 | GTTGAAGTTATCAGGGTGGTGAGG |
| 1055C14-R10 | TGCCAGGAATTATCCAGGTTCCAC |
| 1055C14-F6 | GACCTTGGGATGCCAGATTCACAC |
| 1055C14-R6 | AGGCATCCTCTTGAAGCAGTGCTC |
| 1055C14-F5 | CATCATACATGGTATCAGTGACAC |
| 1055C14-R5 | GTATGAACTGTCTTCACCCAAGGC |
| 1055C14-F3 | CTGGTGGATATCTTCTGACCATAG |
| 1055C14-R3 | TTCTATTTCCGTGGCTATGGCTTG |
| U35G3-F7 | TGCTAGTATTTCCTGAAGAGAGTC |
| U35G3-R7 | TGCAAGGGAACTTGCTTGCCATAG |
| U35G3-F | GATGCCAAATTGAGCCACTTCTC |
| U35G3-R | GTGTCTGACGATGATGTTGCTGAC |
| U35G3-F2 | TTAGACTCTGGGACTCAGGTACTC |
| U35G3-R2 | GAATGTTGGAAAGGAAGGACTCAG |
| U35G3-F3 | GAAGATGGCCAGTTCTAGGGATAG |
| U35G3-R3 | TCTTGGGACCACTGCAAGGGAATG |
| U35G3-F4 | TGTGAAGTTGGCTTCACTTGTGTC |
| U35G3-R4 | ACCAGCCTGTTAGCTTCTTGTCTG |
| U35G3-F5 | GGTTTCTTTCCATGGAACTTGGGC |
| U35G3-R5 | GGGTAAGAGCATAAGAATAACAGC |
| U35G3-F6 | TCTGGTCCTTGTGCATTCAACTGG |
| U35G3-R6 | CTACTGAGGACCTCTTTAGAAGTC |
| 764D10-F | TTCTGACAACTGACTGGATGACTC |
| 764D10-R | TCTGTGTGAACTCTGGCAGTTATC |
| 764D10-F2 | TAGAGACTGTATCATCAGGGCTGC |
| 764D10-R2 | AGGAGGACAGTAGGATCATTGGTG |
| V698D2-F2 | GCAGAGAAGAAGGTGTGTCCATAG |
| V698D2-R2 | TCCGAAATACTGGATGGGTCATAC |
| Prom 6F | AAGGGAGACAGGACTGACAAGGAG |
| Prom 5R | AGTGAGGGTGGAGAGAAGAAACG |
| PLP 5F | GCTTACCCTGCTTGCTTTTTGTGTC |
| PLP 5R | CGGCTAATTCAAAATCCAGCAAAGGG |
| PLP 7F2 | GATGCCTCAGAGACATCGGACTTC |
| PLP 7R | TTAGAGAAAGGGGGATTTCTACGGG |
| PLP 3UTF | AACATAACAAATAACCAACCCTTC |
| PLP 3UTR2 | AAGGTACGATTATTAAG |
| V698D2-F3 | AAGGGGTAGGCAGGGAGACAGG |
| V698D2-R3 | GCTGGGTGACTTTCCACATTGG |
| V698D2-F4 | CCTGCCAAAAACTCCATTGAGAG |
| V698D2-R4 | CCTGATGCTCCTTGTTCTAAGACC |
| V698D2-F6 | CATCTTCCTGATTTATGCCAGACC |
| V698D2-R6 | AGAAATATGTCCCCTGTAGCCTTC |
| V698D2-F | TGAGATCCTGGAACTGACAACCTG |
| V698D2-R | CCACTTACCAGCCTGTAACTTTGG |
| V698D2-F9 | CTGAGCTCTTTGCACTTCCATATC |
| V698D2-R9 | CCTAAATAAACGGAAAGGTGTTCTG |
| V698D2-F8 | AAGCCTGCCTTATCTTCCACATGC |
| V698D2-R8 | AAAGGTAGGACTGACAGAACTAGG |
| V698D2-F7 | TTTGGGCTCTTGCAATGTCTAGGG |
| V698D2-R7 | GATGGCATGGCTCTTTGTCCTAAG |
| 540A13-F5 | AAGAAGGGATTGGATCTAAGCATC |
| 540A13-R5 | ACAGTTCTTGTGATATCATTGGCG |
| 540A13-F3 | TTAGTCTATGGCGGAACACTAACC |
| 540A13-R3 | GCATGATGTCAAATGACTGGTTTG |
| 540A13-F4 | GTGCTGCATTTGAAGCATGGTCTG |
| 540A13-R4 | TATGGACACAAATCTTTCCACAGC |
| 540A13-F | TAACAGCACGAAGACCCTGCTTTG |
| 540A13-R | AGACAAAGAGGATAGGCAAGTGAC |
| V461C10-F | CCTAGAGACTTGCTTTGCTTGGTC |
| V461C10-R | AGCACCAGCCCAAATAAAGTAGG |
| V461C10-F4 | GAAGTGGATACAGGGGCAATTGG |
| V461C10-R4 | GGATCCTGATTTAGCAGGTCAATG |
| V461C10-F8 | TAATCAGTCTTCTTGCCACACCTC |
| V461C10-R8 | CTGAAACTTTCGTATCTGCTTCCC |
| V461C10-F2 | CCTTAGTTCCCTACAGCACCTACC |
| V461C10-R2 | AAGGGAGTGGGTCTTTAATTCTGC |
| V461C10-F3 | CTGGAAGTCATTGGGGTAAAGTTC |
| V461C10-R3 | TCCTATAAACGACCTTAGAAAGCC |
| V461C10-F7 | GGTATTCAGTACACAAATTGAACTG |
| V461C10-R7 | GAAATATTAGCTCACTAGCTGTTGC |
| V461C10-F6 | TCAGCCTCGGTCAAGCAGTAG |
| V461C10-R6 | CATGACAGCCTCCTTGGTAAGTAG |
| 540A13-F2 | GAACCATAGTCTTTGCTTGGCAGC |
| 540A13-R2 | TGACGCTTTCTTACACAGGAGTGC |
| 540A13-F7 | CTCCAAAGTTCTGTTCAAACCCC |
| 540A13-R7 | GATGAATGATTCTCCTGCACTGC |
| 370B6-F | AAGGAATCTATGACATCCACTGCC |
| 370B6-R | CTTCCTCTGAGATGACCTCCAAC |
| 370B6-F2 | ATTGGGGTACACAAATCCTACTTG |
| 370B6-R2 | CTATCTGGGCTGTTCCTGCTG |
| U116E7-F3 | TGTGGAACACCCTCAGTAGACAGC |
| U116E7-R3 | TCACCAAGAAGAACCAGCGG |
| U116E7-F2 | TTCCACCACGGACAGAAGTTGC |
| U116E7-R2 | TTATGAAGCCAGGGGATGTGC |
| U116E7-F | GCCACTTGCTTCCTTCAAAAGC |
| U116E7-R | CTGGAGACTCAGACAAAAACAGGTG |
| V362H12-F | TTCAGCATTGCCTGACAGTCTTGG |
| V362H12-R | ACCTACACAAAGGTCATCCATGGC |
| V362H12-F5 | CAGTGGGATTACATGAAGTGAAAG |
| V362H12-R5 | CTGCAAGGAGCTGAGTGACTTTAC |
| V362H12-F3 | GGGTCTGTGAATCCTGGTTCAATC |
| V362H12-R3 | GGCTACCAGATGCAACTAACTAGG |
| V362H12-F6 | TCAGTGTACATAGGCAGAGATGAC |
| V362H12-R6 | AGACTCCAAAGGCAAGGCATTCTC |
| V362H12-F4* | ATCTTCTGGTTAATAGGTGCCATG |
| V362H12-R4* | TTTGCTGCAGAGATCTCAGTGTTG |
| V362H12-F2* | GAGCACGGGAATGGTTAAATTTCC |
| V362H12-R2* | ACCTTCAAGGCATGGATACCTTAG |
| 839M11-F | CTTGCTAAGCTGCAGTGGGCTATG |
| 839M11-R | GTGGTGTGGCTAAGCAGGTTCTAC |
| 839M11-F4 | CACCCATTCACAGGCATTCACTAG |
| 839M11-R4 | TTGGGCACCTTAGGGTACAATGTG |
| 839M11-F3 | ATGGTAAGCTCTGTGACTGAACTC |
| 839M11-R3 | TTGTGAAGAAAAACGATGTGGCTC |
| 839M11-F2 | CAGTTTGAGGTATTTCCTGTGAGG |
| 839M11-R2 | TATCCCTTCCAGGAACTCGGTATC |
| U240C2-F | ACCTTCAACGCATGAATACGTAGG |
| U240C2-R | GGCATGGTTGAATTTGCTAACTGC |
| U240C2-F2 | GGCTAACTGTCACACCATGATGTG |
| U240C2-R2 | TGGGAACAAGAGGAAACTCTGGG |
| U240C2-F3* | ATGTGGCAGGCACTCGGCTG |
| U240C2-R3* | TCCTGCCAACCACCCTTTCG |
| 467E10-F2* | TGAGCACTTTGAAGGGTGTCACG |
| 467E10-R2* | ATCATAACTGAGGACAGGAGCCG |
| 467E10-F3* | TGGTAAAGTTGAGGACTTAATTGC |
| 467E10-R3* | ACACCAACCAAAATGAGTGTCG |
| 467E10-F | ATCCAGTGCAGAATTCAGGTCCCG |
| 467E10-R | GACCCAACACAGAAGCACCCAAAG |
| 46H11-F | CCCACCCTGATCTCCCTTGAATC |
| 46H11-R | AGGAATTATTGAGGGTGCATTGTG |
| 46H11-F3 | CCTTGATATCATGCAAAAGTCAGAC |
| 46H11-R3 | TCTATGCAGGCTTCTTGGACTCC |
| 46H11-F4 | AGAATGCTGAAGGTGCTGGGG |
| 46H11-R4 | TACCCTCCTCTTCTCTCCAAGAC |
| 46H11-F7 | ATCTACAGTGACAGGGACACAAAG |
| 46H11-R7 | TGGGAGTGATTTTGGAAGGAGTAG |
| 46H11-F6 | GGAACCAGAGAGTCCAAGCTG |
| 46H11-F5 | CATTAGAGGCATCAGTTGGAACAC |
| 46H11-R5 | CTTGGTAGGGATGGAATCACATAG |
| 46H11-F2 | CAGCAGTGCTGACTGTAACTGC |
| 46H11-R2 | ACCATCATTCTTATACCCAGAACC |
| 233G16-F | TAACTCCAGCCTGGGTTGAGTCAG |
| 233G16-R | TTCTGCTGGACAGACTGATCTTGC |
| 233G16-F2 | GTGGTGTAAATCCTACTCCAGAGC |
| 233G16-R2 | TGAGCTGAGCATGATGAGTATTAG |
| 233G16-F3 | AGCATAAATCAGATAGGAGTCCTG |
| 233G16-R3 | AACACATTACTAAGGCAGGTCTG |
| J513M9-F4 | GGCTATGGCAGTTTTGTCCAGTTC |
| J513M9-R4 | AGCTCATCAGAATCTGGATACCGC |
| J513M9-F3 | GATACCCAAATGTCATTGGCAAAAC |
| J513M9-R3 | GCTGAATTGCTCCTCCATACATAG |
| J513M9-F | ACTTGGCTCTTCTGTTCTGAAACC |
| J513M9-R | TGGCTTTGGTTATGTTTAGGCAGG |
| J513M9-F2 | AGGAACCTGCAGACAATCTCTGAG |
| J513M9-R2 | CCCTGATGGGCTTTCTAAAGAATC |
| J513M9-F5 | AGTGCAAACTAGGAGAGGTGAGC |
| J513M9-R5 | CACACCATATCAGCTGGTAAGCTG |
| J513M9-F6 | TAATGGCAGTCGGATTGGACG |
| J513M9-R6 | AGATGTTATCTCATGCAATCTGGC |
| 541I12-F3 | CATACTGAGTAAAGGGCCTACAGC |
| 541I12-R3 | CCTAAAATATCCAGAATACCAGGG |
| 541I12-F5 | AGCTACAACGCTCAATTTCCTGTC |
| 541I12-R5 | AGCTACTCCACTAACCTGTCGCTG |
| 541I12-F4 | TATGGACAGGGAATTACATCTCTG |
| 541I12-R4 | CAAAAGGAAGACCTGAGTGAGAGG |
| 541I12-F | TACTTATCAGGTATTCCTCCCGTC |
| 541I12-R | TATTTGAATGCAGTAGAACAGTAG |
| 541I12-F2 | ACTGTTCTACTGCATTCAAATAAC |
| 541I12-R2 | GTTGAATGGTGTGACTCAGGTAAG |
| J519P24-F | CACCAGCACAATGAACCAATGCAG |
| J519P24-R | AATCTCCAATATCCCACCTCTCCC |
| U173H7-F | TGTCCTGCTTCTCTTAGTTGGGAG |
| U173H7-R | ACACAAGGCTAATAGACTCCTGCG |
| U139A4-F | AAGAGTAAAGTGAGCAGCAGGACC |
| U139A4-R | ATGGTAAGAGGACAGAGTGTGATGG |
| 107D4-F | GACTTGAACAGTAACTCAGCAAGG |
| 107D4-R | TGTCTTAGAAACCTGCTGACACTG |
| 248J6-F | GCAGCTACTGACATTTTGACTGAC |
| 248J6-R | GTGTACAGACTTACTGAGAGTTCC |
| 174H1-F | GAACTATAGAGCACAGCACTTCCG |
| 174H1-R | CAAACTCAATGACTCACTGTTCAG |
| 42H12-F | ACCAAAGCAGTTCACTCCACTAGC |
| 42H12-R | CTACACGCATCGTTTCACATACTC |
| 129B7-F | CGATGAGATGTCAGATAACAAGAG |
| 129B7-R | AGGCATTTGTCAATCCAAAACTGC |
| 129B7-F4 | TCTGTGTTTTGCATTGTGACCCAG |
| 129B7-R4 | AACAAGAACATAACTCTCCTCCAG |
| 129B7-F2 | TAAAGAGGCTGTCCAATGTCTGAC |
| 129B7-R2 | GTGTCATCTGAGAAGGAAACTATC |
| 129B7-F3 | CAACAGACTGAAGTGCAGGTAAAG |
| 129B7-R3 | AGGTTTGCTAAATGCTTTGATGTC |
| U201H11-F | GTCTGGTGTTAATAAGGTCAAGGC |
| U201H11-R | TAGGCACCCAATACATATGTGCTG |
| U92G6-F | TTTATTGCTTCACCACCTCCTCG |
| U92G6-R | TTCACAGACTGCTTGAGCCCTAAG |
| U19H10-F | GCCTGCAAAGATTGTTCGTAAGC |
| U19H10-R | ATTCTGTGTTCCCATCCACCAAGG |
| J3D11-F | ATTTCCTGAGCAAAGGTCCTTGGG |
| J3D11-R | AACCATTCCTAAGTGCTGCATCG |
| 1070B1-F | ACTCATGGGTCACTTTACTCATTG |
| 1070B1-R | GTCCTGTAATGTCAACATCACATC |
| 540N4-F | CAAGCTTGTGGGAATTCAATGAAG |
| 540N4-R | TAAACCAGTTAGAGATGCTATGGC |
| 540N4-F2 | CACATTTCTGTGAGGATACAGTATC |
| 540N4-R2 | AGTTTGTGCTAATGAGGCCCTCTG |
| 540N4-F3 | TTGAATTGCTGTGACAGCCTCCG |
| 540N4-R3 | GCGTTTTCTGACAGCAAGTTTGGG |
| 540N4-F4 | ATCTCTTATGAAGCAGAAGCCCAC |
| 540N4-R4 | TACCTACGCAAAGTCACAGAGCAG |
| 540N4-F5 | AATGCCTTTCTGGGAGAGGAACTC |
| 540N4-R5 | TAAGTCTATGTGGTTTCAAAGGGC |
| 540N4-F6 | CCAGTTAGAAAGACTTAGCCATAC |
| 540N4-R6 | CAGAACAGAGCACACTGAGAGTAG |
| 540N4-F7 | CTCTTTGCTCCTGCTCCTCC |
| 540N4-R7 | GTGACAACACACTTAGAAGCTCTG |
| 364K23-F | CAGTAGGACACCATCCTTCTGTTC |
| 364K23-R | TGTCCACTGAAACGTGAAGTCCTG |
| 820B18-F | AAAGACCCTGCTTTGGAGCTGAGC |
| 820B18-R | AGCATCAGCTCTGTTATGGCATTC |
| A191P20-F2 | CTCACCCTTTCCCTAAATTGGATC |
| A191P20-R2 | ACCAGGTACGACATCATTGTGTAC |
| A191P20-F | GACCATAGAGTGCTTTCAACCACC |
| A191P20-R | TCAAAGGAGTTTACTGGGAAGGG |
| hDys23 F | ATTGAGGGACGCTGGAAGAAGC |
| hDys23 R | GGAATAAGCAAATCGCCATCC |
| hDys26 F | TATGCCAGAAAGGAGGCCTTGAAG |
| hDys23 R | CAACCTCTTCTCTTAGAACCAGG |
| HCFTR-4F | TAGGAAGTCACCAAAGCAGTACAGC |
| HCFTR-4R | AGCTATTCTCATCTGCATTCCAATG |
Primer pairs known to be an exact match to more than one location in the reference genome sequence are indicated by an asterisk (*).
The universal tag sequence is indicated by lowercase letters.
The multiplex PCR approach used in the duPont Hospital laboratory for all breakpoint mapping was amplification of a region in or near PLP1 with a region of the dystrophin gene on Xp (hDys26F and hDys26R or hDys23F labeled with fluorescent dye and hDys23R). The PCR reactions were performed with 50 ng genomic DNA in a 25-μl reaction by adding Taq Buffer (5× Taq Buffer is 83 mM Tris-HCl [pH 8.8], 850 μg/mL BSA, 83 mM (NH4)2SO4, 33.5 mM MgCl2, 34 mM EDTA, and 50 mM β-mercaptoethanol), 5% dimethyl sulfoxide, 12.5 pmol of each primer, 1.5 mM dNTPs, and 0.625 U AmpliTaq (Applied Biosystems). The conditions were as follows: melting at 94°C for 5 min; 25 cycles at 94°C for 30 s, 55°C for 30 s, and 65°C for 1 min; and a final extension at 65°C for 6 min. Detection and quantitation of products amplified with unlabeled primers used ethidium bromide–stained 4% NuSieve 3:1 agarose gels (Cambrex BioScience) and an Eagle Eye II gel documentation system with Eagle Sight software (Stratagene). Alternatively, labeled primers were used, allowing detection by capillary electrophoresis on an ABI Prism 310 Genetic Analyzer with Genescan software (Applied Biosystems). For a male patient, if the region being tested is duplicated, the expected normalized ratio of the region to the dystrophin control is 2, and, if it is not duplicated, the expected ratio is 1.
A third multiplex PCR approach was performed in the duPont Hospital laboratory to validate use of the dystrophin internal control primers in the previous approach and to determine whether the mapping approaches used in the two laboratories gave reproducible results. The Multiplex PCR Kit (Qiagen) was used in accordance with the manufacturer’s instructions for multiplex amplification with unlabeled primers for a region of the PLP1 gene (PLP7F2 and PLP7R), a region of dystrophin (hDys26F and hDys26R), a region of the cystic fibrosis transmembrane receptor gene on chromosome 7 (HCFTR-4F and HCFTR-4R), two regions proximal to PLP1 that are expected to be duplicated in some patients and not in others (421I20-F5 and 421I20-R5; 1055C14-F7 and 1055C14-R7), and two distal regions (540A13-F3 and 540A13-R3; 461C10-F6 and 461C10-R6). Detection and quantitation of products used ethidium bromide–stained 4% NuSieve 3:1 agarose gels (Cambrex BioScience) and a Gel Logic 440 Imaging System with accompanying molecular imaging software (Eastman Kodak).
Amplification and Sequencing of Duplication Junctions
Long-range PCR (LR-PCR) by the Expand High Fidelity PCR System (Roche) was used to amplify junction breakpoints in 11 patients whose proximal and distal breakpoints were mapped by either multiplex PCR or fiber FISH. A forward primer from the distal region and a reverse primer from the proximal region were used to generate a junction product in the patient that was not present in normal control individuals. For breakpoints mapped to within 2–9 kb by multiplex PCR, LR-PCR was performed using proximal and distal primers deduced to be within the duplication. For breakpoints mapped by FISH, several primers were designed from both the proximal and the distal breakpoint regions identified, and these were used in different combinations until a unique product was generated.
LR-PCR was unsuccessful for patients P026 and P134; therefore, a standard protocol was used for inverse PCR (IPCR) (Triglia et al. 1988). This required restriction digestion of genomic DNA with PstI and XbaI for P026 and P134, respectively; DNA ligation at low concentrations; and then PCR amplification using primers designed in a divergent direction near the proximal duplication breakpoint. The LR-PCR and IPCR products were excised from 1% agarose gels and were purified using a gel extraction kit (Qiagen).
Primers used to amplify across the junction by LR-PCR or IPCR are listed in table 2. These fragments were gel purified, and nested primers were used at one or both ends to generate smaller fragments and sequences across the junction. DNA sequencing was performed using the ABI Prism BigDye v3.1 terminator kit and an ABI 377 DNA sequencer (Applied Biosystems).
Table 2.
Primer Used for Amplification of Duplication Junction Fragments by LR-PCR in 13 Patients with PMD
| Patient andPrimer Name | GenBank Accession Number of Clonea | Position in Clone(bp) | Sequence | FragmentSize(kb) |
| P348: | ||||
| 1055C14-R8 | AL049610 | 73504–73481 | AACAGACTGACTGCTACCTTCCAG | |
| V698D2-F5 | Z73964 | 28066–28089 | TGCAGTATGACATTGGCAGTGTTC | 4 |
| P116: | ||||
| 79P11-R5 | AL133348 | 18628–18605 | CAGATCAAGAGTGATAGCTGGAAG | |
| V698D2-F11 | Z73964 | 32874–32896 | CCAATAACAATGTATGAAGGTTG | 5 |
| P015: | ||||
| 1055C14-R11 | AL049610 | 59957–59934 | CTGTACATCTCACACACAATAGGC | |
| V461C10-F4 | Z75896 | 5427–5449 | GAAGTGGATACAGGGGCAATTGG | 6 |
| P224: | ||||
| 421I20-R2 | AL117327 | 32934–32911 | AGACTTTCATCGCTTCACTCTTGC | |
| V461C10-F8 | Z75896 | 6453–6476 | TAATCAGTCTTCTTGCCACACCTC | 3.5 |
| P026: | ||||
| 235H3-F3 | Z93943 | 14075–14096 | ATGACTGGGAAATCACCTGGCG | |
| 235H3-R4 | Z93943 | 13907–13884 | CCTTCAACCAGATGTTTGTGTTCC | ∼2 |
| P083/PMD38: | ||||
| U250H12-F3 | Z69733 | 13198–13221 | GCTTACAGGCATGATATTGAACAG | |
| 540A13-F5 | AL139228 | 899–922 | AAGAAGGGATTGGATCTAAGCATC | 2.5 |
| P134: | ||||
| 349O20-F8 | AL606763 | 35094–35116 | TGAGTCAGTGCCATTTCTATGCG | |
| 349O20-R5 | AL606763 | 34589–34566 | TTCAGATGTACTGATCCGCCTGTG | 3 |
| PMD9: | ||||
| Z81014F14986 | Z81014 | 14986–15015 | TCTGCCTTAATTTCATTATTTACCCAAGTG | |
| AL034405F10069 | AL034485 | 10069–10098 | AGAGAAAGCTGTACGCAATAAAGACATAGG | 3 |
| P255: | ||||
| 1055C14-R10 | AL049610 | 72119–72096 | TGCCAGGAATTATCCAGGTTCCAC | |
| 467E10-F2 | Z74620 | 113–135 | TGAGCACTTTGAAGGGTGTCACG | ∼8 |
| PMD24: | ||||
| R2c105G4 | Z92846 | 31801–31772 | AAATATTCACCAAATTCATAGGTGAACGAG | |
| F8233G16 | AL135959 | 41010–41039 | GAACTACTATACCCACCCACATTTTCAGAG | 3.5 |
| P114: | ||||
| U105G4-R4 | Z92846 | 29781–29758 | GCTGCCTCTTCCTGATGATTAGAG | |
| J513M9-F6 | AL049631 | 116778–116798 | TAATGGCAGTCGGATTGGACG | 7 |
| P176/PMD7: | ||||
| 43H13-R3 | AL035444 | 21234–21211 | TGAGTGAGACTCTGACTCAACTTG | |
| 129B7-F | Z81144 | 1771–1794 | CGATGAGATGTCAGATAACAAGAG | 4 |
| P110: | ||||
| 101D3-F5 | Z85997 | 33396–33419 | CCTCAGACGAGGAATTGCCAAAAG | |
| 540N4-F5 | AL772400 | 62413–62436 | AATGCCTTTCTGGGAGAGGAACTC | ∼9 |
NCBI Web site.
Bioinformatics and Sequence Analysis of the Duplication Breakpoint Regions
Information available from Ensembl, the UCSC Genome Browser, and the NCBI Web site was used for selection of FISH clones, design of PCR primers, and identification of genes in duplicated regions. Information presented is from Ensembl v29.35b, based on NCBI build 35. Data from our FISH studies, multiplex PCR studies, and junction analyses were compiled in a Microsoft Access database, which was used to format the data for uploading and displaying in Ensembl.
Various Web-based sequence-analysis programs were used to investigate the 13 tandem duplication events (see Web Resources). By use of BLASTn (Altschul et al. 1990), with the recombinant junction sequence as the query sequence, the exact positions of the proximal and distal breakpoints were confirmed on Xq22 and within specific genomic clones. The breakpoints represent the boundaries between single-copy and duplicated DNA sequences, and the genomic segments surrounding each breakpoint were investigated and termed “breakpoint regions.” Reference genomic sequences were obtained from Ensembl and UCSC on the basis of breakpoint position. CLUSTALW was used to align the abnormal recombinant junction sequence determined from each duplication event, with the reference genomic sequence from both the proximal and distal breakpoint regions. BLASTn was also used for analysis of sequence homologies, with the use of genomic segments ∼70 bp in size and centered on the DNA breakpoints. Pairwise sequence comparisons of the 2-kb proximal and distal breakpoint regions for each patient were performed using BLAST2 with default parameters (Tatusova and Madden 1999). These 2-kb sequences were also analyzed for repetitive elements and low-complexity sequences by use of RepeatMasker (RepBase database version 7.4). Refined computer-based sequence analysis included the screening for tandem and palindromic inverted repeats in 300-bp breakpoint regions by use of Mreps and Palindrome, respectively. Purine and pyrimidine repeats ⩾10 bp, which equates to roughly one helical turn of DNA (Ussery et al. 2002), were identified, and only perfect uninterrupted tracts were included. We used Mfold version 3.1 (Zuker 2003) for prediction of secondary structures, analyzing 30 bp across each proximal and distal breakpoint and comparing those sequences with the 30-bp recombinant junction sequence. Finally, we used DNA Pattern Find to search for 36 specific sequence motifs elsewhere implicated in DNA rearrangements (Abeysinghe et al. 2003). We compared the results obtained from the 30-bp breakpoint regions (i.e., 15 bp on each side of the breakpoint) with control DNA sequences the same size found 150 bp proximal to each respective breakpoint. The close proximity was chosen to minimize variation in sequence properties due to genomic location.
LCR segments of DNA in a 2-Mb region around the PLP1 gene were investigated using PipMaker (Schwartz et al. 2000). The boundaries of the various repeat elements identified were determined using BLAST, CLUSTALW, and RepeatMasker.
Results
Characterization of 59 Duplication Events on Xq22 and Mapping Breakpoints
The extent of the duplications in 59 patients with PMD was determined by interphase FISH and multiplex PCR using clones and primers throughout a 7.4-Mb region including the PLP1 gene. The combined mapping data for each patient were displayed in Ensembl (fig. 1). Of the 59 duplication events studied, 38 were inherited from carrier female mothers, 20 had unknown inheritance patterns, and 1 was shown by FISH to be a de novo event in the patient (K.J.W. and M.C., unpublished data).
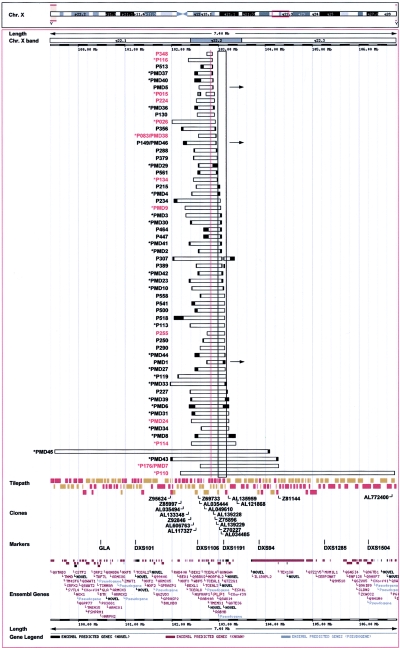
Figure 1.
Breakpoint analysis in 59 patients with PLP1 duplications. Duplication mapping information gained by FISH, multiplex PCR, LR-PCR, and IPCR and uploaded into Ensembl is shown. Horizontal bars indicate duplicated regions in each patient; the unblackened region is duplicated, and breakpoints are located within the blackened ends. The resolution of breakpoint mapping varies among patients because of the different methods used (FISH vs. PCR) and because of the number of PCR primer pairs analyzed in each case. The location of PLP1, which lies at position 102837923–102853693 bp (102.8 Mb) on the X chromosome (Ensembl v29.35b; NCBI build 35), is indicated by a red vertical line. Breakpoint junctions have been sequenced for the 13 patients listed in red (see fig. 2). A block of LCRs distal to PLP1 is indicated by a black-outlined vertical rectangle. Duplications mapped by metaphase FISH within Xq22 are indicated by an asterisk (*). Duplications mapping to other regions are indicated by a horizontal arrow. The location of relevant clones, markers, and Ensembl genes is indicated at the bottom. Patients indicated by “PMD” and a number or by “P” and a number were studied at the Institute of Child Health or the Alfred I. duPont Hospital for Children, respectively. The duplications of three patients—P176/PMD7, P083/PMD38, and P149/PMD46—were mapped by both groups, and the mapping information generated was in agreement, providing evidence that methods in the respective laboratories gave equivalent results. Multiplex PCR amplification performed on DNA from 49 of the patients, by use of primers specific to two proximal and two distal regions flanking PLP1 along with primers for an autosomal region and an X-chromosomal region, verified the reproducibility of the mapping methods used by the two laboratories and validated the use of an X-chromosomal control region in mapping. Families reported elsewhere include PMD1 (Woodward et al. 1998, 2000, 2003; Hodes et al. 2000); PMD2 (Ellis and Malcolm 1994; Harding et al. 1995; Woodward et al. 1998, 2000); PMD9 (Ellis and Malcolm 1994; Woodward et al. 1998, 2000); PMD5 (Hodes et al. 2000; Woodward et al. 2000); PMD3, PMD4, PMD6, and P176/PMD7 (Woodward et al. 2000); PMD24 (Woodward and Malcolm 1999); PMD27 (Sistermans et al. 1998); and P116 (Watanabe et al. 1973; Wilkus and Farrell 1976). In four cases—PMD27, PMD31, PMD6, and PMD8—the extent of the duplication was determined in a female carrier because there was insufficient material from the male patient with the duplication.
Metaphase FISH for 39 patients, with the use of PLP1 and flanking probes, showed that the majority (36/39 cases) had duplicated regions within Xq22. Three patients had a PLP1 duplication located elsewhere in the genome: Xq26 in patient PMD1 (Woodward et al. 2003), Xp22 in PMD5 (Hodes et al. 2000), and the Y chromosome in P149/PMD46 (K.J.W. and M.C., unpublished data). Patient P015 had an additional duplicated region that was proximal to the PLP1 duplication, and patients P307 and P389 had an additional distal duplication. Breakpoint locations for these additional duplications are shown in figure 1.
The duplications including PLP1 ranged in size from 100 kb to 4.6 Mb, with variability at both the proximal and distal breakpoints. Patients PMD45 and P110 had the largest duplications. The duplication in patient PMD45 extended the farthest proximal to PLP1, whereas the duplication in patient P110 extended the farthest distal to PLP1. Interestingly, we found that the duplicated genomic segments in each of the 59 patients were unique. Clustering of the distal duplication breakpoint was apparent. Most patients had breakpoints within a 430-kb region distal to PLP1, and approximately one-half of the patients had a DNA break within a 200-kb genomic segment of distal LCR sequences (fig. 1). The exceptions were patients PMD45, PMD43, P176/PMD7, and P110, who had the largest duplications. A less clustered distribution of proximal breakpoints was observed to lie within a 750-kb region proximal to PLP1. All but one patient (PMD45) had the proximal breakpoint in this region.
In four families examined, the extent of the duplication determined in the patient was the same as that found in a female carrier, suggesting that the duplication had been stably inherited. This justified the assumption about carriers PMD27, PMD6, and PMD8 that the duplication would be identical to that inherited by the patients. However, the possibility of an alteration in the configuration of the rearrangement during meiosis cannot be excluded.
Fine Mapping of Breakpoints and Sequencing of Duplication Junctions
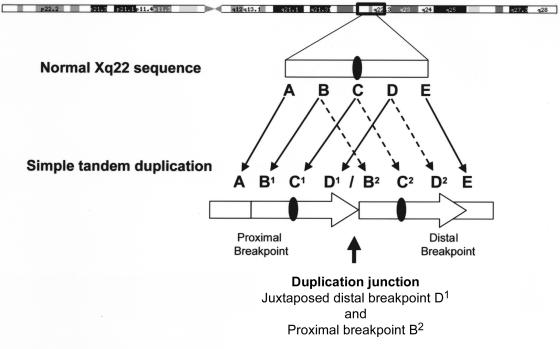
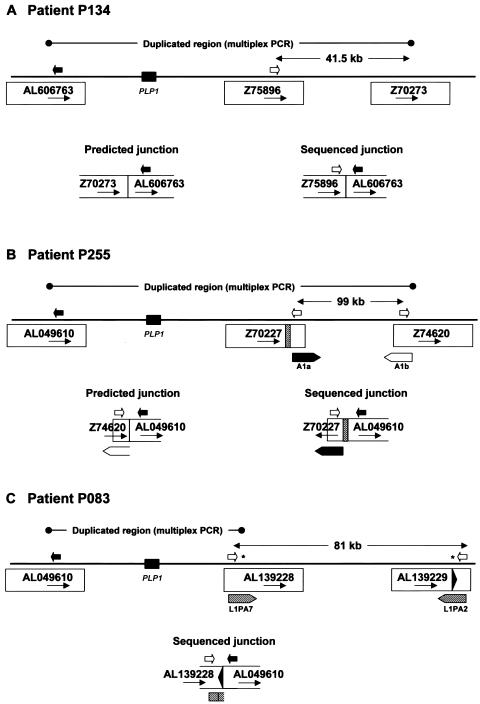
Because it had been demonstrated previously that most PMD duplications are head-to-tail in orientation, we began fine mapping of proximal and distal breakpoints—that is, the regions between single-copy and duplicated sequence (the boundary A/B1 and the boundary D2/E in fig. 2)—with the aim to PCR amplify and sequence across the abnormal junctions formed by the tandem duplications (the boundary D1/B2 in fig. 2). Note that, at these junctions, sequence on the centromeric side is from a more distal portion of the human genome reference sequence (D1 in fig. 2) than is sequence on the telomeric side of the junction (B2 in fig. 2), so that distal clones are abnormally juxtaposed to proximal ones. We mapped and sequenced 13 duplication junctions.
Figure 2.
Generalized duplication structure for tandem PLP1 duplications. Single-copy sequence proximal and distal to the duplication is shown as A and E, respectively. Double-copy sequence is shown as BCD and as an arrow. We have not distinguished between B1C1D1 and B2C2D2 in our patients; one represents the normal Xq22 sequence, and the other is the duplicated sequence. The PLP1 gene is shown as a blackened oval. At the junction, sequences 5′ of the distal duplication breakpoint become adjacent to sequences 3′ of the proximal duplication breakpoint, generating a structure not found in control individuals (region D1/B2). The boundaries of the duplicated and single-copy sequences have been designated as the breakpoint regions; the proximal breakpoint region is A/B1, and the distal breakpoint region is D2/E.
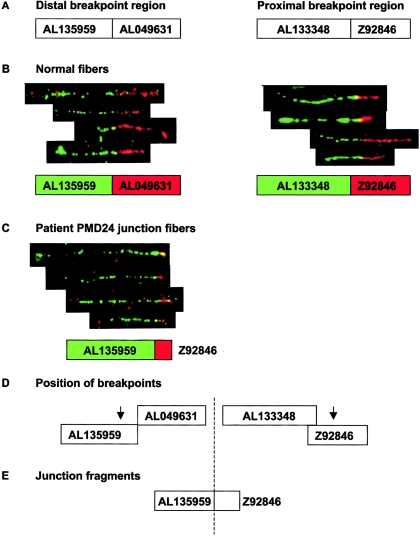
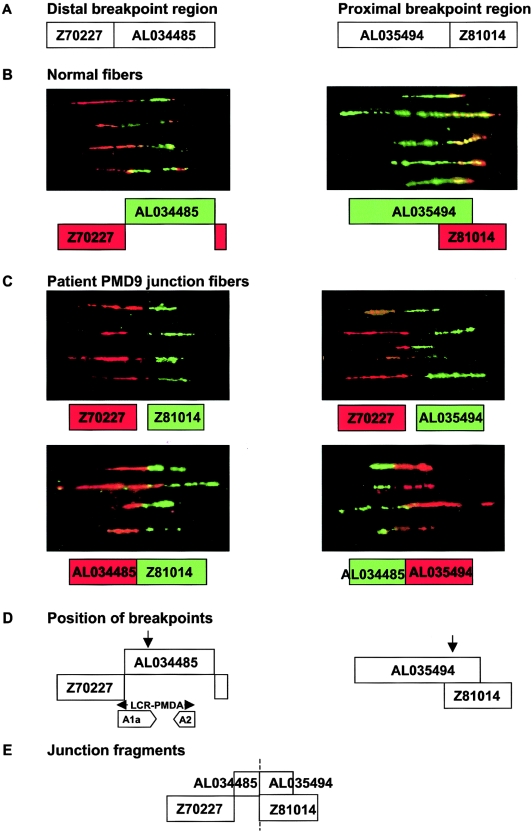
We used fiber FISH for high-resolution mapping of the breakpoints for patients PMD24 and PMD9 (figs. 3 and 4) and LR-PCR for amplification of the duplication junctions. For 11 patients, the proximal and distal breakpoint positions were refined by multiplex PCR and were amplified by LR-PCR or IPCR. The junction sequences obtained (fig. 5) were analyzed by BLASTn searches to confirm the precise breakpoint locations on Xq22 (table 3). Attempts to find junctions in patients P113, P119, and P379 by both LR-PCR and IPCR and in P130, PMD10, PMD34, PMD3, and P290 by LR-PCR failed despite equally well-mapped breakpoints, for unknown reasons. The remaining duplications were not mapped at a resolution high enough to attempt to identify junctions.
Figure 3.
Fiber FISH mapping of duplication breakpoints in patient PMD24. A, Relative sizes and positions of distal and proximal breakpoint clones used for fiber FISH, as shown in Ensembl. Interphase FISH had previously mapped the distal breakpoint within AL135959 and AL049631 and the proximal breakpoint within AL133348 and Z92846. B, Composite image of fiber FISH of normal cell lines, showing the relationship between distal and proximal breakpoint clones. Underneath each fiber FISH image is a representation of the relationship between these clones, as deduced from the fiber FISH data. The color of the box for each clone corresponds to the color in the FISH image. C, Fiber FISH of PMD24 fibers by use of the distal and proximal clones expected to contain the duplication breakpoint. Juxtaposed signals for AL135959 and Z92846 in the patient are shown. Signals for these clones were widely separated in normal controls and showed no relationship (data not shown). These clones are normally located ∼800 kb apart and clearly demonstrate that the duplication was tandem in nature. D, Arrows show the likely position of both breakpoints, on the basis of fiber FISH data. Since only a short red hybridization signal was observed in junction fibers as opposed to normal fibers, we estimated that the breakpoint is in the proximal half of clone Z92846. In contrast, the length of the green signal was almost the same in junction and normal fibers, suggesting that the breakpoint was toward the end of clone AL135959. E, Probable orientation of clones relative to the breakpoint (dashed line). The images shown are composites of several different fibers from the same experiment, but only four individual fibers are shown from each hybridization, for simplicity. The fibers shown are representative of the images captured from each slide.
Figure 4.
Fiber FISH mapping of duplication breakpoints in patient PMD9. A, Relative sizes and positions of distal and proximal breakpoint clones used for fiber FISH, as shown in Ensembl. Interphase FISH and UPQFM-PCR mapped the proximal breakpoint to a 12-kb interval within AL035494 and Z81014 but gave contradictory results for the distal breakpoint, which mapped within a reported LCR (Inoue et al. 2002). B, Fiber FISH of normal cell lines, showing the relationship between distal and proximal breakpoint clones. The color of the box for each clone corresponds to the fluorescent label used in the hybridization and the position indicated by fiber FISH. Genomic clones Z70227 and AL034485 from the distal region identified by UPQFM-PCR showed two signals of similar size lying next to each other without any overlap. However, there was an additional smaller signal from Z70227 (red) present at the other end of the AL034485 signal (green). The majority of the DNA sequence for clone AL034485 and the distal part of Z70227 contains inverted repeat sequences (A1a and A2) as part of LCR-PMDA (see fig. 7B). Therefore, the extra signal probably represents hybridization of Z70227 to part of the A2 repeat unit. The same patterns were observed for both normal and PMD9 fibers. Overlapping signals for the two proximal clones, AL035494 and Z81014, were observed in normal and patient fibers, with the smaller signal from the cosmid Z81014 almost entirely contained within the longer AL035494 signal (green). C, Four fiber FISH experiments on PMD9 fibers, each combining different distal (Z70227 or AL034485) and proximal (Z81014 or AL035494) clones. Underneath each fiber FISH image is a representation of the relationship between these clones, as deduced from the fiber FISH data. The color of the box for each clone corresponds to the color in the FISH image. As expected, no relationship between clones was observed in normal controls (data not shown), but a consistent pattern was seen for patient PMD9. D, The likely position of both breakpoints (arrows), on the basis of fiber FISH data. When distal clone Z70227 was cohybridized with proximal clone Z81014 or AL035494, a gap was seen between the signals, and the Z70227 signal (red) appeared to be longer than the Z81014 signal (green) and of a similar size to the AL035494 signal. Since there is a large overlap between these two proximal clones, and since they both appeared to be the same size as AL034485 at the breakpoint, it seemed likely that the proximal breakpoint was near the sequence junction between AL035494 and Z81014. When distal clone AL034485 was used in the fiber FISH experiments, signals adjacent to either of the two proximal clones were observed. This was consistent with the distal duplication breakpoint lying within the proximal part of AL034485, since the junction signals for this clone appeared relatively short. The fiber FISH data supported the UPQFM-PCR breakpoint mapping. The location of LCR-PMDA and inverted repeat sequences (A1a and A2) are shown with respect to the breakpoint. E, The probable orientation of clones relative to the breakpoint (dashed line). The fiber images shown are composites of several different fibers from the same experiment, but only 4–5 individual fibers are shown from each hybridization, for simplicity. The fibers shown are representative of the images captured from each slide.
Figure 5.
CLUSTALW alignment of 13 tandem duplication breakpoint sequences. Multiple sequence alignments between the 30-bp distal and proximal breakpoint regions and the recombinant junction sequence generated using CLUSTALW are shown. Nucleotides present in all three sequences are indicated by an asterisk (*). Overlap between distal and proximal sequences at the junction breakpoint (red letters) is outlined by a box. Patient P015 had no overlap and a 2-bp insertion (underlined and in black letters). Distal and proximal sequences included in the junction fragment are in green and blue letters, respectively, and those not included are in black letters. Two possibilities for the distal clone of patient P083 are shown, with mismatches indicated in black letters. As indicated, there is no overlap at the junction if AL139229 is at the junction. If AL139228 is at the junction, there is an overlap of 1 base, an A. Tracts of purines (R), pyrimidines (Y), or alternating purines and pyrimidines (RY or YR) >10 bp in length are underlined. Those at the edge of the 30-bp sequence but present within the full-length sequence are included. P348 has Y32 and Y11, P015 has YR14, P083 has RY12 and RY11, PMD9 has R11 and YR10, P255 has Y15 and Y12, P114 has Y12, and P176 has R10. Tandem repeats are indicated by arrows (P348).
Table 3.
Junction Information for 13 Sequenced Tandem Duplication Events
|
Distal |
Proximal |
||||||||||
| Patient | Size ofDuplication(bp) | Clone | Break in Clone | Breakpointon X (bp) | Gene(s) | Repeata | Clone | Break in Clone | Breakpointon X (bp) | Gene(s) | Repeata |
| P348 | 110,706 | Z73964 | 29979–29980 | 102861493 | BC018033 | None | AL049610 | 71628–71629 | 102750787 | BC043382 | AluSx |
| P116 | 509,297 | Z73964 | 33210–33212 | 102864725 | BC018033 | AluSg | AL133348 | 15510–15512 | 102355428 | NT_011651.262 | HERVL |
| P015 | 176,890 | Z75896 | 7582 | 102912383 | None | L1ME3B | AL049610 | 56334 | 102735493 | None | None |
| P224 | 391,975 | Z75896 | 9196–9198 | 102913997 | None | L1PA5 | AL117327 | 32547–32549 | 102522022 | NT_011651.284, AX721149 | AluSg |
| P026 | 926,922 | Z75896 | 24251–24255 | 102929052 | None | L1PA3 | Z95624 | 93–97 | 102002130 | None | None |
| P083/PMD38b | |||||||||||
| 388,154 | AL139229 | 13457 | 102956200 | None | L1PA2 | Z69733 | 14956 | 102568046 | RAB40A, AX721149, AF422143 | L1PA8A | |
| 307,009 | AL139228 | 1637 | 102875055 | BC018033 | L1PA7 | … | … | … | … | … | |
| P134 | 473,896 | Z75896 | 25729–25730 | 102930530 | None | L1PA3 | AL606763 | 34280–34281 | 102456634 | None | AluSx |
| PMD9 | 764,999 | AL034485 | 10807–10809 | 103047142 | None | MLT1H1 | AL035494 | 69208–69210 | 102282143 | NT_011651.261 | L1PA13 |
| P255 | 280,028 | Z70227 | 35116–35117 | 103029562 | MGC39900, BC028039, BC029803 | None | AL049610 | 70375–70376 | 102749534 | AK124135, BC043382 | None |
| PMD24 | 805,457 | AL135959 | 43175 | 103216370 | NT_011651.303 | None | Z92846 | 30511 | 102410913 | None | LTR24 |
| P114 | 954,830 | AL121868 | 6053–6055 | 103364440 | NT_011651.306 | AluJo | Z92846 | 29208–29210 | 102409610 | None | None |
| P176/PMD7 | 1,673,932 | Z81144 | 2858–2860 | 104275619 | IL1RAPL2 | None | AL035444 | 18784–18786 | 102601687 | NT_011651.286 | MLT1C |
| P110 | 4,577,176 | AL772400 | 69335–69340 | 106756530 | NT_011651.351 | AluSg | Z85997 | 33680–33675 | 102179354 | NT_011651.259 | AluYc2 |
DNA breaks in repetitive elements are listed as shown in RepeatMasker.
Two possible distal breakpoints are shown for patient P083/PMD38.
Most of the junction sequences were consistent with the positions predicted by interphase FISH and multiplex PCR and with simple tandem head-to-tail duplication, although subtle rearrangements could not be ruled out. However, sequence analysis of the junctions identified in patients P134 and P255 (fig. 5) revealed sequences that were 41.5 kb and 99 kb proximal, respectively (fig. 6A and 6B), to the multiplex PCR-predicted locations. Interestingly, the distal sequence at the junction in patient P255 was in an inverse orientation compared with what would be expected for a tandem head-to-tail duplication (clone Z70227 in fig. 6B). In addition, alignment of junction sequence from patient P083 with the reference human genome sequence (fig. 5) revealed two potential distal breakpoint sequences (indicated by asterisks [*] in fig. 6C). The distal breakpoint was predicted by multiplex PCR to be in a long interspersed nuclear element (LINE) in clone AL139228, and most of the sequence obtained at the junction matched this clone, but there were five mismatches in the first 55 bases at the junction. These 55 bases matched exactly those in a LINE in the reverse orientation and 81 kb distal in clone AL139229.
Figure 6.
Complex rearrangements in three patients. For each patient, the extent of duplication predicted by multiplex PCR is indicated by a line with circles at each end. The distance between the distal breakpoint predicted by multiplex PCR and the actual distal breakpoint identified by sequence analysis of the junction fragment (A and B) or the distance between two potential distal breakpoints (C) is indicated by a line with arrowheads at each end. The relative position of PLP1 is indicated by a black rectangle; potential breakpoint clones are indicated by accession number; the relative positions of PCR primers used in the analysis are indicated by arrows; and the junctions predicted by multiplex PCR and the actual sequenced junctions are diagrammed. A, Patient P134. B, Patient P255. The relative positions of the A1a and A1b repeats of the LCR (diagrammed in detail in fig. 7) are indicated by thick arrows. A unique region of clone Z70227 (254 bp) that is not present in Z74620 is indicated by a striped rectangle. C, Patient P083. Relative positions of LINEs are indicated by striped arrows, and potential positions of the distal breakpoint are indicated by an asterisk (*). The black arrowhead indicates a 55-bp region of clone AL139229. Not drawn to scale.
Analysis of Breakpoints
Either the proximal or distal boundary between single-copy and duplicated sequence (region A/B1 or D2/E, respectively, in fig. 2) has to be formed during the rearrangement process by which a tandem head-to-tail duplication is formed. Thus, we amplified and sequenced across both of these boundaries by PCR in four patients (P176, P114, an affected relative of P116, and P348). Identical sequences were revealed when the boundary sequences were compared with the reference genomic sequences by use of BLAST2 (not shown), suggesting that one end of the duplication was formed by an error-free homologous recombination mechanism.
Known genes or Genscan gene predictions were found to be interrupted at the junctions in several patients (table 3). No significant phenotypic differences have been noted in these patients during clinical evaluation to date.
Bioinformatics Analysis of Proximal and Distal Breakpoints
No significant homologies were revealed when the recombinant junction sequence from each duplication event was aligned with the reference genomic sequence at the proximal and distal breakpoints. Most patients had minimal sequence identity in the 30 bp surrounding each breakpoint and only short overlaps or microhomologies of 1–6 bp at the junction (fig. 5). One patient (P015) had a 2-bp insertion that was not present in either clone.
A comparison of larger genomic clone segments (2 kb) from the proximal and distal breakpoint regions of each patient identified no sequence similarities for 11 of the 13 tandem duplications. In contrast, sequence similarities between the 2-kb breakpoint regions were identified for P116 and P110. For P116, almost the whole sequence on both sides of the junction was composed of interspersed repetitive elements (see table 3). Similarity was detected between a nearly complete AluSg element at the distal breakpoint and a partial AluJo repeat 65 bp beyond the proximal breakpoint. There were no regions of similarity extending >7 bp beyond the Alu elements. Only a 3-bp overlap was observed at the junction when the two breakpoint sequences were aligned (fig. 5). In P110, we found an exact 6-bp match at the junction and further sequence identity in the flanking region (fig. 5). RepeatMasker revealed that both breakpoints were embedded within Alu elements, with the proximal repeat an AluYc2 and the distal repeat an AluSg. The proximal and distal breakpoint regions showed a high level of homology across the junction, with a chimeric AluYc2-AluSg element formed at the junction, but the homology did not extend beyond the interspersed repetitive elements. A second significant homology within the 2-kb breakpoint region was identified between the AluYc2 proximal breakpoint sequence and an AluSx element immediately 5′ of the distal breakpoint.
Analysis of Breakpoint Features and Flanking Region for 13 Sequenced Duplication Junctions
RepeatMasker (RepBase version 7.4) revealed interspersed repetitive elements, including Alu elements, LINEs, or long terminal repeats (LTRs), either unilaterally or bilaterally in 12 of the 13 sequenced duplication events (table 3). The exception was patient P255, who had unique sequence at both the proximal and the distal ends. Examination of each of the breakpoints individually showed Alu elements at seven junctions, LINEs at seven junctions, LTR elements at four junctions, and unique sequence at eight junctions. Six patients had breaks within Alu elements (table 3), and all but two of these elements (P114 distal and P224 proximal) were almost full length. The breaks appeared to have a random distribution, and none were within the 26-bp core consensus sequence at the 5′ end of Alu elements (Deininger et al. 1981; Rudiger et al. 1995). The nearest were 20 bp away in patient P224 and 16 bp away in patient P134. Six patients had breaks within L1 LINEs (table 3). These elements were mostly partial in length, and the breaks appeared to have a random distribution. Although breakpoints often coincided with Alu elements, LINEs, or LTRs, not all breakpoints mapped to intervals of heavy interspersed repeat content. The percentage of interspersed repetitive DNA within the 2 kb surrounding each breakpoint varied from 0% to 100%, with an average value of 54.6%, corresponding to 65.8% at the distal breakpoints and 43.4% at the proximal breakpoints.
Tandem repeats of purine, pyrimidine, or alternating purine/pyrimidine tracts were found either unilaternally (6/13) or bilaterally (6/13) within 12 of the 13 duplication events. Patient P134 was the exception, with no tracts identified at either breakpoint. Pyrimidine tracts were in the greatest abundance, with more found at the proximal breakpoints (6 distal and 15 proximal). There were equal numbers of purine tracts (7 distal and 3 proximal) and alternating purine/pyrimidine tracts (4 distal and 6 proximal). Seven patients had tracts very close to the junction breakpoints, and these are shown in the CLUSTALW alignments (fig. 5). Patient PMD9 had a purine tract of 11 bp spanning the distal breakpoint, and patient P083 had an alternating purine/pyrimidine tract of 12 bp positioned 2 bp from the proximal breakpoint. The longest tract was a 32-bp purine sequence positioned 5 bp from the distal breakpoint in P348 but not present in the junction. Additional tandem repeats with different nucleotide compositions were identified in five patients (P114, P110, P083, P348, and P026). The longest was 37 bp in length and spanned the proximal breakpoint and junction in patient P348. The repeat was composed of 2.31 copies of a 16-bp sequence (ATCTCAGCTCACTGCA; shown by arrows in fig. 5). The others were 16 or 17 bp in length and were located >30 bp from the breakpoint junction and therefore are not shown in the CLUSTALW alignment (fig. 5).
Palindromes (⩾10 bp) were found in only four duplication events (in patients P176, P114, P255, and P348), and the two found within proximal breakpoint sequences are also present in the junction (P176 and P114). The palindrome closest to the junction breakpoint and therefore included in the CLUSTALW alignment (fig. 5) was within the pyrimidine tract (Y32) in P348. We used Mfold to investigate the role of DNA secondary structure in mediating the duplication event. In all patients except P015, the secondary structures for the junction were less stable than the original breakpoint sequences, which suggests that they are not involved in promoting the rearrangement. Furthermore, our computer-based analysis for 37 short sequence motifs previously associated with chromosomal rearrangements and possibly involved in DNA cleavage and recombination (Abeysinghe et al. 2003) found no differences between the breakpoint and control regions and did not reveal a common mechanism (data not shown).
Topoisomerase I consensus cleavage sites (CAT, CTY, GTY, and RAT) (Been et al. 1984) were frequently found near every breakpoint for each of the 13 duplication events. Since there were two possible distal breakpoints for patient P083, a total of 27 breakpoint and control sequences were investigated. We found an average of six topoisomerase I sites within the breakpoint sequences and seven within the control sequences, 150 bp away, and thereby concluded that the difference was not significant. We found no topoisomerase II consensus cleavage sites and no other sequence motifs that were consistently present at each breakpoint. The DNA polymerase α pause site core sequence (GAG) was found at 21 of the 27 breakpoints but was equally common within the control sequences.
Analysis of the 2-Mb Region Surrounding PLP1 for LCRs
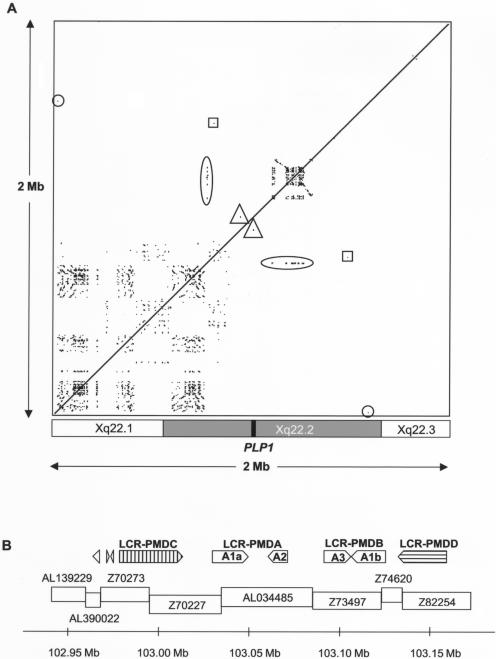
We analyzed the 2-Mb region surrounding PLP1 for LCRs. Three small regions of similarity were identified on each side of PLP1 (shown by triangle, square, and circle symbols in fig. 7A), and these were short (86–3,197 bp), with a low percentage identity (table 4). Investigation using BLASTn revealed no similarities between the repeats and other regions of the human genome. None of the patients had both proximal and distal breakpoints that coincided with the positions of these flanking repeats; therefore, they were not considered to have a major role in generating the duplications. Thus, the PMD duplications are not mediated by homologous LCRs at proximal and distal breakpoints (NAHR).
Figure 7.
Analysis of repeats in the PLP1 genomic region. A, Region-specific repeats in the genomic region surrounding PLP1. A 2-Mb human genomic sequence was masked for interspersed repetitive elements (UCSC Genome Browser, July 2003 release) and was compared against itself by use of PipMaker (BLASTz). The alignments are shown as a dot plot and are plotted on the horizontal axis according to sequence. Alignments—that is, repeated regions—are shown as black lines on the dot plot. Directly repeated sequences are represented as a series of dots forming upward-facing diagonal lines (/), and inverted repeated sequences are represented as a series of dots forming downward-sloping diagonal lines (\). The checkerboard-like pattern proximal to PLP1 represents numerous short, locally repeated sequences. The densely packed lines distal to PLP1 represent a large region of LCRs (further characterized in fig. 7B). There was one small region of similarity to the LCR region found proximal to PLP1 (104 bp with 73.08% identity) (oval). Other small regions of similarity on either side of PLP1 are shown by triangles, squares, and circles (described further in table 4). The approximate locations of the chromosome bands in the region are shown underneath the dot plot, and the position of PLP1 is indicated by a black box. B, Organization of LCRs distal to PLP1. The positions of the various repeat elements are shown relative to genomic clones in the region. LCR-PMDC and LCR-PMDD are novel, and LCR-PMDA and LCR-PMDB (including Ala, A2, A3, and Alb) have been described elsewhere (Inoue et al. 2002). In addition, two small inverted repeats were found in clone Z70273 (277 and 332 bp in size), and another was found in AL390022 (356 bp). Arrows show the direction of each repeat, and the scale bar shows their position on the X chromosome according to Ensembl.
Table 4.
Summary of the Short Regions of Sequence Similarity Identified by BLASTz on Both Sides of PLP1
| Clone | Symbolin Figure 7a | Positionwithin Cloneb | Total Size(bp) | Identityc(%) |
| Z93848 | Triangle | 22975–26171 | 3,197 | 36.1 |
| Z73964 | Triangle | 36996–39898 | 2,903 | 39.8 |
| Z73965 | Square | 9469–9554 | 86 | 68.6 |
| AL049631 | Square | 111144–111231 | 88 | 67.0 |
| AL590407 | Circle | 26638–26549 | 90 | 65.6 |
| AL121860 | Circle | 102438–102527 | 90 | 65.6 |
Symbols corresponding to the ones used in figure 7 to highlight these regions.
The position (bp) of each repeat sequence within the sequence for each genomic clone.
The percentage of identical nucleotides within each alignment.
A large LCR region distal to PLP1 was also found that had a complicated repeat structure (shown by a horizontal oval symbol in fig. 7A). We identified two directly repeated LCRs, LCR-PMDA (comprising Ala [20.3 kb] and A2 [11.7 kb]) and LCR-PMDB (comprising A3 [14.8 kb] and Alb [20.3 kb]), that have been described elsewhere, and both contain a pair of inverted repeats with a high degree of sequence identity (88.1%) (Inoue et al. 2002). In addition, we found a novel pair of inverted repeats, termed “LCR-PMDC” and “LCR-PMDD,” which are 35.6 kb and 27.7 kb in length and lie on either side of LCR-PMDA and LCR-PMDB (fig. 7B), making the entire distal LCR region almost 200 kb in length. The LCR-PMDC and LCR-PMDD repeats are also very similar to each other, since comparison using BLASTz showed 92.4% sequence identity over a region 25 kb in size. They are not similar to LCR-PMDA and LCR-PMDB. In addition, we identified a few small regions of similarity to LCR-PMDA and LCR-PMDB lying proximal to the main LCR region, within genomic clones AL390022 and Z70273 (fig. 7B), and also present ∼164 kb proximal to PLP1 in clone Z73965 (104 bp, with 73.08% identity with LCR-PMDA and LCRPMDB) (see vertical oval symbol in fig. 7A).
The region proximal to PLP1 contained numerous short locally repeated sequences that had an alternating distribution, indicated by the checkerboard-like pattern in figure 7A. Further investigation of these repeated regions found that there were four different types of repeat sequences proximal to PLP1. Although these proximal repeats covered a wider area than the distal repeats, individual repeat units were generally much shorter (mean size ∼6 kb). Between repeat units within each of the four proximal groupings there was less similarity than that found for the distal LCRs. Most showed only 60%–80% similarity in BLASTz comparisons. Many of the repeats included coding sequences, and the majority of the annotated genes in the 1-Mb region proximal to PLP1 were contained within a repeat unit. The relatively low degree of sequence identity between the various copies of the proximal repeats makes it seem unlikely that NAHR-based rearrangements between different copies of these proximal repeats would be likely to occur. However, the presence of numerous repeats in this region suggests that duplications within this region have occurred repeatedly in the past, and it may indicate a tendency toward duplication events in this region.
Discussion
Our molecular dissection of Xq22 rearrangements in 59 patients with PMD has provided insights into the molecular mechanisms of chromosome duplication. We have found unique duplication structures for each patient and widely scattered breakpoints, especially at the proximal end. Interestingly, however, there is some clustering of breakpoints at the distal end. Analysis of the 2-Mb region surrounding PLP1 revealed no large LCRs flanking the gene but a distal LCR region with a complicated structure containing both inverted and direct repeats. We identified two novel LCRs, named “LCR-PMDC” and “LCR-PMDD,” that are similar in sequence to each other but not to LCR-PMDA and LCR-PMDB, which were reported elsewhere (Inoue et al. 2002). The finding of novel LCRs extends the LCR region to ∼200 kb, and almost one-half of the distal breakpoints are within this region. Interestingly, a region with similarity to the distal LCR region was found proximal to PLP1 in Z73965, but it was small, with low identity, and could not account for all the different proximal breakpoints. However, many small repeat sequences with minimal homology and unrelated to the distal LCRs were identified proximal to PLP1.
Sequence analysis of the junctions of 13 tandem duplications (D1/B2 in fig. 2) showed an insertion or microhomology of 1–6 nt in the majority of events but no long stretches of homology. Sequence analysis of the boundaries between single-copy and duplicated sequence at the proximal and distal breakpoints (regions A/B1 and D2/E in fig. 2) of four tandem duplications showed no variation from the reference genomic sequence. Taken together, these data suggest that the mechanism of NAHR between LCRs does not occur in the majority of patients with PMD, in contrast to those with CMT1A and many other genomic diseases (Lupski 1998; Inoue and Lupski 2002), and the data also suggest that PLP1 duplications are formed by a coupled homologous and nonhomologous mechanism. The observation of normal reference genomic sequence at one end of the duplicated region, either the proximal or the distal breakpoint, suggests an error-free homologous repair process. Because both of the sequences are normal at these breakpoints, it is not possible to tell which had been involved in formation of the duplication. Therefore, in our structure of a tandem duplication event illustrated in figure 2, we cannot determine whether B1C1D1 or B2C2D2 is the duplicated sequence. Since PLP1 duplications predominantly occur on the paternal X chromosome (Mimault et al. 1999), the sister chromatid would be used as template for the homologous repair, a common occurrence in mammalian cells (Johnson and Jasin 2000). The abnormal junction with an insertion, a deletion, or microhomology at the other end of the duplicated region suggests a nonhomologous repair process. Such features are reported to occur frequently at nonhomologous junctions (Domer et al. 1995; Super et al. 1997; Megonigal et al. 1998; Gillert et al. 1999; Lovett et al. 2001; Raffini et al. 2002) and have been postulated to play a role in NHEJ (Roth et al. 1985; Roth and Wilson 1986). A model of recombination in which DNA double-stranded breaks (DSBs) are inaccurately repaired by NHEJ has also been described for deletions of PLP1 in patients with PMD (Inoue et al. 2002).
Our data are consistent with a proposed model for generation of tandem duplications that involves coupled homologous and nonhomologous repair of a DSB (Richardson and Jasin 2000; Helleday 2003). In this model, events are initiated by a DSB with ends resected by a 5′→3′ exonuclease, leaving single-stranded 3′ overhangs. At one side of the break, strand invasion of the homologous region on the sister chromatid occurs, and DNA synthesis is initiated at the 3′ invading end. This conservative and usually error-free process would produce the proximal or distal breakpoint of the PMD duplication in which there are no sequence errors. DNA synthesis proceeds beyond the site of the DSB for the length of the duplicated region, and the extended end is then repaired by NHEJ with the free end of the DSB. This error-prone mechanism would generate the abnormal junction that may have microhomology, insertion, or deletion of several bases.
We examined the sequences at the breakpoints of our 13 tandem duplications for elements that might predispose the region to DSB and rearrangement. We found a high incidence of different interspersed repetitive elements at the breakpoints, even though the actual percentage of interspersed repetitive DNA varied considerably between the individual breakpoint regions. We concluded that it is not simply a high density of interspersed repetitive elements that is responsible for the Xq22 instability and that other factors must be involved. Similar findings of interspersed repetitive elements at breakpoints have been reported elsewhere, and their presence has been postulated to be a frequent cause of genomic rearrangement by NAHR, including both deletions and duplications (Graw et al. 2000; Brooks et al. 2001; Batzer and Deininger 2002; Lutskiy et al. 2002). Interestingly, the most comprehensive study of genomewide human segmental duplications found an enrichment of Alu repeats near or within breakpoints (Bailey et al. 2003). In this study, junction analysis was consistent with a model of homologous Alu-Alu nonallelic recombination for the origin and dispersal of duplications both within and between chromosomes, although far fewer tandem rearrangements were analyzed compared with the more frequent interspersed duplications (Bailey et al. 2003). We do not have data suggesting this mechanism for PLP1 duplications in PMD. Our findings are consistent with a mechanism of NHEJ and are more similar to the scattered deletion breakpoints found within the dystrophin gene in patients with Duchenne muscular dystrophy (Nobile et al. 2002; Toffolatti et al. 2002). Interspersed repetitive elements were reported at 42% (16 of 38) of the deletion breakpoints (Nobile et al. 2002; Toffolatti et al. 2002), and we found them at 69% (18 of 26) of our duplication breakpoints. Furthermore, repetitive elements such as Alu and MER5B have been associated with four cases of nonrecurrent deletions on chromosome 17p11.2 that were generated by NHEJ and NAHR (Shaw and Lupski 2005). The significance of repetitive elements located at PLP1 duplication breakpoints remains unclear, but, if we assume the model of coupled homologous and nonhomologous recombination (Richardson and Jasin 2000), their presence does suggest that homologous invasion could also occur within a downstream interspersed repetitive element and that completion of the event could occur by NHEJ or homologous annealing (Richardson and Jasin 2000). Patient P110 had the largest duplication (4.6 Mb) and an atypical pattern of breakpoint sequences. Both were within an Alu repeat, and a chimeric Alu element was formed at the breakpoint junction. This may suggest the possible involvement of either homology-assisted NHEJ or homologous Alu-Alu recombination.
We found no evidence that secondary structures between DNA ends at the duplication junction were involved in mediating the rearrangement or in promoting the NHEJ event. Our analysis of 37 sequence motifs previously associated with DNA rearrangements and possibly involved in DNA cleavage and recombination was unable to reveal a common mechanism for generation of the duplications. Our results supported the apparent random distribution of breakpoints shown in the mapping data in figure 1 and, taken together, may suggest that the duplications are not mediated by specific sequences. A lack of specific features at the DNA sequence level has also been reported in the rearrangements, mainly deletions, in patients with Duchenne muscular dystrophy (Sironi et al. 2003). However, we did observe long stretches of purines and pyrimidines or alternating purines and pyrimidines near many breakpoints and spanning the breakpoint in two cases. Tracts of purines and pyrimidines (>10) can form Z-DNA, which may be recombinogenic (Boehm et al. 1989; Majewski and Ott 2000). These tracts, particularly polypyrimidine, may also have a role in stimulating DNA cleavage by including sites for topoisomerase II (Spitzner et al. 1990) and by being associated with nonhomologous recombination events (Sperry et al. 1989). Our study did not show the presence of topoisomerase II sites near the breakpoints, at least within the 15 bp examined on each side of the breakpoints. Recent analysis of chromosome breakpoints has shown that alternating tracts of purines and pyrimidines and purine tracts of 25–39 nt were significantly overrepresented at deletion breakpoints (Abeysinghe et al. 2003). Also, polypurine tracts (2–23 bp) and polypyrimidine tracts (2–44 bp) were significantly overrepresented at translocation breakpoints (Abeysinghe et al. 2003). Our finding of purine and pyrimidine tracts at many but not all breakpoints suggests that they may assist with but are not necessary for stimulating the PLP1 duplications.
In several cases, a known gene or predicted gene was found to span the duplication breakpoint, and thus the patient would have normal and disrupted copies. Information available about the disrupted genes at most of the junctions was too preliminary to determine what the significance might be. At others, all that remains of the gene is the 3′ end, and there is nothing predicted on the other side to drive expression. However, in patient P083, when the junction is in clone AL139228, there is the potential for formation of a chimeric transcript from the 5′ end of the RAB40A gene on the proximal side and the 3′ end of a transcript of the RAB9 gene (NCBI accession number BC018033) on the distal side of the junction transcribed in telomere-to-centromere orientation. We were unable to obtain tissue from the patient to test for the presence of this transcript. The protein products of these genes are members of the RAB family of small G proteins that are key components in intracellular vesicular transport and have been implicated in oligodendrocyte membrane trafficking (Rodriguez-Gabin et al. 2004). Although the coding region of neither gene was included, the chimeric transcript could potentially have clinical significance if it were to interfere with expression of either normal gene. The 5′ end of the interleukin 1 receptor accessory protein-like–2 gene (IL1RAPL2), including two coding exons, was present at the distal side of the junction for patient P176/PMD7. However, it is unknown how far the transcript would continue on the other side of the junction. No transcripts of the same orientation were found nearby on the other side of the junction. A transcript from this partial gene could affect phenotype by interfering with expression of IL1RAPL2 or by being translated into a truncated protein with a dominant toxic effect. Alternatively, the transcript could be subject to nonsense-mediated decay and not have a phenotypic effect. The IL1RAPL2 gene is specifically expressed in the nervous system and is affected in X-linked mental retardation 34 (Carrie et al. 1999; Ferrante et al. 2001). IL1RAPL2 has been shown to be a Toll receptor–like protein that interacts with the neuronal calcium sensor–1 protein, a modulator of neurotransmitter release (Bahi et al. 2003). However, the patient was not noted to have cognitive defects unusual for PMD.
Although most of the junctions we identified were indicative of simple tandem head-to-tail duplications, at least 9 of the 59 patients in our cohort did not fit this picture. Of 36 patients tested by FISH, 3 had duplications inserted in a region outside Xq22: Xq26 in PMD1 (Woodward et al. 2003), Xp22 in PMD5 (Hodes et al. 2000), and the Y chromosome in P149/PMD46 (K.J.W. and M.C., unpublished data). Thus, an insertion elsewhere is a relatively rare occurrence. Further, the data suggest that six patients may have complex rearrangements. At least three of them—P015, P307, and P389—had a second, nearby duplication in addition to the PLP1-containing duplication when multiplex PCR data were mapped with respect to the reference human genome sequence (fig. 1). These data could be explained by internal deletions within duplicated regions, by rearrangements subsequent to the duplication event, or by duplication events occurring on a substrate that is rearranged with respect to the reference human genome sequence. In patients P134 and P255, a discrepancy between the distal breakpoint predicted by dosage analysis and the actual junction sequence identified (fig. 6A and 6B), and the additional finding that P255 had a distal breakpoint sequence in the inverse orientation from a head-to-tail duplication, also suggested the possibility of complex rearrangements. Patient P134 may have a second junction in addition to the one we identified by IPCR that involves the predicted distal clone. The simplest explanation for the discrepancy in patient P255 is that the duplication event occurred on a substrate that had an inversion of the 99-kb region between the expected and the actual distal breakpoints with respect to the reference human genome sequence (see fig. 6B). In this regard, the expected and actual distal breakpoints are in the A1b and A1a elements, respectively, of the LCR distal to PLP1, which could mediate such an inversion event. The duplication in patient P083, whose distal breakpoint could be in LINEs in one of two clones, could also have occurred on a substrate that already had a rearrangement involving the LINEs. Alternatively, ⩾55 bp from clone AL139229 could have been inverted and inserted at the junction with clone AL139228, as diagrammed in figure 6C. The mechanisms involved in generating these complex rearrangements are largely unknown. The junction sequences suggest that a nonhomologous repair mechanism was involved in their formation, the same as for the tandem head-to-tail duplications. We have no evidence for or against complex rearrangement in any of the remaining patients.
Although PMD differs mechanistically from many other genomic diseases that are mediated by NAHR between LCRs, the region of LCRs distal to the PLP1 gene may be involved in generating the duplications in PMD. In this regard, it may be significant that four of the six patients with complex rearrangements and six of the eight patients in whom we attempted to identify junctions but failed to do so, possibly because they were complex rearrangements, had their distal breakpoint in or near the LCR region. In addition, the three small PLP1 deletions reported elsewhere (Inoue et al. 2002) had distal breakpoints within the region of LCRs, and one of these cases had a complex rearrangement. Thus, the region of LCRs would appear to be the area of greatest instability close to the PLP1 gene. Although we did not identify any further regions of LCRs, our analysis of the 2-Mb region susceptible to duplications identified many proximal-specific repeats that would not be classified as LCRs because of their small size and low homology. It is tempting to speculate that this unique genomic architecture has a role in the nonrecurrent rearrangements that result in PMD by promoting instability in Xq22. More investigation is required to fully understand the mechanisms involved in genomic rearrangements, particularly those of DNA duplication, but this study shows that both homologous and nonhomologous recombination mechanisms are involved. In addition, given that segmental duplications represent ∼5% of the human genome (Bailey et al. 2001), this analysis of duplication events in patients with PMD provides a valuable model for investigating the underlying molecular mechanisms of other genomic duplications.
Acknowledgments
We acknowledge the Wellcome Trust, for generously supporting K.J.W., and the Medical Research Council, for a Ph.D. studentship to M.C. We acknowledge Nemours and the National Institutes of Health (NIH) (grant P20 RR-020173-01 from the National Center for Research Resources) for generously supporting G.M.H. and K.S. We thank the Children’s Research Center of Michigan and the NIH (grant NS043783) for supporting J.Y.G. We are very grateful to the PMD Foundation, the Kylan Hunter Foundation, and all the patients and families for their support and cooperation. The contents of this publication are solely the responsibility of the authors and do not necessarily represent the official views of the granting agencies. We thank Junaid Fukata, Rodger Palmer, Karen Kirtland, Poorvi Patel, Deborah Stabley, Glenn Simon, and Carolyn Ritterson for their help in determining the extent of duplication in some patients. Finally, we pay tribute to Dr. M. E. Hodes, who died in September 2001 and who made a significant contribution to the understanding of PMD.
Web Resources
The accession number and URLs for data presented herein are as follows:
- BLAST, http://www.ncbi.nlm.nih.gov/blast/ (for BLASTn, BLASTz, and BLAST2)
- CLUSTALW, http://align.genome.jp/
- DNA Pattern Find, http://bioinformatics.org/sms/dna_pattern.html
- Ensembl Genome Browser, http://www.ensembl.org/
- Mfold, http://www.bioinfo.rpi.edu/applications/mfold/old/dna/
- Mreps, http://bioweb.pasteur.fr/cgi-bin/seqanal/mreps.pl
- NCBI, http://www.ncbi.nih.gov/ (for clone, primer, and gene information and RAB9 [accession number BC018033])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for PMD) [PubMed]
- Palindrome, http://bioweb.pasteur.fr/seqanal/interfaces/palindrome.html
- PipMaker, http://bio.cse.psu.edu/cgi-bin/pipmaker?basic
- Primer3, http://www.broad.mit.edu/cgi-bin/primer/primer3_www.cgi
- RepeatMasker, http://repeatmasker.org/
- The Wellcome Trust Sanger Institute: DNA Fibers, http://www.sanger.ac.uk/HGP/methods/cytogenetics/fiber_fish.shtml [DOI] [PubMed]
- The Wellcome Trust Sanger Institute: Human X Project, http://www.sanger.ac.uk/HGP/ChrX [DOI] [PubMed]
- UCSC Genome Browser, http://genome.ucsc.edu/
References
- Abeysinghe SS, Chuzhanova N, Krawczak M, Ball EV, Cooper DN (2003) Translocation and gross deletion breakpoints in human inherited disease and cancer. I. Nucleotide composition and recombination-associated motifs. Hum Mutat 22:229–244 10.1002/humu.10254 [DOI] [PubMed] [Google Scholar]
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410 10.1006/jmbi.1990.9999 [DOI] [PubMed] [Google Scholar]
- Bahi N, Friocourt G, Carrie A, Graham ME, Weiss JL, Chafey P, Fauchereau F, Burgoyne RD, Chelly J (2003) IL1 receptor accessory protein like, a protein involved in X-linked mental retardation, interacts with neuronal calcium sensor-1 and regulates exocytosis. Hum Mol Genet 12:1415–1425 10.1093/hmg/ddg147 [DOI] [PubMed] [Google Scholar]
- Bailey JA, Liu G, Eichler EE (2003) An Alu transposition model for the origin and expansion of human segmental duplications. Am J Hum Genet 73:823–834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey JA, Yavor AM, Massa HF, Trask BJ, Eichler EE (2001) Segmental duplications: organization and impact within the current human genome project assembly. Genome Res 11:1005–1017 10.1101/gr.GR-1871R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Batzer MA, Deininger PL (2002) Alu repeats and human genomic diversity. Nat Rev Genet 3:370–379 10.1038/nrg798 [DOI] [PubMed] [Google Scholar]
- Been MD, Burgess RR, Champoux JJ (1984) Nucleotide sequence preference at rat liver and wheat germ type 1 DNA topoisomerase breakage sites in duplex SV40 DNA. Nucleic Acids Res 12:3097–3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehm T, Mengle-Gaw L, Kees UR, Spurr N, Lavenir I, Forster A, Rabbitts TH (1989) Alternating purine-pyrimidine tracts may promote chromosomal translocations seen in a variety of human lymphoid tumours. EMBO J 8:2621–2631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulloche J, Aicardi J (1986) Pelizaeus-Merzbacher disease: clinical and nosological study. J Child Neurol 1:233–239 [DOI] [PubMed] [Google Scholar]
- Brooks EM, Branda RF, Nicklas JA, O’Neill JP (2001) Molecular description of three macro-deletions and an Alu-Alu recombination-mediated duplication in the HPRT gene in four patients with Lesch-Nyhan disease. Mutat Res 476:43–54 [DOI] [PubMed] [Google Scholar]
- Carrie A, Jun L, Bienvenu T, Vinet MC, McDonell N, Couvert P, Zemni R, Cardona A, Van Buggenhout G, Frints S, Hamel B, Moraine C, Ropers HH, Strom T, Howell GR, Whittaker A, Ross MT, Kahn A, Fryns JP, Beldjord C, Marynen P, Chelly J (1999) A new member of the IL-1 receptor family highly expressed in hippocampus and involved in X-linked mental retardation. Nat Genet 23:25–31 [DOI] [PubMed] [Google Scholar]
- Chance PF, Abbas N, Lensch MW, Pentao L, Roa BB, Patel PI, Lupski JR (1994) Two autosomal dominant neuropathies result from reciprocal DNA duplication/deletion of a region on chromosome 17. Hum Mol Genet 3:223–228 [DOI] [PubMed] [Google Scholar]
- Christian SL, Fantes JA, Mewborn SK, Huang B, Ledbetter DH (1999) Large genomic duplicons map to sites of instability in the Prader-Willi/Angelman syndrome chromosome region (15q11-q13). Hum Mol Genet 8:1025–1037 10.1093/hmg/8.6.1025 [DOI] [PubMed] [Google Scholar]
- Chuzhanova N, Abeysinghe SS, Krawczak M, Cooper DN (2003) Translocation and gross deletion breakpoints in human inherited disease and cancer. II. Potential involvement of repetitive sequence elements in secondary structure formation between DNA ends. Hum Mutat 22:245–251 10.1002/humu.10253 [DOI] [PubMed] [Google Scholar]
- Deininger PL, Jolly DJ, Rubin CM, Friedmann T, Schmid CW (1981) Base sequence studies of 300 nucleotide renatured repeated human DNA clones. J Mol Biol 151:17–33 10.1016/0022-2836(81)90219-9 [DOI] [PubMed] [Google Scholar]
- Domer PH, Head DR, Renganathan N, Raimondi SC, Yang E, Atlas M (1995) Molecular analysis of 13 cases of MLL/11q23 secondary acute leukemia and identification of topoisomerase II consensus-binding sequences near the chromosomal breakpoint of a secondary leukemia with the t(4;11). Leukemia 9:1305–1312 [PubMed] [Google Scholar]
- Edelmann L, Pandita RK, Spiteri E, Funke B, Goldberg R, Palanisamy N, Chaganti RS, Magenis E, Shprintzen RJ, Morrow BE (1999) A common molecular basis for rearrangement disorders on chromosome 22q11. Hum Mol Genet 8:1157–1167 10.1093/hmg/8.7.1157 [DOI] [PubMed] [Google Scholar]
- Edelmann L, Spiteri E, Koren K, Pulijaal V, Bialer MG, Shanske A, Goldberg R, Morrow BE (2001) AT-rich palindromes mediate the constitutional t(11;22) translocation. Am J Hum Genet 68:1–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellis D, Malcolm S (1994) Proteolipid protein gene dosage effect in Pelizaeus-Merzbacher disease. Nat Genet 6:333–334 10.1038/ng0494-333 [DOI] [PubMed] [Google Scholar]
- Ferrante MI, Ghiani M, Bulfone A, Franco B (2001) IL1RAPL2 maps to Xq22 and is specifically expressed in the central nervous system. Gene 275:217–221 10.1016/S0378-1119(01)00659-X [DOI] [PubMed] [Google Scholar]
- Gillert E, Leis T, Repp R, Reichel M, Hosch A, Breitenlohner I, Angermuller S, Borkhardt A, Harbott J, Lampert F, Griesinger F, Greil J, Fey GH, Marschalek R (1999) A DNA damage repair mechanism is involved in the origin of chromosomal translocations t(4;11) in primary leukemic cells. Oncogene 18:4663–4671 10.1038/sj.onc.1202842 [DOI] [PubMed] [Google Scholar]
- Graw SL, Sample T, Bleskan J, Sujansky E, Patterson D (2000) Cloning, sequencing, and analysis of Inv8 chromosome breakpoints associated with recombinant 8 syndrome. Am J Hum Genet 66:1138–1144 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding B, Ellis D, Malcolm S (1995) A case of Pelizaeus-Merzbacher disease showing increased dosage of the proteolipid protein gene. Neuropathol Appl Neurobiol 21:111–115 [DOI] [PubMed] [Google Scholar]
- Heath KE, Day IN, Humphries SE (2000) Universal primer quantitative fluorescent multiplex (UPQFM) PCR: a method to detect major and minor rearrangements of the low density lipoprotein receptor gene. J Med Genet 37:272–280 10.1136/jmg.37.4.272 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helleday T (2003) Pathways for mitotic homologous recombination in mammalian cells. Mutat Res 532:103–115 [DOI] [PubMed] [Google Scholar]
- Hodes ME, Pratt VM, Dlouhy SR (1993) Genetics of Pelizaeus-Merzbacher disease. Dev Neurosci 15:383–394 [DOI] [PubMed] [Google Scholar]
- Hodes ME, Woodward K, Spinner NB, Emanuel BS, Enrico-Simon A, Kamholz J, Stambolian D, Zackai EH, Pratt VM, Thomas IT, Crandall K, Dlouhy SR, Malcolm S (2000) Additional copies of the proteolipid protein gene causing Pelizaeus-Merzbacher disease arise by separate integration into the X chromosome. Am J Hum Genet 67:14–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard T, Barker D, Birney E, Cameron G, Chen Y, Clark L, Cox T, et al (2002) The Ensembl genome database project. Nucleic Acids Res 30:38–41 10.1093/nar/30.1.38 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue K, Lupski JR (2002) Molecular mechanisms for genomic disorders. Annu Rev Genomics Hum Genet 3:199–242 10.1146/annurev.genom.3.032802.120023 [DOI] [PubMed] [Google Scholar]
- Inoue K, Osaka H, Imaizumi K, Nezu A, Takanashi J, Arii J, Murayama K, Ono J, Kikawa Y, Mito T, Shaffer LG, Lupski JR (1999) Proteolipid protein gene duplications causing Pelizaeus-Merzbacher disease: molecular mechanism and phenotypic manifestations. Ann Neurol 45:624–632 [DOI] [PubMed] [Google Scholar]
- Inoue K, Osaka H, Thurston VC, Clarke JTR, Yoneyama A, Rosenbarker L, Bird TD, Hodes ME, Shaffer LG, Lupski JR (2002) Genomic rearrangements resulting in PLP1 deletion occur by nonhomologous end joining and cause different dysmyelinating phenotypes in males and females. Am J Hum Genet 71:838–853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson RD, Jasin M (2000) Sister chromatid gene conversion is a prominent double-strand break repair pathway in mammalian cells. EMBO J 19:3398–3407 10.1093/emboj/19.13.3398 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagawa T, Ikenaka K, Inoue Y, Kuriyama S, Tsujii T, Nakao J, Nakajima K, Aruga J, Okano H, Mikoshiba K (1994) Glial cell degeneration and hypomyelination caused by overexpression of myelin proteolipid protein gene. Neuron 13:427–442 10.1016/0896-6273(94)90358-1 [DOI] [PubMed] [Google Scholar]
- Kanaar R, Hoeijmakers JH, van Gent DC (1998) Molecular mechanisms of DNA double strand break repair. Trends Cell Biol 8:483–489 10.1016/S0962-8924(98)01383-X [DOI] [PubMed] [Google Scholar]
- Kurahashi H, Shaikh T, Takata M, Toda T, Emanuel BS (2003) The constitutional t(17;22): another translocation mediated by palindromic AT-rich repeats. Am J Hum Genet 72:733–738 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieber MR, Ma Y, Pannicke U, Schwarz K (2003) Mechanism and regulation of human non-homologous DNA end-joining. Nat Rev Mol Cell Biol 4:712–720 10.1038/nrm1202 [DOI] [PubMed] [Google Scholar]
- Lovett BD, Lo Nigro L, Rappaport EF, Blair IA, Osheroff N, Zheng N, Megonigal MD, Williams WR, Nowell PC, Felix CA (2001) Near-precise interchromosomal recombination and functional DNA topoisomerase II cleavage sites at MLL and AF-4 genomic breakpoints in treatment-related acute lymphoblastic leukemia with t(4;11) translocation. Proc Natl Acad Sci USA 98:9802–9807 10.1073/pnas.171309898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupski JR (1998) Genomic disorders: structural features of the genome can lead to DNA rearrangements and human disease traits. Trends Genet 14:417–422 10.1016/S0168-9525(98)01555-8 [DOI] [PubMed] [Google Scholar]
- Lupski JR, de Oca-Luna RM, Slaugenhaupt S, Pentao S, Guzzetta V, Trask BJ, Saucedo-Cardenas O, Barker DF, Killian JM, Garcia CA, Chakravarti A, Patel PI (1991) DNA duplication associated with Charcot-Marie-Tooth disease type 1A. Cell 66:219–232 10.1016/0092-8674(91)90613-4 [DOI] [PubMed] [Google Scholar]
- Lutskiy MI, Jones LN, Rosen FS, Remold-O’Donnell E (2002) An Alu-mediated deletion at Xp11.23 leading to Wiskott-Aldrich syndrome. Hum Genet 110:515–519 10.1007/s00439-002-0716-4 [DOI] [PubMed] [Google Scholar]
- Majewski J, Ott J (2000) GT repeats are associated with recombination on human chromosome 22. Genome Res 10:1108–1114 10.1101/gr.10.8.1108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Megonigal MD, Rappaport EF, Jones DH, Williams TM, Lovett BD, Kelly KM, Lerou PH, Moulton T, Budarf ML, Felix CA (1998) t(11;22)(q23;q11.2) in acute myeloid leukemia of infant twins fuses MLL with hCDCrel, a cell division cycle gene in the genomic region of deletion in DiGeorge and velocardiofacial syndromes. Proc Natl Acad Sci USA 95:6413–6418 10.1073/pnas.95.11.6413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mimault C, Giraud G, Courtois V, Cailloux F, Boire JY, Dastugue B, Boespflug-Tanguy O, the Clinical European Network on Brain Dysmyelinating Disease (1999) Proteolipoprotein gene analysis in 82 patients with sporadic Pelizaeus-Merzbacher disease: duplications, the major cause of the disease, originate more frequently in male germ cells, but point mutations do not. Am J Hum Genet 65:360–369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobile C, Toffolatti L, Rizzi F, Simionati B, Nigro V, Cardazzo B, Patarnello T, Valle G, Danieli GA (2002) Analysis of 22 deletion breakpoints in dystrophin intron 49. Hum Genet 110:418–421 10.1007/s00439-002-0721-7 [DOI] [PubMed] [Google Scholar]
- Perez Jurado LA, Wang YK, Peoples R, Coloma A, Cruces J, Francke U (1998) A duplicated gene in the breakpoint regions of the 7q11.23 Williams-Beuren syndrome deletion encodes the initiator binding protein TFII-I and BAP-135, a phosphorylation target of BTK. Hum Mol Genet 7:325–334 10.1093/hmg/7.3.325 [DOI] [PubMed] [Google Scholar]
- Raffini LJ, Slater DJ, Rappaport EF, Lo Nigro L, Cheung NK, Biegel JA, Nowell PC, Lange BJ, Felix CA (2002) Panhandle and reverse-panhandle PCR enable cloning of der(11) and der(other) genomic breakpoint junctions of MLL translocations and identify complex translocation of MLL, AF-4, and CDK6. Proc Natl Acad Sci USA 99:4568–4573 10.1073/pnas.062066799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raskind WH, Williams CA, Hudson LD, Bird TD (1991) Complete deletion of the proteolipid protein gene (PLP) in a family with X-linked Pelizaeus-Merzbacher disease. Am J Hum Genet 49:1355–1360 [PMC free article] [PubMed] [Google Scholar]
- Readhead C, Schneider A, Griffiths I, Nave KA (1994) Premature arrest of myelin formation in transgenic mice with increased proteolipid protein gene dosage. Neuron 12:583–595 10.1016/0896-6273(94)90214-3 [DOI] [PubMed] [Google Scholar]
- Richardson C, Jasin M (2000) Coupled homologous and nonhomologous repair of a double-strand break preserves genomic integrity in mammalian cells. Mol Cell Biol 20:9068–9075 10.1128/MCB.20.23.9068-9075.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez-Gabin AG, Almazan G, Larocca JN (2004) Vesicle transport in oligodendrocytes: probable role of Rab40c protein. J Neurosci Res 76:758–770 10.1002/jnr.20121 [DOI] [PubMed] [Google Scholar]
- Roth DB, Porter TN, Wilson JH (1985) Mechanisms of nonhomologous recombination in mammalian cells. Mol Cell Biol 5:2599–2607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth DB, Wilson JH (1986) Nonhomologous recombination in mammalian cells: role for short sequence homologies in the joining reaction. Mol Cell Biol 6:4295–4304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rudiger NS, Gregersen N, Kielland-Brandt MC (1995) One short well conserved region of Alu-sequences is involved in human gene rearrangements and has homology with prokaryotic chi. Nucleic Acids Res 23:256–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Zhang Z, Frazer KA, Smit A, Riemer C, Bouck J, Gibbs R, Hardison R, Miller W (2000) PipMaker—a web server for aligning two genomic DNA sequences. Genome Res 10:577–586 10.1101/gr.10.4.577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw CJ, Lupski JR (2004) Implications of human genome architecture for rearrangement-based disorders: the genomic basis of disease. Hum Mol Genet Spec 1 13:R57–R64 10.1093/hmg/ddh073 [DOI] [PubMed] [Google Scholar]
- ——— (2005) Non-recurrent 17p11.2 deletions are generated by homologous and non-homologous mechanisms. Hum Genet 116:1–7 10.1007/s00439-004-1204-9 [DOI] [PubMed] [Google Scholar]
- Sironi M, Pozzoli U, Cagliani R, Giorda R, Comi GP, Bardoni A, Menozzi G, Bresolin N (2003) Relevance of sequence and structure elements for deletion events in the dystrophin gene major hot-spot. Hum Genet 112:272–288 [DOI] [PubMed] [Google Scholar]
- Sistermans EA, de Coo RF, De Wijs IJ, Van Oost BA (1998) Duplication of the proteolipid protein gene is the major cause of Pelizaeus-Merzbacher disease. Neurology 50:1749–1754 [DOI] [PubMed] [Google Scholar]
- Sperry AO, Blasquez VC, Garrard WT (1989) Dysfunction of chromosomal loop attachment sites: illegitimate recombination linked to matrix association regions and topoisomerase II. Proc Natl Acad Sci USA 86:5497–5501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzner JR, Chung IK, Muller MT (1990) Eukaryotic topoisomerase II preferentially cleaves alternating purine-pyrimidine repeats. Nucleic Acids Res 18:1–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stankiewicz P, Lupski JR (2002) Molecular-evolutionary mechanisms for genomic disorders. Curr Opin Genet Dev 12:312–319 10.1016/S0959-437X(02)00304-0 [DOI] [PubMed] [Google Scholar]
- Super HG, Strissel PL, Sobulo OM, Burian D, Reshmi SC, Roe B, Zeleznik-Le NJ, Diaz MO, Rowley JD (1997) Identification of complex genomic breakpoint junctions in the t(9;11) MLL-AF9 fusion gene in acute leukemia. Genes Chromosomes Cancer 20:185–195 [DOI] [PubMed] [Google Scholar]
- Tatusova TA, Madden TL (1999) BLAST 2 sequences, a new tool for comparing protein and nucleotide sequences. FEMS Microbiol Lett 174:247–250 10.1016/S0378-1097(99)00149-4 [DOI] [PubMed] [Google Scholar]
- Toffolatti L, Cardazzo B, Nobile C, Danieli GA, Gualandi F, Muntoni F, Abbs S, Zanetti P, Angelini C, Ferlini A, Fanin M, Patarnello T (2002) Investigating the mechanism of chromosomal deletion: characterization of 39 deletion breakpoints in introns 47 and 48 of the human dystrophin gene. Genomics 80:523–530 10.1016/S0888-7543(02)96861-8 [DOI] [PubMed] [Google Scholar]
- Triglia T, Peterson MG, Kemp DJ (1988) A procedure for in vitro amplification of DNA segments that lie outside the boundaries of known sequences. Nucleic Acids Res 16:8186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto Y, Ikeda H (1998) Double-strand break repair mediated by DNA end-joining. Genes Cells 3:135–144 10.1046/j.1365-2443.1998.00180.x [DOI] [PubMed] [Google Scholar]
- Ussery D, Soumpasis DM, Brunak S, Staerfeldt HH, Worning P, Krogh A (2002) Bias of purine stretches in sequenced chromosomes. Comput Chem 26:531–541 10.1016/S0097-8485(02)00013-X [DOI] [PubMed] [Google Scholar]
- Watanabe I, Patel V, Goebel HH, Siakotos AN, Zeman W, DeMyer W, Dyer JS (1973) Early lesion of Pelizaeus-Merzbacher disease: electron microscopic and biochemical study. J Neuropathol Exp Neurol 32:313–333 [DOI] [PubMed] [Google Scholar]
- Wilkus RJ, Farrell DF (1976) Electrophysiologic observations in the classical form of Pelizaeus-Merzbacher disease. Neurology 26:1042–1045 [DOI] [PubMed] [Google Scholar]
- Wolf NI, Sistermans EA, Cundall M, Hobson GM, Davis-Williams AP, Palmer R, Stubbs P, Davies S, Endziniene M, Wu Y, Chong WK, Malcolm S, Surtees R, Garbern JY, Woodward KJ (2005) Three or more copies of the proteolipid protein gene PLP1 cause severe Pelizaeus-Merzbacher disease. Brain 128:743–751 10.1093/brain/awh409 [DOI] [PubMed] [Google Scholar]
- Woodward K, Cundall M, Palmer R, Surtees R, Winter RM, Malcolm S (2003) Complex chromosomal rearrangement and associated counseling issues in a family with Pelizaeus-Merzbacher disease. Am J Med Genet A 118:15–24 10.1002/ajmg.a.10103 [DOI] [PubMed] [Google Scholar]
- Woodward K, Kendall E, Vetrie D, Malcolm S (1998) Pelizaeus-Merzbacher disease: identification of Xq22 proteolipid-protein duplications and characterization of breakpoints by interphase FISH. Am J Hum Genet 63:207–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodward K, Kirtland K, Dlouhy S, Raskind W, Bird T, Malcolm S, Abeliovich D (2000) X inactivation phenotype in carriers of Pelizaeus-Merzbacher disease: skewed in carriers of a duplication and random in carriers of point mutations. Eur J Hum Genet 8:449–454 10.1038/sj.ejhg.5200480 [DOI] [PubMed] [Google Scholar]
- Woodward K, Malcolm S (1999) Proteolipid protein gene: Pelizaeus-Merzbacher disease in humans and neurodegeneration in mice. Trends Genet 15:125–128 10.1016/S0168-9525(99)01716-3 [DOI] [PubMed] [Google Scholar]
- Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31:3406–3415 10.1093/nar/gkg595 [DOI] [PMC free article] [PubMed] [Google Scholar]