Abstract
The biological approach to nanotechnology has produced self-assembled objects, arrays and devices; likewise, it has achieved the recognition of inorganic systems and the control of their growth. Can these approaches now be integrated to produce useful systems?
We hear continually that nanoscience and nanotechnology are frontier areas. Everyone is aware that nanotechnology and nanoscience involve the construction and analysis of objects and devices that are very small on the macroscopic scale. Nevertheless, if the ultimate feature sizes of nanoscale objects are about a nanometer or so, we are talking about dimensions an order of magnitude larger than the scale exploited by chemists for over a century. Synthetic chemists have manipulated the constituents, bonding, and stereochemistry of vast numbers of molecules on the angstrom scale, and physical and analytical chemists have examined the properties of these molecules. So what is so special about the nanoscale?
There are many answers to this question, possibly as many as there are people who call themselves nanoscientists or nanotechnologists. A particularly intriguing feature of the nanoscale is that this is the scale on which biological systems build their structural components, such as microtubules, microfilaments, and chromatin. The associations maintaining these and the associations of other cellular components seem relatively simple when examined by high-resolution structural methods, such as crystallography or NMR—shape complementarity, charge neutralization, hydrogen bonding, and hydrophobic interactions.
A key property of biological nanostructures is molecular recognition, leading to self-assembly and the templating of atomic and molecular structures. For example, it is well known that two complementary strands of DNA will pair to form a double helix. DNA illustrates two features of self-assembly. The molecules have a strong affinity for each other and they form a predictable structure when they associate. Those who wish to create defined nanostructures would like to develop systems that emulate this behavior. Thus, rather than milling down from the macroscopic level, using tools of greater and greater precision (and probably cost), they would like to build nanoconstructs from the bottom up, starting with chemical systems.
What are the advantages of building from the bottom up? Dense chemical variety is one advantage. Just as the surfaces of cellular components contain many features per unit area, a complex chemical surface can be used as a building block and, in principle, its orientation and position can be controlled. By contrast, top-down methods work on materials with little chemical diversity. A second advantage is the vastness of the chemical scale. Even a picomole of material is nearly 1012 copies. Thus, one can imagine producing complex components that form well defined structural motifs organized over large areas in two dimensions or volumes in three dimensions.
DNA Nanotechnology
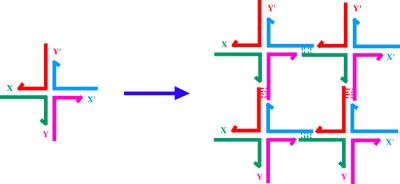
To date, the most successful biomimetic component used for self-assembly has been DNA itself (1). Linear DNA double helices seem to be of limited utility, but one can design synthetic molecules that form stable branched structures, leading to greater structural complexity. Branched DNA molecules can be combined by “sticky-ended” cohesion (2), as shown in Fig. 1. In synthetic systems, sticky ends may be programmed with a large diversity; N-nucleotide sticky ends lead to 4N possible different sequences. Sticky ends of sufficient length cohere by base pairing alone but they can be ligated to covalency. Sticky ends form classic B-DNA when they cohere (3); thus, in addition to the affinity inherent in complementarity, sticky ends also lead to structural predictability. If the positions of the atoms on one component are known near the sticky end, the atoms of the other component are also known. This situation is usefully contrasted with, say, an antibody and its antigen. Although the antigen-combining site may be known, the orientation of the antigen within it cannot be predicted; it must be determined experimentally in each case.
Figure 1.
Formation of a 2D lattice from a junction with sticky ends. X and Y are sticky ends and X′ and Y′ are their complements. Four of the monomers on the left are complexed to yield the structure on the right. DNA ligase can close the gaps left in the complex, which can be extended by the addition of more monomers.
The key static aims of DNA nanotechnology are to use DNA as scaffolding to crystallize biological macromolecules artificially for crystallography (2) and to organize the components of nanoelectronics (4). The first, and likely the second, of these applications entail the assembly of DNA into periodic networks. Thus, the quadrilateral of Fig. 1 would be most useful if extended to form a two-dimensional (2D) or three-dimensional lattice. The branched junctions shown in Fig. 1 are not rigid enough to use as building blocks for a lattice (5). This problem has been solved by combining two branched junctions to produce DNA double-crossover (DX) molecules (6, 7), which consist of two double helices fused by strands that cross between them to tie them together.
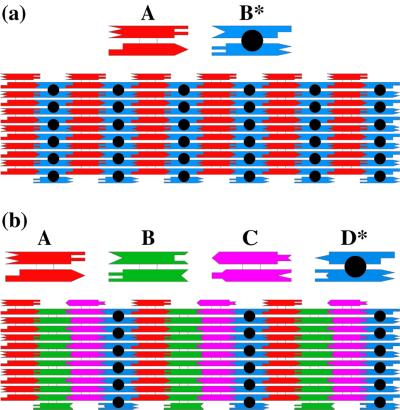
Fig. 2 illustrates two different 2D arrays that have been produced by DX molecules (8). In Fig. 2a, the repeating unit is 2 DX units, and in Fig. 2b, it is 4 DX units. The B* unit in Fig. 2a and the D* unit in Fig. 2b have circles in their centers, representing another helix that is directed out of the plane. This extra helix can serve as a topographic marker for atomic force microscopy. The dimensions of each component are about 4 × 16 nm. Thus, the extra helices produce stripe-like features every 32 nm in the AB* array, and every 64 nm in the ABCD* array (8).
Figure 2.
Arrays assembled from DX molecules. (a) A two-component array. Two DX molecules (A and B*) are illustrated schematically (a Top). The two helices are drawn as rectangles, and the complementary sticky ends are represented by geometrical shapes. A is a conventional DX molecule, but B* contains a DNA hairpin protruding from the plane. Below these molecules is an array that shows the two components fitting together to tile a plane. (b) A four-component array. The same conventions apply as in a. This array uses four tiles, A, B, C, and D*, where A, B, and C are conventional DX molecules and D* contains a hairpin. The stripes are separated by twice the distance seen in a.
Other DNA motifs have been used to produce nanoscale patterns in 2D. Although the individual branched junction is flexible, well structured DNA parallelograms can be prepared from four of them. DNA parallelograms have been used to produce meshworks with tunable cavities (9). Likewise, one can prepare triple-crossover (TX) motifs, containing three fused double helices with coplanar axes. These molecules form 2D arrays that can include TX components rotated out of the plane (10).
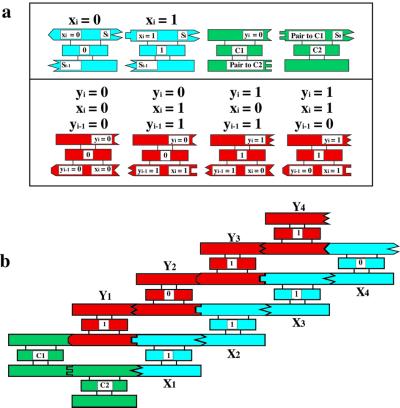
There are several ways to incorporate complexity in DNA arrays. One way is shown in Fig. 2b, where four different units comprise the asymmetric unit of the repeating structure; extension of this approach entails the expense of producing as many components as needed to produce a particular complex pattern in two or three dimensions. Winfree (11) suggested that it is possible to program sticky ends to produce algorithmic assemblies, much like the colored edges of Wang tiles; these tiles form a mosaic in which each tile edge abuts another with the same color. Appropriate sets of Wang tiles can assemble to perform computational operations and to define patterns with much greater complexity than their total number, thereby saving the expense of producing a large number of different tiles. Recently, the feasibility of this approach has been demonstrated in one dimension with triple-crossover molecules, in which a prototype cumulative exclusive OR calculation was executed (12), as shown in Fig. 3. The input and output Boolean values of each step of this calculation are represented by the sticky ends, thus this is a more stringent test of self-assembly than formation of a periodic lattice. The same sticky end on one side of the red answer tiles represents 0, regardless of whether it is on a correct or incorrect tile for a particular position. Despite overall good fidelity, some errors were detected in this experiment.
Figure 3.
A cumulative XOR calculation. The XOR operation takes two Boolean inputs and produces a 0 if they are the same and a 1 if they are different. Shown in a are blue input tiles, Xi which represent 0 or 1, according to the presence of a particular restriction enzyme site. These have been assembled in a particular order in b. The red tiles in a contain the four Boolean possibilities as sticky ends on their lower helices. The input is connected to the output through the green C1 and C2 tiles. At the end of the self-assembly, one strand that runs through the entire system is ligated together, thereby connecting the input to the output. It is read by partial restriction, followed by a denaturing gel, much like a sequencing reaction.
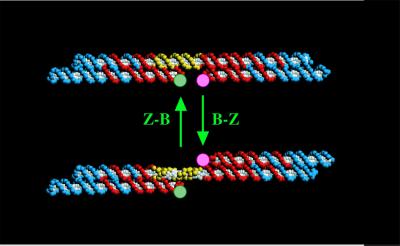
In addition to static structures, DNA can be used to produce nanomechanical devices. An early device (Fig. 4) was based on the transition between right-handed B-DNA and left-handed Z-DNA (13). The system consists of two rigid DX molecules joined by a DNA helix containing a stretch that can undergo the B–Z transition. Yurke et al. (14) have reported a sequence-dependent DNA device that uses branch migration to remove strands from a tweezer-like construct, allowing it to undergo an opening transition. This approach has been used to produce a robust sequence-dependent rotary device (H. Yan, X. Zhang, Z. Shen, and N.C.S., unpublished data). The development of sequence-dependent DNA nanomechanical devices suggests a future for DNA as a controlling element in nanorobotics: N different 2-state devices incorporated in a nanorobotic superstructure could lead to 2N distinct structural states that could be programmed serially to execute mechanical tasks.
Figure 4.
A DNA nanomechanical device based on the B–Z transition. The device consists of two DX molecules connected by a helix (yellow section) that can undergo the B-Z transition. When this occurs, the bottom domain of the right DX molecule swings from the bottom to the top through a rotary motion.
The control over DNA systems seems to be relatively robust. However, to what ends can it be used? The physical properties of DNA are important for understanding biological systems, but their utility in nanoelectronic devices is unproved. To get the maximum use from the organizational capabilities of DNA, it will be necessary to combine DNA with other nanoscale systems, particularly inorganic systems, whose physical properties lend themselves to direct applications. These materials include inorganic nanocrystals and carbon nanotubes, which represent the most exciting potential species to organize in two or three dimensions. The large sizes and abundant functional groups on DNA tiles suggest that multiple functionalities could be attached to a single tile.
The Biological–Inorganic Interface
As noted previously, the use of biological materials offers many advantages over traditional processing methods to construct the next generation of miniaturized electronics devices, particularly including spatial control on the nanometer scale, parallel self-assembly of multiple electronic components on a single device, and correctability. The critical factors in developing a bio-directed self-assembly approach are identifying the appropriate compatibilities and combinations of biological-inorganic materials, synthesis of the appropriate building blocks, and understanding and controlling building block self-assembly processes.
In natural biological systems, macromolecules exert exceptional control over inorganic nucleation, phase stabilization, assembly, and pattern formation (15, 16). Biological systems assemble nanoscale building blocks into complex and functionally sophisticated structures with high perfection, controlled size, and compositional uniformity. These materials are typically soft and consist of a surprisingly simple collection of molecular building blocks (i.e., lipids, peptides, and nucleic acids) arranged in complex architectures. For example, proteins from bones, shells, diatoms, and magnetic bacteria can spatially and temporally nucleate inorganic structures from the nanoscopic to the macroscopic scale. In addition, selectivity and recognition at the molecular scale is a critical feature of living systems. Among the best known examples are antibody–antigen interactions. Unlike the semiconductor industry, which relies on serial lithographic processing to construct the smallest features on an integrated circuit, organisms execute their architectural blueprints by using mostly noncovalent forces acting simultaneously and selectively on many molecular components.
The exquisite selectivity of complementary biological molecules offers a possible avenue to control the formation of complex structures based on inorganic building blocks such as metal or semiconductor nanoparticles. DNA oligomer–nanocrystal complexes, for example, have been examined as building blocks for more complex two- and three-dimensional structures (17, 18). Nanocrystal-labeled proteins have also been used to label biomolecular substrates with increased sensitivity (19, 20). Self-assembled monolayers have been used to template nanocrystal organization and in some cases, covalently bind semiconductor nanocrystals to metal surfaces (21). “Nanonetworks” have been formed with gold nanoclusters by, using dithiol connectors (22), and with iron oxide, using biotin-streptavidin connectors (23). Specific hydrogen-bonding-directed aggregation between nanocrystals, using alkanethiol-modified DNA base pairs, uracil, and 2,6 diaminopyridine, has also been demonstrated (24). Only very modest binding specificity between the biological molecule and the inorganic substrate can be achieved through these grafting chemistries. These approaches show potential for the control and placement of nanoparticles but they have not exploited the atomic composition and plane-specific recognition that a biomolecule can exhibit for an inorganic phase or the nanostructural control and regularity that biomolecules typically impose on crystal phases and crystallographic orientations.
In principle, biological molecules can be used to control the assembly of inorganic nanostructures and hybrid inorganic/organic structures while directing them to self-assemble in the desired manner. Thus, the biological molecules and an unlimited number of different types of nanocrystal building blocks can be mixed in the “pot” and then triggered to self-assemble into their superstructures.
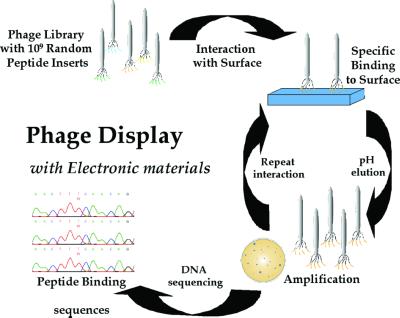
To develop the interactions of biological molecules for inorganic materials, biological selection has been used with the goal of synthesizing technologically important inorganic materials to serve as building blocks for new materials. Because nature has not had the opportunity to produce biomolecular interactions with some of the desired materials, A.M.B. and coworkers (25) used phage display to select peptide sequences, and Brown (26) used repeating polypeptides displayed on the surface of the bacterium Escherichia coli to bind selectively to metal particles. To identify the appropriate compatibilities and combinations of biological-inorganic materials, a combinatorial library of genetically engineered M13 bacteriophage was used to select peptides rapidly that could not only recognize but also control the growth of specific inorganic materials (Fig. 5).
Figure 5.
Peptide selection for electronic materials. The 1.9 × 109 random peptide sequences are exposed to the different crystal substrates; nonspecific peptide interactions are removed with extensive washes. The phage that bind are eluted by lowering the pH and disrupting the surface interaction. The eluted phage are amplified by infecting the E. coli ER2537 host, producing enriched populations of phage, displaying peptides that interact with the specific crystal substrate. The amplified phage are isolated, titered, and reexposed to a freshly prepared substrate surface, thereby enriching the phage population with substrate-specific binding phage. This procedure is repeated three to five times to select the phage with the tightest and most specific binding. The DNA of phage that show specificity is sequenced to determine the peptide-binding sequence.
The phage display library is based on a combinatorial library of random peptides of a given length (e.g., 7- or 12-mers) that are fused to the pIII minor coat protein of the filamentous coliphage M13 (New England Biolabs, www.neb.com). Five copies of the fused random pIII coat protein are located on one end of the phage particle and account for 10–16 nm of the 1 μm viral particle. The library used for the selection consists of 1.9 × 109 random sequences. This peptide combinatorial approach was used to identify proteins that specifically bind to inorganic nanoparticles such as semiconductor nanocrystals. Hence, this is a promising approach for selecting peptides that can recognize specific inorganic crystals and crystallographic orientations of nonbiological origin, including InP, GaAs, and Si (ref. 25; Fig. 6), and can control II-VI (ZnS, CdS, CdSe, ZnSe, and PbS; A.M.B., unpublished data) semiconductor nanocrystal size, crystal structure, shape, and optical properties.
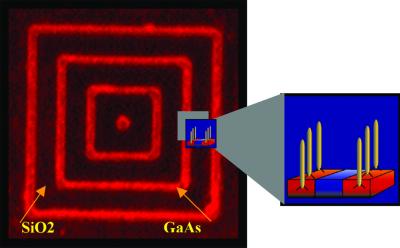
Figure 6.
Peptide selectivity for patterned GaAs. Fluorescently labeled phage displaying a peptide specifically selected to bind to GaAs bind only to the patterned GaAs nested square pattern on a wafer. The red lines (1 μm across) correspond to GaAs and the black spaces (4 μm across) are SiO2. This peptide-specific binding could also be used to deliver nanocrystals to specific locations.
Issues and Prospects
Biomimetic nanotechnology holds great promise as a vehicle to achieve progress in the areas of macromolecular crystallography, nanoelectronics, and nanorobotics. The key issues in the area currently include the following: (i) Can control be obtained over the sizes of crystalline or aperiodic arrays? Can 2D crystals be made larger than their current dimensions on the order of a micrometer or two? (ii) Can crystals be produced that are nearly error-free? Applications in nanoelectronics will require high degrees of perfection, although error-tolerant techniques (27) may alleviate this problem. (iii) Can nanomechanical devices transmit forces in the same way that larger devices do in the macroscopic world? (iv) Can the systems described here and in the related experiments pioneered by Alivisatos et al. (17), Mirkin et al. (18), and Mallouk and coworkers (28) be used to interface the architectural prowess of “wet” biomimetic nanotechnology with the functional potency of the “dry” nanotechnology of nanotubes (29) and nanoelectronic components (30, 31)?
The successes enjoyed by Whitesides and his coworkers (32) working on a slightly larger scale may be taken as an indication that bottom-up assembly and organization can be extended conveniently down through the nanoscale. Current efforts to extend DNA nanotechnology from two- to three-dimensional (3D) arrays can be expected to produce true 3D integration of nanoelectronic components, although this success, when it comes, will lead to other problems of addressing and heat dissipation. Biomolecular recognition and peptide evolution will be used to develop molecular tool kits for the design and synthesis of inorganic nanocrystals with the potential to offer even greater flexibility in materials synthesis and assembly than with current synthetic routes. Biomimetic nanotechnology is just beginning to bloom—the full inflorescence promises to be spectacular.
Acknowledgments
This work has been supported by the National Institute of General Medical Sciences Grant GM-29554 (to N.C.S.), the Office of Naval Research Grant N00014-98-1-0093 (to N.C.S.), the Defense Advanced Research Planning Agency/National Science Foundation Grant CCR-97-25021 (to N.C.S.), National Science Foundation Grants CTS-9986512, EIA0086015, and CTS-0103002 (to N.C.S.), the Army Research Office Grant DADD19–99-0155 (to A.M.B.), and by the Welch Foundation (A.M.B.).
Abbreviations
- DX
double crossover
- 2D
two-dimensional
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Nanoscience: Underlying Physical Concepts and Phenomena,” held May 18–20, 2001, at the National Academy of Sciences in Washington, DC.
References
- 1.Seeman N C. Trends Biotechnol. 1999;17:437–443. doi: 10.1016/s0167-7799(99)01360-8. [DOI] [PubMed] [Google Scholar]
- 2.Seeman N C. J Theor Biol. 1982;99:237–247. doi: 10.1016/0022-5193(82)90002-9. [DOI] [PubMed] [Google Scholar]
- 3.Qiu H, Dewan J C, Seeman N C. J Mol Biol. 1997;267:881–898. doi: 10.1006/jmbi.1997.0918. [DOI] [PubMed] [Google Scholar]
- 4.Robinson B H, Seeman N C. Protein Eng. 1987;1:295–300. doi: 10.1093/protein/1.4.295. [DOI] [PubMed] [Google Scholar]
- 5.Petrillo M L, Newton C J, Cunningham R P, Ma R-I, Kallenbach N R, Seeman N C. Biopolymers. 1988;27:1337–1352. doi: 10.1002/bip.360270902. [DOI] [PubMed] [Google Scholar]
- 6.Fu T-J, Seeman N C. Biochemistry. 1993;32:3211–3220. doi: 10.1021/bi00064a003. [DOI] [PubMed] [Google Scholar]
- 7.Li X, Yang X, Qi J, Seeman N C. J Am Chem Soc. 1996;118:6131–6140. [Google Scholar]
- 8.Winfree E, Liu F, Wenzler L A, Seeman N C. Nature (London) 1998;394:539–544. doi: 10.1038/28998. [DOI] [PubMed] [Google Scholar]
- 9.Mao C, Sun W, Seeman N C. J Am Chem Soc. 1999;121:5437–5443. [Google Scholar]
- 10.LaBean T H, Yan H, Kopatsch J, Liu F, Winfree E, Reif J H, Seeman N C. J Am Chem Soc. 2000;122:1848–1860. [Google Scholar]
- 11.Winfree E. In: DNA Based Computing. Lipton E J, Baum E B, editors. Providence, RI: Am. Math. Soc.; 1996. pp. 199–219. [Google Scholar]
- 12.Mao C, LaBean T H, Reif J H, Seeman N C. Nature (London) 2000;407:493–496. doi: 10.1038/35035038. [DOI] [PubMed] [Google Scholar]
- 13.Mao C, Sun W, Shen Z, Seeman N C. Nature (London) 1999;397:144–146. doi: 10.1038/16437. [DOI] [PubMed] [Google Scholar]
- 14.Yurke B, Turberfield A J, Mills A P, Jr, Simmel F C, Neumann J L. Nature (London) 2000;406:605–608. doi: 10.1038/35020524. [DOI] [PubMed] [Google Scholar]
- 15.Belcher A M, Wu X H, Christensen R J, Hansma P K, Stucky G D, Morse D E. Nature (London) 1996;381:56–58. [Google Scholar]
- 16.Falini G, Albeck S, Weiner S, Addadi L. Science. 1996;271:67–69. [Google Scholar]
- 17.Alivisatos A P, Johnsson K P, Peng X, Wilson T E, Loweth C J, Bruchez M P, Schultz P G. Nature (London) 1996;382:609–611. doi: 10.1038/382609a0. [DOI] [PubMed] [Google Scholar]
- 18.Mirkin C A, Letsinger R L, Mucic R C, Stofoff J J. Nature (London) 1996;382:607. doi: 10.1038/382607a0. [DOI] [PubMed] [Google Scholar]
- 19.Chan W C W, Nie S. Science. 1998;281:2016–2018. doi: 10.1126/science.281.5385.2016. [DOI] [PubMed] [Google Scholar]
- 20.Bruchez M, Moronne M, Gin P, Weiss S, Alivisatos A P. Science. 1998;281:2013–2016. doi: 10.1126/science.281.5385.2013. [DOI] [PubMed] [Google Scholar]
- 21.Colvin V L, Goldstein A N, Alivisatos A P. J Am Chem Soc. 1992;114:5221–5230. [Google Scholar]
- 22.Baum T, Bethell D, Brust M, Schiffrin D J. Langmuir. 1999;15:866–871. [Google Scholar]
- 23.Li M, Wong K K W, Mann S. Chem Mater. 1999;11:23–26. [Google Scholar]
- 24.Cusack L, Rao S N, Wenger J, Fitzmaurice J D. Chem Mat. 1997;9:624–631. [Google Scholar]
- 25.Whaley S R, English D S, Hu E L, Barbara P R, Belcher A M. Nature (London) 2000;405:665–668. doi: 10.1038/35015043. [DOI] [PubMed] [Google Scholar]
- 26.Brown S. Proc Nat Acad Sci USA. 1992;89:8651–8655. doi: 10.1073/pnas.89.18.8651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heath J R, Keukes P J, Snider G, Silliams R. Science. 1998;280:1716–1721. [Google Scholar]
- 28.Mbindyo J K N, Reiss B D, Martin B R, Keating C D, Natan M J, Mallouk T E. Adv Mater. 2001;13:249–254. [Google Scholar]
- 29.Colbert D T, Zhang J, McClure S M, Nikolaev P, Chen Z, Hafner J H, Owens D W, Kotula P G, Carter C B, Weaver J H, et al. Science. 1994;266:1218–1222. doi: 10.1126/science.266.5188.1218. [DOI] [PubMed] [Google Scholar]
- 30.Reed M A, Zhou C, Muller C J, Burgin T P, Tour J M. Science. 1997;278:252–254. [Google Scholar]
- 31.Collier C P, Wong E W, Belohradsky M, Raymo F M, Stoddart J F, Keukes P J, Williams R S, Heath J R. Science. 1999;285:391–394. doi: 10.1126/science.285.5426.391. [DOI] [PubMed] [Google Scholar]
- 32.Trau M, Yao N, Kim E, Xia Y, Whitesides G M, Aksay I A. Nature (London) 1997;390:674–676. [Google Scholar]