Abstract
Despite its remarkable materials properties, the structure of spider dragline silk has remained unsolved. Results from two probe microscopy techniques provide new insights into the structure of spider dragline silk. A soluble synthetic protein from dragline silk spontaneously forms nanofibers, as observed by atomic force microscopy. These nanofibers have a segmented substructure. The segment length and amino acid sequence are consistent with a slab-like shape for individual silk protein molecules. The height and width of nanofiber segments suggest a stacking pattern of slab-like molecules in each nanofiber segment. This stacking pattern produces nano-crystals in an amorphous matrix, as observed previously by NMR and x-ray diffraction of spider dragline silk. The possible importance of nanofiber formation to native silk production is discussed. Force spectra for single molecules of the silk protein demonstrate that this protein unfolds through a number of rupture events, indicating a modular substructure within single silk protein molecules. A minimal unfolding module size is estimated to be around 14 nm, which corresponds to the extended length of a single repeated module, 38 amino acids long. The structure of this spider silk protein is distinctly different from the structures of other proteins that have been analyzed by single-molecule force spectroscopy, and the force spectra show correspondingly novel features.
The last decade has seen a significant increase in the scientific literature on spider dragline silk. This interest is due to the impressive mechanical properties of spider dragline silk, at a time when biomaterials and biomimetics are both exciting interest in the rapidly growing field of materials research. The viscoelastic fibers of spider dragline silk combine both a high tensile strength that is comparable to steel and is only slightly inferior to Kevlar (≈2/3 of its tensile strength), and a high elasticity (≈30% elongation before failure) that is comparable to rubber (1–4). This unique combination makes spider dragline silk mechanically superior to almost any other natural or man-made material. It is apparent that the mechanical properties of the dragline silk protein's intramolecular structure as well as the intermolecular organization of these proteins in the fiber are critical for spider silk performance (2, 5). We report here the partial mechanical and structural characterization of a recombinant dragline silk protein. This recombinant silk protein provides a valuable test system for establishing the relationships between protein structure and mechanical properties in spider silk.
Spider dragline silk can be pictured as a composite material consisting of a semiamorphous matrix filled with tiny (<10 nm) nano-crystalline-like particles (6, 7). The amino acid sequence for spider dragline silk proteins is comprised of poly(A) [poly(alanine)], for some silks substituted by poly(GA) [poly(glycyalanine)], and glycine-rich sequences (2, 8, 9). Despite intensive structural studies on spider dragline silk proteins, their exact structural organization remains to be solved. The 4- to 10-residue-long poly(A) and poly(GA) motifs are thought to be involved in the formation of β-sheet nano-crystalline-like particles. Glycine-rich sequences are thought to fold into some non-α-helical helical structure for GGX or into β-turns for GPGGX, thus forming the semiamorphous matrix (2, 10). On the other hand, a few reports suggest that at least part of these glycine-rich motifs can also fold into β-sheets (11) and/or form an interphase between crystalline-like objects and a semiamorphous matrix (12). NMR and x-ray diffraction experiments show that the crystalline-like particles are well oriented along the silk fiber with polypeptide chains parallel and alanine residues perpendicular to the fiber axis (7, 13–17). These findings suggest that protein molecules are overall well oriented in the silk fiber. This high degree of molecular orientation, together with structural organization of dragline silk proteins, is a prerequisite for the unique mechanical properties of the whole silk fiber (18).
Recombinant spider silk proteins (19–21) have advantages over natural dragline silk for single-molecule research. Recombinant spider silk proteins have a regular, known sequence and can readily be purified in adequate quantities. Natural spider dragline silk proteins are larger and are difficult to solubilize after they have formed silk fibers.
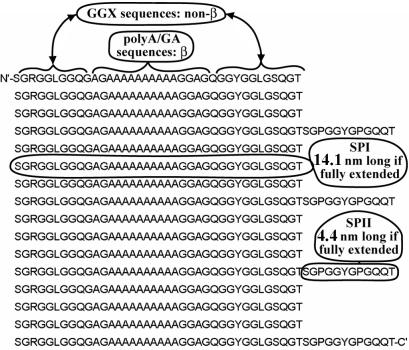
Two consensus sequences, SPI and SPII, represent major repetitive elements from spider dragline silk proteins. A family of recombinant spider silk proteins have been synthesized from these SPI and SPII sequences (22). The SPI sequence consists of 38 aa and includes 16-aa-long poly(A) and poly(GA) stretches, flanked on both sides with a total of 22 aa forming glycine-rich GGX motifs (Fig. 1). The SPII sequence consists of 12 aa representing two GPGGX motifs. All circular dichroism measurements on proteins from this protein family indicate that almost two thirds of the secondary structure of (SPI)n modular proteins is made of β-sheets and the rest is mostly β-turns or similar structures with no significant amount of α-helical structure observed (22). This observation is in good agreement with data obtained previously on native spider dragline silk fibers (9, 16) and suggests that recombinant synthetic modular SPI/SPII proteins have a structural organization very similar (if not identical) to native dragline silk proteins. We used the SPI/SPII modular protein in Fig. 1 for studying the mechanical properties and structural organization of spider silk proteins. The modular protein in Fig. 1 has the formula [(SPI)4 + SPII]4, which we call pS(4+1). With a 52-kDa molecular mass, this is the largest protein available to us from the family of SPI/SPII modular proteins. Its large size is advantageous for both atomic force microscopy (AFM) imaging and single molecule force spectroscopy.
Figure 1.
The sequence of pS(4+1) recombinant silk protein, with selected SPI and SPII modules identified, as well as poly(A/GA) sequences and (GGX)n sequences. N′- and C'- indicate the N-terminal and C-terminal amino acids, respectively.
Materials and Methods
Sample Preparation.
The gene for a pS(4+1) modular recombinant protein was produced as described (22) and inserted in the pET24 expression vector (Novagen). The protein sequence is made of 16 SPI and 4 SPII modules (22) arranged in (SPI)4-SPII-(SPI)4-SPII-(SPI)4-SPII-(SPI)4-SPII pattern (Fig. 1). The pS(4+1) recombinant protein was produced in the Escherichia coli expression strain BL21(DE3) pLysS grown to midlog phase in defined salts medium (23) with 30 μg/ml kanamycin and 34 μg/ml chloramphenicol. Expression was induced with 1 mM isopropyl β-d-thiogalactoside (IPTG). The cells were harvested by centrifugation and lyophilized for purification. The collected cell pellet was lysed with organic acid under denaturing conditions and clarified by centrifugation. The solution was dialyzed into 2 M urea/10 mM Tris, pH 9.9, and loaded on a QAE-Sephadex A50 (Amersham Pharmacia Biotech) column. The flow-through was collected, and the column was washed to recover the silk protein. Protein was dialyzed in a “silk buffer” (160 mM urea/10 mM NaH2PO4/1 mM Tris/10 mM glycine, pH 5.0) and kept at 4°C as a 1 mg/ml stock solution. The silk protein prepared in this way was 99.1% pure, based on amino acid composition analysis.
For AFM observations, the stock solution was diluted in the silk buffer to a desired concentration (100–300 μg/ml) and deposited as a drop on a freshly cleaved mica surface. Protein molecules were allowed to bind to the mica by incubating for 3–5 min, and excess protein was removed by washing with a flow of silk buffer. For observation in air, this procedure was followed by a wash with milliQ-purified water, and the sample was dried under a stream of compressed air purified by passing it through a 0.22-μm filter. For observation under the liquid, the sample was washed with the corresponding solution and was imaged immediately under water or the corresponding buffer to prevent the sample from drying.
For force spectroscopy experiments, samples were prepared essentially as for AFM imaging. No difference was detected between air-dried and non-dried samples. All samples were submerged, just before pulling, under milliQ-purified water with 10 mM CaCl2.
AFM Imaging and Force Spectroscopy.
AFM imaging, both in air and under aqueous solution, was performed by standard procedures in tapping mode on a MultiMode AFM with E scanner and Nanoscope III electronics (Digital Instruments, Santa Barbara, CA). Cantilevers for AFM imaging were also obtained from Digital Instruments. AFM images were captured, processed, and analyzed with nanoscope iii software, versions 4.42r4 and 4.43r8 (Digital Instruments).
Force spectroscopy was performed on a Molecular Force Probe MFP-SA (Asylum Research, Santa Barbara, CA). Silicon nitride cantilevers were obtained from Park Scientific (Stanford, CA). The spring constant for each cantilever was determined by measuring the amplitude of its thermal fluctuations (24), by using the Asylum Research software. The MFP was used to obtain force-vs.-piezo-extension curves, which were calibrated and partially analyzed by a manufacturer-supplied software based on igor pro (WaveMetrics, Lake Oswego, OR) software. To perform worm-like-chain (WLC) curve-fit analyses, this software was correspondingly modified by using the built-in macro language.
Results and Discussion
AFM of Synthetic Silk.
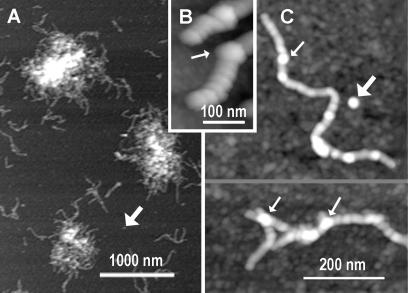
Most of the pS(4+1) silk protein appeared as a fibrous material with a tendency to form nanofiber aggregates on freshly cleaved mica, when imaged in air or in liquid (Fig. 2). The pS(4+1) silk nanofibers were observed not only on bare mica, which is hydrophilic and negatively charged, but also on surfaces that were hydrophobic and either neutral or positively charged (data not shown). The two hydrophobic surfaces were prepared on mica by depositing monolayers of positively charged APTES (3-aminopropyltriethoxysilane), or neutral OcTES (n-octyltriethoxysilane). These findings indicate that nanofiber formation is an intrinsic property of the pS(4+1) silk protein rather than an artifact caused by the interactions of pS(4+1) with the bare mica surface.
Figure 2.
Silk nanofibers formed from pS(4+1) silk protein deposited on mica; AFM height images. (A) pS(4+1) silk protein is present primarily as aggregates of nanofibers. (B and C) Close-up AFM images of the pS(4+1) nanofibers show segmented substructure. Fat arrows indicate isolated blobs that are predicted to be segments of nanofibers, based on their sizes. Thin arrows indicate bulges, which often occur at branch points on nanofibers and may be due to nanofibers overlapping.
Each fiber shows a distinct segmented substructure (Fig. 2 B and C), with an average segment length of 35 ± 9 nm, average height of 3 ± 1.5 nm at the center of the segment, and apparent width of 34 ± 5 nm when imaged in air. Mean volumes of these segments are estimated to be in the range of 1000–1300 nm3. Volumes of biomolecules in AFM images tend to correspond to the volumes predicted by their molecular masses, assuming a molecular density of 1–1.3 g/ml (25–27). Based on this relationship, segments have a mean molecular mass of ≈1000–1600 kDa. This mass is of order 20–35 times the molecular mass of the pS(4+1) protein monomer of 52 kDa.
In addition to pS(4+1) nanofibers, we observed single blobs that are roughly comparable in size and volume to the segments in the nanofibers (Fig. 2, fat arrows). We tentatively identify these blobs as isolated nanofiber segments. There are only a few blobs; the great majority of the material on the surface is either fibrous or what looks like aggregates of fibrous material. If the pS(4+1) blobs bind to the surface as well as the pS(4+1) nanofibers, then nearly 100% of the pS(4+1) molecules were competent for nanofiber formation under imaging conditions.
Bulges were observed on some of the nanofibers (narrow arrows in Fig. 2); their heights were nearly twice as high as the 3-nm heights of typical nanofiber segments. Although observed in different parts of nanofibers, these bulges often coincide with occasions when two nanofibers appear to overlap each other (Fig. 2C Lower). A simple pile-up of the nanofibers can explain such bulges. As seen in Fig. 2, pS(4+1) nanofibers show little branching, and, in fact, we propose that much of the observed nanofiber branching is due to nanofibers overlapping or piling up. Therefore, we propose that the pS(4+1) silk protein possesses a regular and well defined secondary and, possibly, tertiary structure, with well defined fiber-forming structures/signals localized on two opposite ends of each pS(4+1) nanofiber segment.
The nanofibers of pS(4+1) protein display a high degree of flexibility at intersegment contacts as well as a high variability in fiber length. We observed nanofibers as short as 60 nm and as long as 660 nm. The longest nanofibers have more than 20 segments. It is obvious, however, that some factors restrict infinite growth of the nanofibers. We propose that bonds at contact sites are relatively weak and break because of shearing forces on long nanofibers. On the other hand, “shortening” of the nanofibers can be due to misfolding of “fiber-forming” structures/signals at one of the segment ends.
Although the pS(4+1) silk protein is a synthetic protein with a molecular mass (52 kDa) ≈1/6 the mass of native spider dragline silk proteins (250–350 kDa; ref. 5), it is reasonable to expect that the secondary and tertiary structures of these proteins are very closely related and that native silk proteins display similar fiber-forming properties. If this assumption is true, it sheds new light on the silk production picture. Dragline silk is produced by a draw down of a liquid silk dope (28). It seems that, at some stages of the silk dope processing, the protein molecules go from an isotropic state to a nematic liquid-crystalline state (5, 28). This transition is important both for reducing the viscosity of the dope and for reducing the energy needed to align the protein molecules along the silk fiber axis. Fiber formation, similar to the one observed for pS(4+1), can facilitate such isotropic-to-nematic transition for native silk proteins. Transition of monomeric pS(4+1) into fibrous form can lower the minimal necessary flow rates needed to convert from the isotropic state to the nematic state in the ducts of silk-producing ampullate glands and will further reduce the viscosity of silk dope. There is evidence from observations with transmitted polarized light microscopy (29) that the nematic phase of spider silk could be the result of just such a supermolecular assembly of silk polymers as we have observed here.
Molecular Force Spectroscopy of Synthetic Silk.
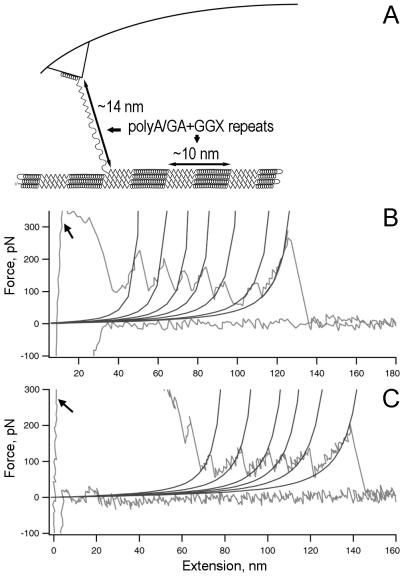
Molecular force spectroscopy gives force-vs.-extension plots for the pS(4+1) silk protein molecules (Fig. 3). The numerous high-force rupture events at the beginning of the pulls are, most likely, because of multiple protein molecules attached to the tip of the AFM probe (arrows, Fig. 3 B and C). As the probe is moved away from the surface, it breaks contact with the surface and with most of the protein molecules. In some cases, only a single molecule will survive the initial pull distances and will stretch with sequential breaking of structural elements, as diagrammed in Fig. 3A. Such a molecule will break off the tip only after being pulled for significantly longer distances. In these cases, a single-molecule force spectrum is recorded at the latter part of the pull as a series of rupture peaks (Fig. 3 B and C).
Figure 3.
Single-molecule force spectroscopy for pS(4+1) silk protein molecules. (A) Diagram of experimental setup, in which rupture peaks are believed to stretch one or more poly(A/GA+GGX) repeating units. (B and C) Force spectra (force-vs.-extension curves) for the unfolding of single molecules of pS(4+1) silk protein. The WLC model curves are fitted to each rupture force peak. The persistence length for each fit is 0.4 nm. Arrows indicate high-force rupture events at the beginning of the pulls, most likely because of multiple protein molecules attached to the tip of the AFM probe.
In numerous previous force spectroscopy studies, the WLC model (30) was used to characterize mechanical unfolding of proteins. The WLC describes the relationship between the protein extension and the entropic force generated as a result of such extension. In particular, the WLC gives peak-to-peak distances that agree closely with the distances expected from the lengths of unfolded polypeptide (31).
We fitted the WLC model to our experimental data by varying two model parameters: persistence length and contour length for the polypeptide backbone chain. The fit was first done to the last rupture event on each selected plot, as a test for a number of pulled molecules. If the fitted persistence length was near 0.4 nm, a typical value for a single polypeptide chain at high force (32), then all preceding rupture events on the plot were fitted with the WLC model for contour length with the persistence length fixed at 0.4 nm. Experimental data and WLC fits were plotted on the same graph (Fig. 3 B and C) to visually assess the quality of the fit. Results from all pulls with single molecule rupture events were analyzed statistically. The average force for rupture peaks was 176 ± 73 pN (Fig. 4 Inset). The high degree of variability in rupture force can be due to both different pulling speeds (refs. 32 and 33; we used from 200 nm/s to 1500 nm/s) and stochastic variability.
Figure 4.
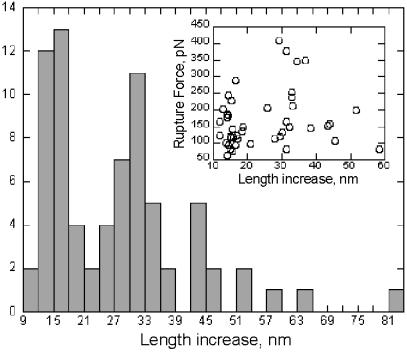
Histogram analysis of length increase (peak-to-peak distances) for rupture events in pS(4+1) silk protein molecules. Note that most of the data coincide with ≈N × 14 nm in length increase, where N = 1, 2, or 3. (Inset) Length increase vs. corresponding rupture force.
These average rupture forces are close to the ones obtained by force spectroscopy for mechanical/structural proteins such as titin (31) and tenascin (31, 34). Both tenascin and titin have Ig-like β-sheet barrel folds made by two antiparallel β-strands, each held together by a number of inter-strand hydrogen bonds (H-bonds) arranged in parallel (35). On the other hand, rupture forces are smaller for α-helix-rich proteins such as spectrin (32), and for proteins that do not normally experience forces, such as the enzyme barnase (36). This difference is attributed to the fact that intrahelix H-bonds in α-helices are arranged in series and that the interhelix hydrophobic or van der Waals interactions between α-helices are significantly weaker than the H-bonds between β-sheets. Thus, the pS(4+1) silk protein seems mechanically to belong to the β-sheet group of proteins. This result correlates well with the predicted β-sheet/β-turn fold for spider dragline silk proteins (2, 10) and with CD data for SPI/SPII modular proteins (22) and dragline silk fibers (9, 16).
The sawtooth pattern of the unfolding force curve for pS(4+1) (Fig. 3 B and C) is similar to other proteins with modular structures (31, 32, 34). This pattern shows that, after the pulling force reaches a certain threshold value, a defined structural unit in the protein breaks in a cooperative way. It is a common observation that the difference in contour length between adjacent rupture peaks corresponds to the length of polypeptide chain released after the preceding rupture event/peak.
We calculated the total length of the extended polypeptide chain for a single pS(4+1) protein molecule to be ≈243 nm, based on the assumption that a single amino acid in the polypeptide chain can be extended up to 0.37 nm in length. The maximum measured total extension for the single-molecule rupture events never exceeded 246 nm. This result indicates that our pulls, as in Fig. 3 B and C, are on single pS(4+1) protein molecules, and that single unfolded pS(4+1) molecules lose contact with the other pS(4+1) molecules in the silk nanofiber after they unfold.
The peak-to-peak distances in pS(4+1) unfolding force curves were not uniform. As seen in the histogram of Fig. 4, the increases in contour length between successive rupture peaks form three distinct groups clustered around ≈14 nm, 28 nm, and 42 nm. The calculated contour length for a fully extended SPI module is 14.1 nm; SPII modules will add an additional 4.4 nm, respectively, and SPI sequences occur four times as frequently as SPII sequences (Fig. 1). These findings indicate that an individual rupture event unfolds a length comparable to 1–3 SPI modules (sometimes together with an SPII module). We only rarely recorded peak-to-peak rupture distances above 45 nm (≈6% of rupture distances were >45 nm). There was no correlation between the force required for a particular rupture event and the length of polypeptide chain released (Fig. 4 Inset).
As in our model of Fig. 3A, we predict that a 14-nm rupture event actually pulls up a poly(A/GA) sequence plus the following (GGX)n sequence. We name these repeating units “poly(A/GA+GGX).”
The peak-to-peak distances are uniform for the other recombinant modular proteins studied by force spectroscopy to date (31, 32, 34). Titin, for example, is composed of tandem Ig domains, and each rupture event corresponds to the unfolding of one independently folded Ig domain. This independent unfolding of titin's Ig domains suggests weak interactions or no interactions between the Ig domains of titin. Titin also differs from pS(4+1) silk protein in that much “hidden” polypeptide length is released when a domain unfolds, because only a few of the amino acids in Ig domains are subjected to the mechanical load during pulling (31).
Unlike titin's ruptures, the ruptures of pS(4+1) silk protein often release two or more repeating units of poly(A/GA+GGX). Furthermore, according to our model in Fig. 3A, poly(A/GA+GGX) silk protein repeats are not independently folded. Instead, these poly(A/GA+GGX) repeats are bonded to distant sequences on the pS(4+1) protein molecule. Specifically, the poly(A/GA) repeats form β-sheets within the protein molecule. We rarely observe a catastrophic unfolding of an entire pS(4+1) protein molecule. Therefore, the intramolecular forces hold the pS(4+1) molecule together even after one or more poly(A/GA+GGX) repeats have been pulled from it.
Model for Silk Structure.
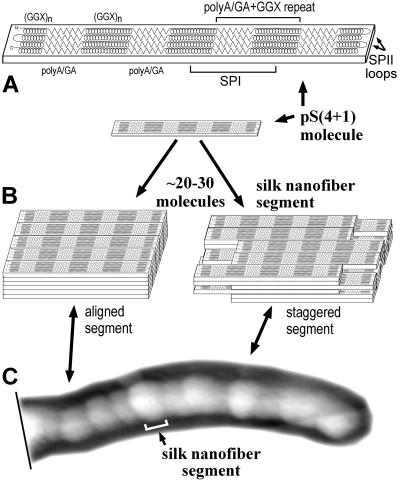
The pS(4+1) nanofiber morphology, as well as its mechanical unfolding pattern, can be explained by the model in Fig. 5. In this model, a single pS(4+1) protein molecule folds into a well defined structure in which β-strands of poly(A/GA) sequences in SPI modules form four H-bond-stabilized β-sheets (Fig. 5A). These β-sheets alternate with the non-β (GGX)n sequences of the SPI modules, which form random coils or non-α-helices. These folded molecules have the shape of an elongated slab with a pattern of alternating hydrophobic and hydrophilic regions formed by β-sheets and by non-β (GGX)n regions, correspondingly. Each slab is a single pS(4+1) molecule, as in Figs. 1 and 5A. The calculated dimensions of a slab are ≈0.54 nm high × 2 nm wide × 40 nm long. The height and width come from the spacing between typical antiparallel β-sheets (estimated as 0.53–0.55 nm) and the interchain distance along the H-bond direction (estimated as 0.47 nm; refs. 37 and 38). The length comes from the length of four folded SPI domains, calculated from their amino acid sequences [0.32–0.35 nm/aa in β-strand poly(A/GA) and ≈0.2 nm/aa in non-α-helical (GGX)n sequences]. The calculated length of the slabs—≈40 nm—is close to the measured segment length in AFM images of pS(4+1) silk nanofibers, which is 35 ± 9 nm.
Figure 5.
A model for pS(4+1) silk nanofiber organization. (A) The single pS(4+1) protein molecule's polypeptide chain folds into a flat slab-like structure in which four hydrophobic β-sheets of poly(A/GA) (zig-zags) are separated by hydrophilic non-α-helical GGX structures (spirals). Four poly(A/GA) β-strands form each of the β-sheets that compose the crystalline-like structures of spider dragline silk. The (GGX)n sequences are random coils or 310-helices or other non-α-helical helical structures. (B) Approximately 30 of these slab-like molecules form a “stack” or “nanofiber segment” because of hydrophobic interactions between β-sheets in aqueous environment. These nanofiber segments are thought to bind to each other through specific “fiber-forming” signals/structures at their ends. Alternate segment models show either perfect or imperfect (staggered) alignment. (C) The whole pS(4+1) protein nanofiber can be viewed as a chain of segments with each segment representing a single pS(4+1) protein “stack.” Under a stretching force (not shown), as in the draw-down step of silk processing, the secondary structure of semiamorphous GGX “matrix” transitions into a more extended form and locks into a 31-helix or β-strand configuration. Numerous inter- and intramolecular H-bonds are formed at this point between these newly formed structures.
In an aqueous environment, these molecular slabs will tend to aggregate, because of intermolecular hydrophobic interactions between their β-sheets. This aggregation is modeled in Fig. 5B as a stack of pS(4+1) molecular slabs. The silk protein molecules are aligned along the axis of the β-strands, as is known from NMR and x-ray diffraction (7, 15). In our diagram of Fig. 5B, these stacks are six slabs high and five slabs wide, based on the dimensions and estimated molecular weight of nanofiber segments in AFM images (Fig. 2 B and C). Because of strong and uniform hydrophobic interactions and H-bonds, the β-sheets in these slabs will form crystalline three-dimensional structures like ones found in native spider dragline silks (6, 7). The H-bonds between β-strands in a β-sheet (and the hydrophobic interactions between β-sheets to a lesser degree) can be viewed as the major stabilizing forces in such structures. The ends of each stack seem to carry signals/structures that facilitate pS(4+1) nanofiber formation. Thus, we suggest that each nanofiber segment corresponds to a separate pS(4+1) stack (Fig. 5 B and C).
The draw-down process, occurring as one of the last steps in a silk fiber production pathway (5), causes major structural rearrangements in silk proteins, resulting in a significant improvement in the fiber mechanical strength (39, 40). During this draw-down step, the glycine-rich part of silk proteins can partially transition from random coil or non-α-helical to the more extended structures, such as 31-helix (10) or β-strand (11). Similar structural rearrangements occur in the silk fibers when they are under a force load (41).
We propose that, during single-molecule force spectroscopy, one or more repeating units [poly(A/GA+GGX)] are pulled from the edge of a slab (Fig. 3A). The spacing between rupture peaks occurs in multiples of ≈14 nm (Fig. 4), which is longer than the 10-nm length of a poly(A/GA+GGX) repeat in the compact “slab” form. This extension from 10 nm to 14 nm is predicted to occur primarily in the non-β GGX sequences, because they have much smaller amino acid spacings in their compact form than in their extended forms.
Spider dragline silk, like lustrin in abalone shell and titin in muscle, thus appears to derive much of its combination of strength and toughness from its modular sacrificial bonds (31, 42). Of course, detailed structural and mechanical studies on spider dragline silk proteins should be performed in the future to validate our model, which remains highly speculative at this point.
Our results show that pS(4+1)-like proteins can be used as a test model for additional studies on recombinant modular SPI/SPII proteins with structurally modified modules. Research on such model systems is enhancing our understanding of the relationship between the spider dragline silk proteins' sequences/structures and their mechanical properties.
Acknowledgments
We thank Nathan Becker for writing valuable macros to analyze pulling curves. This research was supported by National Science Foundation Molecular and Cellular Biosciences (H.G.H., E.O.), National Science Foundation Division of Materials Research-9988640 (J.B.T.), the Materials Research Laboratory (National Science Foundation DMR96-32716 (to J.B.T.)), and Asylum Research.
Abbreviations
- AFM
atomic force microscope/microscopy
- (GGX)n
repeating aa sequences glycine-glycine-X, where X is usually leucine, tyrosine, or glutamine, represented as a loose or tight spiral in Figs. 3A and 5A and B
- H-bond
hydrogen bond
- poly(A/GA)
aa sequences of poly(alanine)/poly(glycylalanine), represented as a zig-zag in Figs. 3A and 5 A and B
- poly(A/GA+GGX)
a poly(A/GA) sequence of SPI plus the (GGX)n sequence of SPI that follows it: GAGAAAAAAAAAAGGAGQGGYGGLGSQGTSGRGGLGGQ
- pS(4+1)
modular recombinant spider silk protein composed of 16 SPI and 4 SPII modules arranged as follows: (SPI)4-SPII-(SPI)4-SPII-(SPI)4-SPII-(SPI)4-SPII (Fig. 1)
- SPI
38-aa-long synthetic spider silk sequence based on sequence from Nephila clavipes: SGRGGLGGQGAGAAAAAAAAAAGGAGQGGYGGLGSQGT
- SPII
12-aa-long synthetic spider silk sequence based on sequence from N. clavipes: SGPGGYGPGQQT
- WLC
worm-like chain (polymer model)
Footnotes
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Nanoscience: Underlying Physical Concepts and Phenomena” held May 18–20, 2001, at the National Academy of Sciences in Washington, DC.
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Hinman M B, Jones J A, Lewis R V. Trends Biotechnol. 2000;18:374–379. doi: 10.1016/s0167-7799(00)01481-5. [DOI] [PubMed] [Google Scholar]
- 2.Hayashi C Y, Shipley N H, Lewis R V. Int J Biol Macromol. 1999;24:271–275. doi: 10.1016/s0141-8130(98)00089-0. [DOI] [PubMed] [Google Scholar]
- 3.Tirrell D A. Science. 1996;271:39–40. doi: 10.1126/science.271.5245.39. [DOI] [PubMed] [Google Scholar]
- 4.Cunniff P M, Fossey S A, Auerbach M A, Song J W, Kaplan D L, Adams W W, Eby R K, Mahoney D, Vezie D L. Polym Adv Technol. 1994;5:401–410. [Google Scholar]
- 5.Vollrath F, Knight D P. Nature (London) 2001;410:541–548. doi: 10.1038/35069000. [DOI] [PubMed] [Google Scholar]
- 6.Grubb D T, Jelinski L W. Macromolecules. 1997;30:2860–2867. [Google Scholar]
- 7.Yang Z T, Liivak O, Seidel A, LaVerde G, Zax D B, Jelinski L W. J Am Chem Soc. 2000;122:9019–9025. [Google Scholar]
- 8.Gatesy J, Hayashi C, Motriuk D, Woods J, Lewis R. Science. 2001;291:2603–2605. doi: 10.1126/science.1057561. [DOI] [PubMed] [Google Scholar]
- 9.Warner S B, Polk M, Jacob K. J Macromol Sci Rev Macromol Chem Phys. 1999;C39:643–653. [Google Scholar]
- 10.Kummerlen J, Vanbeek J D, Vollrath F, Meier B H. Macromolecules. 1996;29:2920–2928. [Google Scholar]
- 11.Fukushima Y. Polym Bull. 2000;45:237–244. [Google Scholar]
- 12.Fossey S A, Tripathy S. Int J Biol Macromol. 1999;24:119–125. doi: 10.1016/s0141-8130(98)00079-8. [DOI] [PubMed] [Google Scholar]
- 13.Seidel A, Liivak O, Calve S, Adaska J, Ji G D, Yang Z T, Grubb D, Zax D B, Jelinski L W. Macromolecules. 2000;33:775–780. [Google Scholar]
- 14.Jelinski L W, Blye A, Liivak O, Michal C, LaVerde G, Seidel A, Shah N, Yang Z. Int J Biol Macromol. 1999;24:197–201. doi: 10.1016/s0141-8130(98)00085-3. [DOI] [PubMed] [Google Scholar]
- 15.Grubb D T, Ji G. Int J Biol Macromol. 1999;24:203–210. doi: 10.1016/s0141-8130(98)00086-5. [DOI] [PubMed] [Google Scholar]
- 16.Shao Z, Vollrath F, Sirichaisit J, Young R J. Polymer. 1999;40:2493–2500. [Google Scholar]
- 17.Shao Z Z, Vollrath F. Polymer. 1999;40:1799–1806. [Google Scholar]
- 18.Termonia Y. Macromolecules. 1994;27:7378–7381. [Google Scholar]
- 19.Lewis R V, Hinman M, Kothakota S, Fournier M J. Protein Expression Purif. 1996;7:400–406. doi: 10.1006/prep.1996.0060. [DOI] [PubMed] [Google Scholar]
- 20.Fahnestock S R, Bedzyk L A. Appl Microbiol Biotechnol. 1997;47:33–39. doi: 10.1007/s002530050884. [DOI] [PubMed] [Google Scholar]
- 21.Arcidiacono S, Mello C, Kaplan D, Cheley S, Bayley H. Appl Microbiol Biotechnol. 1998;49:31–38. doi: 10.1007/s002530051133. [DOI] [PubMed] [Google Scholar]
- 22.Prince J T, McGrath K P, Digiolamo C M, Kaplan D L. Biochemistry. 1995;34:10879–10885. doi: 10.1021/bi00034a022. [DOI] [PubMed] [Google Scholar]
- 23.Riesenberg D, Schulz V, Knorre W A, Pohl H D, Korz D, Sanders E A, Ross A, Deckwer W D. J Biotechnol. 1991;20:17–27. doi: 10.1016/0168-1656(91)90032-q. [DOI] [PubMed] [Google Scholar]
- 24.Hutter J L, Bechhoefer J. Rev Sci Instrum. 1993;64:3342–3342. [Google Scholar]
- 25.Golan R, Pietrasanta L I, Hsieh W, Hansma H G. Biochemistry. 1999;38:14069–14076. doi: 10.1021/bi990901o. [DOI] [PubMed] [Google Scholar]
- 26.Pietrasanta L I, Thrower D, Hsieh W, Rao S, Stemmann O, Lechner J, Carbon J, Hansma H. Proc Natl Acad Sci USA. 1999;96:3757–3762. doi: 10.1073/pnas.96.7.3757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schneider S W, Larmer J, Henderson R M, Oberleithner H. Pflügers Arch Eur J Physiol. 1998;435:362–367. doi: 10.1007/s004240050524. [DOI] [PubMed] [Google Scholar]
- 28.Knight D P, Vollrath F. Proc R Soc London Ser B. 1999;266:519–523. [Google Scholar]
- 29.Viney C, Huber A E, Dunaway D L, Kerkam K, Case S T. In: Silk Polymers: Materials Science and Biotechnology, ACS Symposium Series. Kaplan D, editor. Vol. 544. Washington, DC: Am. Chem. Soc.; 1994. pp. 120–136. [Google Scholar]
- 30.Bustamante C, Marko J F, Siggia E D, Smith S. Science. 1994;265:1599–1600. doi: 10.1126/science.8079175. [DOI] [PubMed] [Google Scholar]
- 31.Fisher T E, Oberhauser A F, Carrion-Vazquez M, Marszalek P E, Fernandez J M. Trends Biochem Sci. 1999;24:379–384. doi: 10.1016/s0968-0004(99)01453-x. [DOI] [PubMed] [Google Scholar]
- 32.Rief M, Pascual J, Saraste M, Gaub H E. J Mol Biol. 1999;286:553–561. doi: 10.1006/jmbi.1998.2466. [DOI] [PubMed] [Google Scholar]
- 33.Evans E. Annu Rev Biophys Biomol Struct. 2001;30:105–128. doi: 10.1146/annurev.biophys.30.1.105. [DOI] [PubMed] [Google Scholar]
- 34.Oberhauser A F, Marszalek P E, Erickson H P, Fernandez J M. Nature (London) 1998;393:181–185. doi: 10.1038/30270. [DOI] [PubMed] [Google Scholar]
- 35.Brändén C-I, Tooze J. Introduction to Protein Structure. New York: Garland; 1999. [Google Scholar]
- 36.Best R B, Li B, Steward A, Daggett V, Clarke J. Biophys J. 2001;81:2344–2356. doi: 10.1016/S0006-3495(01)75881-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Obrien J P, Fahnestock S R, Termonia Y, Gardner K C H. Adv Mat. 1998;10:1185–1195. [Google Scholar]
- 38.Arnott S, Dover D S, Elliott A. J Mol Biol. 1967;30:201–208. doi: 10.1016/0022-2836(67)90252-5. [DOI] [PubMed] [Google Scholar]
- 39.Guess K B, Viney C. Thermochim Acta. 1998;315:61–66. [Google Scholar]
- 40.Knight D P, Knight M M, Vollrath F. Int J Biol Macromol. 2000;27:205–210. doi: 10.1016/s0141-8130(00)00124-0. [DOI] [PubMed] [Google Scholar]
- 41.Sirichaisit J, Young R J, Vollrath F. Polymer. 2000;41:1223–1227. [Google Scholar]
- 42.Smith B L, Schaffer T E, Viani M, Thompson J B, Frederick N A, Kindt J, Belcher A, Stucky G D, Morse D E, Hansma P K. Nature (London) 1999;399:761–763. [Google Scholar]