Abstract
Mycothiol (MSH; 1d-myo-inosityl 2-[N-acetyl-l-cysteinyl]amido-2-deoxy-α-d-glucopyranoside) is the major low-molecular-weight thiol produced by mycobacteria. Mutants of Mycobacterium smegmatis mc2155 deficient in MSH production were produced by chemical mutagenesis as well as by transposon mutagenesis. One chemical mutant (mutant I64) and two transposon mutants (mutants Tn1 and Tn2) stably deficient in MSH production were isolated by screening for reduced levels of MSH content. The MSH contents of transposon mutants Tn1 and Tn2 were found to be less than 0.1% that of the parent strain, and the MSH content of I64 was found to be 1 to 5% that of the parent strain. All three strains accumulated 1d-myo-inosityl 2-deoxy-α-d-glucopyranoside to levels 20- to 25-fold the level found in the parent strain. The cysteine:1d-myo-inosityl 2-amino-2-deoxy-α-d-glucopyranoside ligase (MshC) activities of the three mutant strains were ≤2% that of the parent strain. Phenotypic analysis revealed that these MSH-deficient mutants possess increased susceptibilities to free radicals and alkylating agents and to a wide range of antibiotics including erythromycin, azithromycin, vancomycin, penicillin G, rifamycin, and rifampin. Conversely, the mutants possess at least 200-fold higher levels of resistance to isoniazid than the wild type. We mapped the mutation in the chemical mutant by sequencing the mshC gene and showed that a single amino acid substitution (L205P) is responsible for reduced MSH production and its associated phenotype. Our results demonstrate that there is a direct correlation between MSH depletion and enhanced sensitivity to toxins and antibiotics.
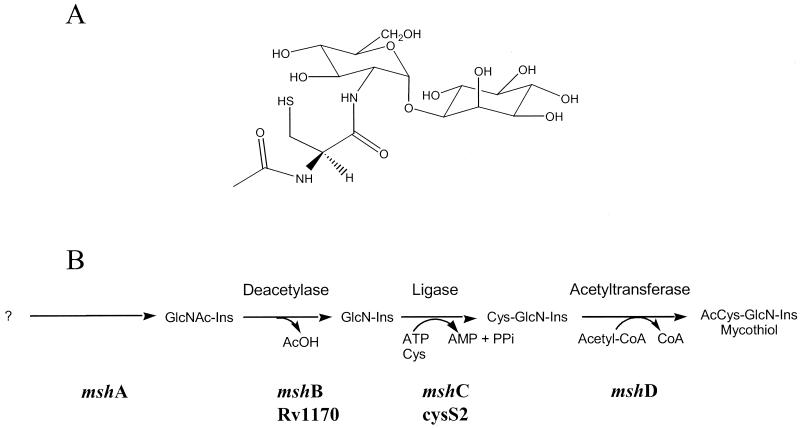
Mycothiol (MSH; Fig. 1A) is a low-molecular-weight thiol that serves functions in mycobacteria analogous to those of glutathione in gram-negative bacteria and eukaryotes (7). These functions include protection against oxidative damage and inactivation of electrophilic toxins (15, 17). The distribution of MSH is limited to gram-positive actinomycetes, and of all the species tested, mycobacteria appear to generate the highest levels of MSH (14). The absence of MSH in mammalian cells suggests that the enzymes involved in the metabolism of MSH may be attractive drug targets for either rational drug design or screening of libraries of inhibitory compounds (25).
FIG. 1.
(A) Structure of MSH; (B) proposed MSH biosynthesis pathway. Ac, acetyl; CoA, coenzyme A.
Although studies of MSH biosynthesis are still at an early stage, as shown in Fig. 1B, elements of the overall pathway have been described (1, 4, 16). Moreover, direct measurements of intermediates in bacterial extracts that support the proposed biosynthetic pathway have been reported (1). The genes for the MSH biosynthesis pathway were designated mshA, mshB, mshC, and mshD (16), with the corresponding enzymes labeled as shown in Fig. 1. It has been shown that 1d-myo-inosityl 2-acetamino-2-deoxy-α-d-glucopyranoside (GlcNAc-Ins) is an intermediate (16), and we presume that its formation is the first dedicated step in MSH biosynthesis. Glucosaminylinositol is formed by deacetylation of GlcNAc-Ins. We have previously identified mshB as the gene that encodes this deacetylase, and it appears that MshB is one of the control points for MSH biosynthesis (16). The next step in MSH biosynthesis is the linking of 1-d-myo-inosityl-2-deoxy-d-glucopyranoside (GlcN-Ins) to the amino acid cysteine by an ATP-dependent ligase to produce Cys-GlcN-Ins. This enzyme is encoded by the mshC gene. In the final step, an acetyltransferase encoded by the mshD gene converts Cys-GlcN-Ins to MSH (acetyl Cys-GlcN-Ins).
To assess the importance of MSH biosynthesis we generated mutants deficient in MSH production. We have previously described a Mycobacterium smegmatis chemical mutant (strain 49) that is blocked in both GlcN-Ins (17) and GlcNAc-Ins (16) production, indicating that it has a defect in the mshA gene or an earlier step. This mutant has increased susceptibility to hydrogen peroxide and rifampin but is highly resistant to isoniazid (17). In the present study we describe the production and characterization of chemical and transposon mutants of M. smegmatis which are deficient in the ligase activity. We further map the mutation in the chemical mutant to the mshC gene and show that a single amino acid substitution from leucine to proline is responsible for reduced levels of MSH production and its associated phenotype.
MATERIALS AND METHODS
Strains and chemicals.
Escherichia coli DH5α [F− recA1 hsdR17 thi-1 gyrA96 supE44 endA1 relA1 recA1 deoR Δ(lacZYA-argF)U169 (φ80 lacZ ΔM15] was grown in Luria-Bertani (LB) medium at 37°C. Kanamycin (100 μg/ml), ampicillin (100 μg/ml), and hygromycin (150 μg/ml) were used when required. M. smegmatis mc2155 was kindly provided by W. R. Jacobs and was grown in 7H10 medium with 10% oleic acid-albumin-dextrose-catalase and 0.05% Tween 20 (PBT). Kanamycin (25 μg/ml) and hygromycin (50 μg/ml) were used when required. Chemicals were purchased from Sigma Chemical Co. (St. Louis, Mo.). Mutagenesis of M. smegmatis mc2155 with N-methyl-N′-nitro-N-nitrosoguanidine (MNNG; Aldrich) to produce mutant I64 was performed as described by Newton et al. (17), except that diamide was not added to the plating medium. M. smegmatis strains mc2155 and I64 were grown at 37°C, while the transposon mutants were grown at 42°C.
Tn611 transposon mutagenesis.
The transposon mutagenesis of M. smegmatis mc2155 was performed as described by Guilhot et al. (8) with the thermosensitive plasmid pCG79 containing Tn611. The plasmid was electroporated into M. smegmatis, and transformants were selected on PBT supplemented with kanamycin at 30°C. Randomly chosen clones were grown at 30°C for 72 h in 5 ml of 7H9 medium supplemented with kanamycin. These cultures were used to inoculate antibiotic-free 7H9 medium, and the inoculated cultures were grown for 24 h at 39°C. Various dilutions were then plated on PBT supplemented with kanamycin and incubated at 39°C.
Molecular biology manipulations.
Genomic DNA was isolated from the M. smegmatis cultures as described by Jacobs et al. (9). Standard recombinant DNA techniques and Southern blotting were carried out as described by Sambrook et al. (20). M. smegmatis electroporation was performed as described by Snapper et al. (22). Mapping of the chemical mutation in the mshC gene was performed by comparing the mshC (cysS2)-cysQ gene region between I64 and mc2155 (unpublished sequence from The Institute for Genome Research database). Genomic DNA from both strains was amplified with the following primers: primer cysS25 (5′-ATGCAATCGTGGTCGGCACCGGCG-3′) and primer cysQ3 (5′-CGTCACAAGGCGGCTCCGAT-3′). The PCR product was cloned into PCR2.1 (Invitrogen), and the mshC gene was sequenced by using the primers described above and the universal T7 and m13 reverse primers.
Assay of MSH and MSH precursors.
Labeling of cell extracts with monobromobimane (mBBr) to determine the thiol content was performed by modifications of previously published protocols (1, 14). Cell pellets from 3 ml of culture were resuspended in 0.5 ml of warm 50% acetonitrile-water containing 2 mM mBBr (Calbiochem, San Diego, Calif.) and 20 mM HEPES (pH 8). The suspensions were incubated for 15 min at 60°C in a water bath and then cooled on ice. A 2- to 5-μl aliquot of 5 M HCl or 5 M trifluoroacetic acid was added to produce a final acidic pH.
Control samples were extracted with 0.5 ml of warm 50% acetonitrile-water containing 5 mM N-ethylmaleimide and 20 mM HEPES (pH 8). The suspensions were incubated for 15 min at 60°C and then cooled on ice. After addition of mBBr to 2 mM, the solution was incubated for a second time for 15 min at 60°C. The control samples were cooled but not acidified. The cell debris in each sample was pelleted by centrifugation (14,000 × g, 5 min) in a microcentrifuge.
High-pressure liquid chromatographic (HPLC) analysis of the thiols was carried out by injecting 25 μl of a 1:4 dilution of the samples in 10 mM HCl onto a Beckman Ultrasphere octyldecyl silane column (5 μm; 250 by 4.6 mm). Elution was accomplished with 0.25% glacial acetic acid (pH 3.6 with NaOH; buffer A) and 95% methanol (buffer B) by using the following gradient: at 0 min, 10% buffer B; at 15 min, 18% buffer B; at 30 min, 27% buffer B; at 32 min, 100% buffer B; at 34 min, 10% buffer B; and at 60 min, 10% buffer B (reinjection). The flow rate was 1 ml/min, and fluorescence detection was as described previously (1).
Ligase activity assay.
The specific activity of ligase was estimated by the ATP-dependent formation of Cys-GlcN-Ins catalyzed by M. smegmatis cell extracts prepared and analyzed in triplicate as described previously (1), except that centrifugation of the extracts was conducted by centrifugation at 14,000 × g for 5 min in a microcentrifuge.
Toxicity studies and antibiotic sensitivity tests.
Hydrogen peroxide toxicity studies were performed as reported by Newton et al. (17). To determine sensitivity to alkylating agents, the bacteria were plated on PBT or LB medium plates supplemented with the appropriate antibiotics and with different concentrations of the alkylating agents mBBr, iodoacetamide, and chlorodinitrobenzene (CDNB), which were poured into the molten agar. To determine sensitivity to redox-cycling agents and antibiotics, disk assays were performed. Briefly, cells were grown to the mid-log phase, and a lawn of cells was plated onto PBT or LB medium plates supplemented with appropriate redox-cycling agents and antibiotics. Various amounts of redox-cycling agents were added to the disk in a 10-μl volume and the disks were allowed to dry. The disks were placed onto the lawn of cells and incubated for 2 to 3 days. To determine antibiotic MICs, isoniazid, erythromycin, and vancomycin Etest strips (Oxoid) were used, and the manufacturer's guidelines for interpretation of the results were followed.
Complementation of mutant I64.
The mshC gene was cloned by PCR from M. smegmatis mc2155. The gene was amplified from genomic DNA with the following primers: primer psodmshCf (5′-GGATCCATGCAATCGTGGTCG-3′) and primer psodmshCr (5′-CATATGTTAGAGGTCCACACCCAG-3′). The forward and reverse primers were designed to contain BamHI and NdeI sites, respectively, for cloning into the pSODIT-2 (kindly provided by K. de Smet) M. smegmatis-E. coli shuttle vector. The corresponding PCR product was cloned into the vector PCR2.1. The mshC gene fragment digested with BamHI and EcoRV (from the cloning vector) was ligated to BamHI- and EcoRV-digested pSODIT-2. The ligation mixture was transformed into E. coli DH5α cells and plated onto LB agar plates containing 100 μg of hygromycin per ml. After verification, the resulting plasmid, designated pSODmhsC, was purified and electroporated into electrocompetent wild-type and mutant M. smegmatis cells.
RESULTS
Production of M. smegmatis chemical mutants.
Chemical mutagenesis with MNNG was used to produce a grided library of 1,600 M. smegmatis mc2155 clones which were subjected to an initial screen for MSH content by a blotting immunoassay protocol (23). A total of 56 colonies were selected for further examination. These were grown in liquid culture, and the cells were extracted for HPLC analysis for their MSH contents. Although several mutants appeared to have reduced MSH contents, one mutant, strain I64, was found to have a significantly lower MSH content (see Table 2) and was selected for further study.
TABLE 2.
Levels of thiol and amine intermediates and ligase activity in MSH-deficient mutants
| M. smegmatis strain | GlcN-Ins concn (μmol/g [dry wt]) | Ligase sp act (nmol/min/mg of protein) | MSH concn (μmol/g [dry wt]) |
|---|---|---|---|
| mc2155 | 0.1 | 0.320 ± 0.010 | 10.0 |
| I64 | 2.5 | 0.004 ± 0.001 | 0.1 |
| Tn1 | 2.6 | 0.004 ± 0.002 | <0.004 |
| Tn2 | 1.9 | 0.004 ± 0.002 | <0.003 |
Production of M. smegmatis MSH transposon mutants.
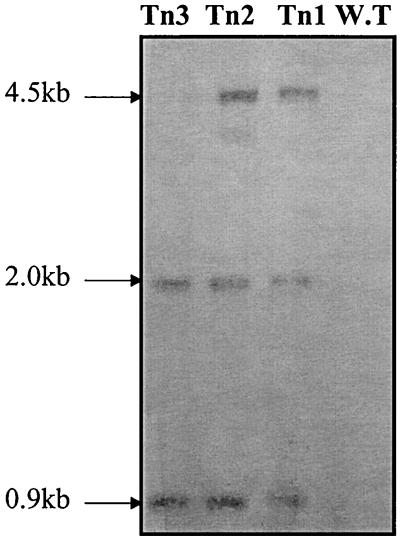
A transposon mutant library of M. smegmatis consisting of 10,000 mutants was created by using thermosensitive plasmid pCG79 (8, 19). One thousand representative mutants were picked, sorted, and cultured individually in enzyme-linked immunosorbent assay (ELISA) plates. The mutants were screened by ELISA with a polyclonal antibody against MSH (24). Although three independent MSH-deficient mutants were identified (Table 1), only two of these mutants (mutants Tn1 and Tn2) were found to stably incorporate the Tn611 transposon into the genome, as judged by Southern analysis (Fig. 2). As observed in Fig. 2, if the insertion is in the genome, three bands are expected when IS6100 is used as a probe for hybridization to PstI-digested genomic DNA of the mutants: two bands (0.9 and 2 kb) identical to those in the parent plasmid and a third one of a different size that in our case turned out to be 4.5 kb for mutants Tn1 and Tn2. A faint band of 4.5 kb was also observed in an initial Southern hybridization in which the genomic DNA of mutant Tn3 was examined. However, subsequent experiments revealed that the transposon insertion in mutant Tn3 is not stable, as this mutant failed to grow at 42°C on selective plates (Table 1). Thus, phenotypic analysis of this mutant was aborted. The Southern hybridization with an IS6110-labeled probe also confirmed that only one transposon was present in the chromosome for each of the two transposon mutants (Fig. 2).
TABLE 1.
Initial identification and characteristics of mutants
| M. smegmatis strain | Growth on isoniazid at 50 μg/ml | A405a | A600 | Signal/ growthb | Growth on isoniazid at 42°C |
|---|---|---|---|---|---|
| mc2155 | − | 1.394 | 0.208 | 4.629 | NDc |
| I64 | + | 0.396 | 0.405 | 0.978 | ND |
| Tn1 | + | 0.256 | 0.279 | 0.918 | + |
| Tn2 | + | 0.278 | 0.320 | 0.869 | + |
| Tn3 | + | 0.286 | 0.288 | 0.993 | − |
Absorbance of the alkaline phosphatase product in the MSH-capture ELISA (23).
Proportional to the MSH contents of the cells.
ND, not determined.
FIG. 2.
Southern blot analysis of the three M. smegmatis::Tn611 clones. The DNA of M. smegmatis mc2155 [the wild type (WT)] was included as a control and, as expected, showed no hybridization signal. Genomic DNAs were digested with PstI and probed for hybridization with IS6100 as described by Perez et al. (19).
MSH-deficient mutants lack Cys-GlcN-Ins ligase activity.
The mutants were analyzed for their levels of MSH and the amine intermediate during exponential growth by sensitive HPLC assays, as described previously (1). All three mutants and strain I64 were found to be deficient in MSH and overproduced GlcN-Ins by about 25-fold (Table 2). In all cases the content of Cys-GlcN-Ins, the ligase product, was undetectable (<0.002 μmol/g [dry weight]). A block in Cys-GlcN-Ins ligase activity would be expected to result in the accumulation of GlcN-Ins. These findings suggest that the ligase gene (mshC) has been interrupted in mutant I64 and the transposon mutants. Specific ligase assays (17) were undertaken with crude extracts from log-phase cells to assess enzyme levels (Table 2). The results show that the levels in mutants Tn1 and Tn2 are similar to the level in chemical mutant I64, with crude extracts of all three mutants having very low levels of l-cysteine-1-d-myo-inosityl-2-amino-2-deoxy-α-d-glucopyranoside ligase(MshC) activity.
MSH-deficient mutants are more sensitive to alkylating agents.
We have previously shown that MSH takes part in the detoxification of alkylating agents (15). Consequently, mutants lacking MSH should be more susceptible to these agents. To test this hypothesis, mutants Tn1, Tn2, and I64 were grown on PBT plates supplemented with increasing amounts of the alkylating agents mBBr, iodoacetamide, and CDNB. The fluorescent alkylating agent mBBr selectively reacts with cellular thiols, and mycobacteria possess an MSH-dependent amidase that can detoxify MSH-monobimane conjugates (15). A lack of MSH would presumably result in increased sensitivity to this toxin. Indeed, as illustrated in Table 3, transposon mutants Tn1 and Tn2 were fourfold more sensitive to mBBr than wild-type strain mc2155. Chemical mutant I64, which has 1% of the wild-type MSH content, was also more susceptible to mBBr, but not to the same extent as the transposon mutants, which have no detectable MSH. Iodoacetamide is another alkylating agent that is commonly used to derivatize proteins. Mutants I64 and Tn1 were twofold more sensitive to iodoacetamide than wild-type strain mc2155, and mutant Tn2 was fivefold more sensitive than wild-type strain mc2155. Mutants Tn1 and Tn2 were also more sensitive to CDNB (two- to threefold), another alkylating agent and a glutathione S-transferase substrate, than wild-type strain mc2155. In the case of the thiol-specific oxidant, diamide, which readily penetrates cells and oxidizes thiols to disulfides (11), we observed more then 10-fold increases in the sensitivities of the transposon mutants. The sensitivity of mutant I64 to diamide was double that of the wild type. In summary, the transposon mutants lacking MSH were more susceptible to alkylating agents than wild-type strain mc2155, although the degree of susceptibility depended on the toxin tested.
TABLE 3.
MICs of various alkylating agents for mc2155 and MSH-deficient mutants
| Agent | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| Wild type | I64 | Tn1 | Tn2 | |
| CDNB | 0.05-0.075 | 0.075 | 0.025 | 0.025 |
| mBBr | 0.1 | 0.075 | 0.025 | 0.025 |
| Iodoacetamide | 0.05 | 0.025 | 0.025 | 0.010 |
| Diamide | >10 | 5 | <1 | <1 |
MSH-deficient mutants are more sensitive to oxidative stress.
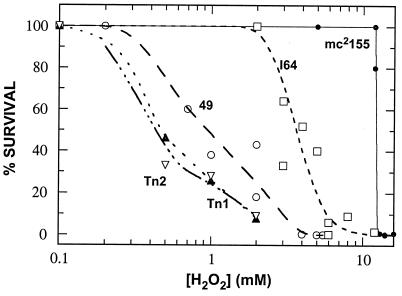
Glutathione-dependent peroxidases and transferases are known to be involved in protecting the cell against oxidative stress (6). M. smegmatis can tolerate high levels of peroxide, while previously reported MSH mutant 49 is very sensitive to hydrogen peroxide (17). As seen in Fig. 3, mutants Tn1 and Tn2 were about 10 times more sensitive to hydrogen peroxide than the wild-type strain, which is unable to tolerate as little as 1 mM H2O2, while the wild-type strain can survive in the presence of 10 mM H2O2. Strain I64 is intermediate between wild-type mc2155 and the transposon mutants in terms of its susceptibility to peroxide stress (Fig. 3).
FIG. 3.
Tests of sensitivities of MSH-deficient mutants versus wild-type strain mc2155 to hydrogen peroxide. Closed circle, mc2155; open squares, mutant I64; open circle, mutant 49; upward closed triangles, mutant Tn1; downward open triangles, mutant Tn2.
When the oxidative stress is in the form of redox-cycling agents that increase the superoxide concentration in the cell, a pattern similar to that described above was seen. As seen in Table 4, mutants Tn1 and Tn2 were markedly more sensitive to plumbagin and the antibiotic nitrofurantoin. The results for the transposon mutants with menadione showed no consistent pattern and are therefore inconclusive. Mutant I64, which contains a low level of MSH, is less sensitive to the agents listed above, undergoing a significant increase in the zone of inhibition only when it was exposed to 0.5 μmol of menadione.
TABLE 4.
Responses of growth of strain mc2155 and MSH-deficient mutants to redox-cycling agents
| Agent (concn [μmol]) | Zone of inhibition (radius [mm])
|
|||
|---|---|---|---|---|
| Wild type | I64 | Tn1 | Tn2 | |
| Plumbagin (0.01) | 13.5 ± 0.5 | 13.3 ± 1.9 | 23.0 ± 1.2 | 23.0 ± 1.0 |
| Plumbagin (0.005) | 11.0 ± 1.0 | 9.3 ± 0.9 | 17.0 ± 0.6 | 15.0 ± 0.0 |
| Menadione (0.5) | 19.7 ± 0.9 | 26.7 ± 0.9 | 31.3 ± 1.3 | 16.4 ± 1.9 |
| Menadione (0.1) | 9.7 ± 0.3 | 8.7 ± 0.7 | 20.7 ± 3.8 | 15.3 ± 2.0 |
| Nitrofurantoin (1.0) | 6.0 ± 0.6 | 8.7 ± 0.3 | 14.0 ± 2.7 | 35.0 ± 3.6 |
| Nitrofurantoin (0.5) | 4.7 ± 0.3 | 5.0 ± 0.6 | 10.7 ± 2.6 | NDa |
ND, not determined.
MSH-deficient mutants are more susceptible to a broad range of antibiotics than the wild-type strain.
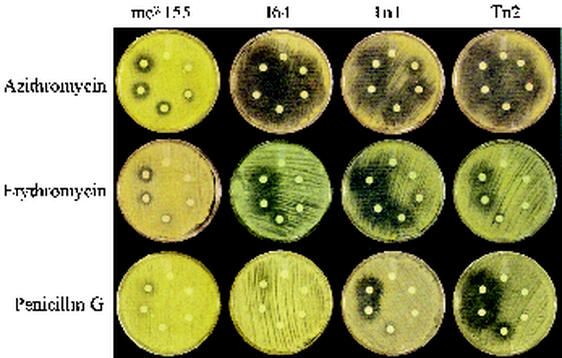
Since the mutants are devoid of MSH they cannot carry out MSH-dependent detoxification via the pathway involving MSH S-conjugate amidase activity (15). If our hypothesis that MSH is involved in protection against antibiotic challenge is correct, then these mutants should be more sensitive to certain antibiotics. Thus, we examined the antibiotic sensitivities of the transposon and chemical mutants. Transposon mutants Tn1 and Tn2 and chemical mutant I64 are 3- to 16-fold more sensitive than the parent strain (mc2155) to a variety of antibiotics, including erythromycin, azithromycin, vancomycin, and penicillin G (Fig. 4 and Table 5). Examination of the drugs used to treat Mycobacterium tuberculosis infection revealed that all the transposon mutants are more sensitive to rifamycin and rifampin than chemical mutant I64 (Table 5). We have not observed any significant difference for ethambutol or pyrazinamide. The opposite effect was observed for isoniazid, to which the mutants possessed over 100-fold higher levels of resistance than the wild type, while I64 exhibited only a 16-fold higher level of resistance (Table 5).
FIG. 4.
Tests of sensitivity MSH-deficient mutants versus wild-type strain mc2155 to antibiotics. Antibiotics were applied to disks with concentrations of 0, 2, 7.8, 32, 125, and 250 μg (clockwise from the top).
TABLE 5.
Antibiotic responses of strain mc2155 and MSH-deficient mutants
| Drug | MIC (μg/ml)
|
|||
|---|---|---|---|---|
| mc2155 | I64 | Tn1 | Tn2 | |
| Isoniazid | 2 | 32 | >250 | >250 |
| Erythromycin | 125 | 32 | 7.8 | 32 |
| Azithromycin | 7.8 | 2 | 2 | 2 |
| Vancomycin | 6 | 1 | 2 | 1.5 |
| Penicillin G | >250 | >250 | 32 | 32 |
| Rifampin | 32 | 32 | 7.8 | 7.8 |
| Rifamycin | 32 | 32 | 7.8 | 2 |
Chemical mutant I64 is mutated in mshC.
Recently, Sareen et al. (21) described the purification of MshC from M. smegmatis mc2155 and showed that the N-terminal sequence matched the ortholog of open reading frame Rv2130c from M. tuberculosis. When a Southern analysis was performed with genomic DNAs from mutant I64 and mc2155, in which the DNAs were probed with the mshC gene from M. smegmatis, there was no difference in the banding pattern between I64 DNA and mc2155 DNA (data not shown), indicating that the lack of MshC activity is probably due to a point mutation rather than the structural disruption of the mshC gene. To identify the mutation in the I64 mutant, mshC genes from both mc2155 and mutant I64 were cloned and sequenced. When the nucleotide sequences were compared, a single difference in the sequences appeared at position 614, where there is a thymine in wild-type strain mc2155 and a cytosine in mutant I64. This single base change results in a change in codon from CTG to CCG and consequently leads to the replacement of a single amino acid: from a leucine in the mc2155 ligase to a proline in the I64 ligase (Fig. 5).
FIG. 5.
Multiple-sequence alignment of partial MshC protein. The region containing the point mutation in mutant I64 (marked with boldface and underlined) was aligned to the MshC proteins of various actinomycetes. Completely identical amino acids are marked by asterisks, and dashes mark identity values of more than 60%.
Wild-type mshC complements mutant I64.
To confirm that this single point mutation in the I64 ligase is responsible for the reduced MshC activity and the reduced levels of MSH production in mutant I64 and the sensitivity of mutant I64 to antibiotics, a complementation approach was performed with wild-type mshC. The intact mshC gene was cloned into pSODIT-2. In this vector, the mshC gene is under the control of the M. tuberculosis superoxide dismutase promoter, which is constitutively expressed. Ten transformants were assayed for MSH, and all the transformants had levels of MSH greater than that detected in the wild type. One transformant was chosen for further characterization (Table 6). This complemented strain had 200% the wild-type level of MSH, and its sensitivity to isoniazid is similar to that of wild-type strain M. smegmatis mc2155. Furthermore, like the wild-type M. smegmatis strain but in contrast to mutant I64, the complement shows resistance to vancomycin and erythromycin, indicating that the mshC gene complements the I64 chemical mutant and reverts its antibiotic susceptibility. The complement was also checked for sensitivity to oxidative agents. As seen in Table 6, the complement was less sensitive to oxidative stress caused by menadione and nitrofurantoin than the I64 mutant and had sensitivity to redox-cycling agents more similar to that of mc2155. These results indicate that the lack of MSH is responsible for the increased sensitivity to antibiotics and redox-cycling agents in mutant I64.
TABLE 6.
Characteristics of chemical mutant I64 complemented with M. smegmatis mshC gene
| M. smegmatis strain | % MSH | MIC (μg/ml)
|
Zone of inhibition (mm)
|
|||
|---|---|---|---|---|---|---|
| Isoniazid | Erythromycin | Vancomycin | Menadione (0.5 μmol) | Nitrofurantoin (1.0 μmol) | ||
| mc2155 | 100 | 6 | 128 | 6 | 12 ± 1 | 5 ± 0 |
| I64 | 1-5 | 32 | 32 | 1.9 | 18 ± 2 | 12 ± 2 |
| I64::psodmshC | 200 | 6 | 128 | 6 | 12 ± 1 | 6 ± 1 |
DISCUSSION
In this study we constructed chemical and transposon mutants deficient in MSH production. We showed that these MSH-deficient mutants were lacking MshC (ligase) activity. Furthermore, we demonstrated that in chemical mutant I64 ligase activity is reduced due to a point mutation in the mshC gene. This point mutation results in an amino acid substitution of a proline for a leucine amino acid residue in chemical mutant I64. The leucine-to-proline substitution at residue 205 is a significant change since proline has restricted rotation about the C—N bond, and this will likely produce a substantial disruption in the surrounding polypeptide region. Since this region is largely conserved in MshC in a wide range of actinomycetes, it is plausible that the L205P substitution in I64 has a disruptive effect on MshC activity (Table 6). By complementing the chemical mutant with the mshC gene, we demonstrated that the mutated mshC gene is responsible for the lack of MSH production in this mutant.
Comparative Southern analysis with genomic DNAs from mutants I64, Tn1, and Tn2 and wild-type strain mc2155, in which the DNAs were probed with the mshC gene from M. smegmatis, revealed that there are no structural differences in the banding patterns between the mutants and mc2155 (data not shown). In the case of mutant I64 we have identified a point mutation in the mshC gene which can be complemented to restore the wild-type phenotype. Contrary to the case for I64, it seems that the mutation is not present in the mshC genes of mutants Tn1 and Tn2. We assume that the transposition occurred in a regulatory gene tightly controlling MshC production. This may explain the phenotypic differences among the mutants in response to various chemical stressors. Further studies are being carried out to identify the nature of these transposon mutations.
We characterized MSH-deficient ligase mutants to identify the role of MSH in the protection of mycobacterial cells against alkylating agents, free radicals, and antibiotics. Members of the family Actinomycetaceae, including the genus Mycobacterium, exhibit innate resistance to many currently available antimicrobial drugs. This phenomenon is generally ascribed to their cell wall impermeability. Here we showed that MSH-deficient mutants have increased sensitivities to a wide variety of agents including antibiotics, oxidizing agents, and alkylating agents.
The demonstration of a direct correlation between antibiotic susceptibility and MSH depletion, together with our earlier identification of the MSH-dependent detoxification system (15), may account for the natural ability of actinomycetes and especially mycobacteria to resist a wide range of antibiotics. Drug inactivation by mechanisms involving low-molecular-weight thiols has been reported before. A plasmid-encoded glutathione S-transferase is able to confer resistance to the antibiotic fosfomycin (3). It seems evident that in actinomycetes MSH serves a role analogous to that of glutathione in eubacteria and eukaryotes.
From the present results one may argue that the accumulation of an intermediate (GlcN-Ins) rather than MSH itself may lead to increased levels of peroxide sensitivity or altered antibiotic sensitivities. We do not think that this is the case, for several reasons. First, we have reported that a mutant, mutant 49, which is blocked at an early MSH biosynthesis step is very sensitive to peroxides, while it has completely different levels of biosynthesis intermediate, failing to produce GlcNAc-Ins, GlcN-Ins, Cys-GlcN-Ins, and MSH (16, 17). Yet, it could still be argued that one of the precursors to MSH is needed in an as yet unidentified process to confer resistance to peroxide. Since the mutants blocked at MshC produce both GlcNAc-Ins and GlcN-Ins but are nevertheless highly sensitive to hydrogen peroxide, we can eliminate the possibility that these intermediates are associated with the peroxide resistance. This leaves the possibility that Cys-GlcN-Ins rather than MSH might be the key to peroxide resistance. However, Cys-GlcN-Ins is present in M. smegmatis at almost undetectable levels (1), which makes it unlikely that it could play a significant role. Thus, we strongly argue that MSH is the key to antibiotic and peroxide sensitivities rather than one of the biosynthesis intermediates.
The main mechanism that detoxified sublethal quantities of oxidants and antibiotics as reported in this study probably consists of the MSH-dependent inactivation mechanisms of alkylating agents that we have reported on previously (15). This mechanism involves cleavage of the amide bond between GlcN-Ins and N-acetylcysteine, leading to the formation of N-acetylcysteine—R, where R is the toxin used, and recycling of GlcN-Ins forms another molecule of MSH (15). It seems likely that this mechanism is also involved in self-detoxification of antibiotics that are produced by fermentation broths of several antibiotic-producing microorganisms and that have been found to be remarkably nontoxic. For example, the polyketide antibiotic derivatives cysfluoretin (2) and WS009A and WS009B (12), produced in Streptomyces spp., were found to be nontoxic compared to the toxicities of compounds with related structures but lacking the N-acetylcysteine moiety when they were tested in vitro and in vivo. Gould and colleagues (5) expressed an ∼40-kb fragment of the polyketide kinamycin biosynthesis gene cluster in the heterologous host Streptomyces lividans ZX7 and found a biosynthetic intermediate, seongomycin, that contained an N-acetylcysteine moiety in the broth. They speculated that MSH was the source of the N-acetyl moiety in this novel polyketide derivative. The source of MSH was probably the host, S. lividans, which is known to produce MSH (14) and likely would have the MSH S-conjugate amidase activity (15) required to produce the mercapturic acid derivative and allow its excretion into the fermentation broth, as was observed (5).
In streptomyces MSH appears to detoxify and protect the cell from a variety of endogenously generated antibiotics and reactive intermediates, as evidenced by the isolation of mercapturic acids from fermentation broths (2, 5, 12). It is proposed that mycobacteria retain this capability and can similarly detoxify exogenously supplied antibiotics using MSH and the MSH S-conjugate amidase. MSH-deficient mutants of M. smegmatis are generally more sensitive to peroxide and antibiotics, which lends support to this proposal.
Interestingly, MSH-deficient mutants of M. smegmatis are extremely resistant to isoniazid. Isoniazid is an antimycobacterial prodrug which needs to be activated inside mycobacteria to form a reactive intermediate of yet undetermined structure (10). If MSH is considered the global reducing low-molecular-weight thiol found in mycobacteria, we suggest that it is involved in activation of isoniazid. MSH disulfide reductase is believed to maintain MSH—and, indirectly, the intracellular milieu of mycobacteria—in a reducing state (18). However, since our MSH mutants have other low-molecular-weight thiols such as cysteine and coenzyme A in their reduced forms (data not shown), these mutants cannot be described as being free of low-molecular-weight thiols. Thus, the isoniazid resistance is probably directly derived from a lack of MSH and not just a lack of low-molecular-weight thiol reductants in general. To add support to the assumption that the reducing activity is not sufficient for drug activation, we have noticed that our mutants do not possess increased levels of resistance to the prodrug ethionamide (data not shown), which is believed to be activated in a manner to similar to that in which isoniazid is activated. We have also shown that another antibiotic, nitrofurantoin, is reduced in our mutants to produce an active compound. It was previously shown that nitrofurantoin is activated by reduction into a nitro ion radical that reacts with molecular oxygen to produce superoxide (13).
The present results demonstrate that MSH is important in a variety of detoxification processes in M. smegmatis but do not provide insight into the details of the mechanisms involved. Highly reactive toxins may react directly with MSH, whereas less reactive compounds may require specific enzymes to promote activation of the toxin or to catalyze its reaction with MSH (e.g., MSH S-transferases). In other cases the effect of MSH could be indirect, occurring through regulation of the expression or activation of enzymes that detoxify a specific agent. Further detailed studies will be needed to elucidate the individual mechanisms involved for each class of toxin.
Our results confirm and expand the correlation between MSH depletion and sensitivity to toxins and antibiotics, supporting the notion that MSH metabolism can be an attractive drug target for novel inhibitory compounds. An inhibitor of MSH biosynthesis may potentially be used in combination with antibiotics which are not in use against mycobacteria at present.
Acknowledgments
M.R. and G.L.N. contributed equally to this work.
We thank Olivia Tang and Mia Unson for help in producing the mutants and William Black and Mabel Rodrigues for early screening of antibiotics. We thank Joseph Aguilera, Firmin Hung, Micah Steffek Stephanie Simmons, and Jennifer Heys for technical assistance and Christophe Guilhot and Brigitte Giquel for providing us with the transposon-carrying plasmids.
This work was supported by grants to Y.A. by the British Columbia Lung Association and the TB Veterans Association and to R.C.F. from the National Institute of Allergy and Infectious Diseases (grant AI49174) and the National Science Foundation (grant 9981850). Both Y.A. and R.C.F. are supported by contracts from eXegenics Pharmaceutics Incorporated and a grant from the Fogarty International Center (grant TW00976). Y.A. is a Canadian Institute of Health Research Foundation-British Columbia Lung Association Scholar.
REFERENCES
- 1.Anderberg, S., G. L. Newton, and R. C. Fahey. 1998. Mycothiol biosynthesis and metabolism: cellular levels of potential intermediates in the biosynthesis and degradation of mycothiol. J. Biol. Chem. 273:30391-30397. [DOI] [PubMed] [Google Scholar]
- 2.Aoyama, T., W. Zhao, F. Kojima, Y. Muraoka, H. Maganawa, T. Takeuchi, and T. Aoyagi. 1993. Cysfluoretin, a new inhibitor of glutathione S-transferase, produced by Streptomyces sp. MI384-DF12. J. Antibiot. 46:1471.. [DOI] [PubMed] [Google Scholar]
- 3.Arca, P., C. Hardisson, and J. E. Suarez. 1990. Purification of a glutathione s-tranferase that mediates fosfomycin resistance in bacteria. Antimicrob. Agents Chemother. 34:844-848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bornemann, C., M. A. Jardine, H. S. C. Spies, and D. J. Steenkamp. 1997. Biosynthesis of mycothiol: elucidation of the sequence of steps in Mycobacterium smegmatis. Biochem. J. 325:623-629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carney, J. R., S.-T. Hong, and S. J. Gould. 1997. Seongomycin: a new sulfur containing benzo[b]fluorene derived from genes clustered with those for kinamycin biosynthesis. Tetrahedron Lett. 38:3139-3142. [Google Scholar]
- 6.Dolphin, D., R. Poulson, and O. Avramovic. 1989. Glutathione: chemical, biochemical, and medical aspects. Parts A and B. Coenzymes and cofactors. John Wiley & Sons, Inc., New York, N.Y.
- 7.Fahey, R. C. 2001. Novel thiols of prokaryotes. Annu. Rev. Microbiol. 55:333-356. [DOI] [PubMed] [Google Scholar]
- 8.Guilhot, C., I. Otal, I. Van Rompaey, C. Martin, and B. Gicquel. 1994. Efficient transposition in mycobacteria: construction of Mycobacterium smegmatis insertional mutant libraries. J. Bacteriol. 176:535-539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jacobs, W. R., Jr., G. V. Kalpana, J. D. Cirillo, L. Pascopella, S. B. Snapper, R. A. Udani, W. Jones, R. G. Barletta, and B. R. Bloom. 1991. Genetic systems for mycobacteria. Methods Enzymol. 204:537-555. [DOI] [PubMed] [Google Scholar]
- 10.Johnsson, K., W. A. Froland, and P. G. Schultz. 1997. Overexpression, purification, and characterization of the catalase-peroxidase KatG from Mycobacterium tuberculosis. J. Biol. Chem. 272:2834-2840. [DOI] [PubMed] [Google Scholar]
- 11.Kosower, N. S., and E. M. Kosower. 1995. Diamide: an oxidant probe for thiols. Methods Enzymol. 251:123-133. [DOI] [PubMed] [Google Scholar]
- 12.Miyata, S., N. Ohhata, H. Murai, Y. Masui, M. Ezaki, S. Takase, M. Nishikawa, S. Kiyoto, M. Okuhara, and M. Kohsaka. 1992. WS009A and B, new endothelin receptor antagonists isolated from Streptomyces sp. no. 89009. I. Taxonomy, fermentation, isolation, physico-chemical properties and biological activities. J. Antibiot. 45:1029-1040. [DOI] [PubMed] [Google Scholar]
- 13.Nachtman, J. P. 1986. Superoxide generation by 1-nitropyrene in rat lung microsomes. Res. Commun. Chem. Pathol. Pharmacol. 51:73-80. [PubMed] [Google Scholar]
- 14.Newton, G. L., K. Arnold, M. S. Price, C. Sherill, S. B. delCardayré, Y. Aharonowitz, and G. Cohen. 1996. Distribution of thiols in microorganisms: mycothiol is a major thiol in most actinomycetes. J. Bacteriol. 178:1990-1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Newton, G. L., Y. Av-Gay, and R. C. Fahey. 2000. A Novel mycothiol-dependent detoxification pathway in mycobacteria involving mycothiol S-conjugate amidase. Biochemistry 39:10739-10746. [DOI] [PubMed] [Google Scholar]
- 16.Newton, G. L., Y. Av-Gay, and R. C. Fahey. 2000. N-Acetyl-1-d-myo-inosityl-2-amino-2-deoxy-α-d-glucopyranoside deacetylase (MshB) is a key enzyme in mycothiol biosynthesis. J. Bacteriol. 24:6958-6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Newton, G. L., M. Unson, S. Anderberg, J. A. Aguilera, N. N. Oh, S. delCardayré, Y. Av-Gay, and R. C. Fahey. 1999. Characterization of Mycobacterium smegmatis mutants defective in 1-d-myo-inosityl-2-amino-2-deoxy-α-d-glucopyranoside and mycothiol biosynthesis. Biochem. Biophys. Res. Commun. 255:239-244. [DOI] [PubMed] [Google Scholar]
- 18.Patel, M. P., and J. S. Blanchard. 2001. Mycobacterium tuberculosis mycothione reductase: pH dependence of the kinetic parameters and kinetic isotope effects. Biochemistry 40:5119-5126. [DOI] [PubMed] [Google Scholar]
- 19.Perez, E., J. A. Gavigan, I. Otal, C. Guilhot, V. Pelicic, B. Giquel, and C. Martin. 1998. Tn6111 transposon mutagenesis in Mycobacterium smegmatis using a temperature-sensitive delivery system. Methods Mol. Biol. 101:187-198. [DOI] [PubMed] [Google Scholar]
- 20.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 21.Sareen, D., M. Steffek, G. L. Newton, and R. C. Fahey. 2002. ATP-dependent l-cysteine:1-d-myo-inosityl 2-amino-2-deoxy-α-d-glucopyranoside ligase, mycothiol biosynthesis enzyme MshC, is related to class I cysteinyl-tRNA synthetases. Biochemistry 41:6885-6890. [DOI] [PubMed] [Google Scholar]
- 22.Snapper, S. B., L. Lugosi, A. Jekkel, R. E. Melton, T. Kieser, B. R. Bloom, and W. R. Jacobs, Jr. 1988. Lysogeny and transformation in mycobacteria: stable expression of foreign genes. Proc. Natl. Acad. Sci. USA 85:6987-6991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Unson, M. D., G. L. Newton, C. Davis, and R. C. Fahey. 1998. An immunoassay for the detection and quantitative determination of mycothiol. J. Immunol. Methods 214:29-39. [DOI] [PubMed] [Google Scholar]
- 24.Unson, M. D., G. L. Newton, K. F. Arnold, C. E. Davis, and R. C. Fahey. 1999. Improved methods for immunoassay of mycothiol. J. Clin. Microbiol. 37:2153-2157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Young, D. B., and K. Duncan. 1995. Prospects for new interventions in the treatment and prevention of mycobacterial disease. Annu. Rev. Microbiol. 49:641-673. [DOI] [PubMed] [Google Scholar]