Abstract
We have identified the simplest RNA binding site for isoleucine using selection-amplification (SELEX), by shrinking the size of the randomized region until affinity selection is extinguished. Such a protocol can be useful because selection does not necessarily make the simplest active motif most prominent, as is often assumed. We find an isoleucine binding site that behaves exactly as predicted for the site that requires fewest nucleotides. This UAUU motif (16 highly conserved positions; 27 total), is also the most abundant site in successful selections on short random tracts. The UAUU site, now isolated independently at least 63 times, is a small asymmetric internal loop. Conserved loop sequences include isoleucine codon and anticodon triplets, whose nucleotides are required for amino acid binding. This reproducible association between isoleucine and its coding sequences supports the idea that the genetic code is, at least in part, a stereochemical residue of the most easily isolated RNA–amino acid binding structures.
Keywords: Selection, SELEX, origin, evolution, minimal, exiguity, translation, genetic code
INTRODUCTION
The method of selection-amplification or SELEX (Ellington and Szostak 1990; Robertson and Joyce 1990; Tuerk and Gold 1990) has added greatly to the known functions of RNA. Most classes of ribozymes now known were selected from randomized sequences (Jaschke 2001) rather than derived from one of the seven (Doudna and Cech 2002) or eight (Nissen et al. 2000) natural ribozyme types. The selection of diverse activities from randomized RNA is relevant to the hypothesis of an RNA World (Gilbert 1986), in which it has been suggested that selection from varied, mostly nonfunctional sequences may have been a principal mode of biological evolution (Yarus 2002). This idea makes the simplest active RNA for all reactions of particular evolutionary interest, as the site requiring the fewest nucleotides should be most numerous in initial pools of randomized sequence (Ciesiolka et al. 1996).
However, even conceding its prominence at the start, it is not logically implied that the most frequent motif after many cycles of selection is the simplest one. More complex, but better-performing motifs can be purified preferentially, particularly if selection conditions are stringent. For example, Geiger et al. (1996) recovered a unique large arginine aptamer of exceptional properties from an exceptionally rigorous selection protocol. Simpler motifs exist, but could not satisfy the selection. A finite sample of sequences late in a selection dominated by the most competent RNAs may, therefore, not even contain a simple, less-active motif.
Even if the simplest site is still prominent in a final selected pool, its smallest form can be difficult to identify. Intercalation of functionless spacers among essential sequence modules is a statistical aid that should make a site more prevalent by orders of magnitude (Yarus and Knight 2002; Knight and Yarus 2003). Therefore, forms of the site with internal spacers will be most easily found. Internal spacer sequences are not trivially distinguishable from the active parts of the motif. Thus, additional experiments are needed to recognize the smallest active structure; for example, random terminal truncation experiments (Pan and Uhlenbeck 1992), selection from remutagenized pools (Bartel et al. 1991), or minimization using random internal deletions (Bittker et al. 2002).
Finally, there is the possibility that among active sequences, the predominant one is the most easily amplified. In a particularly clear experiment on this topic, Coleman and Huang (2002) utilized sequences of 30, 60, 100, and 140 randomized nucleotides together in the same pool. Only the 30- and 60-nt sequences yielded RNAs capable of CoA-thioester synthesis. Longer molecules virtually vanished from the selection when selection for activity became prominent, apparently because of replicative disadvantage. Because RNA active centers detected in the shorter molecules should have existed as subsequences within longer molecules, this experiment suggests that replication rather than activity can be decisive during a selection.
We now demonstrate a method that favors selection-amplification of the smallest active site, by forcing the activity into the smallest nucleotide tract that is able to support activity. We show that selections conducted with unusually short random tracts do yield the simplest motif as a dominant type. In addition, a previously known motif, which we will call the UAUU site, is identified as the simplest RNA structure that binds isoleucine, as well as the most frequently isolated. Conserved and functional anticodon and codon sequences within this most frequent site suggest how they may have been chosen for use in the stereochemical part of the genetic code.
Isoleucine binding selection
The UAUU motif was selected initially from RNA transcribed from DNA templates with 50 randomized positions, using isoleucine-Sepharose affinity and L-isoleucine elution (Majerfeld and Yarus 1998). These original UAUU-containing RNAs comprised 14% of 50 sequenced isolates after 13 rounds of selection-amplification and were derived independently from five parental randomized RNAs. The isolates conserved a UAUUGGGG sequence, disposed in stable structures as an internal loop opposite the sequence AC, the latter initially derived from primer complement (Fig. 1A ▶). Modification-interference assays, mutation, and the synthesis of truncated active derivatives localized the site of isoleucine binding to the loop structure. Of the remaining isolates sequenced, only one other repetitively isolated motif was prevalent, representing 18% of clones. Although it contained a possibly interesting conserved AUAUAUA sequence, this second isolate showed little specificity, having apparently similar affinity for isoleucine, alanine, valine, and methylamine.
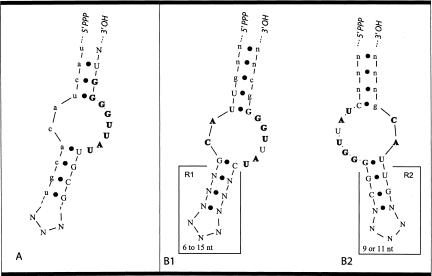
FIGURE 1.
(A) Summary structure of the UAUU motif based on results from the initial selection (Majerfeld and Yarus 1998). Initially, random nucleotides are capitalized and fixed nucleotides are in lowercase. Absolutely conserved nucleotides are in bold. Nonbold capitals designate highly conserved nucleotides (>70% identity in sequence alignment) and nonconserved nucleotides are indicated (N). (B) Refined summary structures of the UAUU motif based on 62 of the 72 sequenced UAUU isolates. Capitalization and boldface are the same as in A, except that lowercase letters were only sometimes derived from the constant regions and could also be selected from randomized nucleotides. Structures B1 and B2 summarize the two permutations of the motif. B1 is the preferred permutation (82%) in which the AC module occurs 5′ of the UAUUGG module.
Therefore, these data suggested the hypothesis that the simplest specific isoleucine site (some amino acid specificity being required for plausible coding) is the UAUU motif. On the basis of the initial isolation (Fig. 1A ▶; Majerfeld and Yarus 1998) and reselection for the motif from 30% doped pools (S. Changayil and M. Yarus, unpubl.), a minimal UAUU motif appeared to require 22–26 nt. This assumes complete conservation of UAUUGGGG and the opposing AC, as well as flanking pairs to stabilize the loop. Accordingly, to determine its robustness and to isolate the simplest binding motif for isoleucine, we repeated isoleucine affinity selection using RNA pools with randomized tracts of 16, 22, and 26 nt instead of the original 50. We chose primer sequences so that no essential sequence of the UAUU motif (or its complement) was present. Thus, conserved loop sequences must come from initially randomized nucleotides.
RESULTS
Selections with limited numbers of randomized nucleotides
RNA pools with 16, 22, and 26 initially randomized positions were applied to carboxyl-linked isoleucine-EAH Sepharose 4B (Majerfeld and Yarus 1994). Bound RNAs were eluted with 1 mL of L-isoleucine containing buffer after washing with 5–7.5 column volumes of selection buffer. The number of template DNA sequences implies a mean of ~60,000 copies of each 16-mer sequence and 19 copies of each possible 22-mer transcribed, so there is a low probability, ~10−9 (Poisson probability of zero copies, P(0) = e−19 ≈ 6 × 10−9; Ciesiolka et al. 1996) that any contiguous 22-mer sequence was absent. Therefore, the design of the experiments implies that any simpler 16-mer or 22-mer isoleucine site, if it exists, should be present at the start of these selections, perhaps many times over. In addition, affinity selection is exceptionally sensitive to ligand affinity – it can be calculated (Ciesiolka et al. 1996) that these conditions should resolve RNAs with Kd ≈ 10 mM. As this seems a reasonable upper limit for biological amino acid concentrations, we argue that the selection applies a minimal plausible criterion for isoleucine binding activity.
RNA pools with 26 initially randomized positions showed an initial peak after five selection cycles and were cloned and sequenced after 6 selection cycles, when 20% of the pool was present in the isoleucine elution peak. RNA pools with 22 initially randomized positions showed a small peak eluted by free isoleucine after seven rounds of selection, indicating purification of isoleucine binding RNAs. Another three rounds of selection further increased isoleucine elution from 7.6% to 17% of the molecules. RNA molecules with 16 initially randomized positions were cloned and sequenced after 11 rounds of selection. At this point, there was still no discernible peak after isoleucine elution. Sequencing results are in Table 1 ▶.
TABLE 1.
Summary of sequences from the 16, 22, and 26-nucleotide selections
| Total no. of clones | ||||||
| Family | 16-mer | 22-mer | 26-mer | Total | Estimated parent RNAs | Consensus sequence |
| UAUU P1 | 0 | 19 | 40 | 59 | 48 | ---N(0-7)- U-U-A-C-G-N(6-15)-C-U-A-U-U-G-G-G- N(0-3) --- |
| UAUU P2 | 0 | 7 | 6 | 13 | 10 | ---N(0-4)- C-U-A-U-U-G-G-G-G-C-N(7-9)-G-U-U-A-C-N(0-4)--- |
| VG1 | 26 | 0 | 0 | 26 | 13 | ---N(9-15)-C-A-C-G-U-A-C--- |
| VG2 | 2 | 2 | 0 | 4 | 4 | ---N(2-4)- C-N-N-G-G-U-A-C-G-C-C-U-N(1-5)--- |
| VG3 | 0 | 7 | 0 | 7 | 1 | ---R-C-G-A-C-C-G-U-A-U-G-M-G-A-A-A-G-U-U-G-G-G--- |
| VG4 | 5 | 2 | 0 | 7 | >3 | ---N(3,9)- C-A-C-G-C-A-U-G-C-C-U-G-G--- |
| VG5 | 0 | 4 | 0 | 4 | 3 | ---N(1-6)- C-A-N-N-A-G-G-A-G-C-G-U-C-G-G-U-N(0-5)--- |
| VG6 | 0 | 0 | 3 | 3 | 1 | ---G-C-C-A-N-U-A-C-A-U-C-U-C-G-C-U-N-U-C-G-U-A-C-U-N-C-G--- |
| VG7 | 2 | 0 | 0 | 2 | 1 | ---U-U-A-C-C-A-C-A-G-U-U-G-G-C-G-G--- |
| VG8 | 2 | 0 | 0 | 2 | 2 | ---N(3-4)- G-A-C-U-G-A-N-A-N-G-U-N(1-2)--- |
| VG9 | 0 | 0 | 1 | 1 | 1 | ---U-A-C-G-C-G-U-U-G-U-C-U-U-C-C-A-G-A-G-C-C-U-G-G-C-C-- |
| Total sequenced | 37 | 41 | 50 | 128 | ||
UAUU P1 and P2 represent the two permutations of the motif (Figure 1B ▶). The consensus sequence lists absolutely conserved positions in underlined bold type. Other listed residues are conserved in at least 70% of the sequences in the family. The dashed lines at the beginning and end of the consensus indicate the 5′ and 3′ constant regions. Comprehensive sequence data is available at bayes.Colorado.EDU/seq.
The UAUU motif was recovered from pools with 22 and 26 randomized positions, but not from the 16-random RNAs. Recovery of the UAUU motif was particularly efficient from 26-random RNAs, with 46 of 50 isolates sequenced (92%) being UAUU examples after six rounds of selection (Table 1 ▶). On the basis of sequence differences between isolates, we estimate that these latter UAUU RNAs were derived independently from 43 different initial RNAs (using three differences as the maximum divergence due to PCR error).
The UAUU motif accounted for 26 of 41, or 61%, of the sequences from the 22-random RNAs after 10 rounds of selection (Table 1 ▶). The 22-random members of the UAUU family were derived from an estimated 17 parents, with more incidences of nearly identical sequences than seen in the 26-randomized selection. Other selected 22-random sequences could be grouped into four families (groups of sequences with similar conserved sequences) of 2–7 isolates (Table 1 ▶).
In contrast, the UAUU motif was not isolated from the 16-random selection, but these sequences also were not random. Instead, the action of selection was clear. The 37 sequenced 16-random RNAs could be divided into five groups, the largest of which contained the conserved sequence CACGUAC in the random region immediately adjacent to the 3′ primer sequence (VG1, Table 1 ▶). This single family, in fact, accounted for 70% of the 16-random sequences.
Characteristics of the UAUU motif
From the 22-mer and 26-mer randomized selections, a total of 72 UAUU isolates were sequenced and aligned to characterize the requirements for isoleucine affinity. Summary structures in Figure 1B ▶ represent the result for 62 of the 72 UAUU isolates. The remaining 10 isolates have the conserved internal loop sequences, but there is variability in loop size (e,g., two isolates are predicted to have an extra U in the loop on both sides of the motif). All 72 UAUU sequences were used to determine sequence conservation in the usual unpaired regions. The 62 isolates represented by Figure 1B ▶ were used to characterize the conservation of helices flanking the loop.
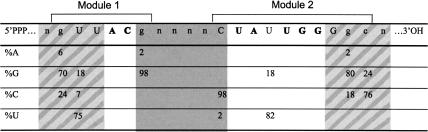
As surmised during its initial isolation (Majerfeld and Yarus 1998), the UAUU motif consists of two sequence modules. Module 1 has the consensus sequence GUUACG. The internal dinucleotide AC is completely conserved and the remaining four nucleotides are conserved in >70% of isolates (Fig. 1B ▶; Table 2 ▶). Module 2 has the consensus CUAUUGGGGC, and has five invariant and five highly preferred nucleotides (>70%; Table 2 ▶). These sequences differ at a few positions from the original selection, because Module 1 was derived initially from a fixed sequence used for amplification. The two conserved modules can be circularly permuted and the preferred permutation is being Module 1 5′ of Module 2 (82%; Fig. 1B1 ▶).
TABLE 2.
Summary of sequence variation within individual UAUU molecules
The sequence order is of permutation 1 (Figure 1B1) through nucleotide frequencies (%) summarize both permutations. Frequencies within the paired regions (shaded) are estimated based on the 62 isolates that are predicted to form the structures in Figure 1B. Bold type, uppercase and lowercase letters have the same meaning as in Figure 1B. All frequencies were determined only from isolates in which nucleotides occur at initially randomized positions.
Within the internal loop, all nucleotides are absolutely conserved except for the second U in the UAUUGG sequence. This nucleotide is G in a minority, 18% of all isolates (Table 2 ▶). The base pairs closing the loop structure are also highly conserved. In the paired region formed by the 5′ end of Module 1 and the 3′ end of Module 2 (R2, Fig. 1B2 ▶); the first base pair outside of the loop structure is usually G:U (74%) and the second is C:G or G:C (94%). The region flanking the conserved internal loop on the other side (R1, Fig. 1B1 ▶) has no apparent sequence conservation, but bases co-vary in a manner suggesting a minimum of one required base pair. The UAUU motif has at least six additional base pairs flanking the loop; an average of 7.9 bp is predicted in the 26-random sequences and 6.7 in 22-random sequences. Up to seven nucleotides involved in flanking base pairs can be derived from the constant region (shown in lowercase in Fig. 1B ▶).
In summary, the UAUU motif has seven invariant and nine highly conserved (>70% identity) nucleotides. Twelve nucleotides shape the loop, including the flanking G:U and G:C pairs. At least 12 additional paired nucleotides are required to stabilize the motif, although more are frequent. An additional three nucleotides are required for the hairpin loop. Thus, a minimum of 27 nucleotides is required to fold the functional motif within a continuous tract of nucleotides. The minimum number required for motif formation is ≈ 20 (27 minus seven paired flanking nucleotides that can be derived from constant regions). This conclusion, based entirely on sequence conservation, is supported by failure to isolate this motif from 16 randomized positions, which is too small, but recovery from 22 nucleotide random pools, which is just large enough (by utilizing at least five constant nucleotides).
Variation within the UAUU motif
We investigated variation of the second U in the UAUUGGGG sequence to G in 18% of isolates (Table 2 ▶). The variant was always a minority among sequences that met the selection. This trend to minority was quite strong in some selections. In the selection with 22 randomized positions and 10 rounds of selection amplification, only one of 26 UAUU sequences (4%) showed the variation. We directly measured the effects of this mutation on binding strength by evaluating the dissociation constant (Kd). Isocratic affinity chromatography has the same logic as equilibrium dialysis, in which one phase is moving and the other immobilized; therefore, it can measure Kd (Ciesiolka et al. 1996). G for U substitution decreases the Kd for L-isoleucine 2.5-fold or 0.55 kcal/mole (UAGU; Table 3 ▶). Thus, mutation to G somewhat compromises isoleucine binding, accounting for its minority status in a selection relying on isoleucine both as a fixed ligand and an eluant.
TABLE 3.
Binding and specificity of the RNAs
Apparent Kd (# measurements averaged) for each sequence family, using isocratic affinity chromatography (Ciesiolka et al. 1996). Median elution volumes from isoleucine-Sepharose were determined for modified selection buffer, and modified selection buffer plus either isoleucine, norleucine, or glycine (1.0 and/or 2.0 mM concentrations). Modified selection buffer contains 0.4 mM ZnCl2 instead of 0.1 mM. The former follows the initial measurements (Majerfeld and Yarus 1998) so that results can be compared. A dash indicates no response to free-L-isoleucine.
Investigating other sequences
Although the UAUU family was by far the most abundant motif in the 22- and 26-random RNA pools, it was not recovered at all from selection with 16 randomized positions. Because 16 nucleotides are not enough to form the UAUU motif defined by sequence conservation, we investigated other isolates to determine whether a simpler binding site was observable when the UAUU motif was not possible. In addition, we characterized every other sequence recovered from the 22- and 26-randomized pools to look for other efficient isoleucine sites. Consensus sequences for these and the number of examples recovered are in Table 1 ▶.
RNAs were tested by chromatography of one or more representative transcripts of each family on isoleucine-Sepharose. Successive elutions used selection buffer followed by 10 mM L-isoleucine and a column sweep with 1 M NaCl (Fig. 2 ▶). Isocratic competitive affinity chromatography with 1 and/or 2 mM isoleucine was also used to estimate the Kd for free amino acids (Table 3 ▶). All sequence families isolated from the 16-mer selection (VG1, VG2, VG4, VG7, and VG8) had some affinity for the Ile-Sepharose matrix (Fig. 2 ▶; Table 3 ▶), but none were detectably eluted by free L-isoleucine (Fig. 2 ▶). Thus, when simplicity of the site is forced, selected RNAs had no significant affinity for free amino acid, but could bind the larger ligand surface presented by isoleucine plus the supporting matrix.
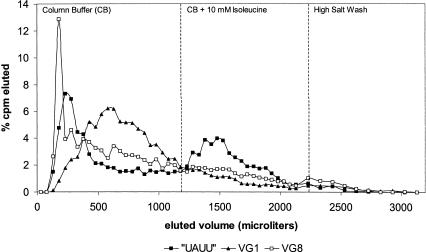
FIGURE 2.
Representative elution profiles for representatives of the VG1, VG8, and UAUU sequence families. VG1 represented ~70% of the final 16-mer pool, and variant VG8 has the strongest Kd for isoleucine-Sepharose among the 16-mer sequence families (Table 3 ▶). Both show affinity for the column (delayed elution), but no peak with isoleucine.
In contrast, most sequences from the 22-mer and 26-mer selections (VG3, VG5, VG6, and VG9) were eluted with L-isoleucine. To compare binding strength and get an index of specificity, isocratic affinity chromatography was used to estimate Kd for isoleucine, glycine, and norleucine (the last has the same sidechain area as isoleucine, but differs in shape; Table 3 ▶).
Among all investigated sequences, the UAUU motif showed the most favorable binding-free energy. The UAUU motif had a Kd of 1.2 mM for free isoleucine, slightly higher than in the initial selection (Majerfeld and Yarus 1998). Measured specificity, however, is comparable with the initial results—the UAUU site binds ~2.3 times (0.5 kcal/mole) better to isoleucine than norleucine and ~5 times (1.0 kcal/mole) better than to glycine. Of non-UAUU sites, VG5 bound free isoleucine best (Kd = 2.0 mM), but was not specific, binding glycine and isoleucine similarly (Table 3 ▶). RNAs VG3 and VG9 bound free isoleucine 2.0- to 2.4-fold more weakly than the UAUU motif. Although VG9 could discriminate sidechains only weakly (Table 3 ▶), VG3 bound isoleucine ~3.3 times (0.7 kcal/mole) better than norleucine and ~16 times (1.6 kcal/mole) better than glycine.
RNA VG3 is therefore unique in being as specific as the UAUU motif, but it shows lower affinity for isoleucine, which may partly explain its low abundance. Also, the appearance of this motif in the 22-random selection only, and also the complete conservation of all but 3 nucleotides throughout the random region (Table 1 ▶) suggest that this site is more complex than UAUU. Interestingly, VG3 loosely resembles the UAUU motif, having a conserved UAU sequence and the conserved string UUGGGGC (Table 1 ▶; the terminal GC are from the constant region). G and U-rich tracts have been associated previously with RNA sites for hydrophobic ligands (Majerfeld and Yarus 1998; Yarus 1998).
DISCUSSION
The selections worked as expected
Below, we interpret the outcome of these selections as typical. In support of this interpretation, we stress that these experiments worked very much as a simple model of selection would have predicted. Isolation of the UAUU motif required the selection of 16 conserved nucleotides plus a minimum of 1 extra base pair from the random region. The required sequences should occur once in 417 = 1010.2 -randomized sequences. One might correct this estimate for the ability of essential sequence modules to shift within the larger random sequence (Yarus and Knight 2002; Knight and Yarus 2003), and for the ability of the motif to fold correctly. However, these corrections are in opposing directions and may have only a small net effect in short random regions. Such refinements are therefore ignored for simplicity.
We performed reconstruction experiments in which individual or multiple radioactive UAUU sequences were mixed with an excess of nonisotopic-randomized RNA and subjected to isoleucine affinity fractionation. These experiments suggest that our affinity chromatography purifies UAUU sequences an average of about 79-fold with respect to randomized RNA. If UAUU sites must be purified ~1010.2-fold starting with completely randomized sequences, this would require about five to six cycles of 79-fold selection on simplest assumptions (i.e., 795.4 ≅ 1010.2). Ignoring the difference between detection of a motif and its complete purification, this agrees well with the outcome of the 22-mer and 26-mer selections, which showed an initial isoleucine-eluted peak at seven and five cycles, respectively. Thus, our affinity selection found UAUU sites at approximately the expected abundance.
Elevating the frequency of the smallest site
The frequency of any motif in an initial randomized pool of different lengths seems fairly well understood. On simple assumptions, any sequence motif should increase monotonically in frequency in an initial pool as the random region is lengthened. Longer tracts give more ways to place required sequence elements (Ciesiolka et al. 1996; Sabeti et al. 1997). This increase is believed to be opposed by an increasing probability of alternative inactive folds as longer arbitrary sequences are added, but the net advantage of longer sequences still remains (Sabeti et al. 1997). Division of the motif into modules with intercalated spacers makes the fold easier to find—this is true for structures of varied complexity, for randomized tracts of varied lengths, and for very disparate numbers of initial molecules (Yarus and Knight 2002). However, the division of the motif into separate pieces is optimally effective only for pieces of approximately equal size (Knight and Yarus 2003). Thus, we expect all active RNAs to decrease in frequency as randomized tracts are shortened, and ultimately to disappear when the active core cannot be formed. At all stages, the active motif requiring the fewest nucleotides should be the most frequent at the start of the experiment, and might, therefore, disappear last.
However, as discussed in the introduction, real selection-amplification experiments may not identify the smallest active motif—because of competitive exclusion by larger but more functional structures, or the intercalation of large functionless spacers, or because differences in replication can be as (or more) important than differences in function. These confounding factors can all be made less significant by progressively increasing selection pressure for small size. Therefore, shrinking the randomized tract (we used the smallest tracts that we are aware of in the small-molecule literature) should be an effective strategy forcing the isolation of an active structure containing the fewest nucleotides. In fact, at least here, where spurious effects should be minimized, the ultimate simplest structure was always also the dominant one.
Thus, the experiments above conform to expectations. The internal loop motif containing CUAUUGGGG (termed the UAUU site for concision) is present at approximately expected frequencies; even so, it decreased in initial abundance (it required more amplification) in the 22-nt pool as compared with 26-nt randomized regions. When only 16 randomized positions were allowed, neither it nor any alternative could be isolated, even by much more extensive selection (although selection among sequences and selection of other functions were evident in the 16-nt pool).
The smallest isoleucine binding RNA
Thus, selection strongly supports the UAUU internal loop as the simplest isoleucine site. The UAUU motif is, by far, the most numerous specific isoleucine site in three independent successful selections for affinity to free isoleucine. The results of the 26-random selection, which yielded 46/50 UAUU RNAs, show that this motif can be truly dominant in frequency. Sequence abundance and motif size have been interrelated previously. Calculations indicate that an RNA active site only 1.6 nucleotides larger requires 10-fold more initial RNA in which to find the motif (Ciesiolka et al. 1996; Yarus and Knight 2002; Knight and Yarus 2003). Thus, the observed reproducible dominance of UAUU among selected sequences provides evidence, on the standard reasoning, that it is the simplest possible binding structure.
However, the argument for the UAUU motif goes beyond dominance interpreted as an indirect consequence of the statistical superiority of smaller motifs (Salehi-Ashtiani and Szostak 2001). When UAUU RNA could not form, the 16-mer random region did not allow a site with affinity for free L-isoleucine. Instead, the RNAs recovered had only a general affinity for the matrix (which then trailed into the fractions harvested; Fig. 2 ▶). Thus, under conditions calculated to cut off competition from larger sites and allow the emergence of smaller active structures, none appear.
Furthermore, if the random tract is lengthened, sites that first are found are dominated by the UAUU motif. By showing that the UAUU site is most frequent throughout multiple selections that compress the site until free isoleucine binding is not recovered, we have shown that simplicity and abundance can be synonymous molecular properties, as calculations suggested. That is, progressive increases in the severity of selection for small size do not displace UAUU RNA, so it likely is the smallest possible free isoleucine binding RNA. Alternative sites bind less well, are infrequent, and may be larger. Moreover, because more than one set of primers was used for amplification (Majerfeld and Yarus 1998), this conclusion is likely to be independent of the particular fixed sequences that necessarily accompany the amino acid site during selection amplification.
The genetic code
We now wish to evaluate these results in the context of a search for the origin of the genetic code (Knight et al. 2003), and particularly to evaluate the possibility of a chemical foundation for the code (Woese 1966). Origin theories called stereochemical usually assert that direct or indirect interactions between codons and/or anticodons and amino acids underlie some codon assignments. These putative interactions should be amenable to laboratory demonstration.
There is strong experimental evidence for one stereochemical hypothesis, that some coding sequences are abstracted from amino acid binding sites. A recent summary shows that for 26 in vitro-selected binding sites for six varied amino acids, the probability of the observed occurrence of codons and anticodons within selected sites was of the order 10−11 or less, supposing that triplets and binding sites are unrelated (Knight et al. 2003), equivalent to tossing ≥35 straight heads with a fair coin. Thus, the code may have emerged in the RNA World (Gilbert 1986), in which larger RNAs of reproducible sequences and binding activities would have probably first been available (Yarus 2001).
Nevertheless, RNA is versatile; often many small RNAs bind the same amino acid. Binding sites can be so diverse that multiple selections fail to reisolate the same sites, as for arginine (Yarus 1998). This makes it difficult to know how sites would be chosen when the genetic code was composed.
However, from this work it appears that that choice of amino acid binding sites may sometimes be strongly constrained. The UAUU motif seems to be uniquely the simplest and the most frequent isoleucine binding structure. This isoleucine binding site is simple enough to occur without selection in tiny randomized pools containing only zeptomol levels of RNA molecules (Yarus and Knight 2002; Knight and Yarus 2003). The UAUU motif also has the most favorable binding-free energy observed, sufficient to participate in isoleucine binding at biological concentrations. Thus, there is a plausible and reproducible chemical connection between isoleucine and its triplets, which appear together in a recurring structure. Postulating evolutionary significance for this result does not require that the triplets have any particular molecular role, only that they must recur because they do something essential within the most abundant binding site. The triplets’ essential nature has been demonstrated many times over by multiple independent reselection (above), as well as by the molecular data on the site (Majerfeld and Yarus 1998). As the exact conditions during the emergence of ribocytes (RNA cells) are unknown and perhaps controversial (Pace 1991), it may also be important that the robustness of the UAUU site (for example, with regard to salt, temperature, and nucleic acid structure) is open to investigation.
MATERIALS AND METHODS
Selections
The isoleucine affinity selection cycle has been described (Majerfeld and Yarus 1998). Selection buffer was 50 mM HEPES (pH 7.0), 300 mM NaCl, 7.5 mM MgCl2, and 0.1 mM ZnCl2. In addition, elution buffer had 10 mM isoleucine. Primer sequences were (5′–3′) TAATACGACTCACTATAGGGCGAAGAAGGAAG AGCG for the 5′ primer and TTCGGCTTCACTTCACACGC for the 3′ primer. Initial template DNA comprised 400, 550, and 1400 pmole of template DNA for 16-random, 22-random, and 26-random selections, respectively.
Isoleucine elution profiles
Profiles were generated by applying 10 pmole of folded internally labeled [32P]RNA to the isoleucine-Sepharose column. RNA was heated at 65°C for 3 min, brought to selection buffer conditions, and allowed to incubate for 5–10 min at room temperature. The 0.2 mL column was washed with 1.2 mL of selection buffer before elution with five column volumes of 10 mM L-isoleucine in selection buffer.
Reconstruction of the affinity purification
A total of 600 pmole of random 26-mer RNA were mixed with either 1.5 pmole of a single 32P-labeled UAUU isolate or 0.9 pmole each of three labeled UAUU isolates. After one round of affinity chromatography under selection conditions, UAUU enrichment was determined from the fraction of the UAUU isolate in the initial versus the final pool. The enrichment was measured as 131-fold and 61-fold for the selection with one and three UAUU isolates, respectively.
Analysis of flanking base pairs
The predicted number of base pairs flanking the UAUU loop were estimated with a Python program. Each sequence was scanned for the conserved loop sequence elements UACG and CUAUUGGG. Sequence between the motifs and extending outward in both directions were examined for covariation indicating required base pairing. Isolates in which the sequence elements were not found were examined manually and with RNA mFold (Walter et al. 1994) to determine whether alternate folding was responsible.
Acknowledgments
We thank members of the laboratory for help in redaction of a draft. This work was supported by NIH and NASA research grants to M.Y.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5114503.
REFERENCES
- Bartel, D.P., Zapp, M.L., Green, M.R., and Szostak, J.W. 1991. HIV-1 Rev regulation involves recognition of non-Watson-Crick base pairs in viral RNA. Cell 67: 529–536. [DOI] [PubMed] [Google Scholar]
- Bittker, J.A., Le, B.V., and Liu, D.R. 2002. Nucleic acid evolution and minimization by nonhomologous random recombination. Nat. Biotechnol. 20: 1024–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesiolka, J., Illangasekare, M., Majerfield, I., Nickles, T., Welch, M., Yarus, M., and Zinnen, S. 1996. Affinity selection-amplification from randomized oligoribonucleotide pools. Methods Enzymol. 267: 315–335. [DOI] [PubMed] [Google Scholar]
- Coleman, T.M. and Huang, F. 2002. RNA-catalyzed thioester synthesis. Chem. Biol. 9: 1227–1236. [DOI] [PubMed] [Google Scholar]
- Doudna, J.A. and Cech, T.R. 2002. The chemical repertoire of natural ribozymes. Nature 418: 222–228. [DOI] [PubMed] [Google Scholar]
- Ellington, A.D. and Szostak, J.W. 1990. In vitro selection of molecules that bind specific ligands. Nature 346: 818–822. [DOI] [PubMed] [Google Scholar]
- Geiger, A., Burgstaller, P., von der Eltz, H., Roeder, A., and Famulok, M. 1996. RNA aptamers that bind L-arginine with sub-micromolar dissociation constants and high enantioselectivity. Nucleic Acids Res. 24: 1029–1036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilbert, W. 1986. The RNA world. Nature 319: 618. [Google Scholar]
- Jaschke, A. 2001. Artificial ribozymes and deoxyribozymes. Curr. Opin. Struct. Biol. 11: 321–326. [DOI] [PubMed] [Google Scholar]
- Knight, R.D. and Yarus, M. 2003. Finding specific RNA motifs: Function in a zeptomole world? RNA 9: 218–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knight, R.D., Landweber, L., and Yarus, M. 2003. Tests of a stereochemical genetic code. In Translation mechanisms (eds. J. Lapointe and L. Brakier-Gingras), pp. 115–128. Landes Biosciences, Houston, TX.
- Majerfeld, M. and Yarus, M. 1994. An RNA pocket for an aliphatic hydrophobe. Nat. Struct. Biol. 1: 287–292. [DOI] [PubMed] [Google Scholar]
- ———. 1998. Isoleucine: RNA sites with associated coding sequences. RNA 4: 471–478. [PMC free article] [PubMed] [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., and Steitz, T.A. 2000. The structural basis of ribosomal activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Pace, N.R. 1991. Origin of life—Facing up to the physical setting. Cell 65: 531–533. [DOI] [PubMed] [Google Scholar]
- Pan, T. and Uhlenbeck, O.C. 1992. In vitro selection of RNAs that undergo autolytic cleavage with Pb2+. Biochemistry 31: 3887–3895. [DOI] [PubMed] [Google Scholar]
- Robertson, D.L. and Joyce, D.F. 1990. Selection in vitro of an RNA enzyme that specifically cleaves single-stranded DNA. Nature 344: 467–468. [DOI] [PubMed] [Google Scholar]
- Sabeti, P.C., Unrau, P.J., and Bartel, D.P. 1997. Accessing rare activities from random RNA sequences: The importance of the length of molecules in the starting pool. Chem. Biol. 4: 767–774. [DOI] [PubMed] [Google Scholar]
- Salehi-Ashtiani, K. and Szostak, J.W. 2001. In vitro evolution suggests multiple origins for the hammerhead ribozyme. Nature 414: 82–84. [DOI] [PubMed] [Google Scholar]
- Tuerk, C. and Gold, L. 1990. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 polymerase. Science 249: 505–510. [DOI] [PubMed] [Google Scholar]
- Walter, A.E., Turner, D.H., Kim, J., Lyttle, M.H., Muller, P., Mathews, D.H., and Zuker, M. 1994. Coaxial stacking of helixes enhances binding of oligoribonucleotides and improves predictions of RNA folding. Proc. Natl. Acad. Sci. 91: 9218–9222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese, C.R., Dugre, D.H., Saxinger, W.C., and Dugre, A.S. 1996. The molecular basis for the genetic code. Proc. Natl. Acad. Sci. 55: 966–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarus, M. 1998. Amino acids as RNA ligands. J. Mol. Evol. 47: 109–117. [DOI] [PubMed] [Google Scholar]
- ———. 2001. On translation by RNAs alone. Cold Spring Harbor Symp. Quant. Biol. LXVI: 207–215. [DOI] [PubMed] [Google Scholar]
- ———. 2002. Primordial genetics: Phenotype of the ribocyte. Annu. Rev. Genet. 36: 125–151. [DOI] [PubMed] [Google Scholar]
- Yarus, M. and Knight, R. 2002. The scope of selection. In The genetic code and the origin of life (ed. L.R. Pouplana). Landes Bioscience, Houston, TX. (http://www.ncbi.nlm.nih.gov/entrez/books)