Abstract
Little is known about the tertiary structure of internal ribosome entry site (IRES) elements. The central domain of foot-and-mouth disease (FMDV) IRES, named 3 or I, contains a conserved GNRA motif, essential for IRES activity. We have combined functional analysis with RNA probing to define its structural organization. We have found that a UNCG motif does not functionally substitute the GNRA motif; moreover, binding of synthetic GNRA stem–loops to domain 3 was significantly reduced in RNAs bearing UCCG or GUAG substitutions. The apical region of domain 3 consists of a four-way junction where residues of the GNRA tetraloop are responsible for the organization of the adjacent stem–loops, as deduced from ribonucleases and dimethyl sulfate accessibility. A single A-to-G substitution in the fourth position of this motif led to a strong RNA reorganization, affecting several nucleotides away in the secondary structure of domain 3. The study of mutants bearing UNCG or GUAG tetraloops revealed lack of protection to chemical attack in native RNA at specific nucleotides relative to the parental GUAA, suggesting that the GNRA motif dictates the organization and stability of domain 3. This effect is likely mediated by the interaction with distant residues. Therefore, the GNRA motif plays a crucial role in the organization of IRES structure with important consequences on activity.
Keywords: GNRA, internal initiation of translation, IRES, RNA structure, UNCG
INTRODUCTION
Internal ribosome entry site (IRES) elements consist of structured mRNA regions that recruit the ribosome to an internal AUG codon, usually with the help of cellular proteins (Jackson 2000). IRES elements, first described in picornavirus, are present in an increasing number of viral and cellular mRNAs whose translation is not compromised at times of cap-dependent inhibition (Holcik et al. 2000; Hellen and Sarnow 2001). During the last years, the knowledge of picornavirus IRES-interacting proteins essential to drive internal initiation of translation has made a great progress (for review, see Belsham and Jackson 2000; Martínez-Salas et al. 2001; Pestova et al. 2001). Translation initiation factors eIF4G, eIF3, eIF4B, and IRES transacting factors (ITAF) polypyrimidine binding protein (PTB) and ITAF45, recognize the foot-and-mouth disease virus (FMDV) and encephalomyocarditis virus (EMCV) IRES at precise locations in RNA structure-dependent manner (Jang and Wimmer 1990; Luz and Beck 1991; Meyer et al. 1995; Kolupaeva et al. 1996, 1998; López de Quinto and Martínez-Salas 2000; Pilipenko et al. 2000; López de Quinto et al. 2001). However, despite the fact that RNA structure is thought to play an essential role in IRES activity, the RNA organization of the picornavirus IRES is far from being understood. The relative large size of IRES elements has imposed a difficulty to enable structural studies similar to those carried out with other RNA molecules (Correll et al. 1997; Kolk et al. 1998; Doherty et al. 2001; Michiels et al. 2001).
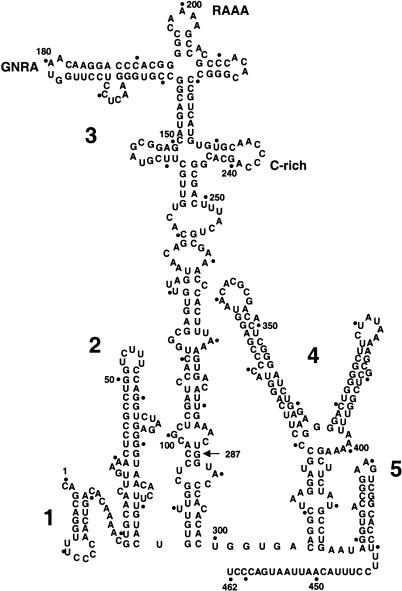
The FMDV IRES is a relatively long RNA region, spanning 462 nucleotides that are predicted to fold in five domains (named 1 to 5 or G to L) containing stable stem–loop structures (see Fig. 1 ▶). Domain 3 (also named I) is the central domain. According to computer prediction programs, its 210 residues acquire a stem–loop structure at its apical region, which contains a conserved, essential GNRA motif in a distal loop (López de Quinto and Martínez-Salas 1997). The proximal part is predicted to fold as a long-stem region interrupted with several bulges. At least four residues encompassing G88 and C297 are involved in the formation of base pairs in a helical structure, as mutational analysis of this region showed the recovery of IRES activity of defective mutants upon restoration of their secondary structure (Martínez-Salas et al. 1996). In terms of protein binding, domain 3 is known to interact with polyC binding protein (PCBP2; Walter et al. 1999; Stassinopoulos and Belsham 2001). However, none of the canonical eukaryotic initiation factors seems to recognize this region of the IRES.
FIGURE 1.
Secondary structure of FMDV IRES. RNA structure was predicted using the Mfold program of GCG package, version 10. Domains are numbered 1 to 5, and span positions 1–85, 84–297, 306–415, and 416–462, respectively; a dot is used to mark positions each 10 nucleotides. The conserved GNRA, RAAA and C-rich motifs in domain 3 are highlighted. An arrow at position 287 points to the 3′ end of the antisense oligonucleotide used in primer extension assays.
We have previously demonstrated long-range RNA–RNA interactions in the FMDV IRES (Ramos and Martínez-Salas 1999). Formation of RNA complexes in vitro depends on ionic strength, temperature, and RNA concentration, suggesting a dynamic tertiary structure that may have a role in modulating IRES biology. Domain 3 is unique, as it is the only IRES region that has the capacity to interact with distal parts of the IRES (Ramos and Martínez-Salas 1999). This property suggests that the central domain may act as a scaffold structure organizing the IRES tertiary architecture. Formation of long-range RNA–RNA interactions mediated by domain 3 is consistent with the presence of structural motifs involved in tertiary folding. A mutational analysis of GNRA conserved motif showed that it is essential for IRES activity in FMDV (López de Quinto and Martínez-Salas 1997) and EMCV (Robertson et al. 1999). Interestingly, the double GCGA substitution mutant was active, whereas AUAA and particularly GUAG mutant bearing a single substitution in the fourth position of the GNRA motif were inactive (López de Quinto and Martínez-Salas 1997). Because GNRA motifs constitute a structural element often involved in organizing RNA structure (Costa and Michel 1997; Doherty et al. 2001) this motif is a candidate to organize IRES architecture.
Little is known about the RNA architecture of IRES elements. Structural studies of small RNA molecules corresponding to IRES fragments is starting to shed light on the organization of hepatitis C (HCV) IRES (Klink et al. 2000; Lukavsky et al. 2000; Kieft et al. 2002; Vos et al. 2002). In the case of c-myc, Apaf-1 and cricket paralysis virus (CrpV) RNAs, the accessibility of probing reagents to the full-length IRES has revealed gross changes relative to the computer assisted predicted structures (Le Quesne et al. 2001; Jan and Sarnow 2002; Mitchell et al. 2003). Thus, there is a clear need to determine RNA structure to establish a valid function–structure relationship.
The RNA structure of FMDV domain 3 deduced from enzymatic and chemical probing studies, in conjunction with the study of mutants affected in the GNRA motif, have underscored the complexity of FMDV IRES. We report here that the absence of GNRA motif-dependent interactions led to inactive elements concomitantly with a reorganization of the RNA structure at the apical region of the central domain. Thus, FMDV IRES structural organization is dependent on the integrity of the GNRA motif.
RESULTS
The UNCG motif does not functionally substitute for the GNRA in IRES activity
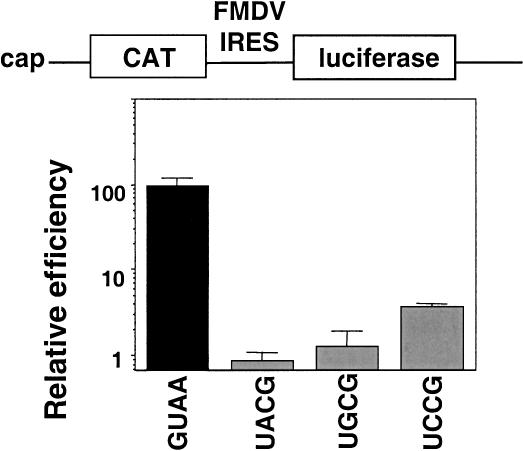
Our previous work has shown that the GNRA motif is essential for FMDV IRES activity (López de Quinto and Martínez-Salas 1997). The GNRA motif is a building block that may contribute to RNA structure mediating in tertiary interactions with distant residues of the molecule (Costa and Michel 1997; Doherty et al. 2001). In some cases in which the GNRA has been involved in hairpin stability, the substitution of GNRA by a UNCG has not altered the activity of the resulting RNA (Selinger et al. 1993). Thus, to gain knowledge about the role of the GNRA motif in IRES activity we studied the effect of a UNCG substitution. The GUAA sequence in domain 3 (Fig. 1 ▶) was substituted by three different versions of UNCG, and the activity of the mutated IRES was examined in transfected BHK-21 cells in the context of bicistronic constructs of the form CAT-IRES-luciferase (Fig. 2 ▶). None of the UNCG IRES mutants were as active as the wild-type sequence. In fact, the mutated IRES were 50- to 100-fold lower active than the GUAA IRES.
FIGURE 2.
A UNCG sequence does not functionally substitute the GNRA motif for IRES activity. Bicistronic constructs containing the FMDV IRES bearing the wild-type GUAA sequence or the indicated UNCG substitution between CAT and luciferase were transfected into BHK-21 cells. Relative IRES activity was calculated as the luciferase/CAT activity in each extract, made relative to that observed in the full-length wild-type IRES.
As mentioned above, the involvement of GNRA sequence in organizing IRES structure was likely. As a first step to assess this possibility, we analyzed the mobility of RNAs bearing the wild-type GUAA motif or the UCCG substitution in native gels. Transcripts corresponding to FMDV domain 3 formed dimers very efficiently following renaturation in native conditions (Ramos and Martínez-Salas 1999). The RNA bearing the UCCG substitution also dimerized, but interestingly, the monomer appeared as a diffuse band (data not shown), suggesting the presence of more RNA conformations than observed in the wild-type GUAA RNA. The nucleotide substitutions introduced at the GNRA motif did not modify the pattern of eIF4G, eIF4B, p110 (eIF3), and PTB binding detected in UV-crosslink assays of labeled mutant IRES (data not shown). These results suggested that the contribution of the GNRA motif to IRES activity could be due to its involvement in RNA organization.
RNA oligonucleotides harboring the GNRA motif interact only with domain 3 or the entire IRES
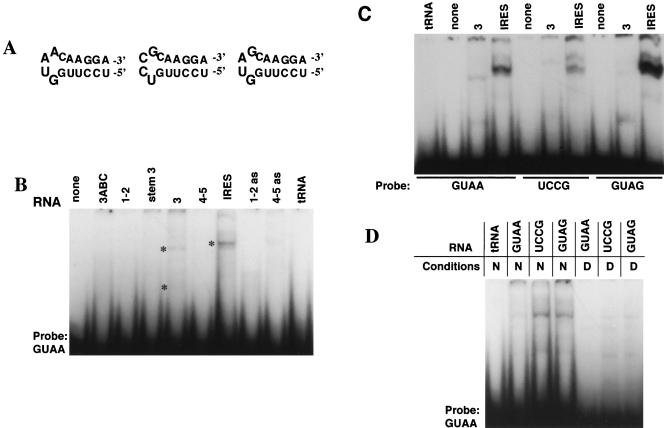
To test the existence of tertiary contacts dependent on the GNRA motif, we performed gel-shift analysis using as probes synthetic RNA oligonucleotides that reproduced the GNRA stem–loop (Fig. 3A ▶). Interaction of the GUAA hairpin was weak but specific for the monomer and the dimer of domain 3, and the full-length IRES. None of the other IRES domains, tRNA, or any of the antisense transcripts used as ligands showed interaction with this probe in gel mobility-shift assays (Fig. 3B ▶). This interaction relied on the presence of Mg2+ ions in the binding buffer, and was dosage-dependent (data not shown).
FIGURE 3.
GNRA-dependent RNA–RNA interactions. (A) Secondary structure of the oligoribonucleotides GUAA, UCCG, and GUAG. (B) Mapping of GUAA stem–loop binding site. The GUAA 32P-labeled oligoribonucleotide (50 nM) was incubated in binding buffer with unlabeled sense IRES domains, antisense, or tRNA (400 nM). Transcripts are named according to the domain they represent; 1–2 (1–85); 3, (84–297); 4–5, (306–462). Transcripts 3ABC (151–227) and stem 3 (84–297, harboring a deletion of residues 152–226) correspond to the apical and basal regions of domain 3, respectively. Complexes were electrophoresed through a 6% acrylamide gel in TBM buffer. Retarded bands corresponding to heterocomplexes are marked with asterisks. (C) Oligoribonucleotides bearing UCCG and GUAG tetraloops modified the efficiency of binding to the entire domain 3 or full-length wild-type FMDV IRES. (D) Substitutions in the GNRA motif of domain 3 alter the efficiency of binding of labeled GUAA stem–loop oligoribonucleotide.
The use of an oligonucleotide bearing the UCCG sequence at the loop significantly decreased the efficiency of binding with domain 3 and the full-length IRES (Fig. 3C ▶). The altered mobility observed in the weak retarded complex may be due to conformational changes in the resulting RNA–hairpin complex. This modification in RNA–RNA interaction may be responsible for the lack of IRES activity observed in UCCG mutant (see Fig. 2 ▶). This result led us to analyze the behavior of a hairpin bearing the sequence GUAG at the loop, a single substitution in the GNRA motif that was one of the most defective IRES in our initial screening of the GNRA mutants activity (López de Quinto and Martínez-Salas 1997). The GUAG oligonucleotide also decreased its efficiency of binding with domain 3 (Fig. 3C ▶). In addition, the GUAG hairpin binds four-fold more strongly with the full-length IRES than the GUAA, likely as a consequence of a change in the binding site. Moreover, binding efficiency of the GUAA oligoribonucleotide to mutants of domain 3, UCCG, or GUAG was enhanced relative to the intensity detected in the wild-type domain 3 (Fig. 3D ▶), suggesting lack of intramolecular competition in the GNRA mutant.
RNA structure mapping of the central region of FMDV IRES by chemical probing
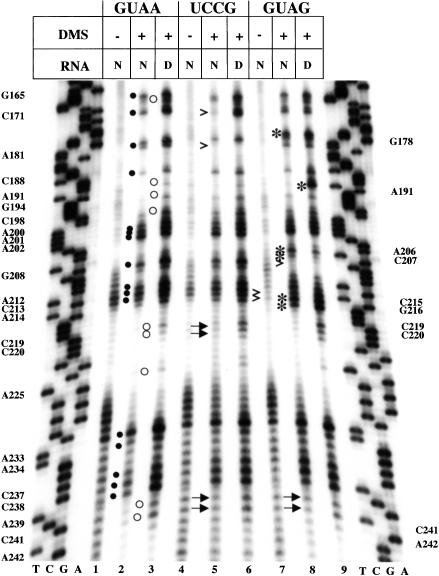
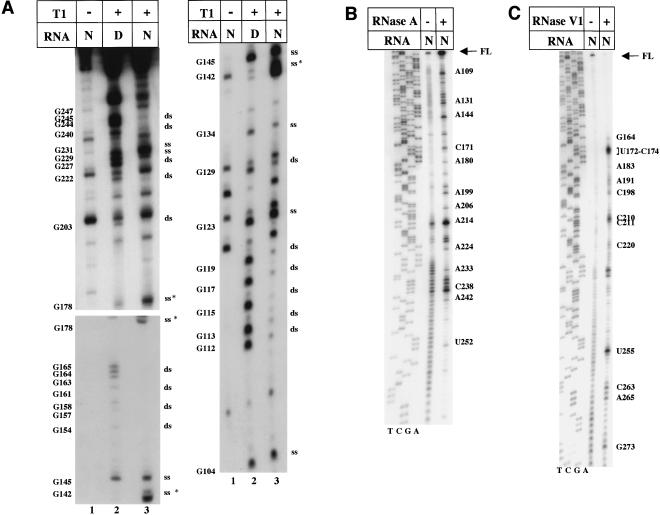
The functional analysis of the GNRA motif together with mapping of the binding site of oligonucleotides mimicking this stem–loop to domain 3 prompted us to study the potential involvement of the GNRA motif in the organization of IRES structure. As a tool to examine the compactness of RNA structure, we performed dimethyl-sulfate (DMS) modification followed of primer extension analysis of the FMDV IRES using a 5′end-labeled antisense oligonucleotide annealed to the base of domain 3. The reverse transcriptase (RT) extension products were analyzed in denaturing 6% acrylamide, 7 M urea gels, in parallel to a sequence ladder prepared with the same 5′ end-labeled primer. Compared to the native, untreated RNA (Fig. 4 ▶, lane 1), methylation of domain 3 in native conditions led to RT-stops at specific residues (Fig. 4 ▶, filled dots in lane 2). These RT-halts should precede one nucleotide to unpaired residues (Brunel and Romby 2000). Interestingly, the native RNA methylated in the presence of Mg2+ ions (N), led to protection of RT-stop at nine positions relative to RNA methylated under denaturing conditions (Fig. 4 ▶, empty circles in lane 3). It has to be noticed that protected residues occurred mostly within the apical region of domain 3, suggesting a reorganization of RNA structure induced by the presence of divalent ions in the solvent.
FIGURE 4.
Chemical probing of mutants in the GNRA motif of FMDV IRES. Domain 3 prepared in vitro was annealed to a 5′ end-labeled antisense primer and used as template in a reverse transcriptase reaction. A sequence ladder generated with the same labeled antisense primer was run in parallel to identify extension products; residues are marked on the left as the complementary sense sequence. Lane 1 shows the cDNA products obtained after primer extension of unmodified RNA. Lanes 2 and 3 show primer extension products of the RNA treated with dimethyl sulfate (DMS) in native (N) or denaturing (D) conditions. Filled dots in lane 2 depict the residues that halted RT elongation in DMS-treated RNA, and residues protected from DMS modification are denoted by open circles in lane 3. Transcripts bearing the UCCG or GUAG domain 3 were incubated with DMS in parallel to the wild type. Filled or empty dots point to the residues, marked on the left, which halted RT-elongation in DMS-treated GUAA RNA. Changes observed in GUAG and UCCG mutants are indicated by asterisks (new RT-stop), empty arrow-heads (lack of RT-stop relative to the GUAA RNA), or filled arrows (lack of protection relative to the GUAA RNA), and are listed on the right side of the autoradiogram.
DMS treatment of UCCG mutant showed that the RT-stop at positions 181 and 171 disappeared in native RNA (Fig. 4 ▶, lane 5) concomitantly with the lack of RT-stop protection at A242, C241, C220, and C219 in UCCG relative to that observed in the wild-type GUAA IRES. The results obtained with a UACG mutant were similar (data not shown). This striking long-range effect of the UNCG substitution in the GNRA motif prompted us to analyze the secondary structure of GUAG mutant. The DMS attacks observed in GUAG mutant revealed significant changes in primer extension, which affected residues spanning positions 206 to 208 and 215–216, in addition to the GNRA motif. The RT-stop at residue G208 was not observed, whereas six new bands were detected (asterisks in Fig. 4 ▶, lanes 8,9). Simultaneously, RT-stops at positions A242, C241 were not protected in GUAG, as it happened in the UCCG mutant. However, in contrast to the UCCG mutant, a strong enhancement of RT-stop was observed at position A191 only under denaturing conditions.
Primer extension analysis carried out using the full-length IRES transcript yielded the same pattern of RT-stops than the isolated domain 3 (data not shown). Therefore, the simultaneity of changes affecting the GNRA motif with those in residues 206–215 and 241–242 suggested that the GNRA motif contributes to distant interactions in the apical region of domain 3.
Enzymatic probing of the central region of FMDV IRES
To complement the structural information obtained by primer extension analysis of chemically modified RNA, we set up the conditions to analyze the accessibility of RNases to native or denatured 5′ end-labeled domain 3. Partial RNase T1 digestion of native RNA revealed the presence of G residues protected from cleavage, which were not protected when digestion was carried out in denaturing conditions. Extensive T1 digestion in native RNA was observed at defined positions (Fig. 5A ▶, lane 3). Conversely, significant protection from T1 cleavage in native RNA indicated involvement in the formation of base pairs (Fig. 5A ▶, cf. positions marked ds in lanes 3 with 2). In some instances, cleavages at G154 through G165 were faint, suggesting that this region may be hidden to T1 in the partial digestion conditions we are using. Enhanced T1 sensitivity at sites 142 and 178 (Fig. 5A ▶, lane 3) was observed in native RNA, suggesting a conformational change induced by Mg2+ ions that lead to overexposure of these residues to the solvent.
FIGURE 5.
(A) RNase T1 probing of FMDV domain 3. [γ-32P]ATP-5′ end-labeled RNA corresponding to domain 3 was incubated with 0.03 U RNase T1 in native (N) or denaturing (D) conditions. Samples were analyzed on 6% acrylamide 7M urea gels run for different amount of time. Asterisks denote the residues with enhanced sensibility to T1; ss and ds stand for single-stranded or double-stranded, respectively. Bands with the same electrophoretic mobility than truncated fragments of the untreated transcript were not considered. (B) RNase A accessibility to domain 3 of the FMDV IRES. Domain 3 was incubated with diluted RNase A (5 × 10−6 mg/mL) to obtain a partial RNA digestion, and then subjected to primer extension analysis. (C) RNase V1 digestion of domain 3, analyzed by primer extension.
To complement the T1 accessibility to G residues, we performed primer extension analysis of partial RNase A digestion that led us to distinguish C and U residues accessible in native RNA (Fig. 5B ▶), indicating therefore that they were unpaired. Then, and to verify the presence of paired nucleotides toward the 3′ end of domain 3, partial RNase V1 digestion followed of primer extension analysis was carried out (Fig. 5C ▶). A discontinuity between RT-stops suggests the presence of bulges interrupting base-paired regions, in agreement with RNase T1 protection on the opposite arm of the stem.
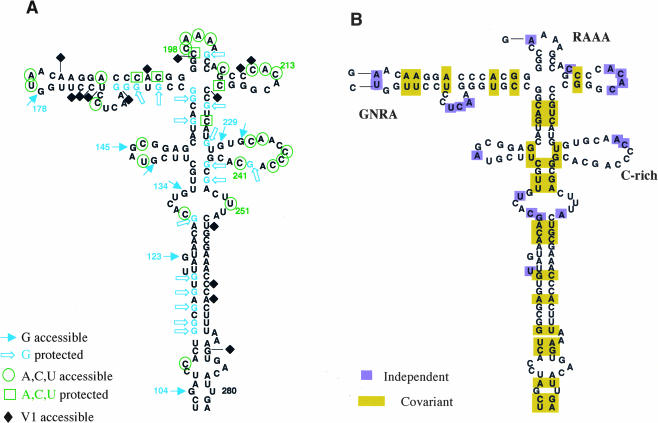
All together, the experimental evidences obtained from RNA probing provide valuable data to define the secondary structure of domain 3. Accessibility of RNase A and T1 to unpaired nucleotides was further verified by addition of DMS-attacked residues that allowed us to identify unequivocally most of the residues exposed in loops (Fig. 6A ▶). On the other hand, protection to RNase digestion and DMS attack in native RNA together with RNAse V1 accessibility has been used to define paired residues (Fig. 6A ▶). The results obtained from chemical probing and RNase accessibility are further supported by the phylogenetic conservation of the secondary structure. Nucleotide substitutions present in 26 different IRES sequences retrieved from databases, including representative examples of the different FMDV serotypes A, O, C, and Sat, were used to assess secondary structure conservation (Fig. 6B ▶). Covariation in these IRES sequences leads to compensatory substitutions, whereas most of the independent substitutions are present in loops. However, and despite the FMDV genetic variability (Martínez-Salas et al. 1993), motifs GNRA, RAAA, or the C-rich are phylogeneticaly conserved.
FIGURE 6.
Model of domain 3 structure in the FMDV IRES. (A) Results from partial T1, RNase A, and V1 digestion, together with DMS attack, have been assembled in a model of the secondary structure. Symbols and residues cited in the text are indicated. (B) Phylogenetic conservation of RNA structure in 26 different IRES sequences retrieved from databases.
Mutants in the GNRA motif showed a reorganization of their secondary structure affecting also distant regions in domain 3
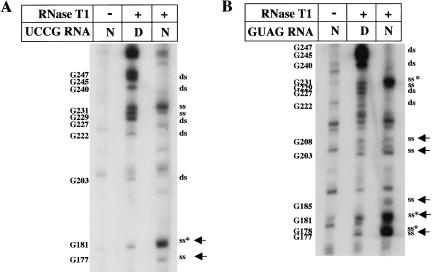
Next, to investigate the role of the GNRA motif in the organization of IRES structure, we have analyzed the secondary structure of mutants in this motif. RNase T1 partial digestion of UCCG mutant demonstrated that this substitution originated three changes in the secondary structure of domain 3 relative to the wild type. First, as expected from sequence modification, a new T1 cleavage appeared at G181, concomitant with absence of digestion at G178 (Fig. 7A ▶). In addition, G177 was also digested. A hypersensitive cleavage at G181 in the substituted sequence confirmed that the UNCG motif is present as a loop in the domain 3. Second, partial RNase A digestion of UCCG IRES showed a change in the pattern of cleavage outside of the mutated region. As summarized in Fig. 8 ▶, position C241 became protected. Thus, the UCCG substitution at the GNRA motif originated three structural modifications, two in the mutated motif, and the other in residues located in a distant region.
FIGURE 7.
Enzymatic probing of UCCG and GUAG mutants. [γ-32P]ATP-5′ end-labeled RNA corresponding to domain 3 substitution mutants bearing the UCCG (A) or GUAG (B) sequence in the GNRA motif were incubated with RNase T1 and analyzed on 6% acrylamide 7 M urea gel electrophoresis. Asterisks denote the residues that showed enhanced sensibility to RNases, ss and ds stands for single stranded and double stranded, respectively. Filled arrows point to new digestion sites relative to the wild-type GUAA sequence.
FIGURE 8.
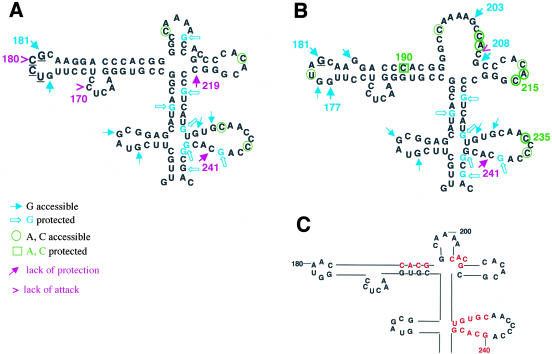
Reorganization of RNA structure in GNRA mutants. Results from DMS-attack have been represented on the secondary structure derived from T1 and RNase A digestion of the RNAs studied, UCCG (A) and GUAG (B). Relevant features of the apical region RNA structure (C).
RNase T1 digestion of GUAG RNA showed a number of differences with the wild-type GUAA IRES, which affected the apical part of domain 3. New cleavages were detected in G177, 181, 185, 203, and 208 (Fig. 7B ▶), indicating a more open conformation than the wild type. Moreover, new RNase A cleavages were detected at residues 205, 215, and 235 (Fig. 8 ▶). Thus, the RNA structure of GUAG mutant bearing a single substitution at the GNRA motif was extensively reorganized, affecting eight distant residues in the apical region of domain 3.
Taken together, these results demonstrated that mutations in the conserved GNRA motif of the apical region of domain 3 lead to changes in RNA structure, which are susceptible to chemical modification analysis. Simultaneous changes in the pattern of accessibility/protection of residues 170, 177, 180, 181, 219, and 241 (Fig. 8A ▶) were detected in the UNCG mutant relative to the wild-type GUAA by the various methods of analysis used. Analysis of the GUAG mutant showed variation in positions 177, 181, 185, 190, 235, and 241, accompanied by a strong change in secondary structure of residues 203–215 in the GUAG mutant (Fig. 8B ▶). Moreover, none of the mutants showed modifications in the proximal region of domain 3 (nt 104–134, and 247–280) by any of the different probing techniques. These data reinforce the role of the GNRA motif as a structural element involved in the organization of the RNA structure of the apical region of domain 3.
DISCUSSION
Structural organization of the central region in the FMDV IRES
Enzymatic and chemical probing results, in conjunction with the phylogenetic conservation and the functional analysis of the GNRA motif, demonstrates that domain 3 of the FMDV IRES is organized as a complex and flexible structure. First, our results support the existence of a four-way junction at the apical region of this domain; more than two base-paired residues are detected at the left, right, and bottom stems (Fig. 6A ▶). In addition, the top stem is consistent with both the protection of G203 to T1 and that of C197 to DMS. On the other hand, we have verified the existence of several loops, which harbor the sequences GUAGC, GUAA, CAAAA, CACACG, and CAACCCCA. Four of these loops consist of phylogeneticaly conserved motifs (Fig. 6B ▶), whereas the ACAC loop admits multiple nucleotide changes. Notably, the GUAA tetraloop and the CAAAA pentaloop accumulate less genetic variability than the adjacent loops.
Second, and to our surprise, the residues G229 and G231, around the C-rich loop, are more accessible to T1 than predicted by computer programs (Fig. 8C ▶). Additionally, DMS attack to C241 is protected in the presence of divalent ions, and G240 is protected from T1 digestion. This finding suggested to us that residues G240CAC243 did not pair permanently to G229UGC232. Thus, as observed in other IRES elements (Jan and Sarnow 2002), it appears that some parts of the FMDV IRES are in a dynamic equilibrium that can be reorganized in response to changing conditions in the environment (Ramos and Martínez-Salas 1999). In agreement with this suggestion, RNase T1 cleavage of G240 in FMDV IRES was enhanced in the presence of PTB and ITAF45 (Pilipenko et al. 2000) and the equivalent region in the closely related GDVII IRES is accessible to RNase One (Pilipenko et al. 2001).
Third, the RNA structure of the middle region of domain 3 has been extensively remodeled. According to T1 and RNase A analysis, an internal bulge included residues G134 as well as U251, but in clear contrast to the chemical probing of the apical region, residues susceptible to DMS attack in the middle region were undetectable (Fig. 6A ▶). This is consistent with a large number of base-paired nucleotides in the secondary structure that do not provide tertiary contacts under the conditions we are using for this study. The five Gs within positions 112–119 conform to a paired region. Presence of paired residues was verified by RNase V1 digestion, which showed arrays of protected residues interrupted by others with strong digestion (Fig. 5C ▶). Besides RNA probing (Fig. 6A ▶), phylogenetic covariation among 26 independent FMDV IRES sequences strongly favors this RNA structure (Fig. 6B ▶).
The GNRA tetraloop is responsible for the organization and stability of RNA structure in the FMDV IRES
The GNRA motif is conserved among picornavirus IRES elements (Jackson and Kaminski 1995). Consistent with this, disruption of the GNRA motif led to IRES inactivation in FMDV and EMCV (López de Quinto and Martínez-Salas 1997; Robertson et al. 1999). We show here that a UNCG motif did not replace functionally the GNRA when tested in vivo in the context of a bicistronic construct. UNCG tetraloops are highly stable structural elements (Tuerk et al. 1988; SantaLucia et al. 1992; Heus and Pardi 2001). Consistent with the presence of a stable loop, G181 in UCCG mutant RNA is hypersensitive to T1 (Fig. 8A ▶). In addition, protection of DMS attack at positions C219 and C241 observed in the GUAA wild-type sequence is lost in UNCG mutants (Fig. 8A ▶), strongly suggesting that changes in the GNRA motif led to structural reorganization of the IRES element.
Our study of the GUAG mutant carrying a single substitution at the fourth position in the GNRA motif confirmed the critical role of this structural element in the organization of domain 3 structure. The G181 change at the GNRA motif, in addition to G178, showed hypersensitivity to RNase T1 (Fig. 8B ▶). Remarkably, the single substitution of A181 to G caused a gross reorganization of region 203–208. RNase digestion of GUAG RNA showed that G203, C205, and G208 are unpaired, consistent with DMS attack to residues 205 and 206 (Fig. 8B ▶).
Results from primer extension of the untreated RNA revealed that the RT-halt induced by secondary structure at positions 214 and 215 in the wild type is relieved in the GUAG mutant (Fig. 4 ▶), consistent with an open configuration of the apical region of domain 3. Thus, a single substitution of A to G at the fourth position in the GNRA loop is responsible for the structural reorganization of the distal stem–loops bearing the RAAA and C-rich conserved motifs (Fig. 8B ▶).
The secondary structure of the proximal region of domain 3, spanning residues 104–134, and 247–280, is not modified in any of the studied mutants according to enzymatic probing; neither is chemical modification-induced changes in RT-stop at this region. Thus, only the apical part of domain 3 is severely reorganized after just a single nucleotide substitution in the GNRA motif to GUAG, or its substitution by a UNCG motif. Remarkably, and in support of long-distance interactions mediated by the GNRA motif, a synthetic oligoribonucleotide bearing the GUAA sequence, but not UCCG or GUAG, specifically interacts in vitro with sequences within domain 3.
Our previous mutational analysis of FMDV GNRA motif showed that GCGA, but not AUAA or GUAG, leads to an active IRES (López de Quinto and Martínez-Salas 1997). Thus regarding a potential GUAA binding site being present in the FMDV IRES, it should be compatible with GYRA receptors. Description of GUAA motif receptors including those selected as high affinity ligands (Costa and Michel 1997), showed a requirement of two consecutive C:G base pairs interacting with the A residues. In the case of the GYRA receptor, the C:G base pair was preceded by a U:A base pair. According to the changes in DMS protection observed in mutants UCCG and GUAG, there are three potential contact sites of the GNRA motif, involving residues C191, C207, and C241. All of them reside in a CACG sequence, potentially forming a double-strand structure (Fig. 8C ▶). It is of interest that primary sequence of G240CAC243 remains invariant among the different FMDV IRES elements retrieved from databases (Fig. 6B ▶). By contrast, many other regions that are paired in the secondary structure show covariation. In addition, residues G229UGC232 are accessible to RNases (Fig. 6A ▶), indicating that nucleotides 241–244 were not paired to 229–232, in contrast to computer program predictions. Thus, despite sequence complementarity, base pairing is prevented opening the possibility of tertiary interaction with distant residues. Further work is required to determine the existence of direct contacts between these regions.
MATERIALS AND METHODS
Plasmids
The IRES region of FMDV Cs8 (Martínez-Salas et al. 1993) was subjected to site-directed mutagenesis to introduce UNCG substitutions at the GNRA motif (Fig. 1 ▶). Mutagenic primer (antisense) 5′-CTTGCGNACAAGGAGGAGTTC-3′, was used in combination with external primers pBIC-s (CGATGAGTGGCAGGGCG GGGC) and pBIC-as (GGCCTTTCTTTATGTTTTTGGCG) as described (Martínez-Salas et al. 1996). Then different mutants of domain 3, nt 84 to 297, were subcloned in pGEM vector as described (Ramos and Martínez-Salas 1999). The nucleotide sequence of the entire length of each IRES under study was obtained using automatic sequencing (ABI PRISM dye terminator cycle sequencing ready reaction kit, Perkin-Elmer).
In vitro transcription
Prior to RNA synthesis, plasmids pGEM-IRES encoding various versions of the FMDV IRES under the control of the T7 RNA polymerase were linearized at a unique SmaI or XhoI restriction sites (domain 3) or the entire IRES, respectively (Ramos and Martinez-Salas 1999). Transcription was performed using 0.5 μg of linear plasmid DNA and the Megashortscript kit (Ambion) during 2 h at 37°C. Reactions were then incubated 15 min with 2 U of DNase I. Newly synthesized RNA was phenol extracted, ethanol precipitated, and resuspended in RNase free water. The integrity of the transcripts as well as efficiency of the synthesis was analyzed in denaturing 6% acrylamide, 7 M urea gel electrophoresis. When required, transcripts were uniformly labeled to a specific activity of ~107 cpm/pmole using (α32P)-CTP (400 Ci/mmole). Template DNA was digested with 1 U of RQ1 DNase (Promega) and unincorporated isotope eliminated by exclusion chromatography in TE-equilibrated columns. RNA was extracted with phenol-chloroform, ethanol precipitated, and resuspended in TE to a concentration of 0.04 pmole/μL (~5 × 105 cpm/μL).
Chemical probing
Chemical modification was performed essentially as described (Brunel and Romby 2000). Monitoring of changes in the RNA folding, dependent on the presence of divalent cations in the solvent, was ensured by comparison of the pattern of DMS attack in native (N) versus denaturing (D) conditions (Peattie and Gilbert 1980). This allows to monitor the appearance of residues protected from methylation in native RNA, which may be indicative of their involvement in the formation of tertiary contacts and stacking of purines in helices (Ehresmann et al. 1987; Watanabe et al. 1994). RNA (about 1.5 μg) was incubated in 20 μL of buffer N (50 mM sodium cacodylate pH 7.5, 300 mM KCl, 10 mM MgCl2) in the presence of 1 μL of DMS (Fluka) freshly diluted 1:5 in ethanol, 15 min at 20°C. To treat the RNA under denaturing conditions, RNA was first denatured during 3 min at 95°C in water, cool down in ice, and adjusted to 20 μL of buffer D (50 mM cacodylate pH 7.5, 1 mM EDTA) prior to addition of 1 μL of DMS and incubation at 20°C 15 min (Brunel and Romby 2000). The methylated RNA was ethanol-precipitated using 2 μg of oyster glycogen as carrier and resuspended in RNase free water, followed of a primer extension analysis.
Primer extension
For primer extension, a 17-mer oligonucleotide 5′CCCGGGTG TGGGTACC3′ complementary to positions 296 to 287 of domain 3, was labeled at its 5′ end with [γ32P]-ATP and T4 polynucleotide kinase (Boehringher Mannheim). Unincorporated isotope was separated by exclusion chromatography using microspin™ columns (Amersham), ethanol precipitated using 2 μg oyster glycogen as carrier and resuspended in water to about 200.000 cpm/μL (0.33 pmole/μL). Then, 1.5 μg of template RNA either, untreated or chemically modified with DMS, was denatured 3 min at 95°C and cool down on ice. Annealing and extension of the labeled antisense primer was carried out in 15 μL of reverse transcriptase (RT) buffer (20 mM Tris-HCl pH 7.5, 100 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 0.01% (v/v) NP-40, 50% (v/v) glycerol in the presence of 100 U de SuperScript II RT (Life Technologies) and 1 mM each dNTPs for 1 h at 45°C. The RNA template was then hydrolyzed in 20 μL of 50 mM Tris-HCl, pH 7.5, 7.5 mM EDTA, 0.5% SDS in the presence of 3.5 μL 3 M KOH at 95° for 3 min followed of incubation at 37°C for 1 h. Hydrolysis was stopped adding 6 μL of 3 M acetic acid. RT-products were precipitated with 2 μg of glycogen carrier and resuspended in 10 μL of water supplemented with denaturing loading buffer (90% formamide, 1 mM EDTA pH 8, 0.1% xylencianol, 0.1% bromophenol). Following denaturation during 3 min at 95°C, the samples were loaded in 6% acrylamide, 7 M urea gels, in parallel to a sequence ladder. The extension products were visualized after autoradiography of the dried gels for about 12 h with intensifying screen.
To prepare the sequence ladder, the Thermo sequenase cycle sequencing kit (Amersham) was used with the 5′-labeled antisense oligonucleotide used for primer extension and plasmid pGEM-IRES as template.
Ribonuclease accessibility
In vitro synthesized RNAs were first dephosphorylated with 1 U of CIP (New England Biolabs) in 100 mM NaCl, 50 mM Tris-HCl, 10 mM MgCl2, 1 mM dithiothreitol pH 7.9, for 30′ at 37°C, and then 5′ end-labeled using [γ-32P]-ATP and T4 polynucleotide kinase (Boehringer Mannheim). Full-length transcripts were identified in 6% acrylamide, 7 M urea gels, and the RNA present in thin slices eluted overnight in 200 μL of 10 mM Tris pH 8, 1 mM EDTA, 0.1% SDS, at 37°C with shaking. Following ethanol precipitation in the presence of oyster glycogen, the eluted RNAs were resuspended in water to about 50,000 cpm/μL.
RNase T1, 0.03 U (Boehringer Manheim) was used to digest partially the labeled RNAs in the presence of 1 μg of unlabeled transcript and 4 μg of tRNA in buffer N (50 mM sodium cacodylate pH 7.5, 300 mM KCl, 10 mM MgCl2) for 30′ at 37°C. For treatment carried out under denaturing conditions, RNAs were incubated in 10 mM Tris-HCl pH 8, 1 mM EDTA (Branch et al. 1989). Similarly, the desired RNAs were treated with 5 × 10−6 mg/mL of RNase A (Aldaz-Carroll et al. 2002) (Boehringer Manheim) under the same buffer conditions used for T1; when required digested RNA was analyzed by primer extension. RNase V1 (Ambion) partial digestions were performed as described (Kolupaeva et al. 2000) using 0.2 U per assay, followed of incubation during 10 min at 37° in buffer N, but then analyzed by primer extension.
RNA–RNA and RNA–protein interaction
For RNA–RNA interactions, [γ-32P]-ATP labeled oligoribonucleotide (GUAA, UCCG, or GUAG) or the indicated FMDV IRES domains were incubated independently at 80°C for 1 min, chilled on ice (Aldaz-Carroll et al. 2002), and then mixed in 50 mM sodium cacodylate pH 7.5, 300 mM KCl, 10 mM MgCl2 (Ferrandon et al. 1997). RNA–RNA complexes were allowed to form for 90 min at 20°C and inmediately analyzed by electrophoresis in native acrylamide gels supplemented with 2.5 mM MgCl2 (Fedor and Uhlenbeck 1990; Paillart et al. 1996). Gels were run at 4°C for 33 min at 180 V in TBM buffer (45 mM Tris, pH 8.3, 43 mM boric acid, 0.1 mM MgCl2). Prior to gel drying, RNA was stained with ethidium bromide and photographed. In separate experiments RNAs were denatured at 65°C for 1 min, and then allowed to renature slowly at room temperature (Kieft et al. 1999) without changes in the pattern of RNA mobility. Dried gels were exposed for autoradiography, as well as to a phosphorimager plate, to quantify the intensity of the retarded bands.
For RNA–protein interaction, uniformly labeled probes (0.04 pmole, ~5 × 105 cpm) were UV-croslinked with soluble proteins present in BHK-21 S10 cell extract (40 μg protein) as described (López de Quinto and Martínez-Salas 2000).
In vivo IRES activity
Relative IRES activity was quantified as the expression of luciferase normalized to that of chloramphenicol acetyltransferase (CAT) from bicistronic mRNAs (López de Quinto and Martínez-Salas 1997). Transfection of 80–90 % confluent BHK-21 monolayers was carried out using cationic liposomes, 1 h after infection with the Vaccinia virus recombinant vTF7-3 expressing the T7 RNA polymerase (Fuerst et al. 1986). Extracts from 1–2 × 105 cells were prepared 20 h postransfection in 100 μL of 50 mM Tris-HCl, pH 7.8, 120 mM NaCl, 0.5% NP40. Experiments were performed on triplicate wells and each experiment was repeated at least three times.
Phylogenetic analysis
Full-length FMDV IRES sequences retrieved from databases, representing the different FMDV serotypes (A, C, O, and Sat), were aligned and then plot in the RNA secondary structure. To remodel the RNA structure of domain 3, the most abundant covariation among the 26 FMDV IRES sequences was always chosen to fit the results of biochemical probing.
Acknowledgments
We thank E. Cano and J. Ramajo for technical assistance. We are also grateful to S. Reigadas, E. Westhof, and C. Gutiérrez for helpful suggestions. This work was supported by grants BMC2002-00983 from MCyT, 07B/0052/2002 from CAM, and an Institutional grant from Fundación Ramón Areces. The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5950603.
REFERENCES
- Aldaz-Carroll, L., Tallet, B., Dausse, E., Yurchenko, L., and Toulmé, J-J. 2002. Apical loop–internal loop interactions: A new RNA–RNA recognition motif identified through in vitro selection against RNA hairpins of the hepatitis C virus mRNA. Biochemistry 41: 5883–5893. [DOI] [PubMed] [Google Scholar]
- Belsham, G.J. and Jackson, R.J. 2000. Translation initiation on picornavirus RNA. In: Translational control of gene expression (eds. N. Sonenberg et al.), pp. 869–900. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Branch, A.D., Benenfeld, B.J., and Robertson, H.D. 1989. RNA fingerprinting. Methods Enzymol. 180: 130–154. [DOI] [PubMed] [Google Scholar]
- Brunel, C. and Romby, P. 2000. Probing RNA structure and RNA–ligand complexes with chemical probes. Methods Enzymol. 318: 3–21. [DOI] [PubMed] [Google Scholar]
- Correll, C.C., Freeborn, B., Moore, P.B., and Steitz, T.A. 1997. Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell 91: 705–712. [DOI] [PubMed] [Google Scholar]
- Costa, M. and Michel, F. 1997. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: Comparison with in vivo evolution. EMBO J. 16: 3289–3302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty, E.A., Batey, R.T., Masquida, B., and Doudna, J.A. 2001. A universal mode of helix packing in RNA. Nat. Struct. Biol. 8: 339–343. [DOI] [PubMed] [Google Scholar]
- Ehresmann, C., Baudin, F., Mougel, M., Romby, P., Ebel, J.P., and Ehresmann, B. 1987. Probing the structure of RNAs in solution. Nucleic Acids Res. 15: 9109–9128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fedor, M.J. and Uhlenbeck, O.C. 1990. Substrate sequence effects on “hammerhead” RNA catalytic efficiency. Proc. Natl. Acad. Sci. 87: 1668–1672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandon, D., Koch, I., Westhof, E., and Nusslein-Volhard, C. 1997. RNA–RNA interaction is required for the formation of specific bicoid mRNA 3′ UTR-STAUFEN ribonucleoprotein particles. EMBO J. 16: 1751–1758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst, T.R., Niles, E.G., Studier, F.W., and Moss, B. 1986. Eukaryotic transient-expression system based on recombinant vaccinia virus that synthesizes bacteriophage T7 RNA polymerase. Proc. Natl. Acad. Sci. 83: 8122–8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellen, C.U. and Sarnow, P. 2001. Internal ribosome entry sites in eukaryotic mRNA molecules. Genes & Dev. 15: 1593–1612. [DOI] [PubMed] [Google Scholar]
- Heus, H.A. and Pardi, A. 1991. Structural features that give rise to the unusual stability of RNA hairpins containing GNRA loops. Science 253: 191–194. [DOI] [PubMed] [Google Scholar]
- Holcik, M., Sonenberg, N., and Korneluk, R.G. 2000. Internal ribosome initiation of translation and the control of cell death. Trends Genet. 16: 469–473. [DOI] [PubMed] [Google Scholar]
- Jackson, R.J. and Kaminski, A. 1995. Internal initiation of translation in eukaryotes: The picornavirus paradigm and beyond. RNA 1: 985–1000. [PMC free article] [PubMed] [Google Scholar]
- Jackson, R.J. 2000. Comparative view of initiation site selection mechanism. In: Translational control of gene expression (eds. N. Sonenberg, J.W.B. Hershey, and M.B. Mathews), pp. 127–184. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY.
- Jan, E. and Sarnow, P. 2002. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 324: 889–902. [DOI] [PubMed] [Google Scholar]
- Jang, S.K. and Wimmer, E. 1990. Cap-independent translation of encephalomyocarditis virus RNA: Structural elements of the internal ribosomal entry site and involvement of a cellular 57-kD RNA-binding protein. Genes & Dev. 4: 1560–1572. [DOI] [PubMed] [Google Scholar]
- Kieft, J.S., Zhou, K., Jubin, R., Murray, M.G., Lau, J.Y. and Doudna, J.A. 1999. The hepatitis C virus internal ribosome entry site adopts an ion-dependent tertiary fold. J. Mol. Biol. 292: 513–529. [DOI] [PubMed] [Google Scholar]
- Kieft, J.S., Zhou, K., Grech, A., Jubin, R., and Doudna, J.A. 2002. Crystal structure of an RNA tertiary domain essential to HCV IRES-mediated translation initiation. Nat. Struct. Biol. 9: 370–374. [DOI] [PubMed] [Google Scholar]
- Klinck, R., Westhof, E., Walker, S., Afshar, M., Collier, A., and Aboul-Ela, F. 2000. A potential RNA drug target in the hepatitis C virus internal ribosomal entry site. RNA 6: 1423–1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolk, M.H., van der Graaf, M., Fransen, C.T., Wijmenga, S.S., Pleij, C.W., Heus, H.A., and Hilbers, C.W. 1998. Structure of the 3′-hairpin of the TYMV pseudoknot: Preformation in RNA folding. EMBO J. 17: 7498–7504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva, V.G., Hellen, C.U., and Shatsky, I.N. 1996. Structural analysis of the interaction of the pyrimidine tract-binding protein with the internal ribosomal entry site of encephalomyocarditis virus and foot-and-mouth disease virus RNAs. RNA 2: 1199–1212. [PMC free article] [PubMed] [Google Scholar]
- Kolupaeva, V.G., Pestova, T.V., Hellen, C.U., and Shatsky, I.N. 1998. Translation eukaryotic initiation factor 4G recognizes a specific structural element within the internal ribosome entry site of encephalomyocarditis virus RNA. J. Biol. Chem. 273: 18599–18604. [DOI] [PubMed] [Google Scholar]
- Kolupaeva, V.G., Pestova, T.V., and Hellen, C.U. 2000. An enzymatic footprinting analysis of the interaction of 40S ribosomal subunits with the internal ribosomal entry site of hepatitis C virus. J. Virol. 74: 6242–6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Quesne, J.P., Stoneley, M., Fraser, G.A., and Willis, A.E. 2001. Derivation of a structural model for the c-myc IRES. J. Mol. Biol. 310: 111–126. [DOI] [PubMed] [Google Scholar]
- López de Quinto, S. and Martínez-Salas, E. 1997. Conserved structural motifs located in distal loops of aphthovirus internal ribosome entry site domain 3 are required for internal initiation of translation. J. Virol. 71: 4171–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ———. 2000. Interaction of the eIF4G initiation factor with the aphthovirus IRES is essential for internal translation initiation in vivo. RNA 6: 1380–1392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- López de Quinto, S., Lafuente, E., and Martínez-Salas, E. 2001. IRES interaction with translation initiation factors: Functional characterization of novel RNA contacts with eIF3, eIF4B, and eIF4GII. RNA 7: 1213–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukavsky, P.J., Otto, G.A., Lancaster, A.M., Sarnow, P., and Puglisi, J.D. 2000. Structures of two RNA domains essential for hepatitis C virus internal ribosome entry site function. Nat. Struct. Biol. 7: 1105–1110. [DOI] [PubMed] [Google Scholar]
- Luz, N. and Beck, E. 1991. Interaction of a cellular 57-kilodalton protein with the internal translation initiation site of foot-and-mouth disease virus. J. Virol. 65: 6486–6494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Salas, E., Sáiz, J.C., Dávila, M., Belsham, G.J., and Domingo, E. 1993. A single nucleotide substitution in the internal ribosome entry site of foot-and-mouth disease virus leads to enhanced cap-independent translation in vivo. J. Virol. 67: 3748–3755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Salas, E., Regalado, M.P., and Domingo, E. 1996. Identification of an essential region for internal initiation of translation in the aphthovirus internal ribosome entry site and implications for viral evolution. J. Virol. 70: 992–998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martínez-Salas, E., Ramos, R., Lafuente, E., and López de Quinto, S. 2001. Functional interactions in internal translation initiation directed by viral and cellular IRES elements. J. Gen. Virol. 82: 973–984. [DOI] [PubMed] [Google Scholar]
- Meyer, K., Petersen, A., Niepmann, M., and Beck, E. 1995. Interaction of eukaryotic initiation factor eIF-4B with a picornavirus internal translation initiation site. J. Virol. 69: 2819–2824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michiels, P.J., Versleijen, A.A., Verlaan, P.W., Pleij, C.W., Hilbers, C.W., and Heus, H.A. 2001. Solution structure of the pseudoknot of SRV-1 RNA, involved in ribosomal frameshifting. J. Mol. Biol. 310: 1109–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell, S.A., Spriggs, K.A., Coldwell, M.J., Jackson, R.J., and Willis, A.E. 2003. The Apaf-1 internal ribosome entry segment attains the correct structural conformation for function via interactions with PTB and unr. Mol. Cell. 11: 757–771. [DOI] [PubMed] [Google Scholar]
- Paillart, J.C., Skripkin, E., Ehresmann, B., Ehresmann, C., and Marquet, R. 1996. A loop-loop “kissing” complex is the essential part of the dimer linkage of genomic HIV-1 RNA. Proc. Natl. Acad. Sci. 93: 5572–5577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peattie, D.A. and Gilbert, W. 1980. Chemical probes for higher-order structure in RNA. Proc. Natl. Acad. Sci. 77: 4679–4682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestova, T.V., Kolupaeva, V.G., Lomakin, I.B., Pilipenko, E.V., Shatsky, I.N., Agol, V.I., and Hellen, C.U. 2001. Molecular mechanisms of translation initiation in eukaryotes. Proc. Natl. Acad. Sci. 98: 7029–7036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pilipenko, E.V., Pestova, T.V., Kolupaeva, V.G., Khitrina, E.V., Poperechnaya, A.N., Agol, V.I., and Hellen, C.U.T. 2000. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes & Dev. 14: 2028–2045. [PMC free article] [PubMed] [Google Scholar]
- Pilipenko, E.V., Viktorova, E.G., Guest, S.T., Agol, V., and Roos, R.P. 2001. Cell-specific proteins regulate viral RNA translation and virus-induced disease. EMBO J. 20: 6899–6908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramos, R. and Martínez-Salas, E. 1999. Long-range RNA interactions between structural domains of the aphthovirus internal ribosome entry site (IRES). RNA 5: 1374–1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robertson, M.E., Seamons, R.A., and Belsham, G.J. 1999. A selection system for functional internal ribosome entry site (IRES) elements: Analysis of the requirement for a conserved GNRA tetraloop in the encephalomyocarditis virus IRES. RNA 5: 1167–1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- SantaLucia Jr., J., Kierzek, R., and Turner, D.H. 1992. Context dependence of hydrogen bond free energy revealed by substitutions in an RNA hairpin. Science 256: 217–219. [DOI] [PubMed] [Google Scholar]
- Selinger, D., Liao, X., and Wise, J.A. 1993. Functional interchangeability of the structurally similar tetranucleotide loops GAAA and UUCG in fission yeast signal recognition particle RNA. Proc. Natl. Acad. Sci. 90: 5409–5413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stassinopoulos, I.A. and Belsham, G.J. 2001. A novel protein-RNA binding assay: Functional interactions of the foot-and-mouth disease virus internal ribosome entry site with cellular proteins. RNA 7: 114–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuerk, C., Gauss, P., Thermes, C., Groebe, D.R., Gayle, M., Guild, N., Stormo, G., d’Aubenton-Carafa, Y., Uhlenbeck, O.C., Tinoco Jr., I., et al. 1988. CUUCGG hairpins: Extraordinarily stable RNA secondary structures associated with various biochemical processes. Proc. Natl. Acad. Sci. 85: 1364–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos, S., Berrisford, D.J., and Avis, J.M. 2002. Effect of magnesium ions on the tertiary structure of the hepatitis C virus IRES and its affinity for the cyclic peptide antibiotic viomycin. Biochemistry 41: 5383–5396. [DOI] [PubMed] [Google Scholar]
- Walter, B.L., Nguyen, J.H., Ehrenfeld, E., and Semler, B.L. 1999. Differential utilization of poly(rC) binding protein 2 in translation directed by picornavirus IRES elements. RNA 5: 1570–1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe. Y., Kawai, G., Yokogawa, T., Hayashi, N., Kumazawa, Y., Ueda, T., Nishikawa, K., Hirao, I., Miura, K., and Watanabe, K. 1994. Higher-order structure of bovine mitochondrial tRNA (SerUGA): Chemical modification and computer modeling. Nucleic Acids Res. 22: 5378–5384. [DOI] [PMC free article] [PubMed] [Google Scholar]