Abstract
During translation, tRNAs cycle through three binding sites on the ribosome: the A, the P, and the E sites. We have determined the structures of complexes between the Haloarcula marismortui large ribosomal subunit and two different E site substrates: a deacylated tRNA acceptor stem minihelix and a CCA-acceptor end. Both of these tRNA mimics contain analogs of adenosine 76, the component responsible for a large proportion of E site binding affinity. They bind in the center of the loop-extension of protein L44e, and make specific contacts with both L44e and 23S rRNA including bases that are conserved in all three kingdoms of life. These contacts are consistent with the footprinting, protection, and cross-linking data that have identified the E site biochemically. These structures explain the specificity of the E site for deacylated tRNAs, as it is too small to accommodate any relevant aminoacyl-tRNA. The orientation of the minihelix suggests that it may mimic the P/E hybrid state. It appears that the E site on the 50S subunit was formed by only RNA in the last common ancestor of the three kingdoms, since the proteins at the E sites of H. marismortui and Deinucoccus radiodurans large subunits are not homologous.
Keywords: Ribosome, X-ray crystallography, E site, 50S ribosomal subunit, tRNA
INTRODUCTION
The ribosome synthesizes proteins encoded by mRNA using aminoacylated tRNA molecules (Maden et al. 1968). In studies of translation in the 1960s, the ribosome was assumed to have two tRNA binding sites, as a donor and an acceptor tRNA appeared to be sufficient for peptidyl transferase (Watson 1964). The site for aminoacyl-tRNA binding was called the A site, and the site for peptidyl-tRNA was called the P site. A third site was proposed later when it was discovered that ribosomes can bind three tRNAs per particle (Rheinberger et al. 1981), and that deacylated tRNA could bind to ribosomes that have occupied A and P sites (Grajevskaja et al. 1982). In vivo, this site receives newly deacylated tRNAs that have been translocated from the P site following peptide bond formation (Kirillov et al. 1983; Lill et al. 1984). It is called the E or exit site (Marquez et al. 2002).
The location, structure, and function of the E site has been investigated by numerous biochemical studies. Chemical footprinting, protection, and cross-linking studies determined that the portion of the E site on the 30S subunit is close to ribosomal protein S11 and the 3′ terminus of 16S rRNA (Wower et al. 1993), and that the component on the 50S subunit is near the L1 stalk and helix 88 of 23S rRNA (Moazed and Noller 1989a; Wower et al. 1993, 2000; Rinke-Appel et al. 1995; Joseph and Noller 1996; Bocchetta et al. 2001). Using modified tRNAs, it was shown that the terminal adenosine of tRNA contributes significantly to E site binding; removal of A76 decreases tRNA affinity for the E site to a level that is below the experimental detection limit (>100-fold; Lill et al. 1988). The functional role of the E site is disputed, as independent studies have concluded that the E site either facilitates translocation (Lill et al. 1986) or, through negative cooperativity with the A site, promotes translational fidelity (Geigenmuller and Nierhaus 1990).
All three ribosomal tRNA binding sites have been studied by structural methods. tRNAs bound to the three sites of the 70S ribosome were first observed at low resolution by electron microscopy (Agrawal et al. 1996; Stark et al. 1997). Subsequently, a model of the Thermus thermophilus 70S ribosome bound with tRNA molecules that had been positioned into a 5.5 Å resolution electron density map provided a more detailed picture of these sites and their interactions with full-length tRNAs (Yusupov et al. 2001). However, it is only at atomic resolution that the specific groups and interactions important for function can be identified. In this regard, the high-resolution crystal structures of isolated subunits with tRNA mimics bound have been effective in illuminating these interactions. For example, the 3 Å resolution structures of the T. thermophilus small subunit complexed with mRNA and mimics of the anticodon tRNA stem-loop bound to the A and P sites of the 30S subunit have identified the components of the small subunit that mediate interactions between mRNA and tRNA (Carter et al. 2000) and have provided insights into the mechanism of decoding (Ogle et al. 2001, 2002). In addition, the atomic resolution structures of Haloarcula marismortui large ribosomal subunit complexed with various peptidyl transferase substrate, intermediate, and product analogs have similarly led to unambiguous identification of the A and P sites in the peptidyl transferase center of the 50S subunit (Nissen et al. 2000), and make it possible to analyze structural aspects the peptidyl transferase reaction (Hansen et al. 2002; Schmeing et al. 2002). Here we describe two atomic resolution structures of the large ribosomal subunit with deacylated tRNA mimics bound in the E site.
RESULTS
The location of the E site on the large ribosomal subunit
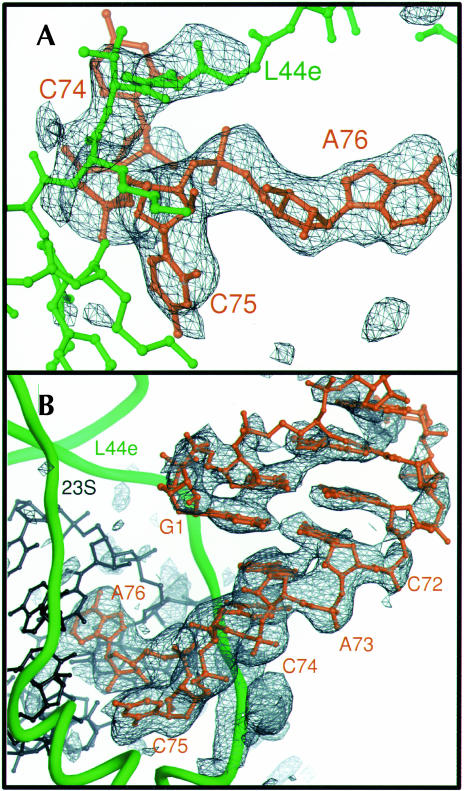
A minihelix that is structurally analogous to the acceptor and TΨC arm of yeast tRNAphe and the RNA oligonucleotide CCA, corresponding to the universally conserved nucleotides 74–76 of tRNA, were soaked into crystals of the H. marismortui large ribosomal subunit and visualized by difference Fourier maps calculated at 3.1 and 2.9 Å resolution, respectively (Fig. 1 ▶; Table 1 ▶). The minihelix binds exclusively to a site in the cleft of 50S subunit about 45 Å away from the P site, near the L1 arm (Fig. 2 ▶). This is the area that had been identified previously as a component of the 50S subunit E site by biochemical studies (Moazed and Noller 1989a; Wower et al. 1993, 2000; Rinke-Appel et al. 1995; Joseph and Noller 1996; Bocchetta et al. 2001) and by lower resolution structures (Agrawal et al. 1996; Stark et al. 1997; Yusupov et al. 2001). Although CCA is a short sequence, it displays significant specificity and binds only at the A, P, and E sites (data not shown). The terminal 3′ CCA nucleotides of the minihelix interact with the E site of the large ribosomal subunit in the same way as the independently bound CCA molecule.
FIGURE 1.
Experimental electron density of E site ligands bound to the H. marismortui 50S ribosomal subunit. (A) Fo(complex)–Fo(parent) electron density map of CCA bound to the 50S subunit, calculated with experimental phases, contoured at 4 σ. Electron density is visible for the portion of ribosomal protein L44e that adopts an altered conformation upon tRNA binding, near the top of the frame. (B) Fo(complex)–Fo(parent) electron density map of the minihelix bound to the 50S subunit, using experimental phases, contoured at 3 σ. Electron density gets weaker further from the CCA end because of increasing disorder. Density for the mobile portion of L44e is at the bottom of the frame.
TABLE 1.
Statistics for data collection and refinement
| Experiment | CCA | Minihelix |
| Resolution (Å)a | 50–2.90 (2.97–2.90) | 50–3.10 (3.18–3.10) |
| λ (Å) | 0.9999 | 1.0015 |
| Observations | 2,687,658 | 1,662,618 |
| Redundancy | 6.7 | 5.1 |
| Completeness (%)a | 99.9 (100) | 99.9 (100) |
| I/σa | 17.6 (2.0) | 13.5 (2.3) |
| Rcyst (%) | 20.8 | 20.4 |
| Rfree (%) | 26.2 | 25.8 |
aNumbers in parentheses indicate the outer resolution bin.
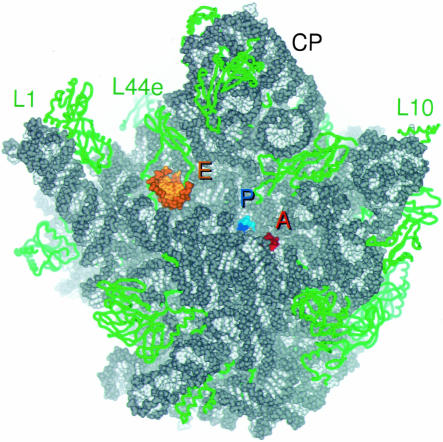
FIGURE 2.
Overview of the tRNA binding sites on the large subunit. CCA oligomers are shown bound at the A site (red) and P site (blue), and the minihelix is shown bound at the E site (orange). 23S rRNA is colored gray and ribosomal proteins are green. Proteins are shown in ribbon format, and RNA all atom, with backbones in darker and bases in lighter colors. The independently solved complex of the L1 arm and L1 protein (Nikulin et al. 2003) was docked into experimental electron density maps of the H. marismortui 50S subunit calculated at 9 Å resolution (Ban et al. 1998).
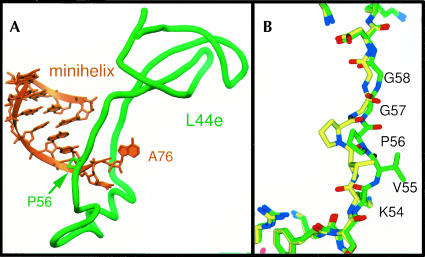
The E site of the 50S subunit contains both protein and rRNA. The duplex region of the minihelix is adjacent to helix 68 of 23S rRNA, whereas the 3′ termini of both substrates contact the stem of helix 88. The protein component of the E site seen interacting with these tRNA mimics is the loop extension of L44e. The CA end of tRNA inserts through the L44e loop, which widens and forms a distinctive lassoed conformation (Fig. 3A ▶). A stretch of seven residues of L44e is displaced up to 5 Å to allow E site binding. It includes two glycines that may facilitate the required conformational change (Fig. 3B ▶).
FIGURE 3.
L44e interacts with the E-site-bound minihelix. (A) The final 2 nt of the minihelix insert through the loop extension of L44e. (B) L44e undergoes a conformational change upon E site tRNA binding, from the conformation seen in yellow to that in green. Proline 56 is shown in both panels A and B to show relative orientation.
Description of E site binding
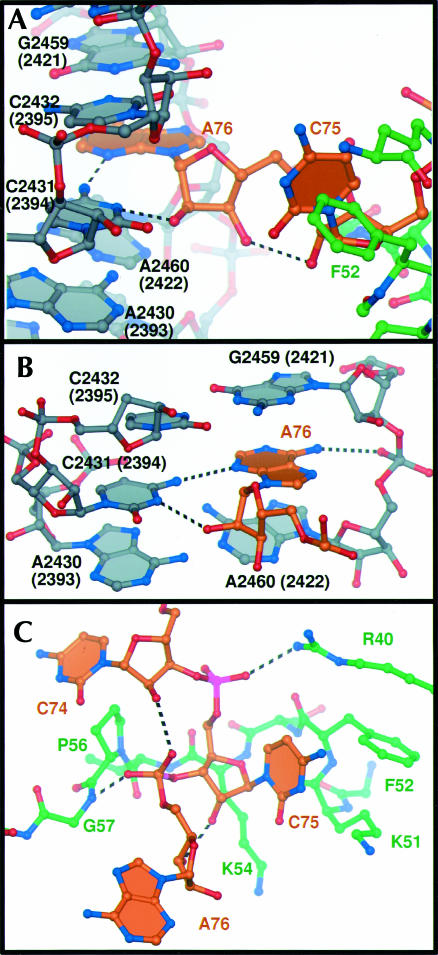
The interaction between the CCA sequence of tRNA and the E site is distinctly different in character from its interactions with the A and P sites. When bound to the A or P sites, the terminal CCA bases are stacked; C75 makes a C-G Watson–Crick base pair with a guanosine from the A or P loop of 23S rRNA, and A76 makes an A-minor interaction with 23S rRNA (Nissen et al. 2000, 2001). In the E site, the terminal bases extend apart and do not base pair with rRNA (Fig. 4 ▶).
FIGURE 4.
Specific interactions of E site binding. (A) The E site ligands interact with both rRNA (gray) and L44e (green). (B) A76 stacks in between 23S rRNA nucleotides G2459 (2421) and A2460 (2422), and hydrogen bonds with universally conserved C2431 (2394). (C) C75 interacts with L44e through hydrogen bonding to Gly 57 and Arg 40, and hydrophobic packing with Lys 54, Lys 51, and Phe 52.
Adenosine 76 inserts into an extension of helix 88, stacking between the splayed apart ribosomal bases G2459 (Escherichia coli numbering 2421) and A2460 (2422; Fig. 4B ▶). It is pinned into position laterally by hydrogen bonds between the N4 of C2431 (2394) and the N3 of A76, as well as by an interaction between a phosphate oxygen of A2460 (2422) and the exocyclic N6 of A76. The sugar phosphate backbone of A76 also makes specific interactions; its 2′ hydroxyl group hydrogen bonds to the N3 of C2431 (2394) and one of its nonbridging phosphate oxygen atoms hydrogen bonds to the backbone amide of L44e glycine 57 (Fig. 4C ▶).
Cytidine 75 is positioned mainly by hydrophobic interactions with L44e (Fig. 4C ▶). Its cytosine base lies on top of the hydrophobic patch created by phenylalanine 52 and the aliphatic carbon chain of lysine 51, and its sugar stacks on the hydrophobic aliphatic carbons of lysine 54. An epsilon nitrogen of arginine 40 donates a hydrogen bond to a nonbridging phosphate oxygen of C75. C74 bends back so that its 2′ hydroxyl group can interact with the phosphate group of A76. In the complex with CCA, the base of C74 is disordered, and only the sugar moiety can be seen. By contrast, in the complex with the minihelix, the base is ordered by stacking with A73 and the double-stranded portion of the minihelix. The duplex portion of the minihelix rises in A-form helical conformation beside helix 68 of 23S rRNA, but only 4 of the 12 bp of the minihelix are seen before the density disappears because of disorder. There is some density for the phosphate backbone continuing several nucleotides further, and one higher phosphate group is more ordered by a hydrogen bond to L44e arginine 87, but the surrounding density is too weak to model. In the 70S structure with tRNAs bound, the acceptor–TΨC helix continues up, and the tRNA elbow contacts protein L1 and the rRNA to which L1 binds. This L1 region is visible in the 9 Å electron density maps of the H. marismortui 50S subunit, but because it is somewhat disordered, it does not appear in the high-resolution maps.
A and P site binding of CCA
The CCA substrate also binds to the peptidyl transferase center (data not shown). Strong electron density is seen at the A site, and CCA binds to it in the same way as the CCA portions of A site substrates previously described (Nissen et al. 2000; Schmeing et al. 2002). The P site density is much weaker, indicating low occupancy binding. Consistent with this observation, the electron density indicates that the highly mobile nucleotide A2637 (2602) of 23S rRNA occupies the conformation seen in the apo structure more than that observed when a P site substrate is bound. The other bases undergoing conformational changes include uridines U2619 (2584) and U2620 (2585), as well as U2541 (2506), which is rotated 180° from any conformation previously seen. However, because the ligands studied here do not include the aminoacyl or peptidyl moieties present when natural substrates occupy these sites, the relevance of these particular conformational differences to the peptidyl transferase process is unclear.
DISCUSSION
Agreement with biochemical results
The structures of these complexes are consistent with and explain the biochemical results concerning the location of the E site on the 50S subunit. The E site was first localized by RNA footprinting studies by Moazed and Noller, who identified 4 nt that are specifically protected by tRNA bound to the E site (Moazed and Noller 1989a). Three of these are near the L1 binding site. The fourth corresponds to H. marismortui cytidine 2431 (2394). These results were corroborated and expanded by later studies. Mankin and colleagues proposed that A76 forms a reverse Hoogsteen base pair with C2431 (2394; Bocchetta et al. 2001). Instead, the N3 and N4 of C2431 (2394) hydrogen bonds to the 2′ hydroxyl and N3 of A76, respectively (Fig. 4B ▶). Also, the nucleotide corresponding to H. marismortui A2460 (C2422) has been cross-linked to deacylated tRNA (Wower et al. 2000). This base stacks directly below A76 (Fig. 4B ▶). In addition, a Fe(II)-EDTA moiety tethered to the end of a tRNA bound in the E site was observed to cleave helix 88 and surrounding RNA (Joseph and Noller 1996), consistent with the finding that A76 stacks onto a base in the first canonical base pair of helix 88, G2459 (2421).
Comparison with tRNA bound to the E site in the 70S structure
The location of the tRNA mimics bound to the E site of the H. marismortui large subunit is largely the same as the 3′ terminus of tRNA modeled into the E site of the 5.5 Å resolution 70S ribosome structure (Yusupov et al. 2001), but some differences in the detailed structures exist. Superimposition of corresponding 23S rRNA phosphorous atoms of T. thermophilus 70S ribosome structure and the H. marismortui 50S subunit structures discussed here allows the comparison of the respective E site substrates (Fig. 5 ▶). The phosphates of the single-stranded ACCA ends are in similar positions, but the bases of C75 and A76 have been modeled to extend in opposite directions in the two structures, with A76 of the tRNA bound to the 70S ribosome protruding away from C2431 (2394), that had been identified by footprinting. This discrepancy is most likely due to the limitations imposed by the 5.5 Å resolution electron density map, which precludes the correct positioning of bases in a single-stranded region.
FIGURE 5.
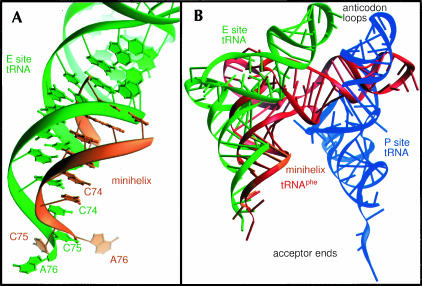
Comparison with tRNA bound to the E site in the 70S structure. (A) The minihelix (orange) is in a position similar to the tRNA (green) bound to the 70S ribosome E site tRNA. Corresponding phosphorous atoms of the H. marismortui 50S subunit and T. thermophilus 70S ribosome were superimposed to allow the comparison. (B) A yeast tRNAphe (red) superimposed onto the duplex portion of the minihelix (orange) has its anticodon stem–loop closer to the anticodon stem loop of a tRNA bound in the 30S P site (blue) than a tRNA bound in the 30S E site (green).
The acceptor stem helices of these two structures occupy similar positions, but are oriented with their helical axes at somewhat different angles. The minihelix observed here can be extended by superimposing the corresponding acceptor helix of a full tRNA on it. The anticodon stem and loop of the superimposed tRNA lies closer to the P site of the 30S subunit than to its E site, which suggests that the orientation of the minihelix stem may be more representative of a tRNA bound in the hybrid P/E state (Moazed and Noller 1989b) than the classic E/E state observed in the T. thermophilus ribosome structure. Alternatively, the orientation of the minihelix may differ because of the absence of the additional constraining interactions that are made by the full tRNA with the 70S ribosome.
Specificity of binding and translocation to E site
In vivo, the acceptor end of tRNA moves to the E site of the 50S subunit subsequent to its deacylation as a result of peptidyl transfer (Moazed and Noller 1989b). To examine the structural basis for the specificity of the E site for deacylated tRNA, we considered what would occur if tRNAs esterified at either the 2′ or 3′ hydroxyl group were introduced into the E site (Fig. 4 ▶). Since the 2′ hydroxyl of A76 is hydrogen bonded to C2431 (2394), any additional atoms added to the 2′ hydroxyl would sterically clash with this base. The binding site of the 3′ hydroxyl group appears to be more accommodating to its modification, despite being within hydrogen bonding distance of C75 2′ hydroxyl group (Fig. 4A,C ▶). The angle for this interaction is not optimal, and there is a small pocket below the 3′ OH. However, when the smallest natural substrate that must differentially bind the E or P site, formyl-methionine-tRNA, is modeled in, steric clashes are predicted to occur between the fMet and a combination of tRNA, rRNA, or L44e, depending on the rotomer angles adopted by the amino acid (not shown). In addition, the presence of any amino acid at this site could disrupt the interactions between the sugars of C75 and A76, rendering the tRNA unable to adopt the conformation required for E site binding.
A survey of the literature reveals that the specificity of the E site for deacylated tRNA has been assessed by comparing the binding of deacylated tRNA with that of a tRNA esterified to a bulky amino acid, usually phenylalanine or N-acetylated phenylalanine (Rheinberger et al. 1981; Lill et al. 1984; Moazed and Noller 1989b). Neither of these aminoacylated tRNAs bind to the E site and, as expected, a phenylalanine at the 3′ hydroxyl of the A76 of the minihelix would clash seriously with the ribosome. There is a small void below the sugar of A76, however, and it is possible that a tRNA acylated with a small amino acid, such as glycine, might have some affinity for the E site.
Both the P site and the E site have some affinity for deacylated tRNA (Rheinberger et al. 1981). What, then, drives the newly deacylated acceptor end from the P site to the E site? Wintermeyer and colleagues have shown that transfer of the peptide chain to the aminoacyl-tRNA lowers its affinity for the A site, driving it into the P site (Semenkov et al. 2000). The newly deacylated acceptor end, having lost the interactions of the peptide with the ribosome, disassociates from the P site and is replaced by the new peptidyl-tRNA. It then can bind to the E site of the large subunit, where it buries the same amount of ribosomal surface area as in the P site, approximately 500 Å2. Additionally, although in some systems the P site has affinity for deacylated tRNAs that is greater than or comparable to that of the E site (Rheinberger et al. 1981), in our system these deacylated moieties have significantly greater affinity for the E site than for the P site, as evidenced by the weak P site density in the CCA soak and the lack of binding of minihelix to the P site.
No evidence for allosteric interactions between the three sites on the large subunit
One model for translation, the “allosteric three site model,” (Geigenmuller and Nierhaus 1990) posits negative allosteric communication between the A and E sites; when the E site is occupied, the A site adopts a low affinity state for binding tRNAs that increases translational fidelity. The A and E sites on the large subunit are over 50 Å apart. If the physical basis for this E to A site communication existed in the 50S subunit, one might expect to see a significant structural shift of some ribosomal component when the E site becomes occupied. No movement of rRNA or protein lying between the A and E sites is seen in these complexes. It is possible, however, that allosteric structural changes occur in the 30S subunit or only in the context of the 70S ribosome with intact tRNAs bound.
Differences between eubacterial and archeal E site ribosomal proteins
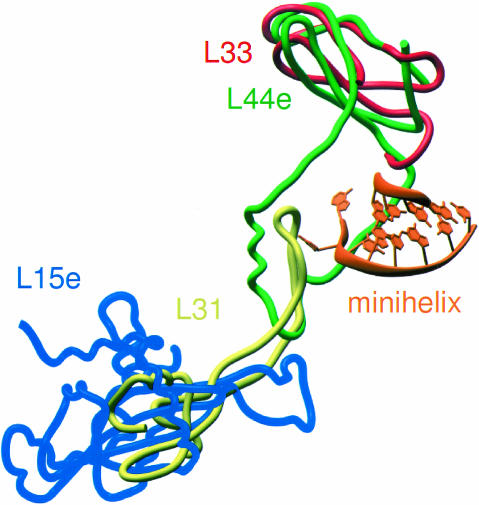
The protein component of the H. marismortui E site is the archeal protein L44e. Eukaryotic ribosomes also contain L44e, but eubacterial ribosomes do not. The structure of the Deinucoccus radiodurans 50S subunit (Harms et al. 2001) reveals that the binding site of the globular domain of L44e is occupied by the eubacterial protein L33, a significantly shorter protein (~57 amino acids) that has no extension (Fig. 6 ▶). The position of the extension of L44e is partially occupied instead by a loop extension of L31, a protein whose globular domain occupies much of the same space as L15e in the H. marismortui structure. Superimposition of the two models reveals that a steric clash would occur between the 3′ terminus of tRNA and the extension of L31, which suggests that a movement of the L31 loop similar to that seen in L44e may occur upon binding of tRNA to the eubacterial E site.
FIGURE 6.
Differences in eubacterial and archeal E site ribosomal proteins. The globular portions of H. marismortui ribosomal proteins L44e (green) and L15e (blue) are replaced by L33 (red) and L31 (yellow), respectively, in the D. radiodurans ribosome (Harms et al. 2001). However, the loop portion of L44e that interacts with the 3′ terminus of the E site-bound minihelix (orange) overlaps with an extension of L31.
The difference between the E sites of archeal and eubacterial 50S subunits indicates that the protein components of the E site were added to the ribosome after bacteria split from eukaryotes and archea. In contrast, the RNA component is conserved: The critical C2431 (2394) is over 99% conserved throughout the three phylogenetic domains of life (Cannone et al. 2002). Thus it appears that the ribosome of the last common ancestor had an E site formed only by RNA, and this site evolved further to include protein components.
MATERIALS AND METHODS
Crystals of the large ribosomal subunit of H. marismortui were grown as described previously (Ban et al. 2000). To prepare complexes with the E site substrates, the crystals were incubated for 5 h in stabilization solution (Schmeing et al. 2002) containing 1 mM minihelix or 10 mM CCA oligonucleotide. Crystals were then flash-frozen in liquid propane. The diffraction data set for the complex of the minihelix and the large ribosomal subunit was collected using beamline 19-ID of the Structural Biology Center (SBC) at the Advanced Photons Source (APS) of Argonne National Laboratory, Argonne, IL, with a 3 × 3 charge-coupled device (CCD) detector, 1.0015 Å wavelength, 80 × 80 μm beam size, and 0.4° oscillations. The diffraction data set for the complex between the 50S subunit and CCA was collected using beamline 8-BM of the Northeast Collaborative Access Team (NE-CAT) at APS, with a Quantum 315 CCD detector, 0.979 Å wavelength, 150 × 150 beam size, and 0.4° oscillations. All data sets were reduced with HKL2000 and scaled with SCALEPACK (Otwinowski and Minor 1987). The structure of the H. marismortui large ribosomal subunit refined at 2.4 Å (Protein Data Bank identification code 1JJ2; Klein et al. 2001) was rigid body refined into each of the E site complex data sets and subjected to energy minimization and individual B-factor refinement using the program CNS (Brunger et al. 1998). The ligands were fit into experimental electron density maps using the program O (Jones et al. 1991). The structures of the complexes underwent successive rounds of modeling, energy minimization, and individual B-factor refinement to yield final models. The atomic coordinates of both complexes have been deposited in the Protein Data Bank (PDB ID codes 1QVF and 1QVG).
To compare the structure of the H. marismortui 50S subunit with the T. thermophilus 70S ribosome (Yusupov et al. 2001) and the D. radiodurans 50S subunit (Harms et al. 2001), phosphorous atoms of the eubacterial 23S rRNA were superimposed onto the corresponding atoms of the H. marismortui 23S, using the least squares function in the program O. The superimposition of the complete structure of yeast tRNAphe (Shi and Moore 2000) onto the minihelix structure was done using all of the backbone atoms of nt 1–4 and 69–72. Buried surface areas were calculated using CNS.
Acknowledgments
The authors are indebted to Larysa Vasylenko for skillful preparation of ribosomes and crystals. We thank Satwik Kamtekar, Scott Bailey, Scott Strobel, Jeff Hansen, and Gregor Blaha for helpful conversations; Dan Klein for modeling the L1 region; and Janice Pata for critical reading of the manuscript. We are grateful for assistance from Craig Ogata, Steve Ealick, Marianne Cuff, and the staff at NE-CAT, SBC, and the Yale Center for Structural Biology. This research was supported by the National Institutes of Health Grant GM22778, and an Agouron Institute grant (to T.A.S and P.B.M.). Use of the Argonne National Laboratory Structural Biology Center and Northeast Collaborative Access beamlines at the Advanced Photon Source was supported by the U.S. Department of Energy, Office of Energy Research, under Contract No. W-31–109-ENG-38.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5120503.
REFERENCES
- Agrawal, R.K., Penczek, P., Grassucci, R.A., Li, Y., Leith, A., Nierhaus, K.H., and Frank, J. 1996. Direct visualization of A-, P-, and E-site transfer RNAs in the Escherichia coli ribosome. Science 271: 1000–1002. [DOI] [PubMed] [Google Scholar]
- Ban, N., Freeborn, B., Nissen, P., Penczek, P., Grassucci, R.A., Sweet, R., Frank, J., Moore, P.B., and Steitz, T.A. 1998. A 9 Å resolution X-ray crystallographic map of the large ribosomal subunit. Cell 93: 1105–1115. [DOI] [PubMed] [Google Scholar]
- Ban, N., Nissen, P., Hansen, J., Moore, P.B., and Steitz, T.A. 2000. The complete atomic structure of the large ribosomal subunit at 2.4 Å resolution. Science 289: 905–920. [DOI] [PubMed] [Google Scholar]
- Bocchetta, M., Xiong, L., Shah, S., and Mankin, A.S. 2001. Interactions between 23S rRNA and tRNA in the ribosomal E site. RNA 7: 54–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger, A.T., Adams, P.D., Clore, G.M., DeLano, W.L., Gros, P., Grosse-Kunstleve, R.W., Jiang, J.S., Kuszewski, J., Nilges, M., Pannu, N.S., et al. 1998. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54: 905–921. [DOI] [PubMed] [Google Scholar]
- Cannone, J.J., Subramanian, S., Schnare, M.N., Collett, J.R., D’Souza, L.M., Du, Y., Feng, B., Lin, N., Madabusi, L.V., Müller, K.M., et al. 2002. The Comparative RNA Web (CRW) Site: An online database of comparative sequence and structure information for ribosomal, intron, and other RNAs. BMC Bioinformatics 3: 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter, A.P., Clemons, W.M., Brodersen, D.E., Morgan-Warren, R.J., Wimberly, B.T., and Ramakrishnan, V. 2000. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 407: 340–348. [DOI] [PubMed] [Google Scholar]
- Geigenmuller, U. and Nierhaus, K.H. 1990. Significance of the third tRNA binding site, the E site, on E. coli ribosomes for the accuracy of translation: An occupied E site prevents the binding of noncognate aminoacyl-tRNA to the A site. EMBO J. 9: 4527– 4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grajevskaja, R.A., Ivanov, Y.V., and Saminsky, E.M. 1982. 70-S ribosomes of Escherichia coli have an additional site for deacylated tRNA binding. Eur. J. Biochem. 128: 47–52. [DOI] [PubMed] [Google Scholar]
- Hansen, J.L., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2002. Structural insights into peptide bond formation. Proc. Natl. Acad. Sci. 99: 11670–11675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harms, J., Schluenzen, F., Zarivach, R., Bashan, A., Gat, S., Agmon, I., Bartels, H., Franceschi, F., and Yonath, A. 2001. High resolution structure of the large ribosomal subunit from a mesophilic eubacterium. Cell 107: 679–688. [DOI] [PubMed] [Google Scholar]
- Jones, T.A., Zou, J.Y., Cowan, S.W., and Kjeldgaard, M. 1991. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A 47: 110–119. [DOI] [PubMed] [Google Scholar]
- Joseph, S. and Noller, H.F. 1996. Mapping the rRNA neighborhood of the acceptor end of tRNA in the ribosome. EMBO J. 15: 910– 916. [PMC free article] [PubMed] [Google Scholar]
- Kirillov, S.V., Makarov, E.M., and Semenkov, Yu.P. 1983. Quantitative study of interaction of deacylated tRNA with Escherichia coli ribosomes. Role of 50 S subunits in formation of the E site. FEBS Lett. 157: 91–94. [DOI] [PubMed] [Google Scholar]
- Klein, D.J., Schmeing, T.M., Moore, P.B., and Steitz, T.A. 2001. The kink-turn: A new RNA secondary structure motif. EMBO J. 20: 4214–4221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lill, R., Robertson, J.M., and Wintermeyer, W. 1984. tRNA binding sites of ribosomes from Escherichia coli. Biochemistry 23: 6710–6717. [DOI] [PubMed] [Google Scholar]
- ———. 1986. Affinities of tRNA binding sites of ribosomes from Escherichia coli. Biochemistry 25: 3245–3255. [DOI] [PubMed] [Google Scholar]
- Lill, R., Lepier, A., Schwagele, F., Sprinzl, M., Vogt, H., and Wintermeyer, W. 1988. Specific recognition of the 3′-terminal adenosine of tRNAPhe in the exit site of Escherichia coli ribosomes. J. Mol. Biol. 203: 699–705. [DOI] [PubMed] [Google Scholar]
- Maden, B.E., Traut, R.R., and Monro, R.E. 1968. Ribosome-catalysed peptidyl transfer: The polyphenylalanine system. J. Mol. Biol. 35: 333–345. [DOI] [PubMed] [Google Scholar]
- Marquez, V., Wilson, D.N., and Nierhaus, K.H. 2002. Functions and interplay of the tRNA binding sites of the ribosome. Biochem. Soc. Trans. 30: 133–140. [DOI] [PubMed] [Google Scholar]
- Moazed, D. and Noller, H.F. 1989a. Interaction of tRNA with 23S rRNA in the ribosomal A, P, and E sites. Cell 57: 585–597. [DOI] [PubMed] [Google Scholar]
- ———. 1989b. Intermediate states in the movement of transfer RNA in the ribosome. Nature 342: 142–148. [DOI] [PubMed] [Google Scholar]
- Nikulin, A., Eliseikina, I., Tishchenko, S., Nevskaya, N., Davydova, N., Platonova, O., Piendl, W., Selmer, M., Liljas, A., Drygin, D., et al. 2003. Structure of the L1 protuberance in the ribosome. Nat. Struct. Biol. 10: 104–108. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Hansen, J., Ban, N., Moore, P.B., and Steitz, T.A. 2000. The structural basis of ribosome activity in peptide bond synthesis. Science 289: 920–930. [DOI] [PubMed] [Google Scholar]
- Nissen, P., Ippolito, J A., Ban, N., Moore, P.B., Steitz, T.A. 2001. RNA tertiary interactions in the large ribosomal subunit: The A-minor motif. Proc. Natl. Acad. Sci. 98: 4899–4903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogle, J.M., Brodersen, D.E., Clemons Jr., W.M., Tarry, M.J., Carter, A.P., and Ramakrishnan, V. 2001. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science 292: 897– 902. [DOI] [PubMed] [Google Scholar]
- Ogle, J.M., Murphy, F.V., Tarry, M.J., and Ramakrishnan, V. 2002. Selection of tRNA by the ribosome requires a transition from an open to a closed form. Cell 111: 721–732. [DOI] [PubMed] [Google Scholar]
- Otwinowski, Z. and Minor, W. 1987. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 276: 307–326. [DOI] [PubMed] [Google Scholar]
- Rheinberger, H.J., Sternbach, H., and Nierhaus, K.H. 1981. Three tRNA binding sites on Escherichia coli ribosomes. Proc. Natl. Acad. Sci. 78: 5310–5314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinke-Appel, J., Junke, N., Osswald, M., and Brimacombe, R. 1995. The ribosomal environment of tRNA: Crosslinks to rRNA from positions 8 and 20:1 in the central fold of tRNA located at the A, P, or E site. RNA 1: 1018–1028. [PMC free article] [PubMed] [Google Scholar]
- Schmeing, T.M., Seila, A.C., Hansen, J.L., Freeborn, B., Soukup, J.K., Scaringe, S.A., Strobel, S.A., Moore, P.B., and Steitz, T.A. 2002. A pre-translocational intermediate in protein synthesis observed in crystals of enzymatically active 50S subunits. Nat. Struct. Biol. 9: 225–230. [DOI] [PubMed] [Google Scholar]
- Semenkov, Y.P., Rodnina, M.V., and Wintermeyer, W. 2000. Energetic contribution of tRNA hybrid state formation to translocation catalysis on the ribosome. Nat. Struct. Biol. 7: 1027–1031. [DOI] [PubMed] [Google Scholar]
- Shi, H. and Moore, P.B. 2000. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: A classic structure revisited. RNA 6: 1091–1105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stark, H., Orlova, E.V., Rinke-Appel, J., Junke, N., Mueller, F., Rodnina, M., Wintermeyer, W., Brimacombe, R., and van Heel, M. 1997. Arrangement of tRNAs in pre- and posttranslocational ribosomes revealed by electron cryomicroscopy. Cell 88: 19–28. [DOI] [PubMed] [Google Scholar]
- Watson, J.D. 1964. The synthesis of proteins upon ribosomes. Bull. Soc. Chim. Biol. 11: 1399–1425. [PubMed] [Google Scholar]
- Wower, J., Scheffer, P., Sylvers, L.A., Wintermeyer, W., and Zimmermann, R.A. 1993. Topography of the E site on the Escherichia coli ribosome. EMBO J. 12: 617–623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wower, J., Kirillov, S.V., Wower, I.K., Guven, S., Hixson, S.S., Zimmermann, R.A. 2000. Transit of tRNA through the Escherichia coli ribosome. Cross-linking of the 3′ end of tRNA to specific nucleotides of the 23 S ribosomal RNA at the A, P, and E sites. J. Biol. Chem. 275: 37887–37894. [DOI] [PubMed] [Google Scholar]
- Yusupov, M.M., Yusupova, G.Z., Baucom, A., Lieberman, K., Earnest, T.N., Cate, J.H., and Noller, H.F. 2001. Crystal structure of the ribosome at 5.5 Å resolution. Science 292: 883–896. [DOI] [PubMed] [Google Scholar]