Abstract
Small nucleolar RNAs (snoRNAs) play important roles in ribosomal RNA metabolism. In Saccharomyces cerevisiae, box C/D snoRNAs are synthesized from excised introns, polycistronic precursors, or independent transcription units. Previous studies have shown that only a few independently transcribed box C/D snoRNAs are processed at their 5′ end. Here we describe 12 additional independently transcribed box C/D snoRNAs that undergo 5′-end processing. 5′ Extensions found in the precursors of these snoRNAs contain cleavage sites for Rnt1p, the S. cerevisiae homolog of RNase III, and unprocessed precursors accumulate in vivo in the absence of Rnt1p. Rnt1p cleavage products were identified in vivo when the 5′ → 3′ exonucleases Xrn1p and Rat1p are inactivated (xrn1Δ rat1-1) and in vitro using model RNA substrates and recombinant Rnt1p. Some of these snoRNAs show increased levels of unprocessed precursors when the rnt1Δ deletion is combined to the xrn1Δ rat1-1 mutation, suggesting that these exonucleases participate in the 5′ processing or the degradation of the snoRNA precursors. Unprocessed precursors are not significantly destabilized in the absence of the trimethylguanosine capping enzyme Tgs1p, suggesting that a 5′ monomethyl cap is sufficient to ensure stabilization of these precursors. These results demonstrate that the majority of independently transcribed box C/D snoRNAs from the yeast genome undergo 5′-end processing and that the Rnt1p endonuclease and the Xrn1p and Rat1p 5′ → 3′exonucleases have partially redundant functions in the 5′-end processing of these snoRNAs.
Keywords: Endoribonucleases, exoribonuclease, Rnt1p, RNase III, stem–loop
INTRODUCTION
Small nucleolar RNAs (snoRNAs) are essential cofactors in ribosomal RNA (rRNA) metabolism. A few snoRNAs are necessary for cleavage steps in the maturation of the 35S rRNA precursor, but most of them are required to guide bases or sugar modifications within the rRNA precursor (Tollervey and Kiss 1997; Kiss 2001). Small nucleolar RNAs are subdivided in two major structural families (Balakin et al. 1996; Ganot et al. 1997b). Box C/D snoRNAs guide the methylation of the ribose 2′ hydroxyl groups of nucleotides in the 35S rRNA precursor (Cavaille et al. 1996; Kiss-Laszlo et al. 1996). H/ACA snoRNAs guide the conversion of uridines to pseudouridines in the rRNA precursor (Ganot et al. 1997a; Ni et al. 1997).
Small nucleolar RNAs are usually produced by posttranscriptional processing from precursors species (Tollervey and Kiss 1997). Although most mammalian snoRNAs are encoded within intron sequences and processed from either unspliced precursors or lariat species, only a few yeast snoRNAs are intron encoded (U18, U24, snR34, snR38, snR39, snR44, snR54, and snR59). Most yeast small RNAs are either generated from independent transcription units or initially transcribed as polycistronic units. In the case of polycistronic transcription units, processing intermediates are generated by RNase III cleavage in the spacer separating each snoRNA from the other (Chanfreau et al. 1998a,b; Qu et al. 1999). The resulting intermediates carrying only one snoRNA sequence are further processed by exonucleases (Petfalski et al. 1998; Qu et al. 1999) to generate the mature ends.
The initial transcripts of independently transcribed box C/D snoRNAs all possess a trimethylguanosine (TMG) cap. The snoRNAs that are not processed at their 5′ end such as U3, snR4, and snR13 will retain the TMG cap structure (Samarsky and Fournier 1999). Those that undergo 5′-end processing, such as snR39b, snR40, snR47, and snR79 (Z9), will lose the TMG cap structure (Chanfreau et al. 1998a). In these documented cases, the yeast ortholog of RNase III, Rnt1p, cleaves the 5′ extension found in the precursor of these snoRNAs, and provides an entry site for exonucleolytic digestion. The exonucleases responsible for the final trimming step have not been identified, but have been speculated to be Xrn1p and/or Rat1p (Chanfreau et al. 1998a).
Because of the limited number of independently transcribed box C/D snoRNA genes described, it is not clear whether 5′-end processing is a common pathway or limited to a few snoRNAs. In this study, we examine the question of the processing of recently identified independently transcribed box C/D snoRNAs. We show that most of them undergo 5′-end processing by a combination of endonucleolytic cleavage by Rnt1p and exonucleolytic digestion by Xrn1p and/or Rat1p. These results show that the majority of independently transcribed box C/D snoRNAs undergo 5′-end processing, and that the enzymes involved in their processing have partially redundant functions.
RESULTS
In silico detection of putative Rnt1p target sites upstream from recently identified box C/D snoRNA genes
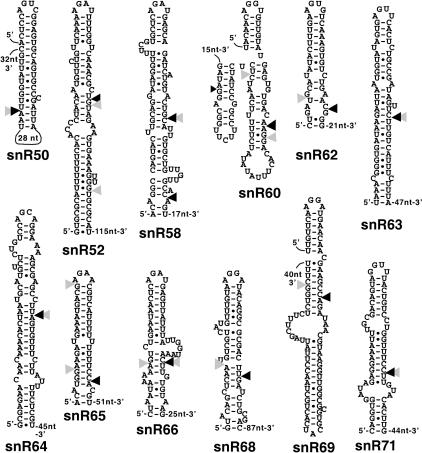
We analyzed the predicted secondary structure of sequences found upstream of box C/D snoRNA gene sequences identified through computational screening (Lowe and Eddy 1999; Samarsky and Fournier 1999). This analysis revealed the presence of putative stem–loop structures upstream from the independently transcribed box C/D snoRNAs snR50, snR52, snR58, snR60, snR62, snR63, snR64, snR65, snR66, snR68, snR69, and snR71 (Fig. 1 ▶). Most of these stem–loop structures are capped by tetraloops showing the consensus sequence AGNN. One exception was snR71, which showed a GGUU tetraloop sequence. In three cases (snR50, snR60, and snR69), the predicted secondary structures were formed by coaxial stacking of a short stem–loop carrying the AGNN tetraloop onto a longer stem (Fig. 1 ▶). The Saccharomyces cerevisiae homolog of RNase III, Rnt1p, specifically cleaves double-stranded structures capped by tetraloop with the sequence AGNN, and the enzyme selects the phosphodiester bond to be cleaved between 13 and 16 bp from the tetraloop (Chanfreau et al. 2000). Thus, the presence of predicted dsRNA structures carrying AGNN terminal tetraloops upstream from these novel independently transcribed box C/D snoRNA sequences suggested that these snoRNAs undergo 5′-end processing through Rnt1p cleavage.
FIGURE 1.
Secondary structure prediction of RNA sequences found upstream of box C/D snoRNAs. Shown are secondary structures corresponding to one of the 5% suboptimal structures calculated by Mfold (Mathews et al. 1999). The distance to the mature snoRNA sequence is indicated on the 3′ end of the structure. Cleavage sites mapped in vitro are indicated by black triangles and cleavage sites mapped in vivo are indicated by gray triangles.
5′-end processing of box C/D snoRNAs by Rnt1p cleavage and exonucleolytic digestion
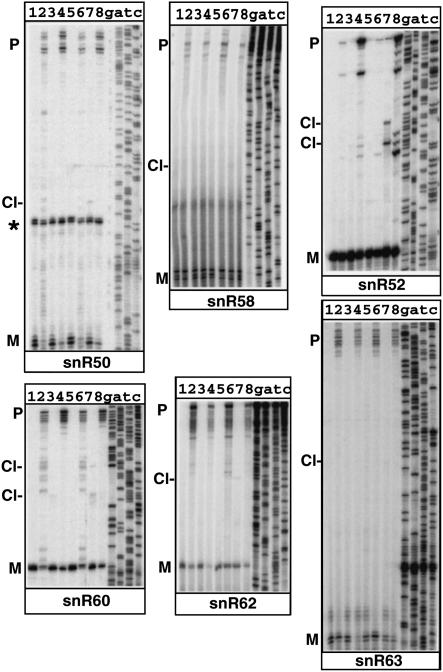
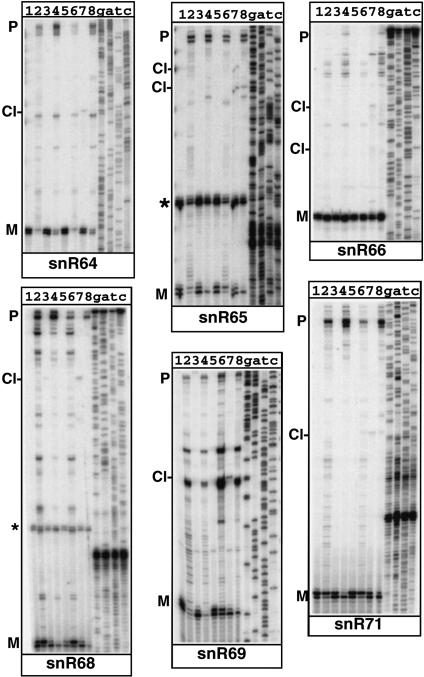
Theoretical RNA folding of sequences found upstream of these snoRNAs suggested that Rnt1p might play a role in the 5′-end processing of these C/D snoRNA species. To test this hypothesis, we analyzed the 5′ end of these snoRNAs by primer extension of RNAs extracted from a wild-type strain and from a yeast strain in which the RNT1 gene was deleted (rnt1Δ). To get conclusive evi- dence for Rnt1p cleavage in vivo, we also sought to detect Rnt1p cleavage products in vivo. These cleaved intermediates are normally unstable because they are rapidly degraded by exonucleases. To detect these species, we used yeast strains carrying deletion or mutant versions of the 5′ → 3′ exonucleases Xrn1p and Rat1p. Xrn1p is cytoplasmic and is not essential (Johnson 1997), whereas Rat1p is nuclear and is essential (Amberg et al. 1992; Johnson 1997). Although Xrn1p is in majority cytoplasmic, full inactivation of 5′ → 3′ exonuclease activities in the yeast nucleus requires inactivation of both Rat1p and Xrn1p, possibly because of a small nuclear fraction of Xrn1p that can functionally complement some of the roles of Rat1p (Petfalski et al. 1998; Danin-Kreiselman et al. 2003). A yeast strain carrying both the xrn1Δ disruption and a rat1 thermosensitive mutation (xrn1Δ rat1-1, provided by D. Tollervey, University of Edinburgh) was analyzed by primer extension. If snoRNA precursors are cleaved by Rnt1p, cleaved intermediates should become detectable upon shift of this strain to a nonpermissive temperature (Petfalski et al. 1998; Qu et al. 1999; Danin-Kreiselman et al. 2003). We also used a triple mutant strain in which the rnt1 knockout was combined to the xrn1Δ rat1-1 mutation (rnt1Δ xrn1Δ rat1-1). Analysis of this strain served two purposes. First, we wanted to demonstrate that any product detected upon inactivation of the Xrn1p and Rat1p exonucleases was dependent upon Rnt1p cleavage (Danin-Kreiselman et al. 2003). Second, this triple mutant strain allowed us to analyze whether 5′-end processing was more affected by inactivation of both Rnt1p and the 5′ → 3′ exonucleases than by inactivation of Rnt1p alone. Primer extension analysis was performed on RNAs extracted from each of these four strains grown at 25°C or shifted to 37°C. This analysis revealed several features observed for all snoRNAs examined (Fig. 2 ▶). For all of them, 5′-extended species were detected in the rnt1Δ strain (lanes 2 and 6), showing that Rnt1p plays a role in the 5′-end processing of these RNAs, as predicted from the secondary structure analysis described in Figure 1 ▶. In most cases, we could clearly detect a product in RNAs extracted from the xrn1Δ rat1-1 strain shifted at 37°C (labeled as Cl-, Fig. 2 ▶, lane 7) that was absent in samples extracted from the rnt1Δ xrn1Δ rat1-1 strain (lanes 8) shifted in the same conditions. These species are likely to correspond to Rnt1p cleavage products that are stabilized in vivo, because of the inactivation of the 5′ → 3′ exonucleases. In the cases of snR58 and snR63, cleaved intermediates were found in very low abundance but were still detectable. The precise location of these cleavage sites was mapped at the nucleotide level using sequencing lanes run in parallel and generated with the same 5′-end-labeled oligonucleotide (Fig. 2 ▶). The location of these cleavage sites is indicated by gray triangles on the secondary structures on Figure 1 ▶. In most cases, cleavage was observed in the 13- to 16-bp range to the terminal tetraloop structure, in agreement with the cleavage site selection rules of the enzyme. These results show that depletion of Rnt1p affects 5′-end processing of these snoRNAs, and that cleaved processing intermediates can be identified in vivo using exonucleases mutant strains.
FIGURE 2.
Primer extension analysis of 5′-end processing of box C/D snoRNAs in wild-type and rnt1Δ, xrn1Δ rat1-1, and rnt1Δ xrn1Δ rat1-1 mutant strains. (Lane 1) Wild-type strain, 25°C. (Lane 2) rnt1Δ strain, 25°C. (Lane 3) xrn1Δ rat1-1 strain, 25°C. (Lane 4) rnt1Δ xrn1Δ rat1-1 strain, 25°C. (Lane 5) Wild-type strain, shifted to 37°C. (Lane 6) rnt1Δ strain, shifted to 37°C. (Lane 7) xrn1Δ rat1-1 strain, shifted to 37°C. (Lane 8) rnt1Δ xrn1Δ rat1-1 strain, shifted to 37°C. In some cases, the oligonucleotide used cross-hybridized to another RNA species (indicated by an asterisk) on the figure. The mature species are indicated by “M”, and the precursors are indicated by “P”. Cleaved intermediates detected in the xrn1Δ rat1-1 strain, shifted to 37°C (lanes 7) are indicated by Cl-.
The amount of mature species accumulating in the rnt1Δ strain was strikingly variable depending on snoRNAs. Some snoRNAs such as snR50, snR64, and snR69 showed almost no accumulation of mature snoRNA in the absence of Rnt1p (Fig. 2 ▶, cf. lanes 1 and 2). Others, such as snR52 and snR58, showed no perturbation in the level of mature snoRNA produced in the absence of Rnt1p. These results show that the major mechanism to ensure mature snoRNA production differs significantly depending on individual snoRNA species. Some snoRNAs are strictly dependent on Rnt1p cleavage for mature snoRNA production, whereas some others seem to be able to overcome the lack of Rnt1p cleavage through exonucleolytic digestion (although we cannot rule out that another, yet unidentified, endonuclease may play a role as well).
We also noticed some differences in the mature 5′ end generated through the endonucleolytic cleavage pathway followed by exonucleolytic digestion compared to the 5′ end generated exclusively through the exonucleolytic pathway (identified in the rnt1Δ strain). In the latter case, the mature end was sometimes shifted by 1 nt upstream compared to the end generated through Rnt1p endonucleolytic cleavage followed by exonucleolytic digestion (Fig. 2 ▶). The basis for this observation remains unclear.
The analysis of the triple mutant strain revealed that in some cases, the fully extended unprocessed precursors were more abundant in the rnt1Δ xrn1Δ rat1-1 triple mutant strain than in the rnt1Δ strain. This phenotype was often detectable at 25°C (compare lanes 2 and 4 for snR52, snR60, snR64, snR66, snR68, and snR71), even before complete inactivation of Rat1p. Although fully unprocessed precursors are hardly detectable in the xrn1Δ rat1-1 mutant strain at any temperature, this result suggests that the role of the Xrn1p and/or Rat1p exonuclease is not limited to digesting the Rnt1p cleavage product, but that they also function in processing full-length precursors when these species accumulate in the absence of Rnt1p. This is consistent with the fact that some mature snoRNA levels are barely affected by Rnt1p absence (e.g., snR52 and snR66). These results show that the endonucleolytic and the exonucleolytic pathways have partially redundant functions in the 5′ processing of some of these snoRNAs.
Recombinant Rnt1p cleaves model substrates containing box C/D snoRNA 5′ extensions
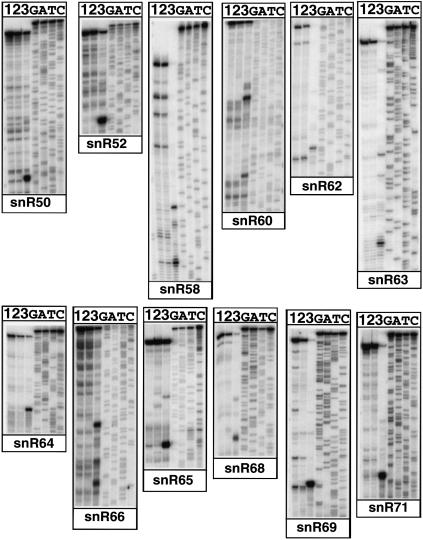
To demonstrate directly that the cleaved intermediates identified in vivo by primer extension in the xrn1Δ rat1-1 strain at a nonpermissive temperature correspond to bona fide Rnt1p cleavage sites, we generated model transcripts of ~200 nt containing the stem–loop structures, and we incubated these unlabeled transcripts with bacterially expressed purified recombinant Rnt1p or with a catalytically inactive mutant (E320K). Cleavage site(s) were mapped by primer extension analysis with reverse transcriptase in parallel to a sequence ladder generated with the same 5′-end-labeled oligonucleotide. In all cases, incubation with recombinant Rnt1p resulted in major cleavage products that were not observed upon incubation with buffer alone or with the mutant enzyme (Fig. 3 ▶). Mapping of the cleavage site with a sequencing ladder revealed that cleavage occurred 13 to 16 bp downstream from the AGNN tetraloops, in agreement with the cleavage site selection rules of the enzyme (Chanfreau et al. 2000) and in agreement with the results observed in vivo (Fig. 1 ▶). Some sites mapped in vitro were located on the 3′ side of the stem–loop, whereas the corresponding sites mapped in vivo were located on the 5′ side of the loop (Fig. 1 ▶, compare gray and black arrowheads) in a staggered location. Although the staggered cut on both sides of the double-stranded RNA is consistent with properties of RNase III endonucleases, it is possible that for these species, 5′ extended products are stabilized in vivo due to the presence of the stem–loop, as described previously (Henry et al. 1994), whereas concerted double cleavage in vitro leads to a most prominent primer extension block on the 3′ side of the molecules. For two snoRNAs (snR52 and snR58), we could detect a downstream cleavage product 12–14 bp from the cleavage site located closer to the tetraloop. This second cleavage was detected either in vivo (snR52) or in vitro (snR58). These observations suggest that Rnt1p may cleave sequentially dsRNA when the duplex following the tetraloop is long. We also noticed that the sequence of the tetraloop found in the target stem–loop upstream of snR71 differs from the AGNN consensus described for yeast RNase III. However, a GGUU tetraloop is likely to adopt the same tetraloop conformation described for an AGUU sequence (Wu et al. 2001). In addition, we recently identified Rnt1p cleavage sites in the introns of RPS22B and RPL18A that contain GGUU terminal tetraloop structures (Danin-Kreiselman et al. 2003). These results show that yeast RNase III can accommodate a variety of sequences at the first position, as long as the tetraloop can adopt an AGNN-type conformation. In conclusion, the data obtained from the in vitro cleavage experiments were usually consistent with the cleavage sites observed in vivo, and further confirmed that the stem–loops located upstream of these snoRNA sequences correspond to Rnt1p cleavage sites. These data also confirmed that the primer extension stop detected in the xrn1Δ rat1-1 strain shifted to 37°C correspond to the cleaved intermediates.
FIGURE 3.
In vitro cleavage of snoRNA precursors model substrates by recombinant Rnt1p. In vitro transcribed unlabeled RNAs were incubated with buffer alone (lane 1), recombinant wild-type (lane 3), or mutant (E320K, lane 2) Rnt1p, and used as primer extension templates.
Inactivation of the trimethylguanosine capping enzyme Tgs1p does not strongly destabilize the snoRNA precursors observed in the rnt1 knockout strain
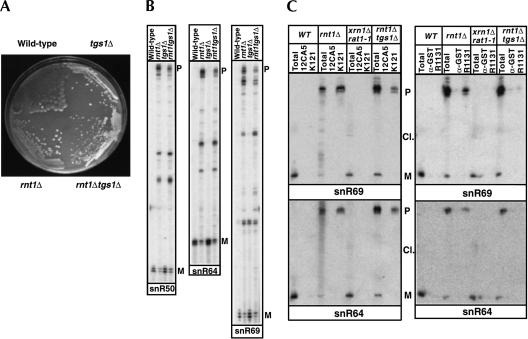
In previous studies, unprocessed precursors detected in the rnt1Δ strain were shown to possess a TMG cap (Chanfreau et al. 1998a,b). One hypothesis proposed to explain the stability of these species was that the 5′ TMG cap protects these precursors against degradation by exonucleases (Chanfreau et al. 1998a,b). The TGS1 gene has recently been shown to encode the trimethylguanosine capping enzyme (Mouaikel et al. 2002). To test the hypothesis that unprocessed precursors are stabilized because of the presence of the TMG cap, we generated a rnt1Δ tgs1Δ double knockout strain. A heterozygote tgs1Δ::BHIS disruption was obtained in a heterozygote rnt1Δ::BTRP deletion diploid strain, and haploid single and double mutant spores were obtained by diploid sporulation and tetrad dissection. Interestingly, the rnt1Δ tgs1Δ double knockout exhibited slower growth than the rnt1Δ knockout, but was still viable at 25°C (Fig. 4A ▶). We analyzed by primer extension the levels of unprocessed and mature snoRNAs in the four spores obtained from this tetrad analysis (WT, rnt1Δ, tgs1Δ, rnt1Δ tgs1Δ). The primer extension profile of three snoRNAs for which the rnt1 deletion significantly affected mature snoRNA levels (snR50, snR64, snR69) is shown in Figure 4B ▶. We did not observe a strong destabilization of these snoRNAs precursors, in contrast to the stated hypothesis. The levels of precursors were slightly reduced, but not as dramatically as one could expect if the TMG cap was solely responsible for stabilizing these precursors.
FIGURE 4.
Analysis of snoRNAs in wild-type, rnt1Δ, tgs1Δ, and rnt1Δ tgs1Δ strains. (A) Growth of wild-type, rnt1Δ, tgs1Δ, and rnt1Δ tgs1Δ strains. Shown are sister spores obtained after dissection of a tetrad. Spores were streaked on YPD medium and grown at 23°C for 6 days. (B) Analysis of the 5′ end of the snR50, snR64, and snR69 snoRNAs by primer extension in wild-type, rnt1Δ, tgs1Δ, and rnt1Δ tgs1Δ strains. Legend as in Figure 2 ▶. (C) Immunoprecipitation analysis of the TMG cap status of precursors and mature snoRNAs in wild-type, rnt1Δ, xrn1Δ rat1-1, and rnt1Δ tgs1Δ strains using the K121 and RR131 antibodies. Total RNAs, RNAs selected after binding to protein G beads coupled to K121 anti-TMG antibodies or to 12CA5 (anti-HA) antibodies, and RNAs selected after binding to protein A beads coupled to polyclonal R1131 anti-TMG antibodies or anti-GST antibodies were loaded on a 6% polyacrylamide sequencing gel, transferred to a Hybond N+ membrane, and probed with snR64-Rev and snR69-Rev antisense oligonucleotide probes hybridizing to the indicated snoRNAs.
To confirm the TMG cap status of these unprocessed precursors, mature snoRNAs, and cleaved intermediates, we performed immunoprecipitation experiments on total RNAs extracted from wild-type, rnt1Δ, xrn1Δ rat1-1 strain shifted to 37°C, and rnt1Δ tgs1Δ strains, using a monoclonal K121 anti-TMG antibody coupled to protein G sepharose beads. We used the monoclonal antibody 12CA5 as negative control. After incubation with beads and extensive washing, RNAs were loaded on polyacrylamide gels and detected by Northern blot and hybridization with oligonucleotide probes (Fig. 4C ▶). In RNAs extracted from the rnt1Δ sample, extended species of snR64 and snR69, could be immunoprecipitated by anti-TMG antibodies, but not by the control 12CA5 antibody. The mature snoRNAs detected in the wild-type and xrn1Δrat1-1 strains were not immunoprecipitated by the anti-TMG antibody, further confirming that these mature snoRNAs are generated through 5′-end processing. The cleaved intermediates detected in the xrn1Δ rat1-1 strain shifted to 37°C were also not immunoprecipitated by the anti-TMG antibody, confirming that these species correspond to cleavage products. Precursors of snR64 and snR69 accumulating in the rnt1Δ tgs1Δ double knockout strain were immunoprecipitated less efficiently compared to the input samples than the precursors accumulating in the rnt1Δ strain. Quantification of these Northern blots showed that between 20 and 25% of the precursors accumulating in the rnt1Δ tgs1Δ strain were immunoprecipitated by the K121 anti-TMG antibody, whereas immunoprecipitation of precursors accumulating in the rnt1Δ strain was quantitative. The same results were obtained with snR50 (data not shown). This result shows that inactivation of the TMG capping enzyme results in an inhibition of TMG capping, as expected (Mouaikel et al. 2002). We note however that a significant fraction of unprocessed precursors is immunoprecipitated by the anti-TMG antibody in the rnt1Δ tgs1Δ strain.
Because trimethylation is expected to be absent in the rnt1Δ tgs1Δ strain, the level of immunoprecipitation observed for the precursors in this strain might be explained by the cross-reactivity of the K121 anti-TMG antibody with monomethylated cap structures. We therefore used another antibody, R1131 (a kind gift of R. Lührmann, Max Planck Institute), which reacts more specifically with trimethylguanosine cap structures (Fig. 4C ▶). Unprocessed snoRNA precursors of snR64 and snR69 were not precipitated by the R1131 antibody in the rnt1Δ tgs1Δ strain, whereas the precursors found in the rnt1Δ strain were precipitated. The same results were obtained with snR50 (data not shown). Because the precursors accumulating in the rnt1Δ tgs1Δ strain are precipitated by the K121 antibodies, but not by the R1131 antibodies, we conclude that these precursors are monomethylated at their 5′ end, and that the precipitation of unprocessed precursors observed in the rnt1Δ tgs1Δ strain is due to a cross-reactivity of the K121 antibody with the monomethyl cap structure. Because the unprocessed precursors detected in the rnt1Δ tgs1Δ strain are not strongly destabilized, we conclude that the monomethyl cap is sufficient to ensure their stabilization.
DISCUSSION
A large number of independently transcribed box C/D snoRNAs undergo 5′-end processing
In this study, we demonstrate that 12 novel box C/D snoRNAs undergo 5′-end processing. Only four independently transcribed box C/D snoRNAs had been shown to be processed at the 5′ end so far, snR39b, snR40, snR47, and snR79 (Chanfreau et al. 1998a). Other known box C/D snoRNAs are either TMG capped at their 5′ end or processed from intronic lariats or polycistronic precursors (Samarsky and Fournier 1999). The 12 additional examples reported here show that the majority of the 23 monocistronic independently transcribed box C/D snoRNAs from S. cerevisiae undergo 5′-end processing. Some of the stem–loops that constitute these processing signals are conserved in Hemiascomycetes species other than S. cerevisiae, suggesting that they correspond to functionally important processing signals (Chanfreau 2003). Processing of box C/D snoRNAs at the 5′ end is therefore much more widespread than previously thought. It is worthwhile to note that for the other major family of snoRNAs (H/ACA), 5′-end processing is not very common, with only three independently transcribed snoRNAs undergoing 5′-end processing (Chanfreau et al. 1998a). Most other H/ACA snoRNAs are either TMG capped or intron encoded. The basis for this difference in the processing pathways of these molecules is intriguing. Because the proteins that assemble onto these snoRNAs differ, and because it is thought that processing and RNP biogenesis are coupled (see below), the differences in snoRNP assembly pathways may have dictated a difference in the requirement for processing of these molecules.
Partial redundance of endonucleolytic and exonucleolytic digestion pathways
In all the cases reported here, we found evidence of Rnt1p cleavage sites in the 5′ extension. The sizes of the 5′ extensions fall within a range of 150 to 200 nt. The location of the Rnt1p cleavage site in these extensions is variable, ranging from 21 nt upstream of the mature sequence (e.g., snR62) to more than 100 nt for snR52. The functional importance of Rnt1p cleavage is variable depending on the snoRNAs, ranging from being essential to generate the mature 5′ end (e.g., snR50) to almost dispensable (e.g., snR52). The basis for this differential requirement for Rnt1p cleavage is unknown. It is possible that some of the Rnt1p target stem–loops are not very stable and do not form very efficiently in vivo. For these snoRNAs, 5′-end processing would rely more heavily on exonuclease digestion, and depletion of Rnt1p would not profoundly affect the level of mature snoRNA production. We have tried to correlate the requirement for Rnt1p with the stability of the predicted stem–loops as estimated by Mfold or with the length of the 5′ extensions, but no correlation could be found. Another unexpected finding of this study was that the pathway relying exclusively on exonucleolytic digestion is partially redundant with the pathway that combines endonucleolytic cleavage and exonucleolytic digestion. In some cases, we observed an increase of accumulation of unprocessed precursors when both Rnt1p and the 5′ → 3′ exonucleases are inactivated. This suggests that these exonucleases can digest full-length precursors and that this pathway contributes to production of at least a fraction of the mature snoRNAs in the cell. Thus, the function of these exonucleases is not restricted to the digestion of Rnt1p cleavage products. In addition, these exonucleases are probably also involved in the degradation of unprocessed precursors that accumulate in the absence of Rnt1p. Because these exonucleases can probably not attack full-length TMG capped precursors, it is likely that a decapping activity must be active for these exonucleases to function in the processing of unprocessed precursors.
Is 5′-end processing a quality control step for proper snoRNP assembly?
Finally, it is interesting to question why such a large number of independently transcribed snoRNAs undergo 5′-end processing. In this study, we show that the majority of independently transcribed box C/D snoRNAs undergo 5′-end processing. It has been suggested that 3′-end processing of extensions of the precursors of noncoding small RNAs provides a way to complete assembly of RNPs before degradation of the transcripts by cellular exonucleases (Kufel et al. 2000). It is possible that 5′-end processing and the presence of 5′ extensions in the precursors of these small RNAs serves a similar purpose, and provides a way to avoid 5′ → 3′ degradation of the transcripts before complete snoRNP assembly. All the unprocessed precursors of snoRNA molecules identified in the rnt1 deletion strain are TMG capped (Chanfreau et al. 1998a; this study). This observation suggests that TMG-cap synthesis rapidly follows transcription of the snoRNA sequence. Rapid hypermethylation of the precursor would be consistent with the fact that in contrast to most vertebrates sn(o)RNAs, hypermethylation of the 5′ end of yeast small RNAs and of U3 vertebrate snoRNA seems to occur in the nucleus (Terns and Dahlberg 1994; Mouaikel et al. 2002). The acquisition of this TMG cap was proposed to prevent premature 5′ → 3′ degradation and protects the snoRNA until snoRNP assembly is completed, or at least delays the action of the exonucleases (Chanfreau et al. 1998a; this study). We note that in the absence of the TMG capping enzyme, the snoRNA precursors are not strongly destabilized (Fig. 4B ▶). The results obtained by immunoprecipitation with the K121 and R1131 antibodies (Fig. 4C ▶) suggest that the precursors accumulating in the rnt1Δ tgs1Δ strain are partially methylated. This result shows that monomethylcaps are sufficient to ensure stabilization of these precursors, and that complete destabilization would require demethylation and/or decapping. After snoRNP assembly, the 5′ extensions are removed by cleavage and/or decapping and exonucleolytic digestion, and the exonucleases will stop at the boundaries of the snoRNAs, which are already marked by the presence of the snoRNP proteins. This mechanism would ensure that most of the snoRNAs produced by the transcriptional machinery are properly packaged into snoRNPs, and adds to the list of quality-control mechanisms provided by RNA-processing reactions.
MATERIALS AND METHODS
Yeast strains
The xrn1Δ rat1-1 strain was provided by D. Tollervey (Petfalski et al. 1998). The rnt1Δ xrn1Δ rat1-1strain is described in Danin-Kreiselman et al. (2003). The rnt1Δ tgs1Δ double knockout strain was obtained by disrupting the TGS1 open reading frame using a PCR-generated HIS marker as described (Longtine et al. 1998) in a diploid strain heterozygote for the rnt1 disruption (Chanfreau et al. 1998b). After sporulation and tetrad dissection, several tetrads were obtained that showed that the rnt1Δtgs1Δdouble knockout strains, prototrophs for both histidine and tryptophan are viable.
RNA analysis
Secondary structure folding using Mfold (Mathews et al. 1999) was performed on M. Zuker’s Web site (Zuker 2003). Total RNA extraction from yeast strains was performed as described (Chanfreau et al. 1998b). Yeast strains were grown on YPD at 25°C, unless otherwise stated. For temperature shifts to 37°C, the strains were shifted for 2 h 30 min to 3 h in medium that was pre-equilibrated at 37°C. Primer extension on total RNAs was performed as described (Chanfreau et al. 1998b), using reverse oligonucleotides specific for each snoRNA (indicated by snR_-Rev on the list). Unlabeled RNA precursors for in vitro cleavage assays were synthesized by in vitro transcription of templates carrying a T7 RNA polymerase promoter obtained by PCR using an upstream oligonucleotide containing the T7 promoter (indicated by snR_-T7) on the list, and the reverse oligonucleotide (snR_-Rev). PCR templates were transcribed in vitro using the Megashortscript kit (Ambion). In vitro cleavage using recombinant Rnt1p and primer extension mapping on gel-purified RNAs was performed as described (Chanfreau et al. 2000; Danin-Kreiselman et al. 2003). Immunoprecipitations using monoclonal K1212 anti-TMG and 12CA5 antibodies (Calbiochem–Oncogene Research) and acrylamide Northern blot analysis were performed as described (Chanfreau et al. 1998b). The R1131 antibody was a kind gift from R. Lührmann. Immunoprecipitations using the R1131 and the anti-GST antibody were performed as described (Kwan et al. 2000).
Oligonucleotides sequences
snR50-Rev: GATTCAATCACGAAAAATCTGCTGC
snR50-T7: CAGGGATCCTAATACGACTCACTATAGGACCTTT CCCTCCTTCC
snR52-Rev: GGAAGGCAACATAAGTTTTTCTAATCC
snR52-T7: CAGGGATCCTAATACGACTCACTATAGGTGATTA CATGTACG
snR58-T7: CGGAATTCTAATACGACTCACTATAGGCGAGGAA GCTAACAGGCC
snR58-Rev: GCGAATTCGGCAATTAACCCTCACTAAAGGTCT AACGTCTAAAAGTTCG
snR60-Rev: CAGATAGGAGCGAAAGACTAATTTCGATGG
snR60-T7: CAGGGATCCTAATACGACTCACTATAGGCTTGTCC TTGTCCACG
snR62-Rev: CGGAATTCGGCAATTAACCCTCACTAAAGGGAATT GTTGATAGTCGTATATC
snR62-T7: CAGGATCCTAATACGACTCACTATAGGGGAGTTTT AAAGAACTCATGTTGC
snR63-Rev: GAAGTACTTCTGTTCTTAATAGACC
snR63-T7: CGCGGATCCTAATACGACTCACTATAGGTAACAT AACTCTTCTACTGCAGTC
snR64-Rev: GGCGCCCTAAGCTCTACTAAAAACTGGGC
snR64-T7: CAGGGATCCTAATACGACTCACTATAGGTGTTGTT ACGCATCTC
snR65-Rev: GTTTGAATGCTTTCAGATACTATCTAGC
snR65-T7: CAGGGATCCTAATACGACTCACTATAGGTGGAA AAACTTCCCTCCG
snR66-Rev: GGTCAGTAATAAAAAAGCAATCTCGTCC
snR66-T7: CAGGGATCCTAATACGACTCACTATAGGAGCTT ACTATTCTTGGTCC
snR68-Rev: ACAGCCCCCGTCAATACGATAACGC
snR68-T7: CAGGGATCCTAATACGACTCACTATAGGAGGTT ACGATCAAGTATC
snR69-Rev: CGAATCGAAGAGCTGGGTTTATAGC
snR69-T7: CAGGGATCCTAATACGACTCACTATAGGAAGAG AAAATTGGTCC
snR71-Rev: CAATCATATCAAAAGATCTGAGTGAGC
snR71-T7: CAGGGATCCTAATACGACTCACTATAGGCCCAA TTTATGGCAGC
Acknowledgments
We thank R. Lührmann (Max Planck Institute) for the generous gift of R1131 antibodies and D. Tollervey (University of Edinburgh) for providing the xrn1Δ rat1-1 strain. A.L. was supported by a fellowship from the Chemistry–Biology Interface Training Program (USPHS National Research Service Award GM0896). C.Y.L. is supported by a fellowship from the Cellular and Molecular Biology Training Program (USPHS National Research Service Award GM07185). This work was supported by grant R01-GM61518 from NIH.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Article and publication are at http://www.rnajournal.org/cgi/doi/10.1261/rna.5126203.
REFERENCES
- Amberg, D.C., Goldstein, A.L., and Cole, C.N. 1992. Isolation and characterization of RAT1: An essential gene of Saccharomyces cerevisiae required for the efficient nucleocytoplasmic trafficking of mRNA. Genes & Dev. 6: 1173–1189. [DOI] [PubMed] [Google Scholar]
- Balakin, A.G., Smith, L., and Fournier, M.J. 1996. The RNA world of the nucleolus: Two major families of small RNAs defined by different box elements with related functions. Cell 86: 823–834. [DOI] [PubMed] [Google Scholar]
- Cavaille, J., Nicoloso, M., and Bachellerie, J.P. 1996. Targeted ribose methylation of RNA in vivo directed by tailored antisense RNA guides. Nature 383: 732–735. [DOI] [PubMed] [Google Scholar]
- Chanfreau, G. 2003. Conservation of RNase III processing pathways and specificity in Hemiascomycetes. Eukaryotic Cell (in press). [DOI] [PMC free article] [PubMed]
- Chanfreau, G., Legrain, P., and Jacquier, A. 1998a. Yeast RNase III as a key processing enzyme in small nucleolar RNAs metabolism. J. Mol. Biol. 284: 975–988. [DOI] [PubMed] [Google Scholar]
- Chanfreau, G., Rotondo, G., Legrain, P., and Jacquier, A. 1998b. Processing of a dicistronic small nucleolar RNA precursor by the RNA endonuclease Rnt1. EMBO J. 17: 3726–3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanfreau, G., Buckle, M., and Jacquier, A. 2000. Recognition of a conserved class of RNA tetraloops by Saccharomyces cerevisiae RNase III. Proc. Natl. Acad. Sci. 97: 3142–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danin-Kreiselman, M., Lee, C.Y., and Chanfreau, G. 2003. RNase III-mediated degradation of unspliced pre-mRNAs and lariat introns. Mol. Cell 11: 1279–1289. [DOI] [PubMed] [Google Scholar]
- Ganot, P., Bortolin, M.L., and Kiss, T. 1997a. Site-specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809. [DOI] [PubMed] [Google Scholar]
- Ganot, P., Caizergues-Ferrer, M., and Kiss, T. 1997b. The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes & Dev. 11: 941–956. [DOI] [PubMed] [Google Scholar]
- Henry, Y., Wood, H., Morrissey, J.P., Petfalski, E., Kearsey, S., and Tollervey, D. 1994. The 5′ end of yeast 5.8S rRNA is generated by exonucleases from an upstream cleavage site. EMBO J. 13: 2452–2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson, A.W. 1997. Rat1p and Xrn1p are functionally interchangeable exoribonucleases that are restricted to and required in the nucleus and cytoplasm, respectively. Mol. Cell. Biol. 17: 6122–6130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss, T. 2001. Small nucleolar RNA-guided post-transcriptional modification of cellular RNAs. EMBO J. 20: 3617–3622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss-Laszlo, Z., Henry, Y., Bachellerie, J.P., Caizergues-Ferrer, M., and Kiss, T. 1996. Site-specific ribose methylation of preribosomal RNA: A novel function for small nucleolar RNAs. Cell 85: 1077–1088. [DOI] [PubMed] [Google Scholar]
- Kufel, J., Allmang, C., Chanfreau, G., Petfalski, E., Lafontaine, D.L., and Tollervey, D. 2000. Precursors to the U3 small nucleolar RNA lack small nucleolar RNP proteins but are stabilized by La binding. Mol. Cell. Biol. 20: 5415–5424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwan, S., Gerlach, V.L., and Brow, D.A. 2000. Disruption of the 5′ stem-loop of yeast U6 RNA induces trimethylguanosine capping of this RNA polymerase III transcript in vivo. RNA 6: 1859–1869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine, M.S., McKenzie 3rd, A., Demarini, D.J., Shah, N.G., Wach, A., Brachat, A., Philippsen, P., and Pringle, J.R. 1998. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast 14: 953–961. [DOI] [PubMed] [Google Scholar]
- Lowe, T.M. and Eddy, S.R. 1999. A computational screen for methylation guide snoRNAs in yeast. Science 283: 1168–1171. [DOI] [PubMed] [Google Scholar]
- Mathews, D.H., Sabina, J., Zuker, M., and Turner, D.H. 1999. Expanded sequence dependence of thermodynamic parameters improves prediction of RNA secondary structure. J. Mol. Biol. 288: 911–940. [DOI] [PubMed] [Google Scholar]
- Mouaikel, J., Verheggen, C., Bertrand, E., Tazi, J., and Bordonne, R. 2002. Hypermethylation of the cap structure of both yeast snRNAs and snoRNAs requires a conserved methyltransferase that is localized to the nucleolus. Mol. Cell 9: 891–901. [DOI] [PubMed] [Google Scholar]
- Ni, J., Tien, A.L., and Fournier, M.J. 1997. Small nucleolar RNAs direct site-specific synthesis of pseudouridine in ribosomal RNA. Cell 89: 565–573. [DOI] [PubMed] [Google Scholar]
- Petfalski, E., Dandekar, T., Henry, Y., and Tollervey, D. 1998. Processing of the precursors to small nucleolar RNAs and rRNAs requires common components. Mol. Cell. Biol. 18: 1181–1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qu, L.H., Henras, A., Lu, Y.J., Zhou, H., Zhou, W.X., Zhu, Y.Q., Zhao, J., Henry, Y., Caizergues-Ferrer, M., and Bachellerie, J.P. 1999. Seven novel methylation guide small nucleolar RNAs are processed from a common polycistronic transcript by Rat1p and RNase III in yeast. Mol. Cell. Biol. 19: 1144–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samarsky, D.A. and Fournier, M.J. 1999. A comprehensive database for the small nucleolar RNAs from Saccharomyces cerevisiae. Nucleic Acids Res 27: 161–164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Terns, M.P. and Dahlberg, J.E. 1994. Retention and 5′ cap trimethylation of U3 snRNA in the nucleus. Science 264: 959–961. [DOI] [PubMed] [Google Scholar]
- Tollervey, D. and Kiss, T. 1997. Function and synthesis of small nucleolar RNAs. Curr. Opin. Cell. Biol. 9: 337–342. [DOI] [PubMed] [Google Scholar]
- Wu, H., Yang, P.K., Butcher, S.E., Kang, S., Chanfreau, G., and Feigon, J. 2001. A novel family of RNA tetraloop structure forms the recognition site for Saccharomyces cerevisiae RNase III. EMBO J. 20: 7240–7249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker, M. 2003. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]