Abstract
Hand-foot-genital syndrome (HFGS) is a rare, dominantly inherited condition affecting the distal limbs and genitourinary tract. A nonsense mutation in the homeobox of HOXA13 has been identified in one affected family, making HFGS the second human syndrome shown to be caused by a HOX gene mutation. We have therefore examined HOXA13 in two new and four previously reported families with features of HFGS. In families 1, 2, and 3, nonsense mutations truncating the encoded protein N-terminal to or within the homeodomain produce typical limb and genitourinary abnormalities; in family 4, an expansion of an N-terminal polyalanine tract produces a similar phenotype; in family 5, a missense mutation, which alters an invariant domain, produces an exceptionally severe limb phenotype; and in family 6, in which limb abnormalities were atypical, no HOXA13 mutation could be detected. Mutations in HOXA13 can therefore cause more-severe limb abnormalities than previously suspected and may act by more than one mechanism.
Hand-foot-genital syndrome (HFGS [MIM 140000]) is a rare, dominantly inherited condition characterized by distal limb and distal genitourinary tract malformations. To date, only eight affected families and one sporadic case have been reported (Stern et al. 1970; Poznanski et al. 1975; Giedion and Prader 1976; Elias et al. 1978; Goeminne 1981; Verp et al. 1983; Halal 1988; Hennekam 1989; Verp 1989; Cleveland and Holmes 1990; Donnenfeld et al. 1992; Fryns et al. 1993). In the limbs, the most striking abnormality is first-digit hypoplasia, comprising short, proximally placed thumbs with hypoplastic thenar eminences and short, medially deviated halluces. There is also ulnar deviation of the second fingers, clinodactyly/brachydactyly of the fifth fingers, brachydactyly of the second to fifth toes, and delayed ossification, fusion, and shortening of the carpals and tarsals. These abnormalities appear to be fully penetrant, bilateral, and symmetrical, with little variation in severity. By contrast, the genitourinary tract abnormalities are incompletely penetrant and variably severe. Genital abnormalities include hypospadias in males (Poznanski et al. 1975; Giedion and Prader 1976; Goeminne 1981; Halal 1988; Donnenfeld et al. 1992; Fryns et al. 1993) and Müllerian duct fusion defects in females (Stern et al. 1970; Halal 1988; Donnenfeld et al. 1992). The latter range from isolated longitudinal vaginal septum to double uterus with double cervix and have resulted in fetal loss or neonatal death in at least three families. Urinary abnormalities include ectopic ureteric orifices, vesicoureteric reflux and pelviureteric junction obstruction (Poznanski et al. 1975; Verp et al. 1983; Halal 1988; Donnenfeld et al. 1992), and can lead to chronic pyelonephritis, renal insufficiency, and renal transplant. HFGS has been shown in one affected family to be caused by a nonsense mutation in the homeobox of HOXA13 (Mortlock and Innis 1997), thus making it only the second human malformation syndrome known to be caused by a HOX gene mutation. (The first was synpolydactyly [MIM 186000], which is caused by mutations in HOXD13 [Akarsu et al. 1996; Muragaki et al. 1996; Goodman et al. 1997, 1998]). In this report, we describe five novel mutations in HOXA13 identified during a study of two new and four previously reported families with features of HFGS.
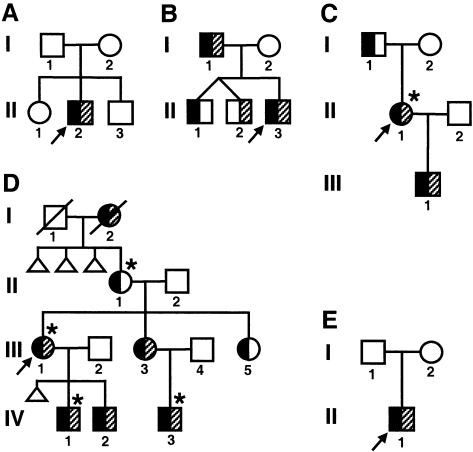
Families 1 and 5 were ascertained after referral for genetic counseling to the Kennedy-Galton Center at Northwick Park Hospital, London, and the Department of Medical Genetics at the University Medical Center, Utrecht, respectively. In family 1 (fig. 1A), the proband (II.2) had hand and foot abnormalities typical of HFGS (fig. 2) as well as a short penis tethered inferiorly to the scrotal sac. His parents and two sibs were clinically unaffected. In family 5 (fig. 1E), the proband (II.1) had severe hand and foot abnormalities, including extremely short thumbs, absent halluces, and marked hypoplasia of all middle phalanges (fig. 3), as well as glandular hypospadias. He was the only child of clinically normal parents. Families 2 (Fryns et al. 1993), 3 (Cleveland and Holmes 1990), 4 (Elias et al. 1978; Verp 1989; Verp et al. 1983; Donnenfeld et al. 1992), and 6 (Hennekam 1989) were reported elsewhere. Follow-up of family 3 (fig. 1C) revealed that II.1, who was known to have typical limb abnormalities, also had a septate uterus and bilateral vesicoureteric reflux, resulting in recurrent urinary tract infections; her newborn son, III.1, had typical limb abnormalities with penile hypospadias and absence of the distal foreskin. Another affected male, IV.2, was also ascertained in family 4 (fig. 1D); he had typical limb abnormalities, penoscrotal hypospadias, severe chordee, and a bifid scrotum.
Figure 1.
Pedigree drawings. A, Family 1; B, family 2; C, family 3; D, family 4; and E, family 5. Black shading of the left half of symbols indicates hand and foot abnormalities. Gray shading of the right half of symbols indicates genital abnormalities (hypospadias or other penile abnormality in males; uterine and/or vaginal abnormalities in females). An asterisk (*) above a symbol indicates that the individual had a urinary tract abnormality on diagnostic imaging.
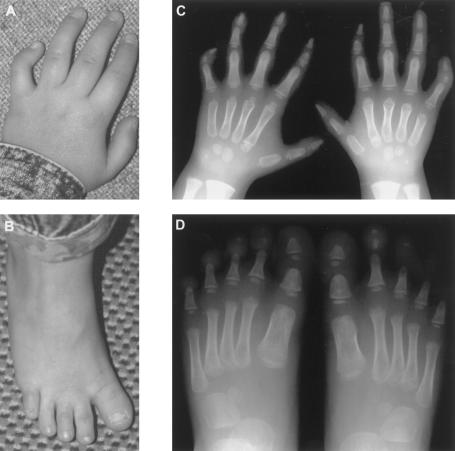
Figure 2.
Limb abnormalities in II.2 from family 1 at age 8 years. Photographs of left hand (A), showing short, proximally placed thumb, ulnar deviation of the second finger, and clinodactyly of the fifth finger; and left foot (B), showing short, medially deviated hallux. Radiographs of both hands (C), showing short first metacarpals, small pointed first distal phalanges, hypoplastic second and fifth middle phalanges, proximal pseudoepiphyses of the second metacarpals and delayed ossification of the carpal centers; and both feet (D), showing short first metatarsals, small triangular first proximal and distal phalanges, absent ossification of the third to fifth middle phalanges, and hypoplasia/absence of the distal phalanges.
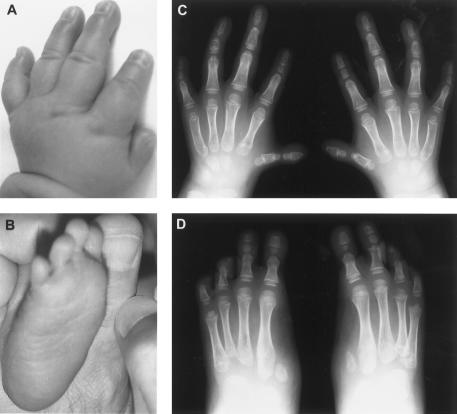
Figure 3.
Limb abnormalities in II.1 from family 5. Photographs at age 1 mo of left hand (A), showing extremely small thumb; and right foot (B ), showing absence of hallux. Radiographs at age 5 years of both hands (C), showing extremely short first metacarpals, small pointed first distal phalanges, marked hypoplasia of all middle phalanges, especially the second and fifth, pseudoepiphyses of the third and fourth middle phalanges and the first and second metacarpals, and delayed ossification of the carpal centers; and both feet (D), showing absence of first digits apart from rudimentary bases of the first metatarsals, hypoplasia/absence of all middle phalanges, lack of epiphyses associated with the middle and distal phalanges, and abnormal tarsals with absent or fused cuneiforms.
Blood samples were obtained from consenting individuals (family 1: I.1, I.2, and II.2; family 2: I.1, I.2, II.1, II.2, and II.3; family 3: I.1, I.2, and II.1; family 4: I.2, III.1, III.3, and III.5; family 5: I.1, I.2, and II.1; and family 6: the proband), with the approval of the local ethical review boards. To search for mutations in HOXA13, the coding sequence of the gene (GenBank) was amplified by PCR in four overlapping segments. Amplified fragments were subcloned into pCR-Script (Stratagene), cycle sequenced (Applied Biosystems Prism Dye Terminator Kit), and analyzed on an ABI 377 sequencer (Applied Biosystems). The sequence obtained differs at nine positions from that previously reported, and these corrections have been submitted to GenBank. Segregation of the mutations identified was assessed by restriction digestion of amplified fragments if possible and direct sequence analysis if not.
In family 1, II.1 was heterozygous for an A→C substitution in exon 1 at base 407 (fig. 4A; base numbers refer to the coding sequence, starting at the first residue of the initiator codon). His unaffected parents did not carry the mutation, confirming that it had arisen de novo; it converts a serine residue to a stop codon and is predicted to result in a truncated protein containing only the first 135 amino acids of the wild-type protein (fig. 4C). In family 2, I.1 was heterozygous for a C→T substitution in exon 1 at base 586 (fig. 4A). The same mutation was identified in II.1 and II.3 but not in II.2; it converts a glutamine residue to a stop codon and is predicted to result in a truncated protein containing only the first 195 amino acids of the wild-type protein (fig. 4D). In family 3, II.1 was heterozygous for a C→T substitution in exon 2 at base 1093 (fig. 4A). The same mutation was present in I.1; it too converts a glutamine residue to a stop codon and is predicted to result in a truncated protein containing only the first 364 amino acids of the wild-type protein (fig. 4E). In family 4, III.1 was heterozygous for a 24-bp in-frame insertion in exon 1 after base 387 (fig. 4A). This insertion occurs in a stretch of 18 imperfect trinucleotide repeats encoding the third of three N-terminal polyalanine tracts. It probably arose by duplication of repeats 7–14 and expands the tract from 18 to 26 alanines. I.2, III.3, and III.5 carried the same expansion, which must thus have remained stable over at least three generations. In family 5, II.1 was heterozygous for an A→C substitution in exon 2 at base 1114 (fig. 4A). Neither of his unaffected parents carried the substitution, confirming that it had arisen de novo. This substitution converts the 51st residue of the homeodomain from asparagine to histidine.
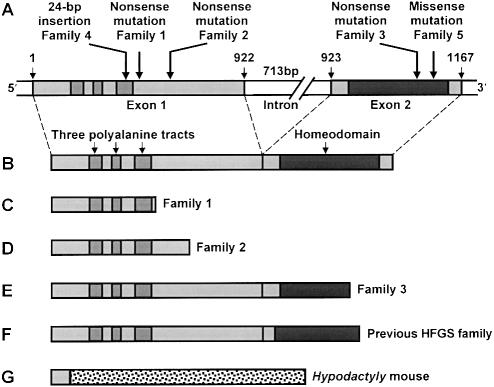
Figure 4.
Mutations in HOXA13. A, Genomic structure of HOXA13, showing sites of the five mutations identified in this study. B, Wild-type HOXA13 protein. C–F, Predicted truncated mutant proteins in family 1 (C), lacking the last 253 amino acids; family 2 (D), lacking the last 193 amino acids; family 3 (E), lacking the last 24 amino acids; and family previously investigated by Mortlock and Innis (1997) (F), lacking the last 20 amino acids. G, Stable truncated mutant protein in the Hypodactyly mouse (Post et al. 2000), caused by a frameshift mutation in exon 1 of Hoxa13; the first 25 amino acids are followed by 275 amino acids with no wild-type counterpart and a premature stop codon.
In all five families, the limb abnormalities were fully penetrant, bilateral, and symmetrical, with analogous involvement of the hands and feet. In particular, II.2 from family 2, who was previously reported as affected but whose limbs were normal, turned out not to carry the familial mutation, highlighting the diagnostic importance of limb abnormalities. By contrast, even when our new data are included, genital abnormalities have been noted in only 16 of 30 males and in 9 of 16 females, and urinary abnormalities have been noted in only 2 males and 6 females.
In the proband from family 6, no mutation was found in the entire coding region, the last 33 bases of the 5′ UTR, the first 31 and last 39 bases of the intron, or the first 34 bases of the 3′ UTR, although a regulatory mutation outside these areas cannot be excluded. A routine karyotype was also normal. Unfortunately, stored metaphase spreads were not available to test by FISH for a microdeletion involving 7p14, where the HOXA cluster is located, nor were blood samples from additional family members available to test for linkage to 7p14. In this family, there were no limb abnormalities other than short halluces, and no males had hypospadias, although three females had Müllerian duct fusion defects (Hennekam 1989). Family 6 thus appears to exhibit a unique “foot-uterus” syndrome, which is dominantly inherited but only partially penetrant and is probably not caused by a HOXA13 mutation.
The five HOXA13 mutations identified in this study fall into three distinct classes: three would truncate the encoded protein, one would expand a polyalanine tract, and one would alter an amino acid in the homeodomain. The effects of the first two classes are clinically indistinguishable, whereas those of the third are exceptionally severe.
The truncated proteins predicted in families 1 and 2 lack the entire homeodomain, and those in family 3 lack four key homeodomain residues responsible for contacting target DNA (fig. 4C, D, and E). Three of these residues are likewise removed by the only HOXA13 mutation that has previously been described (Mortlock and Innis 1997; fig. 4F). In no case has the stability of the truncated protein yet been examined. Limb abnormalities virtually identical to HFGS also occur in heterozygous Hypodactyly mice (Hd/+), which carry a 50-bp deletion after the 25th codon of Hoxa13, resulting in a frameshift followed by a long tract of novel sequence and a premature stop (Mortlock et al. 1996). Although this truncated protein (fig. 4G) lacks the last 363 amino acids of Hoxa13, including the homeodomain, it is nevertheless stable (Post et al. 2000).
The same limb phenotype has recently been reported in a patient with a de novo interstitial deletion at 7p14, which encompasses the entire HOXA cluster (Devriendt et al. 1999), suggesting that the truncation mutations, too, result in haploinsufficiency for HOXA13. Mice heterozygous for a targeted disruption or deletion of Hoxa13, however, have far milder limb abnormalities (Fromental-Ramain et al. 1996). In the forelimbs the phalanges of digit one are occasionally fused, whereas in the hindlimbs the claw and distal phalanx of digit one are often malformed, with occasional 2/3 soft-tissue syndactyly. Mice homozygous for these mutations die in utero at 11.5–15.5 days postconception. Their first digits are absent, and the remaining digits are hypoplastic and webbed, but these abnormalities are again far milder than those in Hd/Hd mice, which have only a single incompletely formed digit on each paw (Mortlock et al. 1996).
The phenotypic differences between these mice and humans with truncation mutations can be explained in several ways. First, the truncated human HOXA13 proteins, although unable to bind DNA specifically, may nevertheless exert a deleterious functional effect. If so, however, all four truncated proteins must be stably expressed, and the effect must be mediated by the first 135 amino acids of HOXA13 (the only region they all share). Strong overexpression of the Hypodactyly mutant protein (which retains only the first 25 of these amino acids but has an additional 275 amino acids with no wild-type counterpart) has indeed been shown to cause limb-reduction defects but in only 3 of 15 transgenic mice (Post et al. 2000). More plausibly, the human truncation mutations may act as null alleles. If so, one possibility is that the effects of haploinsufficiency for HOXA13 on limb development are more severe in humans than in mice. In that case, however, expression of the Hypodactyly mutant protein in mice must have a very similar effect to haploinsufficiency for HOXA13 in humans. Alternatively, the human and mouse truncation mutations may all act as null alleles, and the targeted mouse mutations may not produce a straightforward loss of Hoxa13 function, perhaps because insertion of the selectable marker cassette used to make the mice disrupts expression of neighboring genes in the cluster (Olson et al. 1996).
The polyalanine tract expansion in family 4 makes HOXA13 the second HOX protein and the fourth transcription factor in which such expansions have been shown to cause congenital malformations (Muragaki et al. 1996; Mundlos et al. 1997; Brown et al. 1998). In all four proteins, the polyalanine tracts are normally short (15–18 residues) and show little or no polymorphism in length. The expansions are also short (7–15 extra residues) and meiotically stable. They thus differ sharply from other pathological trinucleotide repeat expansions and are probably caused by unequal crossing over during replication (Warren 1997). Although polyalanine tracts are common in homeodomain and other transcription factors, neither their normal function nor the effects of expansions are understood. In HOXD13 at least, such expansions do not perturb the protein’s stability (F.R.G., unpublished data), but the penetrance and severity of the associated phenotype increase with increasing expansion size (Goodman et al. 1997), which suggests a progressive gain of function, and the mutant protein may act as a dominant negative (Zákány and Duboule 1996). The HOXA13 polyalanine tract expansion identified in this study produces the same phenotype as truncation or deletion of HOXA13; it may therefore simply inactivate the protein or alternatively confer a subtle gain of function, perhaps resulting in a dominant negative effect.
The missense mutation in family 5—the first identified in a human HOX protein—alters an invariant asparagine residue in the homeodomain’s recognition helix that directly contacts target DNA (Wolberger et al. 1991; Gehring et al. 1994). Replacing this residue with histidine would probably not affect the resultant protein’s synthesis or stability but would almost certainly perturb its interactions with DNA. Although the side chain of histidine is bulky, it is partially positive at neutral pH and may permit DNA binding. The mutant protein might therefore recognize new targets or fail to recognize some of its normal ones while nevertheless retaining its capacity to affect transcription once bound. This would result in a novel gain of function, perhaps explaining the exceptionally severe limb phenotype.
Acknowledgments
We are most grateful to the families who participated in this study, as well as to Dr. Christine Hall for her help in assessing the radiographs. This work was supported by grants from the Medical Research Council, UK (Clinical Training Fellowship and Clinician Scientist Fellowship to F.R.G.); the Peabody Foundation (L.B.H.); and the National Institutes of Health (NIH) (HD34059) and the University of Michigan Multipurpose Arthritis and Musculoskeletal Diseases Center (UM-MAC; NIH P60 AR20557) (J.W.I.).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Web/GenBank (for the sequence of HOXA13 [U82827])
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for HFGS [MIM 140000] and for synpolydactyly [MIM 186000])
References
- Akarsu AN, Stoilov I, Yilmaz E, Sayli BS, Sarfarazi M (1996) Genomic structure of HOXD13 gene: a nine polyalanine duplication causes synpolydactyly in two unrelated families. Hum Mol Genet 5:945–952 [DOI] [PubMed] [Google Scholar]
- Brown SA, Warburton D, Brown LY, Yu C, Roeder ER, Stengel-Rutkowski S, Hennekam RCM, et al (1998) Holoprosencephaly due to mutations in ZIC2, a homologue of Drosophila odd-paired. Nat Genet 20:180–183 [DOI] [PubMed] [Google Scholar]
- Cleveland RH, Holmes LB (1990) Hand-foot-genital syndrome: the importance of hallux varus. Pediatr Radiol 20:339–343 [DOI] [PubMed] [Google Scholar]
- Devriendt K, Jaeken J, Matthijs G, Van Esch H, Debeer P, Gewillig M, Fryns J-P (1999) Haploinsufficiency of the HOXA gene cluster, in a patient with hand-foot-genital syndrome, velopharyngeal insufficiency, and persistent patent ductus Botalli. Am J Hum Genet 65:249–251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnenfeld AE, Schrager DS, Corson SL (1992) Update on a family with hand-foot-genital syndrome: hypospadias and urinary tract abnormalities in two boys from the fourth generation. Am J Med Genet 44:482–484 [DOI] [PubMed] [Google Scholar]
- Elias S, Simpson JL, Feingold M, Sarto GE (1978) The hand-foot-uterus syndrome: a rare autosomal dominant disorder. Fertil Steril 29:239–240 [Google Scholar]
- Fromental-Ramain C, Warot X, Messadecq N, LeMeur M, Dolle P, Chambon P (1996) Hoxa-13 and Hoxd-13 play a crucial role in the patterning of the limb autopod. Development 122:2997–3011 [DOI] [PubMed] [Google Scholar]
- Fryns JP, Vogels A, Decock P, van den Berghe H (1993) The hand-foot-genital syndrome: on the variable expression in affected males. Clin Genet 43:232–234 [DOI] [PubMed] [Google Scholar]
- Gehring WJ, Qian YQ, Billeter M, Furukubo-Tokunaga K, Schier AF, Resendez-Perez D, Affolter M, et al (1994) Homeodomain-DNA recognition. Cell 78:211–223 [DOI] [PubMed] [Google Scholar]
- Giedion A, Prader A (1976) Hand-foot-uterus-(HFU) syndrome with hypospadias: the hand-foot-genital-(HFG) syndrome. Pediatr Radiol 4:96–102 [DOI] [PubMed] [Google Scholar]
- Goeminne L (1981) Syndroom van Poznanski-Stern. Tijdschrift voor Geneeskunde 37:1461–1465 [Google Scholar]
- Goodman FR, Giovannucci-Uzielli ML, Hall C, Reardon W, Winter R, Scambler P (1998) Deletions in HOXD13 segregate with an identical, novel foot malformation in two unrelated families. Am J Hum Genet 63:992–1000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman FR, Mundlos S, Muragaki Y, Donnai D, Giovannucci-Uzielli ML, Lapi E, Majewski F, et al (1997) Synpolydactyly phenotypes correlate with size of expansions in HOXD13 polyalanine tract. Proc Natl Acad Sci USA 94:7458–7463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halal F (1988) The hand-foot-genital (hand-foot-uterus) syndrome: family report and update. Am J Med Genet 30:793–803 [DOI] [PubMed]
- Hennekam RCM (1989) Acral-genital anomalies combined with ear anomalies. Am J Med Genet 34:454–455 [DOI] [PubMed]
- Mortlock DP, Innis JW (1997) Mutation of HOXA13 in hand-foot-genital syndrome. Nat Genet 15:179–180 [DOI] [PubMed] [Google Scholar]
- Mortlock DP, Post LC, Innis JW (1996) The molecular basis of hypodactyly (Hd): a deletion in Hoxa13 leads to arrest of digital arch formation. Nat Genet 13:284–289 [DOI] [PubMed] [Google Scholar]
- Mundlos S, Otto F, Mundlos C, Mulliken JB, Aylsworth AS, Albright S, Lindhout D, et al (1997) Mutations involving the transcription factor CBFA1 cause cleidocranial dysplasia. Cell 89:773–779 [DOI] [PubMed] [Google Scholar]
- Muragaki Y, Mundlos S, Upton J, Olsen BR (1996) Altered growth and branching patterns in synpolydactyly caused by mutations in HOXD13. Science 272:548–551 [DOI] [PubMed] [Google Scholar]
- Olson EN, Arnold HH, Rigby PW, Wold BJ (1996) Know your neighbors: three phenotypes in null mutants of the myogenic bHLH gene MRF4. Cell 85:1–4 [DOI] [PubMed] [Google Scholar]
- Post LC, Margulies EH, Kuo A, Innis JW (2000) Severe limb defects in Hypodactyly mice result from the expression of a novel mutant HOXA13 protein. Dev Biol 217:290–300 [DOI] [PubMed] [Google Scholar]
- Poznanski AK, Kuhns LR, Lapides J, Stern AM (1975) A new family with the hand-foot-genital syndrome; a wider spectrum of the hand-foot-uterus syndrome. Birth Defects Original Article Series 11:127–135 [PubMed] [Google Scholar]
- Stern AM, Gall JCJ, Perry BL, Stimson CW, Weitkamp LR, Poznanski AK (1970) The hand-foot-uterus syndrome. J Pediatr 77:109–116 [DOI] [PubMed] [Google Scholar]
- Verp MS (1989) Urinary tract abnormalities in hand-foot-genital syndrome. Am J Med Genet 32:555 [DOI] [PubMed] [Google Scholar]
- Verp MS, Simpson JL, Elias S, Carson SA, Sarto GE, Feingold M (1983) Heritable aspects of uterine anomalies. I. Three familial aggregates with Müllerian fusion anomalies. Fertil Steril 40:80–85 [DOI] [PubMed] [Google Scholar]
- Warren ST (1997) Polyalanine expansion in synpolydactyly might result from unequal crossing-over of HOXD13. Science 275:408–409 [DOI] [PubMed] [Google Scholar]
- Wolberger C, Vershon AK, Liu B, Johnson AD, Pabo CO (1991) Crystal structure of a MAT alpha 2 homeodomain-operator complex suggests a general model for homeodomain-DNA interactions. Cell 67:517–528 [DOI] [PubMed] [Google Scholar]
- Zákány J, Duboule D (1996) Synpolydactyly in mice with a targeted deficiency in the HoxD complex. Nature 384:69–71 [DOI] [PubMed] [Google Scholar]