Abstract
We examined a large French family with autosomal dominant cerebellar ataxia (ADCA) that was excluded from all previously identified spinocerebellar ataxia genes and loci. The patients—seven women and a 4-year-old boy—exhibited slowly progressive childhood-onset cerebellar gait ataxia associated with cerebellar dysarthria, moderate mental retardation (IQ 62–76), and mild developmental delays in motor acquisition. Nystagmus and pyramidal signs were also observed in some cases. This unique association of clinical features clearly distinguishes this new entity from other previously described ADCA. Cerebral magnetic-resonance imaging showed moderate cerebellar and pontine atrophy in two patients. We performed a genomewide search and found significant evidence for linkage to chromosome 19q13.3-q13.4, in an ∼8-cM interval between markers D19S219 and D19S553.
The autosomal dominant cerebellar ataxias (ADCAs) comprise a wide spectrum of diseases with variable onset and different clinical and neuropathological profiles, reflecting the degree of cerebellar and brain-stem dysfunction or degeneration (Harding 1993). Molecular genetic studies have revealed that ADCAs are genetically heterogeneous, and 11 different loci have been mapped so far (Stevanin et al. 2000). Polyglutamine-coding (CAG)n-repeat expansions are responsible for the disease in five of its forms (SCA1 [MIM 164400], SCA2 [MIM 183090], SCA3/MJD [MIM 109150], SCA6 [MIM 183086], and SCA7 [MIM 164500]), whereas untranslated CAG tracts are suspected in SCA8 (MIM 603680) and SCA12 (MIM 604326 [Holmes et al. 1999; Koob et al. 1999]). The phenotypes associated with the different loci overlap, so that the locus cannot be deduced on the basis of clinical expression alone (Stevanin et al. 2000). In the French population, ∼40% of ADCAs remain unexplained by any of the known loci.
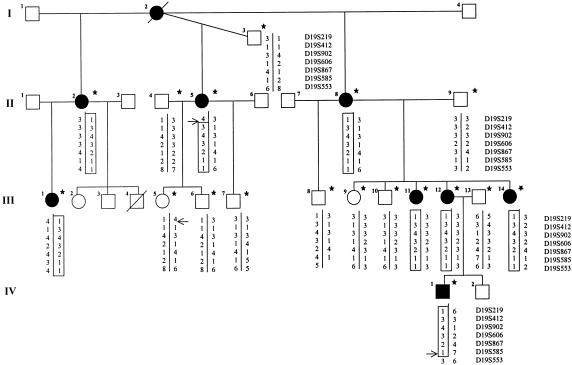
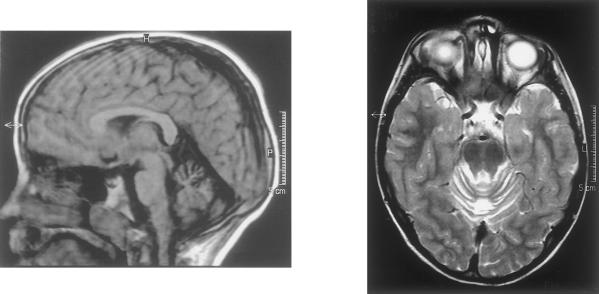
A large French family with cerebellar ataxia was selected. Blood samples of eight patients, six at-risk individuals, and four spouses were obtained after informed consent was signed (fig. 1). The patients were examined at home, except for the youngest affected individual (IV-1), who was also examined by cerebral magnetic-resonance imaging (MRI) and electroencephalography in pediatric clinics. The mini mental-state (MMS) examination was performed at home, and IQ was assessed during childhood in patients II-11, III-12, III-14, and IV-1. A summary of the clinical profile of the eight patients is given in table 1. The most striking features were onset in early childhood in all but one patient and the mild progression of the disease. Cerebellar ataxia was associated with petit mal epilepsy in the only affected boy. Motor development was delayed: patient III-1 walked alone only at age 12 years, patient III-12 at age 3 years, and patients IV-1 and III-14 at age 4 years. No reliable information on either the age at onset or developmental delay could be obtained for the three affected women in generation II, since they did not complain of unsteadiness before adulthood. Mental retardation was obvious although mild in generations III and IV. MMS examination scores were below the pathological limit of 24/30 in five affected women, except for one who had a score of 25/30. Intellectual impairment was mild and global, with IQs of 62–76. There was no evidence of progressive deterioration. Slight dysmorphia and short stature were noted in one patient (III-1). Cerebral MRI in patient IV-1 showed cerebellar vermian atrophy, normal hemispheres, an enlarged fourth ventricle, atrophy of the posterior pons and bulbar thinning (fig. 2). In patient III-1, electromyogram, brain-stem auditory- and somatosensory-evoked potentials, fundoscopy, audition, and karyotype were normal, but a horizontal gaze–evoked nystagmus with a vertical downbeat component was detected by electro-oculography. The presence of petit mal epilepsy in patient IV-1 was confirmed by the presence of a typical spike-wave complex at 3 cycles/s on electroencephalography.
Figure 1 .
Partial pedigree of family AAD-045. Haplotypes for seven microsatellite markers spanning ∼8 cM on chromosome 19q are shown. The haplotype segregating with the disease is boxed. Recombination events are indicated by horizontal arrows. Stars indicate sampled individuals.
Table 1.
Clinical Features in Family AAD-045
| Patient (Sex) | Age atExamination(years) | Age atOnset(years) | Disability | CerebellarAtaxia (Upper Limbs) | Dysarthria | Increased Reflexes | Nystagmus | Mental Status | Additional Signs |
| II-2 (F) | 61 | Childhood | Cannot walkalone (age 38 years) | Moderate (mild) | Severe | +, Plantar flexor | Horizontal | MMS 25/30 | Syncinesias |
| II-5 (F) | 59 | 45 | Wheelchair-bound (age 55 years) | Severe (mild) | Severe | + Except at ankles, unilateral extensor | Horizontal | MMS 23/30 | Torticollis, swallowing difficulties, urinary urgency |
| II-8 (index case) (F) | 53 | Childhood | Cannot run | Moderate (none) | Mild | +, Plantar flexor | Horizontal | MMS 23/30 | Generalized bradykinesia, upward gaze palsy, swallowing difficulties, urinary urgency |
| III-1 (F) | 29 | <1 | Cannot run | Moderate (none) | Moderate | +, Increased muscle tone | Horizontal, Vertical, square waves | Mental retardation | Short stature, slight facial dysmorphia |
| III-11 (F) | 31 | <4 | Cannot run | Mild | None | +, Plantar extensor | Horizontal | MMS 20/30,IQ 71 | |
| III-12 (F) | 28 | Childhood | Cannot run | Mild | Moderate | +, Plantar flexor | Horizontal | MMS 25/30,IQ 68 | |
| III-14 (F) | 24 | <3 | Cannot run | Mild | Mild | +, Unilateral plantar extensor | None | IQ 62 | |
| IV-1 (M) | 4 | <1 | Cannot run | Moderate | Moderate | + | None | IQ 76 | Petit mal epilepsy |
Figure 2 .
Cerebral MRI of patient IV-1 at age 5 years. The T1-weighted midsagital (left) and T2-weighted axial (right) sequences show atrophy in the dorsal part of the cerebellar vermis but normal hemispheres. The fourth ventricle is enlarged. Atrophy is visible in the posterior part of the pons and the medulla. The cerebral cortex is normal.
The striking clinical features in this family are onset during childhood, very slow disease progression, and delayed acquisition of motor and cognitive skills. Delayed attainment of motor milestones has been reported in some autosomal recessive cerebellar ataxias, such as in the carbohydrate-deficiency syndrome (Barone et al. 1999), but without progression. Mental retardation is associated with cerebellar ataxia in the autosomal recessive syndrome of the Cayman Islands (Nystuen et al. 1996) but has never been reported in ADCA. Subcortical dementia is, however, present in a small subgroup (<10%) of patients with ADCA with large CAG repeats at the SCA1, SCA2, and SCA7 loci (Dubourg et al. 1995; Cancel et al. 1997; David et al. 1998; Trojano et al. 1998; Burk et al. 1999). In addition, the neurological phenotype in this kindred is a relatively pure cerebellar ataxia, with dysarthria, nystagmus, and discrete pyramidal features. However, additional neurological signs, such as swallowing difficulties, urinary urgency, and bradykinesia, are observed in patients at age ⩾50 years. Since epilepsy is present in most patients from families with SCA10 (Matsuura et al. 1999; Zu et al. 1999), the petit mal epilepsy in only one member of family AAD-045 may be coincidental. Pontocerebellar atrophy on cerebral MRI is common in ADCA and is consistent with the progressive course of this neurodegenerative disorder (Klockgether et al. 1998).
Trinucleotide-repeat expansions in the SCA1, SCA2, SCA3/MJD, SCA6, SCA7, SCA8, SCA12, and DRPLA genes were excluded by molecular analysis of the index case (II-2) (fig. 1). Linkage analyses also excluded the loci for SCA4 (MIM 600223), SCA5 (MIM 600224), SCA10 (MIM 603516), and SCA11 (MIM 604432) (Stevanin et al. 1999; authors' unpublished data). A genomewide search was performed with the ABI PRISM version 2 linkage-mapping set (PE Biosystems). PCR amplifications of microsatellites were performed either according to the manufacturer's conditions or as described in the Genome Database. Pairwise and multipoint LOD scores were calculated using the MLINK and LINKMAP programs of the computer package FASTLINK version 3.0P (Cottingham et al. 1993; Lathrop et al. 1985) as described elsewhere (Stevanin et al. 1994), with penetrance set at 98% and with equal allele frequencies. Allele frequencies in white subjects (Genome Database) were used for markers of the candidate region. The results obtained by the two modes of calculation differed only slightly.
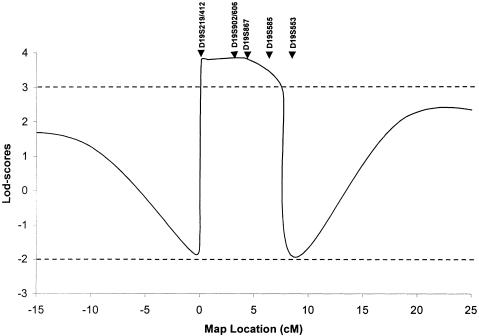
Since no father-to-son transmissions were observed, we began the genomewide search with chromosome X markers, all of which generated significant negative results, excluding the possibility of a mutation on this chromosome in this family. After excluding ∼93% of the autosomes, we obtained positive LOD scores for several consecutive markers on chromosome 19q. Ten additional markers from this region, selected from the Généthon (Dib et al. 1996) and Center for Medical Genetics, Marshfield Medical Research Foundation genetic maps (Broman et al. 1998), were then tested to confirm and refine the genetic localization. A maximum LOD score (Zmax) of 3.50 at recombination fraction (θ)=.00 was obtained for marker D19S867 (table 2). Multipoint analyses with seven markers spanning the D19S219–D19S553 interval localized the disease-causing gene to a region between these markers and generated Zmax=3.85 at marker D19S867 (fig. 3). A single haplotype segregated with the disease. Recombination events in patients II-5 and IV-1 excluded markers D19S219 and D19S553, respectively, delimiting an ∼8-cM region on chromosome 19q13.3-q13.4 (fig. 1).
Table 2.
Pairwise LOD Scores for the Disease Locus and 13 Chromosome 19 Markers[Note]
| LOD Score at θ = |
|||||||||
| Markers | .0 | .01 | .05 | .1 | .2 | .3 | .4 | θ at Zmax | Zmax |
| D19S420 | −1.84 | .29 | 1.16 | 1.44 | 1.44 | 1.15 | .67 | .15 | 1.49 |
| D19S900 | −1.96 | .18 | 1.05 | 1.33 | 1.36 | 1.09 | .63 | .15 | 1.40 |
| D19S219 | −1.88 | .25 | 1.11 | 1.39 | 1.40 | 1.11 | .63 | .14 | 1.45 |
| D19S412 | 2.37 | 2.33 | 2.17 | 1.95 | 1.51 | 1.03 | .51 | .00 | 2.37 |
| D19S902 | 2.97 | 2.92 | 2.71 | 2.44 | 1.86 | 1.24 | .59 | .00 | 2.97 |
| D19S606 | 3.10 | 3.05 | 2.84 | 2.58 | 2.00 | 1.37 | .70 | .00 | 3.10 |
| D19S866 | 2.54 | 2.49 | 2.28 | 2.02 | 1.45 | .84 | .24 | .00 | 2.54 |
| D19S867 | 3.50 | 3.44 | 3.21 | 2.91 | 2.26 | 1.55 | .79 | .00 | 3.50 |
| D19S907 | 2.55 | 2.50 | 2.31 | 2.06 | 1.53 | .97 | .38 | .00 | 2.55 |
| D19S904 | .59 | .57 | .52 | .46 | .33 | .21 | .10 | .00 | .59 |
| D19S585 | .36 | .35 | .31 | .26 | .17 | .08 | .02 | .00 | .36 |
| D19S553 | −3.61 | .06 | .62 | .75 | .69 | .49 | .23 | .10 | .75 |
| D19S571 | −3.61 | −.09 | .77 | 1.04 | 1.05 | .78 | .40 | .14 | 1.10 |
Note.— Calculations were performed using allele-specific frequencies in White control. The use of equal allele frequencies only slightly modified the values in this family.
Figure 3 .
Multipoint linkage analysis with seven chromosome 19q markers: D19S219/D19S412–3 cM–D19S902/D19S606–1 cM–D19S867–2 cM–D19S585–2 cM–D19S553. Because of computer limitations, three overlapping multipoint calculations, using D19S219-D19S412-D19S902, D19S902-D19S606-D19S867, and D19S867-D19S585-D19S553, were performed to generate this curve. The LOD scores obtained in the overlapping regions were similar in all analyses.
Onset occurred during childhood in all generations; unlike the situation for other ADCAs, there was no evidence of anticipation. In ADCAs caused by polyglutamine expansions, childhood onset is always associated with large CAG-repeat expansions and rapid disease progression. To determine whether the mutation in this family is a CAG/polyglutamine-repeat expansion, rapid expansion detection (RED) analyses were performed on patients III-12 and IV-1, as described elsewhere (Zander et al. 1998). Furthermore, proteins were extracted, according to two different protocols (Stevanin et al. 1996), from immortalized lymphoblasts of four patients with early onset and/or marked severity of the disease (II-5, II-8, III-11, and III-14). The extracts were electrophoresed and blotted onto cellulose membranes, then hybridized with the 1C2 antibody, which selectively recognizes large polyglutamine-repeat tracts (Trottier et al. 1995). Consistent with the clinical observations, we found no large CAG/polyglutamine-repeat sequences in the members of the family tested. The 1C2 antibody labeled no specific proteins containing a polyglutamine expansion on the Western blots. RED analysis of genomic DNA revealed no ligation products corresponding to CAG/CTG repeats >40 units (data not shown). Both methods, however, detect only relatively large expansions; therefore, a small CAG repeat, as in SCA6 (Zhuchenko et al. 1997), or another type of trinucleotide-repeat expansion cannot be excluded.
In conclusion, we have mapped, on human chromosome 19, the locus responsible for a new phenotype of ADCA with mental retardation: a phenotype that we propose to designate as “spinocerebellar ataxia 13” (SCA13). This family extends the spectrum of clinical features associated with ADCA. Since the clinical syndrome in this family does not correspond to any of the three types of ADCA described by Harding, it therefore might represent a new clinical and genetic entity.
Several genes have been identified in the 8-cM region on 19q, including those for Bcl-2–associated X protein (BAX), phospholipase A2 (PLA2G4C), and calmodulin (CALM3), which are potential candidates. However, the clinical course of the disorder, in which delayed motor and mental development are observed before the onset of progressive ataxia, suggests that the responsible gene might be involved in both the development and maintenance and/or survival of specific neuronal populations. The 8-cM candidate region defined by haplotype reconstruction and multipoint analysis is still large. Additional families with linkage to this locus, as well as the development of new markers, are needed to refine the candidate interval.
Acknowledgments
The authors are grateful to the family members for participating; to Drs. Merle Ruberg and Ayman Tourbah for constructive comments on the manuscript; to Drs. P. Bejjani and D. Héron for clinical examination; to Y. Trottier for providing the 1C2 antibody; and to Dr. G. Cancel, C. Penet, I. Lagroua, Y. Pothin, and J. Bou for technical assistance. This study was supported financially by the Association Française contre les Myopathies, the VERUM Foundation, and Biomed Concerted Action grant BMH4 CT96 0244. G.S. was the recipient of fellowships from the Société de Secours des Amis des Sciences and the Fondation pour la Recherche Médicale.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Center for Medical Genetics, Marshfield Medical Research Foundation, http://www.marshmed.org/genetics
- Généthon, http://www.genethon.fr/
- Genome Database, The, http://www.gdb.org/
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nim.nih.gov/omim (for SCA1 [MIM 164400], SCA2 [MIM 183090], SCA3/MJD [MIM 109150], SCA4 [MIM 600223], SCA5 [MIM 600224], SCA6 [MIM 183086], SCA7 [MIM 164500], SCA8 [MIM 603680], SCA10 [MIM 603516], SCA11 [MIM 604432], and SCA12 [MIM 604326])
References
- Barone R, Pavone L, Fiumara A, Bianchini R, Jaeken J (1999) Developmental patterns and neuropsychological assessment in patients with carbohydrate-deficient glycoconjugate syndrome type IA (phosphomannomutase deficiency). Brain Dev 21:260–263 [DOI] [PubMed] [Google Scholar]
- Broman KW, Murray JC, Sheffield VC, White RL, Weber JL (1998) Comprehensive human genetic maps: individual and sex-specific variation in recombination. Am J Hum Genet 63:861–869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk K, Globas C, Bosch S, Graber S, Abele M, Brice A, Dichgans J, et al (1999) Cognitive deficits in spinocerebellar ataxia 2. Brain 122:769–777 [DOI] [PubMed] [Google Scholar]
- Cancel G, Durr A, Didierjean O, Imbert G, Burk K, Lezin A, Belal S, et al (1997) Molecular and clinical correlations in spinocerebellar ataxia 2: a study of 32 families. Hum Mol Genet 6:709–715 [DOI] [PubMed] [Google Scholar]
- Cottingham RW Jr, Idury RM, Schäffer AA (1993) Faster sequential genetic linkage computations. Am J Hum Genet 53:252–263 [PMC free article] [PubMed] [Google Scholar]
- David G, Durr A, Stevanin G, Cancel G, Abbas N, Benomar A, Belal S, et al (1998) Molecular and clinical correlations in autosomal dominant cerebellar ataxia with progressive macular dystrophy (SCA7). Hum Mol Genet 7:165–170 [DOI] [PubMed] [Google Scholar]
- Dib C, Faure S, Fizames C, Samson D, Drouot N, Vignal A, Millasseau P, et al (1996) A comprehensive genetic map of the human genome based on 5,264 microsatellites. Nature 380:152–154 [DOI] [PubMed] [Google Scholar]
- Dubourg O, Durr A, Cancel G, Stevanin G, Chneiweiss H, Penet C, Agid Y, et al (1995) Analysis of the SCA1 CAG repeat in a large number of families with dominant ataxia: clinical and molecular correlations. Ann Neurol 37:176–180 [DOI] [PubMed] [Google Scholar]
- Giunti P, Stevanin G, Worth P, David G, Brice A, Wood NW (1999) Molecular and clinical study of 18 families with ADCA type II: evidence for genetic heterogeneity and de novo mutation. Am J Hum Genet 64:1594–1603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harding AE (1993) Clinical features and classification of inherited ataxias. Adv Neurol 61:1–14 [PubMed] [Google Scholar]
- Holmes SE, O'Hearn EE, McInnis MG, Gorelick-Feldman DA, Kleiderlein JJ, Callahan C, Kwak NG, et al (1999) Expansion of a novel CAG trinucleotide repeat in the 5′ region of PPP2R2B is associated with SCA12. Nat Genet 23:391–392 [DOI] [PubMed] [Google Scholar]
- Klockgether T, Skalej M, Wedekind D, Luft AR, Welte D, Schulz JB, Abele M, et al (1998) Autosomal dominant cerebellar ataxia type I: MRI-based volumetry of posterior fossa structures and basal ganglia in spinocerebellar ataxia types 1, 2 and 3. Brain 121:1687–1693 [DOI] [PubMed] [Google Scholar]
- Koob MD, Moseley ML, Schut LJ, Benzow KA, Bird TD, Day JW, Ranum LP (1999) An untranslated CTG expansion causes a novel form of spinocerebellar ataxia (SCA8). Nat Genet 21:379–384 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1985) Multilocus linkage analysis in humans: detection of linkage and estimation of recombination. Am J Hum Genet 37:482–498 [PMC free article] [PubMed] [Google Scholar]
- Matsuura T, Madhureeta A, Mehrdad K, Bachinski LL, Zoghbi HY, Ashizawa T (1999) Mapping of the gene for a novel spinocerebellar ataxia with pure cerebellar signs and epilepsy. Ann Neurol 45:407–441 [DOI] [PubMed] [Google Scholar]
- Nystuen A, Benke PJ, Merren J, Stone EM, Sheffield VC (1996) A cerebellar ataxia locus identified by DNA pooling to search for linkage disequilibrium in an isolated population from the Cayman Islands. Hum Mol Genet 5:525–531 [DOI] [PubMed] [Google Scholar]
- Stevanin G, Dürr A, Brice A (2000) Clinical and molecular advances in autosomal dominant cerebellar ataxias: from genotype to phenotype and physiopathology. Eur J Hum Genet 8:4–18 [DOI] [PubMed] [Google Scholar]
- Stevanin G, Herman A, Brice A, Dürr A (1999) Clinical and MRI findings in spinocerebellar ataxia type 5. Neurology 53:1355–1357 [DOI] [PubMed] [Google Scholar]
- Stevanin G, LeGuern E, Ravise N, Chneiweiss H, Durr A, Cancel G, Vignal A, et al (1994) A third locus for autosomal dominant cerebellar ataxia type I maps to chromosome 14q24.3-qter: evidence for the existence of a fourth locus. Am J Hum Genet 54:11–20 [PMC free article] [PubMed]
- Stevanin G, Trottier Y, Cancel G, Durr A, David G, Didierjean O, Burk K, et al (1996) Screening for proteins with polyglutamine expansions in autosomal dominant cerebellar ataxias. Hum Mol Genet 5:1887–1892 [DOI] [PubMed] [Google Scholar]
- Trojano L, Chiacchio L, Grossi D, Pisacreta AI, Calabrese O, Castaldo I, De Michele G, et al (1998) Determinants of cognitive disorders in autosomal dominant cerebellar ataxia type 1. J Neurol Sci 157:162–167 [DOI] [PubMed] [Google Scholar]
- Trottier Y, Lutz Y, Stevanin G, Imbert G, Devys D, Cancel G, Saudou F, et al (1995) Polyglutamine expansion as a pathological epitope in Huntington's disease and four dominant cerebellar ataxias. Nature 378:403–406 [DOI] [PubMed] [Google Scholar]
- Zander C, Thelaus J, Lindblad K, Karlsson M, Sjöberg K, Schalling M (1998) Multivariate analysis of factors influencing repeat expansion detection. Genome Res 8:1085–1094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhuchenko O, Bailey J, Bonnen P, Ashizawa T, Stockton DW, Amos C, Dobyns WB, et al (1997) Autosomal dominant cerebellar ataxia (SCA6) associated with small polyglutamine expansions in the alpha 1A-voltage-dependent calcium channel. Nat Genet 15:62–69 [DOI] [PubMed] [Google Scholar]
- Zu L, Figueroa KP, Grewal R, Pulst SM (1999) Mapping of a new autosomal dominant spinocerebellar ataxia to chromosome 22. Am J Hum Genet 64:594–599 [DOI] [PMC free article] [PubMed] [Google Scholar]