Abstract
Hereditary motor and sensory neuropathies, to which Charcot-Marie-Tooth (CMT) disease belongs, are a common cause of disability in adulthood. Growing awareness that axonal loss, rather than demyelination per se, is responsible for the neurological deficit in demyelinating CMT disease has focused research on the mechanisms of early development, cell differentiation, and cell-cell interactions in the peripheral nervous system. Autosomal recessive peripheral neuropathies are relatively rare but are clinically more severe than autosomal dominant forms of CMT, and understanding their molecular basis may provide a new perspective on these mechanisms. Here we report the identification of the gene responsible for hereditary motor and sensory neuropathy–Lom (HMSNL). HMSNL shows features of Schwann-cell dysfunction and a concomitant early axonal involvement, suggesting that impaired axon-glia interactions play a major role in its pathogenesis. The gene was previously mapped to 8q24.3, where conserved disease haplotypes suggested genetic homogeneity and a single founder mutation. We have reduced the HMSNL interval to 200 kb and have characterized it by means of large-scale genomic sequencing. Sequence analysis of two genes located in the critical region identified the founder HMSNL mutation: a premature-termination codon at position 148 of the N-myc downstream-regulated gene 1 (NDRG1). NDRG1 is ubiquitously expressed and has been proposed to play a role in growth arrest and cell differentiation, possibly as a signaling protein shuttling between the cytoplasm and the nucleus. We have studied expression in peripheral nerve and have detected particularly high levels in the Schwann cell. Taken together, these findings point to NDRG1 having a role in the peripheral nervous system, possibly in the Schwann-cell signaling necessary for axonal survival.
Introduction
Hereditary motor and sensory neuropathy–Lom (HMSNL [MIM 601455]), which is an autosomal recessive form of Charcot-Marie-Tooth disease, occurs in divergent Romani (Gypsy) groups descended from a small founder population—the Vlax, or Danubian Roma. The disorder was first described in affected families from Bulgaria (Kalaydjieva et al. 1996) and was subsequently diagnosed in families in Italy (Merlini et al. 1998), Slovenia (Butinar et al. 1999), Germany (Baethmann et al. 1998), Spain (Colomer et al. 2000), France, and Rumania. HMSNL is an early-onset peripheral neuropathy that progresses to severe disability in adulthood. Clinically, it presents with muscle weakness and wasting, tendon areflexia, skeletal and foot deformities, sensory loss affecting all modalities, and severe reduction in nerve conduction velocities (Baethmann et al. 1998; Kalaydjieva et al. 1998; Merlini et al. 1998; Butinar et al. 1999). Neural deafness develops during the second or third decade of life, with abnormalities in the brain-stem auditory-evoked potentials suggesting involvement of the entire tract, including the central auditory pathways (Kalaydjieva et al. 1998; Butinar et al. 1999). The neuropathologic observations in HMSNL (Baethmann et al. 1998; Kalaydjieva et al. 1998; Butinar et al. 1999; King et al. 1999) include Schwann-cell dysfunction, which is manifested by hypomyelination and demyelination/remyelination, failure of compaction of the innermost myelin lamellae, and poor hypertrophic response (onion-bulb formation) to the demyelination process. At the same time, axonal involvement is documented by early, severe, and progressive axonal loss and by the presence of curvilinear intra-axonal inclusions that are similar to those seen in the dying-back type of distal axonopathy in experimental vitamin E deficiency.
Findings from a number of recent clinical and experimental studies (Killian et al. 1996; Garcia et al. 1998; Robertson et al. 1999; Sahenk 1999; Sancho et al. 1999) of the common autosomal dominant demyelinating forms of Charcot-Marie-Tooth (CMT) disease have indicated that the neurological deficit in demyelinating neuropathies is related to axonal loss, rather than to demyelination per se. The neuropathologic features of HMSNL make it impossible to attribute the primary defect to either Schwann cells or neurons, and they strongly suggest that impairment of Schwann cell–axonal interaction is a major component of the pathogenesis of this disease. The molecular basis of HMSNL may thus be of relevance to the general understanding of the pathogenetic mechanisms and causes of disability in demyelinating neuropathies.
The disease gene was mapped to a 3-cM interval on 8q24.3, where closely related disease haplotypes and strong linkage-disequilibrium values suggested a single founder mutation (Kalaydjieva et al. 1996). Similar polymorphic haplotypes were subsequently identified in HMSNL chromosomes in affected families across Europe, supporting the assumption of genetic homogeneity and founder effect (Chandler et al. 2000). We now report the identification of the HMSNL gene and the founder mutation causing the disease.
Subjects and Methods
Physical Mapping of the HMSNL Region
A contig of genomic clones spanning the HMSNL interval was assembled by screening of bacterial-artificial-chromosome (BAC) and PAC libraries for the known sequence-tagged sites (STSs) in the region and for the end sequences of clones identified in our previous rounds of library screening. The screening was performed by means of PCR amplification (Research Genetics Human BAC DNA Pools, California Institute of Technology, B&C libraries, cell line 978K) or filter hybridization (PAC library #709, RPCI 6, Roswell Park Cancer Institute; created by Pieter de Jonge and obtained through the German Human Genome Project Center, Max Planck Institute). Clone orientation was obtained by STS content mapping and by halo-FISH (Raap and Wiegant 1994). Nonoverlapping clone ends were used as STSs in the next round of library walking.
Refined Genetic Mapping
For the identification of new polymorphic microsatellites, BAC and PAC contig clones were digested with frequent cutter restriction endonucleases and were shotgun cloned into pBluescript. A replica membrane of the gridded colonies was hybridized with a cocktail of γ[32P]-ATP end-labeled di-/tri-/tetranucleotide repeat sequences, and positive clones were sequenced. Markers (D8S558, D8S529, D8S378, and D8S256) available from public databases were PCR amplified with the use of fluorescently labeled primers (Research Genetics map pair set), were length-separated on a 373 XL DNA analyzer (PE Biosystems), and were analyzed using GENOTYPER software (PE Biosystems). AFM116yh8 and all newly identified microsatellites were analyzed through incorporation of α[32P]-dCTP into the PCR product during amplification. The PCR primers for the newly identified markers were those described by Chandler et al. (2000). Vertical-gel electrophoresis, which was performed in a Hoeffer Pokerface II apparatus, was followed by autoradiography for 2–12 h. Allele calling was performed manually. Haplotypes were constructed manually and were examined for recent and historical recombinations. The marker positions were determined by STS content mapping of the contig clones.
A total of 174 individuals were genotyped for 24 markers in the HMSNL region. Informed consent was obtained from all participants in the study.
Sequencing
BAC/PAC DNA isolation and purification with the QIAfilter Plasmid Midi kits were performed according to the manufacturer’s protocols (Qiagen). End sequencing was performed using universal primers T7 and SP6.
Sequence analysis of the WISP1 and NDRG1 genes included all coding regions and ⩾100 bp of flanking intronic sequences. PCR amplification was performed using the primers shown in table 1. The PCR products were purified with Qiagen QIAquick spin columns. Both strands were sequenced with the same primers used for PCR amplification.
Table 1.
PCR Primers for Sequencing Analysis of NDRG1 and WISP1
|
Primer |
||
| Gene and Exon | Forward | Reverse |
| NDRG1: | ||
| 1 | GACTGCGAGGGTCTGGGAG | CTTACTCCTGGAGTACGC |
| 2 | CTTCTTGCCATTGGTCTTG | GCATGCCCATAAGTACAAG |
| 3 | GATTCAGGTCATAGAAAGG | AGAGAAGACGGGATGAGG |
| 4 | CACGCGGATGCCATGAAC | GCATTTCTGGCTTTTCCAG |
| 5 | CTTTGCCACCGAGACACC | GAGCAAAGCACCTGAACC |
| 6 | CTAATGGCTTCTCTGTGTC | GTCAGTCCAGATCAAAGC |
| 7 | AGGCTCCCGTCACTCTG | GTCTTCCTTCATCTTAAAATG |
| 8 | CCTAGTGTTTCAGATTGCTG | GAGAGCTCGTAGCTCCAG |
| 9 | GGAGTCCAGCAATGCCAC | CTGAGCACCACACAATGC |
| 10 | GAGTAGTGACCAGCTCAG | CAAACTCAGAGCCTGCCTCTTC |
| 11 | ACAGGGCCTCTCTCAAGTTG | CTGGGTAATGCTCAGTCTC |
| 12 | CAGGCCTGGGAGTGGGACAATC | GCAGGCAGGGCCACTTCAAC |
| 13 | CAAGCCACATCTGCTGAATCC | CTTTGCAGCCTCAGATCACC |
| 14 | GACACCAGCAGCCTTGCCTG | CCTAGGGAATCAGAGTCCTC |
| 15 | GGAAACTGGCTCAGACAGG | CATGCCCTCCACACACCTAAC |
| 16 | GTGGACATGGAGAGGACG | GTCTCCACCAGAGCTCACTC |
| WISP1: | ||
| 1 | CATATCTGGTGCTCCTGATGG | GTAGCAGGACCCAGTAGAGAAG |
| 2 | GACAGGAATGCAATGGCAG | GGTGTATCTCCTGCTGAAC |
| 3 | GCATGGTCCACATGGAGCC | GGTGGTCAGAGTTCCAGG |
| 4 | GTGTGGTGAAAGTGAGGGTTG | GCTTGTGAAGTCTAGACATCC |
| 5 | GTAAGGTGGAATGCTCCCAC | CAGATCAGGGTAACTAAGGC |
Sequencing of end clones and PCR products was performed using Big Dye Terminator Cycle Sequencing Ready Reaction Kit reagents (PE Biosystems). The reactions were run on an ABI 377 sequencer (PE Biosystems) and were analyzed using Sequence Navigator software, version 1.0.1.
For large-scale genomic sequencing, BAC/PAC DNA was isolated using the double-acetate method (Birnboim and Doly 1979). The closed-circle band was sonicated, and 1.5–2-kb fragments were size-selected by agarose electrophoresis and were ligated into the SmaI site of the M13mp18 vector. M13 templates were prepared by means of the Triton method (Mardis 1994). Shotgun sequencing was performed using ThermoSequenase (Amersham) and Big Dye Terminator Cycle Sequencing Ready Reaction Kit chemistry (PE Biosystems). Data were collected using ABI 377 automated sequencers and were assembled with the program PHRAP sequence-assembly program (University of Washington Genome Center.
Computational Analysis
The genomic-sequence data were analyzed using the RUMMAGE-DP program (Genome Sequencing Centre, Institute of Molecular Biotechnology, Jena, Germany), which combines >25 different programs (references available at the Web site of the Genome Sequencing Centre, Institute of Molecular Biotechnology, Jena, Germany), including five programs for exon prediction; RepeatMasker, for tagging repetitive sequences; programs for prediction of CpG islands; and homology searches using BLAST, version 1.4, and FASTA, version 2.0. Recognition of promoter regions and transcription-start positions was performed using both Ghosh/Prestridge (TSSG) and Wigender (TSSW) motif databases.
Screening for the R148X Mutation
Exon 7 of NDRG1 was PCR amplified as a 176-bp product, by use of the following primers: 5′-AGGCTCCCGTCACTCTG-3′ (forward) and 5′-GTCTCCTTCATCTTAAAATG3′ (reverse). Restriction digests were performed for 4 h at 65°C in a mix containing 1 × TaqI buffer, 10 μl PCR product, and 10 U TaqI (Promega). The restriction products (lengths 104 bp and 72 bp) were separated in 4% agarose gels stained with ethidium bromide and were visualized under UV light.
Expression Studies
S.A.G.E. library data were obtained through screening of our own libraries constructed from peripheral nerve, glioblastoma, and fetal-brain RNA (Michiels et al. 1999) and through searching of S.A.G.E. public databases. The sequence of the NDRG1 S.A.G.E. tag is GGACTTTCCT. Expression levels are given as the number of transcript tags/106 transcripts in the S.A.G.E. library.
Northern blot analysis was conducted on RNA extracted, according to standard protocols, from total peripheral nerve and from cultured nonmyelinating Schwann cells and hNT2 cells (Sambrook et al. 1989). Reverse transcriptase–PCR (RT-PCR) of NDRG1 from RNA derived from the same sources mentioned above was performed using primers 5′-AACCCACACAGTCACCCT-3′ (forward) and 5′-GAAGTACTTGAAGGCCTC-3′ (reverse). The 189-bp products were run on a 1% agarose gel in 1 × Tris-borate EDTA, were blotted, and were hybridized with the PCR product obtained with the same primers.
Analysis of tissue-specific transcripts was performed by 5′ rapid amplification of cDNA ends (5′-RACE) and by RT-PCR of two fragments spanning the entire coding region of NDRG1. 5′-RACE (Boehringer Mannheim 5′/3′ RACE kit) was performed on total RNA from human fetal brain, adult peripheral nerve, and lymphocytes, by use of the following NDRG1-specific primers: NDRG1-R1 (5′-ACACAGCGTGACGTGAACAG-3′), for the reverse-transcription step); and NDRG1-R2 (5′-CAGAGCCATGTAAAGTCTCG-3′) and NDRG1-R3 (5′-ATGTCCTGCTCCTGGACATC-3′), for the 5′-RACE reactions. The products were tested on agarose gels and were sequenced with primer NDRG1-R3. One-step RT-PCR was performed on the same sources of RNA as was 5′-RACE, by use of the following two primer pairs: NDRG1 5′ UTR-F (5′-GAAGCTCGTCAGTTCACC-3′) and NDRG1 exon 4-R (5′-GTGATCTCCTGCATGTCCTC3′), and NDRG1 exon 4-F (5′-GAGGACATGCAGGAGATCAC-3′) and NDRG1 exon 15-R (5′-CCAGAGGCTGTGCGGGACC-3′).
Radiation-Hybrid Mapping
The chromosomal location of NDRG2 was determined by radiation-hybrid (RH) mapping. PCR screening of the GeneBridge RH panel was performed using primers selected from the unique 3′ UTR sequence of KIAA1248, showing no homology to NDRG1 or NDRG3. The primer sequences were as follows: NDRG2RH-F2 (5′-CTGGGGCTCCATTCACCAAAGC-3′) and NDRG2RH-R2 (5′-AGCCCAGCCCAAGCTTAGCTC-3′). The results were submitted to the RH server of the Whitehead Institute/MIT Center for Genome Research, for calculation.
Results
Physical and Refined Genetic Mapping
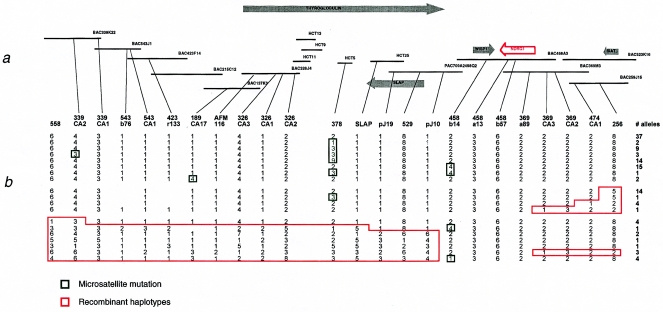
We have assembled a 1-Mb contig of genomic BAC, PAC, and cosmid clones, with a minimum tiling path shown in figure 1. The contig spans the entire HMSNL region, as defined by the recombinations identified in the initial study (Kalaydjieva et al. 1996). The contig was anchored to the four known markers in this region on 8q24, following the order provided by public databases (cen-D8S529/D8S378-AFM116yh8-D8S256-tel). Our subsequent findings have shown the correct orientation to be as follows: cen-AFM116yh8-D8S378-D8S529-D8S256-tel. The contig clones were used for physical mapping of expressed-sequence tags (ESTs) roughly positioned in this region and for identification of new polymorphic markers.
Figure 1.
a, Diagram of the BAC, PAC, and cosmid (HCT) clone contig of the HMSNL region. Known genes are represented by arrows pointing in the direction of their transcription. b, Positions of polymorphic microsatellites used to construct haplotypes on disease chromosomes. The HMSNL critical interval is flanked by markers pJ10 and 369CA3 and contains the WISP1 and NDRG1 genes.
Refined genetic mapping included 174 individuals (60 patients and 114 unaffected relatives) from seven European countries; the individuals were genotyped for 24 polymorphic microsatellites, 19 of which were identified in our study (Chandler et al. 2000). Ten recombinant haplotypes, whose distribution differed between disease chromosomes originating from the diverse Romani groups, helped to narrow the HMSNL region (fig. 1b). In five of the seven centromeric recombinations (fig. 1b, bottom), the breakpoints mapped to the same 90-kb interval between markers pJ10 and 458b14, thus placing the centromeric boundary of the region at pJ10. Haplotype analysis of the telomeric recombinants placed the distal boundary at marker 369CA3 (fig. 1b, right).
Within the pJ10–369CA3 interval, all HMSNL chromosomes shared an identical haplotype for markers 458a13-458b57-369a89. This haplotype was not found in any of the 88 normal chromosomes studied. Marker 458b14 presented with three different alleles in the disease chromosomes; however, on the basis of the conserved flanking haplotypes, this variation was assumed to result from microsatellite mutations (similar to those observed in 339CA2, 189CA17, and, especially, D8S378; fig. 1b, green boxes).
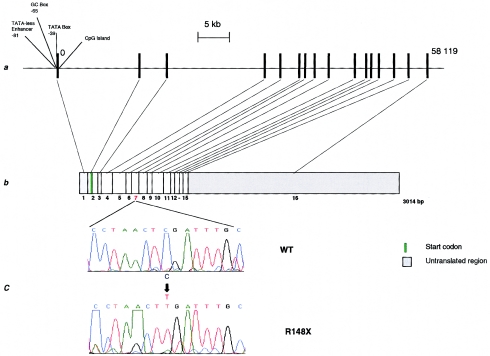
The critical HMSNL gene interval was thus defined, on the basis of recombination and homozygosity mapping, as being located between newly identified markers pJ10 and 369CA3. The entire region was contained in three overlapping genomic clones—PAC 709A2498Q2 and BACs 458A3 (GenBank accession number AF192304) and 369M3 (GenBank accession number AF186190) (fig. 1). Large-scale sequencing of these clones identified the final exons of thyroglobulin in PAC 709A2498Q2 and the full length of two known genes: the Wnt1-inducible signaling protein 1, WISP1 (Pennica et al. 1998), in BAC 458A3, and NDRG1 (aliases RTP, NDR1, DRG1, and CAP43) (Kokame et al. 1996; Van Belzen et al. 1997; Kurdistani et al. 1998; Zhou et al. 1998; Xu et al. 1999) in BACs 458A3 and 369M3 (fig. 1a). WISP1 and NDRG1 are located tail to tail, in opposite orientations, and are separated by a small distance of ∼38 kb. The WISP1 gene spans ∼38 kb of genomic DNA, with the coding regions split into five exons. NDRG1 is spread over 60 kb of genomic DNA and consists of 16 exons, including an untranslated first exon (fig. 2).
Figure 2.
Scale diagram of the genomic and cDNA organization of NDRG1, with an illustration of the HMSNL mutation. a, Genomic organization. The NDRG1 gene consists of 16 exons spanning 60 kb of genomic sequence. A CpG island overlaps with the first exon and the 5′ end of intron 1. A potential promoter with a TATA box, a GC box similar to the N-myc binding region of mouse Ndr1, and a TATA-less enhancer were located 39 bp, 65 bp, and 81 bp upstream, respectively, of the first exon. The 5′ UTR is split between the first two exons. b, cDNA structure. Translation starts from nucleotide position 123. c, HMSNL mutation: C→T substitution at base 564 results in a stop signal (TGA) at codon 148 in exon 7 (R148X). All nucleotide positions are given relative to the published sequence (GenBank accession number D87953).
The HMSNL Mutation
The search for the mutation was conducted by sequencing of the untranslated and promoter regions, all exons, and ⩾100 nucleotides of the flanking introns of WISP1 and NDRG1 in a panel of DNA samples from patients with HMSNL and unaffected controls from the same population.
This analysis revealed a total of 13 single-nucleotide polymorphisms (SNPs) in the two genes (table 2); only one of the 13 SNPs was in WISP1. The difference is due to the fact that sequence variation in NDRG1 was investigated more extensively in individuals of diverse ethnic background, whereas WISP1 was analyzed only in the Roma. So far, our results have shown that SNPs in the NDRG1 gene occur with a frequency of ⩾1/423 nucleotides. In patients with HMSNL, the WISP1 gene sequence was identical to the published wild-type sequence.
Table 2.
SNPs Identified in NDRG1 and WISP1
| Gene and SNP | Nucleotide Positiona | Ethnic Background |
| NDRG1: | ||
| T/G | 5′ UTR, nt 15b | Afro-American |
| C/T | 5′ UTR, nt 3c | Dutch; Roma |
| C/T | Intronic, IVS1+48 | Dutch; Roma |
| C/T | Intronic, IVS2−5 | Afro-American |
| C/T | Intronic, IVS6−33 | Afro-American; Dutch; Roma |
| A/G | Intronic, IVS10 + 83 | Dutch |
| A/C | Intronic, IVS10−50 | Afro-American; Dutch; Roma |
| C/T | Intronic, IVS11−7 | Roma |
| C/T | Intronic, IVS13+147 | Afro-American; Dutch; Roma |
| A/G (293Pro→Pro) | Exon 14, 989c | Afro-American |
| A/C | Intronic, IVS14−124 | Afro-American |
| A/G | 3′ UTR, 1395c | Afro-American |
| WISP1: | ||
| C/T (307Asn→Asn) | Exon 5, 1009d | Roma |
Designated as proposed by Antonarkis et al. (1998), with the positive IVS (intronic) numbers starting from the G of the donor-site invariant GT and with the negative IVS numbers starting from the G of the acceptor-site invariant AG. nt = nucleotide position.
Analysis of NDRG1 in individuals affected with HMSNL identified a C→T transition in exon 7, at nucleotide position 564 (according to the numbering of the reducing agents and tunicamycin-responsive protein (RTP) sequence [GenBank accession number D87953]). This substitution results in the replacement of arginine by a translation-termination signal at codon position 148 (fig. 2). The R148X mutation was found in the homozygous state in all 60 patients with HMSNL that were included in the present study.
The C→T substitution abolished a TaqI site, and a restriction assay was designed as a screening test for the R148X mutation. In families with HMSNL, the mutation segregated in 100% agreement with the carrier status predicted by haplotype analysis. Analysis of 69 additional unaffected members of the extended kindred (the Lom pedigree) in which the disease was first described resulted in the detection of 24 carriers.
Screening for the R148X mutation also included 10 Romani families from Rumania who had unspecified autosomal recessive peripheral neuropathies. The R148X mutation was found in six of these families, in whom it cosegregated with the disease phenotype and occurred in the homozygous state in the affected patients.
We did not find the R148X mutation among 101 unrelated unaffected control individuals, including 68 non-Romani Bulgarians and 33 Roma who originate from the same groups as do the patients with HMSNL but who belong to kindreds with other genetic disorders.
The NDRG Family
NDRG1 is a known gene that has previously been identified in several independent in vitro studies of human cell lines (Kokame et al. 1996; Van Belzen et al. 1997; Kurdistani et al. 1998; Zhou et al. 1998; Xu et al. 1999). The encoded protein is highly conserved in evolution (Kräuter-Canham et al. 1997; Shimono et al. 1999; Yamauchi et al. 1999). The genomic organization of NDRG1, as revealed in the present study (fig. 2), is also conserved and is closely related to that of the mouse gene (Shimono et al. 1999).
The results of previous experiments (Van Belzen et al. 1997; Piquemal et al. 1999) have suggested that NDRG1 is a unique gene; however, a recent study (Van Belzen et al. 1997; Piquemal et al. 1999) has demonstrated the existence of an Ndr gene family in the mouse. Since the existence of homologous genes in humans could affect the specificity and, hence, the reliability of expression studies, we have used the novel mouse sequences to search the human-genome databases. This search has confirmed the existence of related human genes, which we will refer to as NDRG2 and NDRG3, respectively, for the genes homologous to mouse Ndr2 and Ndr3.
NDRG2 was found to be represented by 147 ESTs and two cDNA sequences. To determine its chromosomal localization, we have performed RH mapping with use of the GeneBridge panel. NDRG2 was localized to chromosome 14q11.2, at 6.72 cR from D14S264, with LOD score = 15.0.
The NDRG3 gene was represented by 86 ESTs and a genomic clone from chromosome 20q11.21-q11-23. This provisional chromosomal localization was confirmed by electronic PCR. In the same genomic clone, this search identified four STSs (three STSs flanking NDRG3 and one located in its 3′ UTR) that have also been independently localized to chromosome 20 by means of RH mapping.
The BLAST comparison showed considerable homology between the three human NDR genes, with greater divergence in the terminal parts of the sequences. At the protein level, the identity (similarity) is 54% (70%) between NDRG1 and NDRG2, 67% (81%) between NDRG1 and NDRG3, and 58% (71%) between NDRG2 and NDRG3. These values are very similar to the percent homology reported for the members of the mouse Ndr family (Okuda and Kondoh 1999). Both mouse Ndr2 and Ndr3 (Okuda and Kondoh 1999) and human NDRG2 and NDRG3 lack the highly hydrophilic amino-acid-sequence motif (GTRSRSHTSE) that is typical of NDRG1 and that is repeated three times at its C-terminus.
Expression Analysis of NDRG1
The ubiquitous expression of NDRG1 is documented by 343 entries in the UniGene cluster (GenBank accession number Hs 75789) and by previous studies (Kokame et al. 1996; Van Belzen et al. 1997; Kurdistani et al. 1998; Zhou et al. 1998; Piquemal et al. 1999; Xu et al. 1999) using various experimental systems. To date, no information on the peripheral nervous system has been published.
To obtain a quantitative comparison of the levels of NDRG1 expression in different tissues, we have performed S.A.G.E. library screening and database searches. The following results, presented as the number of transcript tags/106 transcripts in the S.A.G.E. library, were obtained: peripheral nerve, 400; colorectal cancer (HCT116), 213; glioblastoma multiformae libraries, 210 and 99; brain, 146; normal colon and some primary colon tumors, 81–105; and prostate cancer 139 and 158. The aforementioned values indicate that NDRG1 is abundantly expressed in peripheral nerve, where the levels of expression are significantly in excess of those in the other tissues examined.
Northern blot analysis comparing total adult peripheral nerve RNA, cultured nonmyelinating Schwann cells, and hNT2 cells, which can be induced to neuronal differentiation, showed strong signal in total peripheral nerve and Schwann cells. Expression was lower in undifferentiated hNT2 cells, and no signal was obtained in differentiated hNT. In view of the high sequence homology between the genes of the NDRG family and the possibility of cross-hybridization, these results were verified and confirmed by RT-PCR using NDRG1-specific primers (fig. 3). Our preliminary immunocytochemistry findings in peripheral nerve point to NDRG1 localization in the Schwann-cell cytoplasm, with no evidence of axonal expression (not shown).
Figure 3.
RT-PCR using specific NDRG1 primers in hNT2 cells that are not differentiated (lane 1), hNT2 cells induced to neuronal differentiation (lane 2), in vitro cultured nonmyelinating Schwann cells (lane 3), total adult peripheral nerve (lane 4), and fetal brain (lane 5). NDRG1 specificity was confirmed by transfer of the RT-PCR products to a membrane and by back-hybridization with the PCR product.
We have used 5′-RACE and RT-PCR to check for the presence of tissue-specific NDRG1 transcripts in peripheral nerve, fetal brain, and lymphocytes. The results of 5′-RACE did not provide evidence of different transcription-start sites: these experiments identified a short (15-nucleotide) novel additional sequence immediately upstream of the 5′ UTR of the longest published NDRG1 sequence (Kokame et al. 1996), which, however, was common to all three transcripts. RT-PCR and, subsequently, sequencing of the entire coding region of NDRG1 in peripheral nerve, fetal brain, and lymphocytes revealed a single transcript, identical to the published cDNA sequence, with no evidence of tissue-specific alternatively spliced forms.
Discussion
The heterogeneous category of hereditary motor and sensory neuropathies consists of a large number of clinically and genetically distinct conditions (recently reviewed in Keller and Chance [1999] and Schenone and Mancardi [1999]), including autosomal recessive forms, some of which have been placed on the human genetic map (Ben Othmane et al. 1993; Bolino et al. 1996; Casaubon et al. 1996; LeGuern et al. 1996; Bouhouche et al. 1999). Relative to autosomal dominant CMT disease, these conditions are rare. However, they are clinically more severe (Harding and Thomas 1980) and are less likely to result from mutations in structural myelin proteins; therefore, understanding their genetic basis may provide insight into hitherto unknown molecular mechanisms of peripheral-nervous-system development and axon-glia interactions. The genetic heterogeneity of autosomal recessive peripheral neuropathies and the limited number and size of families affected by any single disorder have presented a major obstacle to molecular research and gene identification. In the case of HMSNL, positional cloning was facilitated by the substantial number of patients identified over a short period of time after the initial description of the disease as well as by the history of the disease-causing mutation. HMSNL occurs in an ethnic group that is marginalized by most health-care systems; therefore, ascertainment can be predicted to be limited. The number of affected individuals in whom a diagnosis has already been made suggests that the disease is relatively common and may be the prevalent form of peripheral neuropathy among the Roma. On the other hand, the origin of the HMSNL mutation has been estimated to pre-date the arrival of the proto-Roma in Europe (Kalaydjieva et al. 1996), indicating that the mutation was present in the ancestral population before it split into numerous small groups separated by geographic dispersal, social pressures, and rules of endogamy. The independent evolution and diversification of disease haplotypes in the different Romani groups across Europe have provided a powerful tool for the refined mapping of the HMSNL gene.
The molecular defect shared by all affected individuals was found to be a truncating mutation in NDRG1. This gene encodes a highly conserved protein with a high degree of homology to the proteins in other species. The amino acid similarity to the Drosophila protein is 44%; to sunflower, 48% (Kräuter-Canham et al. 1997); to rat Bdm1, 75% (Yamauchi et al. 1999); and to mouse Ndr1, 96% (Shimono et al. 1999). These proteins show no homology to known motifs, except for a putative phosphopantetheine-binding site (Kokame et al. 1996; Van Belzen et al. 1997; Piquemal et al. 1999) and a 46% similarity to the ligand-binding domain of the inositol 1,4,5-triphosphate receptor (Kräuter-Canham et al. 1997).
The evolutionary conservation of NDRG1-related proteins points to an important biological role. The previously proposed functions of human NDRG1 are based on studies of non-neural tissues. NDRG1 has been shown to be repressed in cell transformation (Van Belzen et al. 1997; Kurdistani et al. 1998) and up-regulated in growth-arrested differentiating cells (Van Belzen et al. 1997; Kurdistani et al. 1998; Piquemal et al. 1999) and under conditions of cellular stress (Kokame et al. 1996; Zhou et al. 1998; Xu et al. 1999). Inducing agents include p53 (Kurdistani et al. 1998), increased intracellular Ca2+ and forskolin (Zhou et al. 1998), retinoic acid, and vitamin D (Piquemal et al. 1999). NDRG1 expression has been shown to cycle with cell division (Kurdistani et al. 1998), and studies of the intracellular localization of the protein suggest translocation between the cytoplasm and the nucleus (Van Belzen et al. 1997; Kurdistani et al. 1998; Piquemal et al. 1999). A role as a developmental gene has been documented for Ndr1, which, in the mouse embryo, is repressed by N-myc and is up-regulated in cells committed to terminal differentiation (Shimono et al. 1999). The accumulated data suggest involvement in growth arrest and cell differentiation during development and in the maintenance of the differentiated state in the adult, possibly as a signaling protein shuttling between the cytoplasm and the nucleus.
In terms of patterns of expression and proposed general functions, NDRG1 clearly resembles PMP22/gas3. PMP22 is also widely expressed in embryonic and adult tissues (Patel et al. 1992; Baechner et al. 1995) and is believed to be involved in growth arrest and cell differentiation (Manfioletti et al. 1990; Zoidl et al. 1997). The highest levels of expression are found in the myelinating Schwann cell, where PMP22 is a component of compact myelin (Snipes et al. 1992). PMP22 is now known to be responsible for CMT type 1A, hereditary neuropathy with liability to pressure palsies, and some forms of Dejerine-Sottas syndrome in humans (Patel et al. 1992; Timmerman et al. 1992; Valentijn et al. 1992; Chance et al. 1993; Roa et al. 1993), and for naturally occurring models of peripheral neuropathy in the mouse (Suter et al. 1992a, 1992b). A number of studies of affected humans as well as of natural and transgenic rodent models have pointed to the complex pathogenesis of these disorders where altered myelin stability and demyelination are only one aspect. The observed significant phenotypic changes in both Schwann cells and axons (Hanemann et al. 1997; Garcia et al. 1998; Sahenk 1999; Sancho et al. 1999; Robertson et al. 1999) have suggested that, in addition to its function as a myelin protein, PMP22 plays a role in early peripheral-nervous-system development and differentiation and in Schwann cell–axonal interactions (reviewed in Naef and Suter 1998).
Axons and glia in the peripheral nervous system are involved in a most complex system of communication, the integrity of which is essential for the differentiation, survival, and normal function of both types of cells (Snipes and Suter 1994; Jessen and Mirsky 1998, 1999). Both the involvement of NDRG1 in these mechanisms and a possible functional link to PMP22 remain to be investigated in functional studies as well as through the identification of NDRG1 mutations in other peripheral neuropathies. The high levels of NDRG1 expression in peripheral nerve and, specifically, in the Schwann cell, together with the characteristics of the HMSNL phenotype, point to a possible involvement of NDRG1 in Schwann-cell differentiation and the signaling necessary for axonal survival. The role of NDRG1 in growth arrest and cell differentiation, proposed for other tissues, may thus be conserved in the peripheral nervous system and may be related to the complex developmental transitions marking the stages of differentiation of the Schwann-cell lineage and Schwann cell–axonal interactions (Jessen and Mirsky 1998, 1999). At the same time, the abundant expression in adult peripheral nerve and the putative phosphopantetheine-binding domain present in the NDRG1 protein point to its possible dual role and additional involvement in the lipid biosynthetic pathways operating in the myelinating Schwann cell.
Acknowledgments
We thank all affected families, for their participation in the study; clinical colleagues, for referring patients for genetic analysis; Jeroen Vreijling, Danielle Dye, and Anthony Akkari, for expert technical assistance; and Garth Nicholson, for providing normal peripheral nerve tissue. The study was supported by the Australian National Health Medical Research Council, the Muscular Dystrophy Association of the United States of America, and The Wellcome Trust.
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- Baylor College of Medicine Gene Finder, http://dot.imgen.bcm.tmc.edu:9331/gene-finder/gf.html/
- Electronic PCR, National Center for Biotechnology Information, http://www.ncbi.nlm.nih.gov/STS/elecpcr.cgi/
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/ (for BAC 458A3 [accession number AF192304]; BAC 369M3 [accession number AF186190]; NDRG1 (RTP) mRNA [accession number D87953]; NDRG1 5′ UTR novel sequence [accession number AF230380]; NDRG1 UniGene cluster [accession number Hs 75789]; NDRG1 LocusLink [accession number ID 10397]; sunflower SF21 [accession number AF189148]; Drosophila melanogasterBcDNA.GH02439 [accession number AF145594]; Rattus norvegicusBdm1 [accession number AF045564]; mouse Ndr1 [accession number U60593]); Ndr2 [accession number AB033921]; Ndr3 [accession number AB033922]; and sequences representing human NDRG2 [accession number AF159092 and AB033074] and NDRG3 [accession number AL031662])
- Genome Sequencing Centre, Institute of Molecular Biotechnology, http://genome.imb-jena.de/ (for the RUMMAGE-DP program)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for HMSNL [MIM 601455])
- RepeatMasker Documentation, http://ftp.genome.washington.edu/RM/RepeatMasker.html
- Serial Analysis of Gene Expression Tag to Gene Mapping (S.A.G.E. map), http://www.ncbi.nlm.nih.gov/sage/
- University of Washington Genome Center, http://www.genome.washington.edu/UWGC/analysistools/phrap.htm (for the PHRAP sequence-assembly program)
- Whitehead Institute for Biomedical Research/MIT Center for Genome Research, http://www.genome.wi.mit.edu/
References
- Antonarakis S (1998) Recommendations for a nomenclature system for human gene mutations. Nomenclature Working Group. Hum Mutat 11:1–3 [DOI] [PubMed] [Google Scholar]
- Baechner D, Liehr T, Hameister H, Altenberger H, Grehl H, Suter U, Rautenstrauss B (1995) Widespread expression of the peripheral myelin protein-22 gene (PMP22) in neural and non-neural tissues during murine development. J Neurosci Res 42:733–741 [DOI] [PubMed] [Google Scholar]
- Baethmann M, Göhlich-Ratmann G, Schröder JM, Kalaydjieva L, Voit T (1998) HMSNL in a 13-year-old Bulgarian girl. Neuromuscul Disord 8:90–94 [DOI] [PubMed] [Google Scholar]
- Ben Othmane K, Hentati F, Lennon F, Ben Hamida C, Blel S, Roses AD, Pericak-Vance MA, et al (1993) Linkage of a locus (CMT4A) for autosomal recessive Charcot-Marie-Tooth disease to chromosome 8q. Hum Mol Genet 2:1625–1628 [DOI] [PubMed] [Google Scholar]
- Birnboim HC, Doly J (1979) A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res 7:1513–1523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bolino A, Brancolini V, Bono F, Bruni A, Gambardella A, Romeo G, Quattrone A, et al (1996) Localization of a gene responsible for autosomal recessive demyelinating neuropathy with focally folded myelin sheaths to chromosome 11q23 by homozygosity mapping and haplotype sharing. Hum Mol Genet 5:1051–1054 [DOI] [PubMed] [Google Scholar]
- Bouhouche A, Benomar A, Birouk N, Mularoni A, Meggouh F, Tassin J, Grid J, et al (1999) A locus for an axonal form of autosomal recessive Charcot-Marie-Tooth disease maps to chromosome 1q21.2-q21.3. Am J Hum Genet 65:722–777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butinar D, Zidar J, Leonardis L, Popovic M, Kalaydjieva L, Angelicheva D, Sininger Y, et al (1999) Hereditary auditory, vestibular, motor, and sensory neuropathy in a Slovenian Roma (Gypsy) kindred. Ann Neurol 46:36–44 [DOI] [PubMed] [Google Scholar]
- Casaubon LK, Melanson M, Lopes-Cendes I, Marineau C, Andermann E, Andermann F, Weissenbach J, et al (1996) The gene responsible for a severe form of peripheral neuropathy and agenesis of the corpus callosum maps to chromosome 15q. Am J Hum Genet 58:28–34 [PMC free article] [PubMed] [Google Scholar]
- Chance PF, Alderson MK, Leppig KA, Lensch MW, Matsunami N, Smith B, Swanson PD, et al (1993) DNA deletion associated with hereditary neuropathy with liability to pressure palsies. Cell 72:143–151 [DOI] [PubMed] [Google Scholar]
- Chandler D, Angelicheva D, Heather L, Gooding R, Gresham D, Yanakiev P, de Jonge R, et al (2000) Hereditary motor and sensory neuropathy–Lom (HMSNL): refined genetic mapping in Romani (Gypsy) families from several European countries. Neuromuscul Disord (in press) [DOI] [PubMed] [Google Scholar]
- Colomer J, Iturriaga C, Kalaydjieva L, Angelicheva D, King RHM, Thomas PK (2000) Hereditary motor and sensory neuropathy–Lom (HMSNL) in a Spanish family: clinical, pathological and genetic studies. Neuromuscul Disord (in press) [DOI] [PubMed] [Google Scholar]
- Garcia A, Combarros O, Calleja J, Berciano J (1998) Charcot-Marie-Tooth disease type 1A with 17p duplication in infancy and early childhood: a longitudinal clinical and electrophysiologic study. Neurology 50:1061–1067 [DOI] [PubMed] [Google Scholar]
- Hanemann CO, Gabreëls-Festen AA, Stoll G, Müller HW (1997) Schwann cell differentiation in Charcot-Marie-Tooth disease type 1A (CMT1A): normal number of myelinating Schwann cells in young CMT1A patients and neural cell adhesion molecule expression in onion bulbs. Acta Neuropathol (Berl) 94:310–315 [DOI] [PubMed] [Google Scholar]
- Harding A, Thomas PK (1980) Autosomal recessive forms of hereditary motor and sensory neuropathy. J Neurol Neurosurg Psychiatry 43:669–678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R (1998) Origin and early development of Schwann cells. Microsc Res Tech 41:393–402 [DOI] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R (1999) Schwann cells and their precursors emerge as major regulators of nerve development. Trends Neurosci 22:402–410 [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, Hallmayer J, Chandler D, Savov A, Nikolova A, Angelicheva D, King R, et al (1996) Gene mapping in Gypsies identifies a novel demyelinating neuropathy on chromosome 8q24. Nat Genet 14:214–217 [DOI] [PubMed] [Google Scholar]
- Kalaydjieva L, Nikolova A, Turnev I, Petrova J, Hristova A, Ishpekova B, Petkova I, et al (1998) Hereditary motor and sensory neuropathy–Lom, a novel demyelinating neuropathy associated with deafness in Gypsies: clinical, electrophysiological and nerve biopsy findings. Brain 121:399–408 [DOI] [PubMed] [Google Scholar]
- Keller MP, Chance PF (1999) Inherited peripheral neuropathy. Semin Neurol 19:353–362 [DOI] [PubMed] [Google Scholar]
- Killian JM, Tiwari PS, Jacobson S, Jackson RD, Lupski JR (1996) Longitudinal studies of the duplication form of Charcot-Marie-Tooth polyneuropathy. Muscle Nerve 19:74–78 [DOI] [PubMed] [Google Scholar]
- King RH, Tournev I, Colomer J, Merlini L, Kalaydjieva L, Tomas PK (1999) Ultrastructural changes in peripheral nerve in hereditary motor and sensory neuropathy–Lom. Neuropathol Appl Neurobiol 25:306–312 [DOI] [PubMed] [Google Scholar]
- Kokame K, Kato H, Miyata T (1996) Homocysteine-respondent genes in vascular endothelial cells identified by differential display analysis: GRP78/BiP and novel genes. J Biol Chem 271:29659–29665 [DOI] [PubMed] [Google Scholar]
- Kräuter-Canham R, Bronner R, Evrard J-L, Hahne G, Friedt W, Steinmetz A (1997) A transmitting tissue- and pollen-expressed protein from sunflower with sequence similarity to the human RTP protein. Plant Science 129:191–202 [Google Scholar]
- Kurdistani SK, Arizti P, Reimer CL, Sugrue MM, Aaronson SA, Lee SW (1998) Inhibition of tumor cell growth by RTP/rit42 and its responsiveness to p53 and DNA damage. Cancer Res 58:4439–4444 [PubMed] [Google Scholar]
- LeGuern E, Guilbot A, Kessali M, Ravise N, Tassin J, Maisonobe T, Grid D, et al (1996) Homozygosity mapping of an autosomal recessive form of demyelinating Charcot-Marie-Tooth disease to chromosome 5q23-q33. Hum Mol Genet 5:1685–1688 [DOI] [PubMed] [Google Scholar]
- Manfioletti G, Ruaro ME, Del Sal G, Philipson L, Schneider C (1990) A growth arrest-specific (gas) gene codes for a membrane protein. Mol Cell Biol 10:2924–2930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mardis ER (1994) High-throughput detergent extraction of M13 subclones for fluorescent DNA sequencing. Nucleic Acids Res 22:2173–2175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlini L, Villanova M, Sabatelli P, Trogu A, Malandrini A, Yanakiev P, Maraldi NM, et al (1998) Hereditary motor and sensory neuropathy Lom type in an Italian Gypsy family. Neuromuscul Disord 8:182–185 [DOI] [PubMed] [Google Scholar]
- Michiels EMC, Oussoren E, Van Groenigen M, Pauws E, Bossuyt PMM, Voute PA, Baas F (1999) Genes differentially expressed in medulloblastoma and fetal brain. Physiol Genomics 1:83–91 [DOI] [PubMed] [Google Scholar]
- Naef R, Suter U (1998) Many facets of the peripheral myelin protein PMP22 in myelination and disease. Microsc Res Tech 41:359–371 [DOI] [PubMed] [Google Scholar]
- Okuda T, Kondoh H (1999) Identification of new genes Ndr2 and Ndr3 which are related to Ndr1/RTP/Drg1 but show distinct tissue specificity and response to N-myc. Biochem Biophys Res Commun 266:208–215 [DOI] [PubMed] [Google Scholar]
- Patel PI, Roa BB, Welcher AA, Schoener-Scott R, Trask BJ, Pentao L, Snipes GJ, et al (1992) The gene for the peripheral myelin protein PMP-22 is a candidate for Charcot-Marie-Tooth disease type 1A. Nat Genet 1:159–165 [DOI] [PubMed] [Google Scholar]
- Pennica D, Swanson TA, Welsh JW, Roy MA, Lawrence DA, Lee J, Brush J, et al (1998) WISP genes are members of the connective tissue growth factor family that are up-regulated in Wnt1-transformed cells and aberrantly expressed in human colon tumors. Proc Natl Acad Sci USA 95:14717–14722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquemal D, Joulia D, Balaguer P, Basset A, Marti J, Commes T (1999) Differential expression of the RTP/Drg1/Ndr1 gene product in proliferating and growth-arrested cells. Biochim Biophys Acta 1450:364–373 [DOI] [PubMed] [Google Scholar]
- Raap A, Wiegant J (1994) Use of DNA-halo preparations for high-resolution DNA in situ hybridization. Methods Mol Biol 33:123–130 [DOI] [PubMed] [Google Scholar]
- Roa BB, Dyck PJ, Marks HG, Chance PF, Lupski JR (1993) Dejerine-Sottas syndrome associated with point mutation in the peripheral myelin protein 22 (PMP22) gene. Nat Genet 5:269–272 [DOI] [PubMed] [Google Scholar]
- Robertson AM, Huxley C, King RHM, Thomas PK (1999) Development of early postnatal peripheral nerve abnormalities in Trembler-J and PMP22 transgenic mice. J Anat 195:331–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahenk Z (1999) Abnormal Schwann cell–axon interactions in CMT neuropathies: the effects of mutant Schwann cells on the axonal cytoskeleton and regeneration-associated myelination. Ann NY Acad Sci 883:415–426 [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T (1989) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- Sancho S, Magyar JP, Aguzzi A, Suter U (1999) Distal axonopathy in peripheral nerves of PMP22-mutant mice. Brain 122:1563–1577 [DOI] [PubMed] [Google Scholar]
- Schenone A, Mancardi GL (1999) Molecular basis of inherited neuropathies. Curr Opin Neurol 12:603–616 [DOI] [PubMed] [Google Scholar]
- Shimono A, Okuda T, Kondoh H (1999) N-myc–dependent repression of Ndr1, a gene identified by direct subtraction of whole mouse embryo cDNAs between wild type and N-myc mutant. Mech Dev 83:39–52 [DOI] [PubMed] [Google Scholar]
- Snipes GJ, Suter U, Welcher AA, Shooter EM (1992) Characterization of a novel peripheral nervous system myelin protein (PMP-22/SR13). J Cell Biol 117:225–238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snipes J, Suter U (1994) Signaling pathways mediating axon–Schwann cell interactions. Trends Neurosci 17:399–401 [DOI] [PubMed] [Google Scholar]
- Suter U, Moskow JJ, Welcher AA, Snipes GJ, Kosaras B, Sidman RL, Buchberg AM, et al (1992a) A leucine-to-proline mutation in the putative first transmembrane domain of the 22-kDa peripheral myelin protein in the Trembler-J mouse. Proc Natl Acad Sci USA 89:4382–4386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suter U, Welcher AA, Ozcelik T, Snipes GJ, Kosaras B, Sidman RL, Buchberg AM, et al (1992b) Trembler mouse carries a point mutation in a myelin gene. Nature 356:241–244 [DOI] [PubMed] [Google Scholar]
- Timmerman V, Nelis E, Van Hul W, Nieuwenhuijsen BW, Chen KL, Wang S, Othman KB, et al (1992) The peripheral myelin protein gene PMP-22 is contained within the Charcot-Marie-Tooth disease type 1A duplication. Nat Genet 1:171–175 [DOI] [PubMed] [Google Scholar]
- Valentijn LJ, Bolhuis PA, Zorn I, Hoogendijk JE, van den Bosch N, Hensels GW, Stanton VP Jr, et al (1992) The peripheral myelin gene PMP-22/GAS-3 is duplicated in Charcot-Marie-Tooth disease type 1A. Nat Genet 1:166–170 [DOI] [PubMed] [Google Scholar]
- Van Belzen N, Dinjens WNM, Diesveld MPG, Groen NA, van der Made ACJ, Nozawa Y, Vliestra R, et al (1997) A novel gene which is up-regulated during colon epithelial cell differentiation and down-regulated in colorectal neoplasms. Lab Invest 77:85–92 [PubMed] [Google Scholar]
- Xu B, Lin L, Rote NS (1999) Identification of a stress-induced protein during human trophoblast differentiation by differential display analysis. Biol Reprod 61:681–686 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y, Hongo S, Ohashi T, Shioda S, Zhou C, Nakai Y, Nishinaka N, et al (1999) Molecular cloning and characterization of a novel developmentally regulated gene, Bdm1, showing predominant expression in postnatal rat brain. Brain Res Mol Brain Res 68:149–158 [DOI] [PubMed] [Google Scholar]
- Zhou D, Salnikow K, Costa M (1998) Cap43, a novel gene specifically induced by Ni2+ compounds. Cancer Res 58:2182–2189 [PubMed] [Google Scholar]
- Zoidl G, D’Urso D, Blass-Kampmann S, Schmalenbach C, Kuhn R, Müller HW (1997) Influence of elevated expression of rat wild-type PMP22 and its mutant PMP22Trembler on cell growth of NIH3T3 fibroblasts. Cell Tissue Res 287:459–470 [DOI] [PubMed] [Google Scholar]