Abstract
Insulin-dependent diabetes mellitus (IDDM) is a polygenic disease caused by progressive autoimmune infiltration (insulitis) of the pancreatic islets of Langerhan, culminating in the destruction of insulin-producing β cells. Genome scans of families with diabetes suggest that multiple loci make incremental contributions to disease susceptibility. However, only the IDDM1 locus is well characterized, at a molecular and functional level, as alleleic variants of the major histocompatibility complex (MHC) class II HLA-DQB1, DRB1, and DPB1 genes that mediate antigen presentation to T cells. In the nonobese diabetic (NOD) mouse model, the Idd1 locus was shown to be the orthologous MHC gene I-Ab. Inheritance of susceptibility alleles at IDDM1/Idd1 is insufficient for disease development in humans and NOD mice. However, the identities and functions of the remaining diabetes loci (Idd2–Idd19 in NOD mice) are largely undefined. A crucial limitation in previous genetic linkage studies of this disease has been reliance on a single complex phenotype—diabetes that displays low penetrance and is of limited utility for high-resolution genetic mapping. Using the NOD model, we have identified an early step in diabetes pathogenesis that behaves as a highly penetrant trait. We report that NOD-derived alleles at both the Idd5 and Idd13 loci regulate a T lymphocyte–dependent progression from a benign to a destructive stage of insulitis. Human chromosomal regions orthologous to the Idd5 and -13 intervals are also linked to diabetes risk, suggesting that conserved genes encoded at these loci are central regulators of disease pathogenesis. These data are the first to reveal a role for individual non-MHC Idd loci in a specific, critical step in diabetes pathogenesis—T cell recruitment to islet lesions driving destructive inflammation. Importantly, identification of intermediate phenotypes in complex disease pathogenesis provides the tools required to progress toward gene identification at these loci.
Introduction
Type 1 or insulin-dependent diabetes mellitus (IDDM) is a complex trait that is conferred by allelic variation at multiple loci. However, efforts to define the genetics of common polygenic disorders have been hampered by the low penetrance of individual loci, and IDDM is an intensively studied example. Genetic analyses of families with IDDM have generated discrepancies likely reflecting variability among study populations, which limits consensus on disease linkage to only a few loci (Concannon et al. 1998; Mein et al. 1998). Genetic dissection of IDDM has been enabled by the nonobese diabetic (NOD) mouse model (Makino et al. 1980), which closely recapitulates the human disease. Nineteen Idd susceptibility loci have been identified in the NOD mouse, and human-genome scans reveal similarly complex inheritance (Davies et al. 1994; Concannon et al. 1998) in mice (Makino et al. 1980; Ikegami et al. 1995; McAleer et al. 1995; Wicker et al. 1995; Vyse and Todd 1996; Podolin et al. 1997, 1998). The major diabetes-susceptibility locus, IDDM1 in humans (MIM 222100) and Idd1 in mice, maps to variation in the major histocompatibility complex (MHC) class II genes. The remarkable sequence similarity between human HLA-DQB1 and mouse H-2 I-AB susceptibility alleles lies in residues crucial for antigen presentation to T cells (Hattori et al. 1986; Acha-Orbea and McDevitt 1987; Todd et al. 1987). However, the molecular basis of MHC-linked diabetes susceptibility is more complex than was originally appreciated. For example, different HLA-DQB alleles are associated with diabetes risk in distinct populations, and evidence for association of both DRB1 (Sheehy et al. 1989) and DPB1 alleles has been reported elsewhere (Erlich et al. 1996; Noble et al. 1996a, 1996b, 2000). Beyond the critical role of class II alleles, evidence for linkage to extended MHC haplotypes includes variants in the peptide transporter genes in some human populations (Caillat-Zucman et al. 1995; Deng et al. 1995). In the NOD mouse, recent data support at least two additional diabetes-associated genes in the MHC region near the transporter gene Lmp2 (Hattori et al. 1999). Thus, MHC markers are unique in displaying strong linkage to the diabetes phenotype in humans and mice, but a susceptible MHC haplotype alone is insufficient to confer disease.
In marked contrast to the strong effects of MHC haplotype, whole-genome scans in NOD mice (Cordell and Todd 1995) and in humans (Davies et al. 1994; Concannon et al. 1998; Mein et al. 1998) suggest that many non-MHC Idd loci make incremental contributions to disease susceptibility. Only two human loci outside the MHC have been clearly established in independent studies. IDDM2 on chromosome 11p15 maps to a VNTR sequence upstream of the insulin gene (Bennett et al. 1995) and has been suggested to affect insulin expression and T cell tolerance to the protein (Bennett and Todd 1996; Vafiadis et al. 1997). A region on human chromosome 2q has been linked to IDDM, by several groups (Davies et al. 1994; Copeman et al. 1995; Owerbach and Gabbay 1995; Morahan et al. 1996; Esposito et al. 1998; Mein et al. 1998) and is syntenic to the Idd5 locus on mouse chromosome 1 (Todd et al. 1991; Ghosh et al. 1993; Garchon et al. 1994; Cordell and Todd 1995; Cordell et al. 1998). However, identification of non-MHC susceptibility genes in humans and mice has been difficult, and neither the identities nor the pathogenic contributions of most of the remaining loci are known.
Previous efforts to map diabetes-susceptibility genes in humans and rodents share a crucial limitation—the reliance on a single complex phenotype, diabetes. Since this clinical endpoint reflects the actions of many genes acting in complex pathways, it is not surprising that the influence of single loci is hard to resolve. We reasoned that the identification of intermediate (preclinical) diabetes phenotypes in islet inflammation might better reflect the action of one or a subset of essential genes common to IDDM-susceptible humans and rodents. One such phenotype was provided by identification of a pivotal early step in the islet inflammatory process.
Infiltration of the pancreatic islets of Langerhans by T cells and antigen-presenting cells (APCs) is a protracted process that precedes overt diabetes by months to years (Hanninen et al. 1992; Itoh et al. 1993). Infiltration begins as a leukocyte accumulation around the islet perimeter (peri-insulitis), which then invades the islet interior (invasive insulitis), ultimately resulting in diabetes in 70%–90% of female NOD mice by age 6 mo (Signore et al. 1989; Castano and Eisenbarth 1990; Jarpe et al. 1991; Waters et al. 1992; Jansen et al. 1994; Signore et al. 1994; Fox and Danska 1997). T cells are critical for invasive insulitis, since protracted peri-insulitis occurs in NOD mice depleted of T cells by administration of antibodies directed against T cell surface markers (Charlton et al. 1988; Sempe et al. 1991; Chatenoud et al. 1994, 1997). Considerable evidence indicates that destruction of islet β cells is also T cell–dependent (reviewed in Toyoda and Formby 1998). Although T cell function is clearly central to destructive islet inflammation, the Idd loci that control this aspect of autoimmune pathogenesis remain undefined.
A clue to the genetic control of T cell pathogenesis was provided by our previous study, in which we compared insulitis progression in both NOD and the nonobese, diabetes-resistant (NOR) strain of mice (Fox and Danska 1998). NOR is a recombinant inbred strain that inherits 88% of the genome from NOD and the remaining 12% from the C57BLKS/J strain (Prochazka et al. 1992). The NOR and NOD strains share the diabetogenic MHC haplotype H-2g7 (Idd1) but differ at 5 of the remaining 18 Idd loci (Prochazka et al. 1992; Serreze et al. 1994). Thus, the inheritance of C57BLKS/J-derived alleles at Idd4, -5, -9, -11, and -13 is sufficient to protect NOR from IDDM. We found that NOR diabetes resistance correlated with a protracted peri-insulitis involving minimal T cell contribution (Fox and Danska 1998). Thus, transition from peri- to invasive insulitis appeared to be dependent on T cell infiltration and the Idd loci that are allelically variable between NOR and NOD mice. Importantly, these results implicated distinct genetic control of different immune-cell types in early islet inflammation. Here, we have investigated the genetic control of this step in diabetes development by performing a linkage analysis in (NOD×NOR) F2 progeny. Progression from peri- to invasive insulitis behaved as a highly penetrant trait controlled by an interaction between 2 of the 19 Idd loci, Idd5 and -13. Remarkably, chromosomal regions syntenic to Idd5 and -13 are the only known non-MHC diabetes-susceptibility loci that are shared by humans and mice. Our data reveal for the first time that these two loci regulate a critical early step in diabetes pathogenesis, orchestrated through T cell recruitment to nascent autoimmune islet lesions.
Material and Methods
Mice
All mice used in these studies were maintained in a barrier facility at the Hospital for Sick Children (Toronto). In our colony, diabetes incidence at age 6 mo in NOD animals is 83% in females, 9% in males, and 0 in NOR and (NOD×NOR) F1 mice.
Immunohistochemistry
Pancreata were removed from 80-d-old NOD and NOR (NOD×NOR) F1 female mice and from (NOD×NOR) F2 intercross male and female mice, were immersed in TissueTec (Bayer Labs), snap-frozen in liquid nitrogen, and stored at −70° C. Preparation of serial frozen sections was performed with a Leica CM 3050 Cryostat (Leica Canada). To maximize analysis of independent islet infiltrates, serial 5-μm sections were prepared from each of three depths of the 5-mm-thick tissue block, in which each series of sections examined was sampled at intervals of ⩾300 μm apart, as described elsewhere (Fox and Danska 1998). Previous morphometric studies demonstrate that 2–3-mo-old mice have islets averaging 100 μm in diameter and that each pancreas contains ∼1,000 islets (Bonnevie-Nielsen et al. 1983). Pancreatic sections were stained with Mayer's hematoxylin and eosin Y (H+E; Sigma), to visualize leukocyte infiltration. For immunohistochemical analysis, serial sections were then incubated with biotin–anti-CD3 (145-2C11; [Leo et al. 1987]) or biotin–anti-MHC class II (10.2-16 [Oi et al. 1978]) antibodies and ExtrAvidin-Horseradish peroxidase (Sigma). Antibody staining was visualized with AEC substrate (Sigma). Antibodies were prepared by protein G purification from tissue-culture supernatants and were biotin conjugated (Cedarlane Labs), as described elsewhere (Fox and Danska 1997). Insulitis severity was scored in H+E–stained pancreatic sections. Approximately 250 islets from three different tissue depths >300 μm apart were examined in each animal and were graded by assignment of the following scores: 0, no visible infiltrates; 1, peri-insulitis, indicated by perivascular and peri-islet infiltrates; 2, <50% of the islets displaying invasive infiltrates (defined as islet interior occluded by leukocytes); 3, >50% of the islets displaying invasive insulitis when <50% of the islet is occluded by leukocytes; or 4, complete infiltration, virtually all islets displaying invasive insulitis of 50%–100% of the islet. This approach and scoring system has been used extensively for analysis of autoimmune insulitis, by many laboratories (Wicker et al. 1987, 1992; Stein et al. 1992; Yui et al. 1996).
Genomic DNA Preparation
Splenic DNA from C57BLKS/J mice was purchased from the Jackson Labs DNA Resource (Jackson Laboratory). Genomic DNA was also prepared from tail snips of 30 d-old DBA/2J, C57BL/6J, NOD, NOR (NOD×NOR) F1, and (NOD×NOR) F2 intercross mice, by standard methods. Each preparation was diluted 1:100 for use in PCR amplification.
Genotyping
Genotyping of NOR and (NOD×NOR) F2 intercross progeny was performed using microsatellite markers (Whitehead Institute for Biomedical Research/MIT Center for Genome Research). PCR primer pairs were obtained from Research Genetics. (NOD×NOR) F2 intercross animals were typed for microsatellite alleles at Idd4 (D11Mit30 and D11Mit320), Idd5 (D1Mit3, D1Mit8, D1Mit24, D1Mit46, D1Mit77, and D1Mit122), Idd9/11 (D4Mit13, D4Mit16, and D4Mit72), and Idd13 (D2Mit17, D2Mit135, D2Mit338, D2Mit395, D2Mit411, D2Mit423, and D2Mit490). These mice were also typed at C57BLBKS-derived regions in the NOR genome that do not contain known Idd loci on chromosomes 7 (D7Mit105), 12 (D12Mit230), and 18 (D18Mit4 and D18Mit197). NOR mice were typed for NOD-, DBA/2J- or C57BL/6J-derived alleles at multiple markers on chromosome 1 (D1Mit3, D1Mit8, D1Mit11, D1Mit18, D1Mit22, D1Mit24, D1Mit46, D1Mit48, D1Mit65, D1Mit66, D1Mit72, D1Mit77, D1Mit122, D1Mit178, D1Mit212, D1Mit213, D1Mit231, D1Mit245, D1Mit279, D1Mit305, D1Mit306, D1Mit322, D1Mit383, D1Mit411, D1Mit414, and D1Mit430) and chromosome 2 (D2Mit17, D2Mit22, D2Mit62, D2Mit135, D2Mit144, D2Mit147, D2Mit206, D2Mit229, D2Mit256, D2Mit333, D2Mit338, D2Mit343, D2Mit393, D2Mit395, D2Mit411, D2Mit423, D2Mit447, D2Mit452, D2Mit480, D2Mit490, and D2Mit493). The order of these markers derives from the Whitehead Institute for Biomedical Research/MIT Center for Genome Research. For most primer pairs, 30 ng of genomic DNA was amplified for 35–40 cycles, with the following conditions: 30 s at 94°C, 30 s at 55°C, and 1 min at 72°C. For D11Mit30, D4Mit11, D12Mit51, and D18Mit21, PCR amplification was performed with 30 ng of genomic DNA for 40 cycles (30 s at 94°C, 40 s at 50°C, and 1 min 10 s at 72°C in 2.5 mM MgCl2). The PCR amplification products were electrophoresed through 2% NuSieve (American Bioanalytical) and 2% agarose (Gibco BRL) gels and were visualized with ethidium bromide.
Statistical Analysis
Quantitative-trait linkage analysis was performed as follows: a genetic map was built on the basis of the Whitehead Institute for Biomedical Research/MIT Center for Genome Research and EUCIB (UK Human Genome Project Mapping Center) marker orders, and markers were analyzed for trait linkage, by the MAPMAKER EXP v3.0 and QTL v1.1 suite of programs (Lander et al. 1987; Genome Center Software Information and Documentation). Multipoint analysis was conducted using recessive, dominant, and additive models. Linkage to chromosome 1 (Idd5) was maximal under an additive model, and linkage to chromosome 2 (Idd13) was maximal under a dominant model (see the Results section). The LOD scores, proportion of the trait variance explained, and 1-LOD confidence intervals of the position of the peaks were determined for all animals and separately for females and males. Haplotype construction was performed, to detect close double recombinants, and microsatellite typing was reexamined, to correct any errors. For chromosome 2 markers, several mice still typed as close double recombinants and were excluded from the study. Furthermore, since the marker order on chromosome 2 differs markedly between different reference maps, we also analyzed chromosome 2 by using pairwise linkage.
Results
T Cell Dependence of Transition from Peri- to Invasive Insulitis
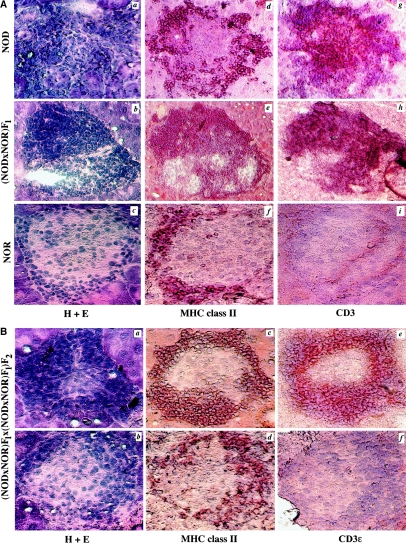
To determine whether progression from peri- to invasive insulitis was invariably associated with T cell infiltration, we analyzed T cell and APCs presence in serial pancreatic sections of 80-d-old NOD, NOR, (NOD×NOR) F1, and (NOD×NOR) F2 intercross mice. Invasive insulitis in NOD and (NOD×NOR) F1 animals (fig. 1A, panels a and b) was characterized by coincident infiltration of MHC class II+ APCs (fig. 1A, panels d and e) and CD3ɛ+ T cells (fig. 1A, panels g and h). In contrast, only mild peri-insulitis was seen in NOR mice (fig. 1A, panel c). Although robust MHC class II+ staining was evident in NOR peri-islet infiltrates (fig. 1A, panel f), few or no CD3ɛ-expressing T cells were observed in these lesions (fig. 1A, panel i), a result concordant with our previous study (Fox and Danska 1998). In (NOD×NOR) F2 intercross progeny (n=19), both the NOD and NOR parental phenotypes were observed. Progression to invasive insulitis (fig. 1B, panel a) was associated with islet infiltration by APCs and T cells (fig. 1B, panels c and e, respectively). In contrast, other (NOD×NOR) F2 mice displayed peri-insulitis (fig. 1B, panel b) characterized by MHC class II+ APCs in the absence of CD3ɛ+ T cells (fig. 1B, panels d and f, respectively). These results suggested that the transition from peri- to invasive insulitis depends on T cell recruitment and is regulated by loci that are allelically variable between NOD and NOR mice.
Figure 1 .
Invasive insulitis is T cell dependent. A, Serial pancreatic cross-sections from 80-d-old female NOD, (NOD×NOR) F1, and NOR mice were stained with H+E, to visualize infiltrating cells (panels a–c); with the anti-MHC class II antibody 10.2-16, to visualize APC (panels d–f); and with the anti-CD3e antibody 145-2C11, to visualize T cells (panels g–i). Panels d–i were counterstained in Mayer's hematoxylin. B, Serial sections from two representative (NOD×NOR) F2 intercross progeny were stained with H+E (panels a and b), anti-MHC class II (panels c and d), and anti-CD3 (panels e and f). Positive protein expression is indicated by dark brown coloration. Original magnification, ×250.
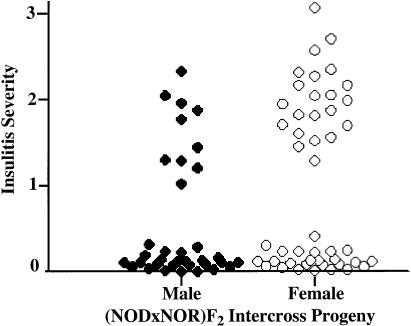
Next, we examined whether the progression from peri- to invasive insulitis behaved as a preclinical trait suitable for genetic linkage analysis. Insulitis severity was measured in 82 male and female (NOD×NOR) F2 progeny at age 80 d. Two distinct phenotypes were evident in the F2 animals: 39% had invasive insulitis (insulitis score ⩾1.5) and 61% had mild peri-insulitis (insulitis score ⩽0.3), a result characteristic of a bimodal trait (fig. 2). Sex dimorphism is well established in NOD diabetes, in which disease incidence is higher in female, compared with male, mice (Makino et al. 1980; Pozzilli et al. 1993). In 80-d-old (NOD×NOR) F2 animals, only a marginal increase in insulitis severity was observed in female, compared with age-matched male, mice (fig. 2; Mann-Whitney test for significance, U=590, P=.03). We previously used quantitative reverse transcriptase–coupled PCR to examine inflammatory progression in purified male and female NOD islets. At age 30 d, T cell cytokine profiles in islet lesions were already biased toward T helper 1 in females and T helper 2 in males. Sex differences in the abundance of T cells and APCs were not evident until age 70 d (Fox and Danska 1997). Examination of the insulitis phenotype at age 80 d in the current study focused on an early stage in autoimmune disease progression, a stage prior to the appearance of substantial differences in severity of male and female islet inflammation.
Figure 2 .
Insulitis severity in (NOD×NOR) F2 intercross progeny. The average insulitis severity was calculated in 82 80-d-old (NOD×NOR) F2 intercross mice by the scoring of H+E–stained pancreatic cross-sections. As shown, a scatter plot comparing individual insulitis severity in male (blackened circles) and female (unblackened circles) animals clearly indicates that insulitis severity behaves as a bimodal trait.
NOD-Derived Alleles at Idd5 and Idd13 Control Invasive Insulitis
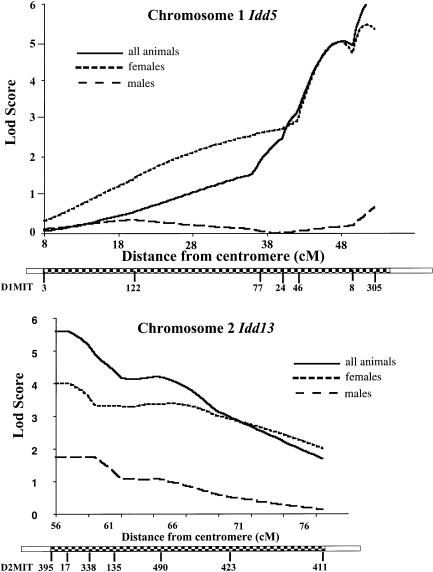
All (NOD×NOR) F2 intercross progeny were genotyped for NOD- or BKS-derived alleles at microsatellite markers spanning the Idd4, -5, -9/11, and -13 loci and at markers in C57BLKS/J-derived regions of the NOR genome that lacked previously identified Idd loci. Pairwise (table 1) and multipoint (fig. 3) statistical analyses of the insulitis-severity scores and genotypes revealed significant linkage (Lander and Kruglyak 1995) of invasive insulitis to markers of Idd5 on chromosome 1 and Idd13 on chromosome 2. LOD scores were calculated separately for males and females, as well as for all the (NOD×NOR) F2 animals. A significant LOD score for all animals was observed at the marker D1Mit305 (LOD, 5.7; fig. 3A) when an additive model was used, with the females displaying strong linkage (LOD, 5.3) whereas the males produced virtually no linkage across this region when the same model was used (LOD, <1.0). We also observed significant linkage to a second locus with maximal linkage at the marker D2Mit395, with LOD scores of 5.7 for all animals, 4.0 for females, and 1.7 for males, under the assumption of a dominant model (fig. 3B and table 1). In contrast, all other markers examined in regions of allelic variation between the NOD and NOR strains did not show significant linkage (LOD, ⩽2.6; data not shown). D1Mit305 and D2Mit395 accounted for 31% and 27%, respectively, of the genetic variation in insulitis phenotype and generated scores 2 log greater than the benchmark defined for significant linkage (Lander and Kruglyak 1995).
Table 1.
Pairwise Linkage Analysis of Chromosome 2[Note]
|
Position |
||||
| Marker | % Variance | LOD Score | MIT | JAX |
| D2Mit395 | 26.9 | 5.3 | 55.7 | 66.9 |
| D2Mit17 | 16.4 | 3.1 | 56.8 | 69.0 |
| D2Mit338 | 23.3 | 4.5 | 59.0 | 57.5 |
| D2Mit135 | 20.2 | 3.9 | 61.2 | 73.0 |
| D2Mit490 | 19.7 | 3.8 | 64.5 | 64.5 |
| D2Mit423 | 14.3 | 2.7 | 68.9 | 68.9 |
| D2Mit411 | 7.9 | 1.4 | 77.6 | 77.6 |
Note.— The (NOD×NOR) F2 data for markers of chromosome 2 were also analyzed by pairwise linkage analysis. The marker name, percentage of variance, and LOD score are shown. As is also shown, the relative order of these markers differs according to information available at the Whitehead Institute for Biomedical Research/MIT Center for Genome Research (MIT) and the Jackson Laboratory (JAX). For both databases, the position of each marker from the centromere of chromosome 2 is shown in centimorgans.
Figure 3 .
NOD-derived alleles at Idd5 and Idd13 confer invasive insulitis. The LOD scores for the linkage of invasive insulitis in females, males, and all animals are shown vs. the distance in centimorgans from the centromeres of A, chromosome 1 and B, chromosome 2. The relative order of the microsatellite markers along each chromosome (according to Whitehead Institute for Biomedical Research/MIT Center for Genome Research) is shown beneath the x axis of each graph. C57BL/6J-derived regions (checkered boxes) are shown on NOR chromosomes 1 and 2. The remaining regions (unblackened boxes) are NOD-derived. *(NOD×NOR) F2 intercross animals could not be scored for linkage telomeric to D1Mit305 or centromeric to D2Mit395 since these markers very close to the boundaries of these C57BL/6J-derived chromosome segments in NOR mice.
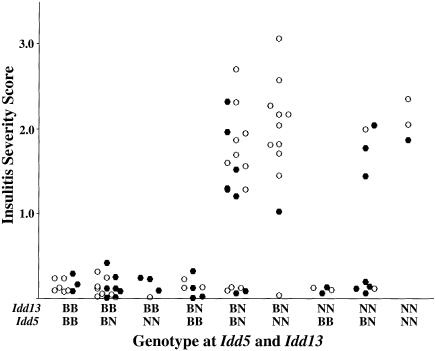
These findings suggest that NOD-derived alleles at both Idd5 and Idd13 drive invasive insulitis, providing evidence of a genetic interaction controlling this phenotype (Frankel and Schork 1996). To analyze this interaction, we compared the degree of insulitis in (NOD×NOR) F2 progeny with all possible combinations of NOD and NOR alleles at the two loci (fig. 4). Clearly, homozygosity for BKS-derived alleles at either Idd5 (D1Mit305) or Idd13 (D2Mit395) conferred potent resistance to invasive insulitis; all animals of these genotypes displayed insulitis scores <0.3 (fig. 4). The powerful role of NOD alleles at these two loci was also evident in this study. Insulitis progression (severity score >1.0) was observed only in the (NOD×NOR) F2 animals that carried at least one NOD-derived allele at both Idd5 and Idd13 (fig. 4). (NOD×NOR) F2 mice carrying NOD alleles at Idd5 and Idd13 provided evidence for the penetrance of the insulitis-progression phenotype. Seventy-four percent (32/43) of F2 animals carrying at least one NOD-derived allele at both Idd5 and Idd13 displayed insulitis scores >1.0. Evidence for genetic interaction between Idd5 and Idd13 is provided by comparison of insulitis values of mice with at least one NOD allele at each locus (n=38) to the other animals (n=44). Using a t test for nonequal variances in the two groups yielded t=9.3, 44 df; two-tailed P<.0001. Thus, a genetic interaction between NOD-derived alleles at the Idd5 and Idd13 loci promotes the transition from peri- to invasive insulitis, an obligate step in early islet inflammation. The effect could be blocked by homozygosity for BKS-derived resistance alleles at either locus. Interestingly, although the phenotype distributions were very similar between male and female (NOD×NOR) F2 progeny (fig. 2), males contributed little to the Idd5 linkage (fig. 3). This observation likely reflects the relatively small numbers of animals with each of the nine possible Idd5 and Idd13 genotype combinations when they are subdivided by sex (fig. 4).
Figure 4 .
Histogram scatter plots of the insulitis severity in male (blackened circles) and female (unblackened circles) (NOD×NOR) F2 intercross progeny distributed by combination of NOD (N) and C57BKS/J (B) alleles at Idd5 (D1Mit305) and Idd13 (D2Mit395).
Idd5 and Idd13 Regulate T Cell Influx into Islet Lesions
Progression to invasive insulitis was associated with T cell invasion of islet lesions (fig. 1A and B), suggesting that the two features are controlled by the same loci. We used immunohistochemistry to examine T cells and APCs in 19 randomly selected (NOD×NOR) F2 intercross mice. Eight of these animals (3 male and 5 female) had inherited at least one NOD-derived allele at Idd5 and Idd13; all displayed invasive insulitis (severity scores 1.2–2.0), and 98% of their affected islets showed T cell infiltration (table 2, top). In contrast, 11 animals (2 male and 9 female) were homozygous for BKS-derived alleles at either or both of the Idd5 and Idd13 loci; all of these mice displayed mild peri-insulitis (severity scores 0.08–0.23), and T cells were observed in only 4.8% of the islet lesions scored in these animals (table 2, bottom). Strikingly, these results suggest that the mechanism by which Idd5 and Idd13 drive insulitis progression is T cell recruitment to, or retention in, islet lesions.
Table 2.
T Cell Infiltration of (NOD×NOR) F2 Islets, Regulated by Idd5 and Idd13[Note]
| Idd5/Idd13 | Insulitis Severity | CD3+Islets/Total Islets | Sex |
| NN/NN | 1.30 | 15/15 | Male |
| BN/NN | 1.81 | 12/12 | Female |
| NN/BN | 1.87 | 9/11 | Male |
| NN/BN | 1.71 | 10/10 | Female |
| NN/BN | 1.62 | 21/21 | Female |
| BN/BN | 2.04 | 18/18 | Female |
| BN/BN | 1.82 | 18/18 | Female |
| BN/BN | 1.20 | 13/13 | Male |
| Average or total | 1.67±.29 | 116/118 | |
| BB/BB | .09 | 1/5 | Female |
| BB/BB | .23 | 0/2 | Female |
| BB/BB | .08 | 0/4 | Male |
| BN/BB | .31 | 0/0 | Female |
| BN/BB | .11 | 0/5 | Male |
| BN/BB | .09 | 0/3 | Female |
| BB/NN | .12 | 0/2 | Female |
| BB/NN | .10 | 1/2 | Female |
| BB/NN | .13 | 0/3 | Female |
| BB/BN | .13 | 0/5 | Female |
| BB/BN | .12 | 0/4 | Female |
| Average or total | .14±.07 | 2/35 |
Note.— Frozen pancreatic sections were prepared from 19 (NOD×NOR) F2 intercross animals. Each animal was typed as heterozygous (BN) or homozygous for NOD-derived (NN) or C57BLKS/J-derived (BB) alleles at Idd5 (D1Mit305) and Idd13 (D2Mit395), and insulitis severity was assessed. Serial frozen pancreatic sections were examined for the presence of CD3ɛ+ T cells by immunohistochemistry. Top, Eight (NOD×NOR) F2 mice displayed invasive insulitis and T cell infiltration. Bottom, Eleven (NOD×NOR) F2 mice displayed benign peri-insulitis and minimal T cell infiltration. Mean insulitis severity ±SD in each group is shown. The number of T cell–infiltrated islets relative to the total number of islets examined is also shown.
Genetic Maps of NOR Chromosomes 1 and 2
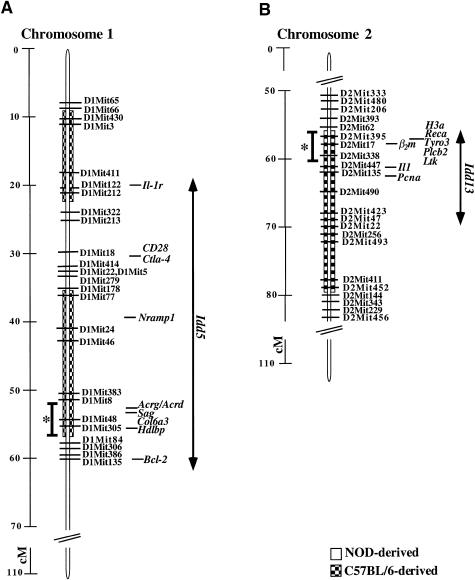
Progress toward the identification of the genes at Idd5 and Idd13 depends on high-resolution genetic mapping. NOR is a recombinant inbred strain providing an opportunity to define the C57BLKS/J-derived segments, of chromosomes 1 and 2, in which the genes regulating insulitis progression reside. The C57BLKS/J strain was derived from two parental strains, C57BL/6J (B6) and DBA/2J (Naggert et al. 1995). Consequently, NOR mice harbor chromosomal segments of NOD, DBA/2J, and B6 origin. To refine the boundaries of the NOR chromosome 1 and 2 segments that were of NOD, DBA/2J or B6 origin, NOR mice were typed with microsatellite markers, and these markers were ordered according to current genetic maps (see the Materials and Methods section). Using 25 microsatellite markers on chromosome 1, two B6-derived segments—one of 16 cM and another of 21 cM in length, located between the D1Mit66 and D1Mit322 loci and between the D1Mit178 and D1Mit84 loci, respectively—were resolved in NOR (fig. 5A). Control of invasive insulitis was most closely linked to D1Mit305, near the telomeric boundary of the latter segment (fig. 3). Analysis of 22 microsatellite markers on chromosome 2 showed that NOR mice contained a 25-cM B6-derived segment extending between D2Mit62 and D2Mit144 (fig. 5B). On chromosome 2, maximal linkage of T cell–dependent invasive insulitis was observed at D2Mit395, near the centromeric boundary of the B6-derived segment (fig. 3 and 5B). As mentioned in the context of multipoint linkage analysis, there is not yet a clear consensus on the order of some chromosome 2 markers. On the basis of these marker positions and the linkage analysis, we computed a 5-cM 1-LOD confidence interval for Idd5 on chromosome 1 (fig. 5A) and a 4-cM 1-LOD confidence interval for Idd13 on chromosome 2 (fig. 5B).
Figure 5 .
Chromosomes 1 and 2 in NOR mice. NOR genomic DNA was typed for NOD- (unblackened boxes) or C57BL/6J-derived (checkered boxes) alleles at each microsatellite marker shown on chromosomes 1 (A) and 2 (B). The distance in centimorgans between markers is shown to the far left. Several candidate genes that map to each region are also shown (in italics). * = 1-LOD confidence intervals were calculated with MAPMAKER QTL. The gene locations and distances in centimorgans are adapted from published studies (Seldin 1996, 1997; Siracusa et al. 1996, 1997), the Whitehead Institute for Biomedical Research/MIT Center for Genome Research, and the Jackson Laboratory. The original location of Idd5 encompasses an interval 20–60 cM from the centromere of chromosome 1 (Cornall et al. 1991; Garchon et al. 1991). Idd13 was originally mapped to a 4-cM interval of chromosome 2 that spans the β2m, Il1, and Pcna genes (Serreze et al. 1994), within which the 1-LOD confidence interval for insulitis progression resides.
Discussion
Studies in humans, NOD mice, and BioBreeding (BB) rats have demonstrated that the generation and function of islet-reactive T cells depends on MHC haplotype (Colle et al. 1981; Hattori et al. 1986; Todd et al. 1987; Wicker et al. 1987). Clearly, β islet–cell destruction requires the coinheritance of multiple susceptibility loci (reviewed in Cordell and Todd 1995). Unraveling how and when non-MHC Idd genes regulate IDDM pathogenesis is key to the development of therapies to modulate disease progression in individuals at high-risk. The success of identification of MHC class II as a major diabetes gene, as well as the remarkable sequence similarity between human DQβ and mouse I-Aβ susceptibility alleles, established a precedent that will not be easily fulfilled by other Idd genes. Unlike the high diabetes risk conferred by MHC class II alleles, it has been suggested that non-MHC genes contribute a small, incremental influence on disease susceptibility. Whole-genome scans in humans and in mice (Cordell and Todd 1995) have supported this idea, although the former have yielded disparate results in different study populations (Concannon et al. 1998; Mein et al. 1998). To gain greater insight into the genetic regulation of diabetes, it is critical to identify molecular and cellular events in pathogenic islet inflammation and to link these preclinical phenotypes to the genetic pathways that regulate IDDM susceptibility.
Previously, we defined an early step in disease pathogenesis, the transition from peri-insulitis, involving APC migration, to T cell–dependent invasive insulitis, which distinguishes NOD mice from the diabetes-resistant NOR strain (Fox and Danska 1998). Here, we report that NOD alleles at two loci, Idd5 and Idd13, cooperatively regulate both the recruitment/retention of T cells to islet lesions and the progression to invasive insulitis. Importantly, the absence of a NOD-derived allele at either locus results in complete resistance to invasive insulitis, despite the presence of NOD alleles at other known Idd loci, including the MHC (Idd1). Consistent with our results, both male and female NOD mice congenic for a BKS-derived chromosome 2 segment that includes the Idd13 interval defined here do not display invasive insulitis (Serreze et al. 1998), and (NOD×CBA) recombinant inbred animals homozygous for CBA alleles on chromosome 1 or 2 are insulitis resistant (Reifsnyder et al. 1999). Remarkably, human and rat chromosomal regions orthologous to murine Idd5 and -13 regions are the only identified non-MHC loci that are also linked to IDDM susceptibility in humans and rats. Thus, the Idd5 and -13 loci are likely to encode regulators of diabetes pathogenesis in all three species. Although it remains to be determined in what cell types (T cells, APCs, and/or islets) Idd5 and -13 exert their effects, our study provides a biological rationale to inform the search for candidate susceptibility genes at these loci.
Idd5 in Diabetes Pathogenesis
Idd5 was originally identified as a chromosome 1 locus that affects the timing of peri-insulitis onset (Garchon et al. 1991) and subsequently was shown to affect both the kinetics and the severity of insulitis (Ghosh et al. 1993). Multiple diabetes-susceptibility regions may map within the 40-cM Idd5 interval (Yui et al. 1996; Cordell et al. 1998). As displayed in table 3 and figure 5A, a chromosome segment containing murine Idd5 is orthologous to four human diabetes-susceptibility regions, including IDDM7 (Copeman et al. 1995; Esposito et al. 1998), IDDM12 (Owerbach and Gabbay 1995; Esposito et al. 1998), and IDDM13 (Morahan et al. 1996; Esposito et al. 1998), all on chromosome 2q, and IDDM6 on chromosome 18q (Merriman et al. 1997, 1998), as well as to the Iddm5 region of rat chromosome 13 (Martin et al. 1999). In the data presented here, T cell–dependent invasive insulitis maps to a 1-LOD confidence interval near D1Mit305, a position that likely excludes several candidate genes. Interleukin-1 receptor (Il1-r [Cornall et al. 1991; Garchon et al. 1991]) and the natural resistance–associated macrophage protein (Nramp-1) involved in the clearance of intracellular pathogens (Govoni and Gros 1998) both map to regions of chromosome 1 that are well outside the confidence interval for the insulitis phenotype (fig. 4A). Furthermore, the high-resolution map of chromosome 1 indicated that Bcl-2 and Ctla-4 are NOD-derived in the NOR strain (fig. 4A), excluding them as candidates for the T cell–dependent invasive insulitis phenotype. Bcl-2 was suggested as a candidate gene for the enhanced resistance of NOD T cells to apoptosis after interleukin-2 withdrawal (Garchon et al. 1994). Enhanced resistance to cyclophosphamide-induced death by NOD compared with B6 T cells was linked to a region of chromosome 1, and Ctla-4 was suggested as a candidate gene (Colucci et al. 1996, 1997). Allelic variants in Bcl-2 and Ctla-4 may contribute to diabetes susceptibility but apparently are not responsible for the phenotype described here. A number of genes without obvious links to diabetes pathogenesis map proximal to D1Mit305 (fig. 5A), including those for the nicotinic acetylcholine receptor, Acrg (Heidmann et al. 1986; Cohen-Haguenauer et al. 1989); collagen type VI, Col6a3 (Schurr et al. 1990); retinal S-antigen, Sag (Danciger et al. 1989); and the high-density lipoprotein-binding protein, Hdlbp (Xia et al. 1993; LeBoeuf et al. 1994). At Idd5, as at all other Idd regions, the mapping of expressed sequence tags and defined coding regions is far from complete, so it remains likely that these regions contain genes that have yet to be identified and may include novel genes involved in diabetes susceptibility.
Table 3.
Chromosomal Regions Orthologous to Murine Idd5 and Idd13 Contain Diabetes-Susceptibility Genes in the Rat and Human Genomes[Note]
| Mouse | Rat | Human | ||||
| Name (Chromosome) | Locations | Phenotype (References) | Name (Chromosome) | Location (References) | Name | Location (References) |
| Idd5 (1) | Bcl2- D1Nds1 | Peri-insulitis (Garchon et al. 1991) | Iddm5 (13) | D13Mit1 diabetes progession (Martin et al. 1999) | IDDM7 | Chromosome 2q (Coperman et al. 1995; Esposito et al. 1998) |
| Idd5 (1) | D1Nds4 | Spontaneous and cyclophos-accelerated diabetes (Cornall et al. 1991) | IDDM12 | Chromosome 2q31 (Owerbach and Gabby 1995; Esposito et al. 1998) | ||
| Idd5 (1) | D1Mit5 | Insulitis kinetics and severity (Ghosh et al. 1993), insulitis susceptibility (Yui et al. 1996) | IDDM13 | Chromosome 2q D2S164 (Morahan et al. 1996; Esposito et al. 1998) | ||
| Idd5 (1) | D1Mit4–Bcl2, D1Mit5–Mit15, D1Mit305 | T cell recruitment and insulitis progression, interaction with Idd13 (present study) | IDDM6 | Chromosome 18q21 (Merriman et al. 1997, 1998) | ||
| Idd13 (2) | D2Mit490–Mit144, H3a-IL-1, IL-1-Pcna | IDDM (Serezze et al. 1998) | Iddm6 (3) | D3Mgh10, diabetes; D3Mit4, insulitis severity, interaction with Iddm5 (Martin et al. 1999) | IDDM3 | Chromosome 15q26 D15S107 (Field et al. 1994; Zuberi et al. 1996) |
| Idd13 (2) | D2Mit395 | T cell recruitment and insulitis progression, interaction with Idd5 (present study) | ||||
|
Mouse |
Rat |
Human |
||||||
| Name (Chromosome) | Locations | Phenotype (References) | Name (Chromosome) | Location (References) | Name | Location (References) | ||
| Idd5 (1) | Bcl2- D1Nds1 | Peri-insulitis (Garchon et al. 1991) | Iddm5 (13) | D13Mit1 diabetes progession (Martin et al. 1999) | IDDM7 | Chromosome 2q (Coperman et al. 1995; Esposito et al. 1998) | ||
| Idd5 (1) | D1Nds4 | Spontaneous and cyclophos-accelerated diabetes (Cornall et al. 1991) | IDDM12 | Chromosome 2q31 (Owerbach and Gabby 1995; Esposito et al. 1998) | ||||
| Idd5 (1) | D1Mit5 | Insulitis kinetics and severity (Ghosh et al. 1993), insulitis susceptibility (Yui et al. 1996) | IDDM13 | Chromosome 2q D2S164 (Morahan et al. 1996; Esposito et al. 1998) | ||||
| Idd5 (1) | D1Mit4–Bcl2, D1Mit5–Mit15, D1Mit305 | T cell recruitment and insulitis progression, interaction with Idd13 (present study) | IDDM6 | Chromosome 18q21 (Merriman et al. 1997, 1998) | ||||
| Idd13 (2) | D2Mit490–Mit144, H3a-IL-1, IL-1-Pcna | IDDM (Serezze et al. 1998) | Iddm6 (3) | D3Mgh10, diabetes; D3Mit4, insulitis severity, interaction with Iddm5 (Martin et al. 1999) | IDDM3 | Chromosome 15q26 D15S107 (Field et al. 1994; Zuberi et al. 1996) | ||
| Idd13 (2) | D2Mit395 | T cell recruitment and insulitis progression, interaction with Idd5 (present study) | ||||||
Note.— Mouse, human, and rat homology maps (NCBI Human/Mouse Homology Relationships) establish that regions of the human and rat genomes previously found to be linked to type 1 diabetes risk, are orthologous to the murine Idd5 and Idd13 intervals identified in this and prior studies of the NOD mouse. For Idd5 on chromsome 1, multiple diabetes-susceptibility regions were identified over a broad region of human chromosome 2q and on human 18q. Interestingly, the region of the rat genome orthologous to murine Idd13 on chromosome 2 (55–60 cM), is Iddm6, which demonstrates genetic interaction with Iddm5 on rat chromosome 13. The latter region is orthologous to murine chromosome 1 (56–62 cM) which overlaps the 1-LOD confidence interval for the Idd5 linkage to insulitis progression in this study.
Idd13 in Diabetes Pathogenesis
The second locus to which we mapped T cell–dependent invasive insulitis is D2Mit395 in the Idd13 locus. Idd13 originally was mapped to a 4-cM region of chromosome 2, flanked by the genes for beta-2-microglobulin (β2m) and proliferating-cell nuclear antigen (Pcna; Serreze et al. 1994), although recent evidence suggests that it is ⩾30 cM in length (Serreze et al. 1998). It is likely that the linkage reported here for the insulitis progression lies within the previously published Idd13 interval (Serreze et al. 1998), although discrepancies between current chromosome 2 maps (Whitehead Institute for Biomedical Research/MIT Center for Genome Research, Jackson Laboratory [Siracusa et al. 1996, 1997]) remain to be clarified. Importantly, murine Idd13 is orthologous to the Iddm6 region of rat chromosome 3 (Martin et al. 1999) and may be orthologous to a human chromosome 15 segment that contains the IDDM3 locus (Field et al. 1994; Zamani et al. 1996), but there is some disagreement on this point (Mein et al. 1998; table 3). Of particular interest, insulitis severity in the BB rat model was shown to be influenced by genetic interaction between the Iddm6 locus and the Iddm5 locus (Martin et al. 1999), a result consistent with our findings of strong interaction between the syntenic murine Idd13 and Idd5 loci, respectively. In all three species, this chromosome region includes β2m, a subunit of the MHC class I complex. In multiple studies, allelic variation between NOD and NOR mice has been reported to affect both the structure and the expression of MHC class I. For example, the interval containing the NOD-derived Idd13 encodes a β2m variant that affects an MHC class I serological determinant (Serreze et al. 1998). In addition, interferon-γ–mediated up-regulation of cell-surface MHC expression is reportedly absent in cultured NOD macrophages (Leiter and Serreze 1992), although the Idd13 locus does not appear to control this effect (Serreze et al. 1998). Allelic variation in the β2m sequence also exists between normal mouse strains (Michaelson 1983), so it remains unresolved how functional variants in this protein may contribute to autoimmune pathogenesis. Additional genes within the confidence interval computed from our analysis include a cluster encoding minor histocompatibility antigens (Graff et al. 1994), recombinase A (Reca [Takahasi et al. 1994; Roca and Cox 1997]), lymphocyte tyrosine kinase (Ltk [Snijders et al. 1997]), and receptor tyrosine kinase (Tyro3 [Liao et al. 1996]) involved in bone resorption (Nakamura et al. 1998). The proinflammatory cytokine interleukin-1 (Il1 [Siracusa et al. 1996, 1997]) also has been considered a candidate gene for mouse Idd13 and for human IDDM13 on chromosome 2q (Esposito et al. 1998). Considerable work remains to isolate and analyze the function of gene(s), at the Idd13 and Idd5 loci, that regulate the progression from benign to invasive insulitis.
Conclusions
Rodent models of polygenic disease provide a unique opportunity to coordinate genetic analyses with the study of cellular pathogenesis. By comparing NOD and NOR mice, we have identified a discrete step in insulitis progression that is conferred by two susceptibility loci and, by inference, two (sets of) genes that are common to IDDM-susceptible humans and rodents. NOR mice inherit genes from the NOD background that promote the peri-islet and perivascular infiltration by APCs, the earliest detectable stage in diabetes pathogenesis. However, this stage is apparently benign, since these lesions contain few T cells (Fox and Danska 1998), and NOR animals rarely progress to overt diabetes. Here we have shown that NOD alleles at Idd5 and Idd13 together regulate T cell influx into islet lesions, which is an obligate step in autoimmune pathogenesis and evident months before overt disease onset. Our identification of punctuated steps in the insulitis process suggested a “simpler” preclinical phenotype that enabled genetic linkage to two of many susceptibility loci and insight into the cellular biology that these loci control. Genetic analysis of other polygenic diseases underscores the utility of this strategy. In (NZB×NZW) F1 mice, B cell phenotypes that are markers of the autoimmune events in systemic lupus erythematosus have greatly assisted in the identification of susceptibility loci (Vyse et al. 1996; Mohan et al. 1997). In type 2 diabetes, physiological and genetic studies in rat models has allowed linkage of multiple loci with specific facets of the disease, including impaired insulin secretion, increased body-mass index, and elevated blood-glucose levels (Galli et al. 1996; Gauguier et al. 1996). The genetic variants that confer diabetes risk in rats or mice may well differ from those in humans, and, similarly, different constellations of genes may mediate diabetes in genetically diverse human populations. Although these genes may differ between species and populations, they are likely to act on common biological pathways that control islet inflammation and β cell destruction. Importantly, the identification of rodent genes within these shared pathways has promise for resolving the molecular basis of diabetes pathogenesis, thereby illuminating targets for therapeutic intervention in high-risk or newly diagnosed individuals.
Acknowledgments
We thank Drs. C. Guidos and R. McInnes for critical comments on the manuscript and Dr. J. Rommens and members of the Danska and Guidos labs for valuable discussions. These studies were supported by grants from the Canadian Diabetes Association and the Juvenile Diabetes Foundation International (to J.S.D.). J.S.D. is a Research Scientist of the National Cancer Institute of Canada. C.J.F. was supported by the Hospital for Sick Children Research Training Center and the Novo-Nordisk Studentship/Banting and Best Diabetes Center. A.D.P. is a Fellow of the Medical Research Council of Canada.
Electronic-Database Information
The accession number and URLs for data in this article are as follows:
- Genome Center Software Information and Documentation, http://waldo.wi.mit.edu/genome_software
- Jackson Laboratory, http://www.jax.org
- NCBI Human/Mouse Homology Relationships, http://www.ncbi.nlm.gov/Homology
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for IDDM1 [MIM 222100]
- Research Genetics, http://www.resgen.com
- UK Human Genome Mapping Project Resource Centre, http://www.hgmp.mrc.ac.uk
- Whitehead Institute for Biomedical Research/MIT Center for Genome Research, http://www-genome.wi.mit.edu
References
- Acha-Orbea H, McDevitt HO (1987) The first external domain of the non-obese diabetic mouse class II I-Ab chain is unique. Proc Natl Acad Sci USA 84:2435–2439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett ST, Lucassen AM, Gough SC, Powell EE, Undlien DE, Pritchard LE, Merriman ME, et al (1995) Susceptibility to human type 1 diabetes at IDDM2 is determined by tandem repeat variation at the insulin gene minisatellite locus. Nat Genet 9:284–292 [DOI] [PubMed] [Google Scholar]
- Bennett ST, Todd JA (1996) Human type 1 diabetes and the insulin gene: principles of mapping polygenes. Annu Rev Genet 30:343–370 [DOI] [PubMed] [Google Scholar]
- Bonnevie-Nielsen V, Skovgaard LT, Lernmark A (1983) β-Cell function relative to islet volume and hormone content in the isolated perfused mouse pancreas. Endocrinology 112:1049–1056 [DOI] [PubMed] [Google Scholar]
- Caillat-Zucman S, Daniel S, Djilali-Saiah I, Timsit J, Garchon HJ, Boitard C, Bach JF (1995) Family study of linkage disequilibrium between TAP2 transporter and HLA class II genes: absence of TAP2 contribution to association with insulin-dependent diabetes mellitus. Hum Immunol 44:80–87 [DOI] [PubMed] [Google Scholar]
- Castano L, Eisenbarth GS (1990) Type-I diabetes: a chronic autoimmune disease of human, mouse, and rat. Annu Rev Immunol 8:647–679 [DOI] [PubMed] [Google Scholar]
- Charlton B, Bacelj A, Mandel TE (1988) Administration of silica particles or anti-Lyt2 antibody prevents β-cell destruction in NOD mice given cyclophosphamide. Diabetes 37:930–935 [DOI] [PubMed] [Google Scholar]
- Chatenoud L, Primo J, Bach J-F (1997) CD3 antibody–induced dominant self tolerance in overtly diabetic NOD mice. J Immunol 158:2947–2954 [PubMed] [Google Scholar]
- Chatenoud L, Thervet E, Primo J, Bach J-F (1994) Anti-CD3 antibody induces long-term remission of overt autoimmunity in nonobese diabetic mice. Proc Natl Acad Sci USA 91:123–127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen-Haguenauer O, Barton PJ, Buonanno A, Cong NV, Masset M, De Tand MF, Merlie J, et al (1989) Localization of the acetylcholine receptor gamma subunit gene to human chromosome 2q32-pter. Cytogenet Cell Genet 52:124–127 [DOI] [PubMed] [Google Scholar]
- Colle E, Guttman RD, Seemayer TA (1981) Spontaneous diabetes mellitus in the rat. I: association with the major histocompatibility complex. J Exp Med 154:1237–1242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colucci F, Bergman M-F, Penha-Goncalves C, Culjo CM, Holmberg D (1997) Apoptosis resistance of nonobese diabetic peripheral lymphocytes linked to the Idd5 diabetes susceptibility region. Proc Natl Acad Sci USA 94:8670–8674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colucci G, Cilio CM, Leijon K, Penha-Goncalves C, Bergman M-L, Holmberg D (1996) Programmed cell death in the pathogenesis of murine IDDM: resistance to apoptosis induced in lymphocytes by cyclophosphamide. Proc Natl Acad Sci USA 9:217–276 [DOI] [PubMed] [Google Scholar]
- Concannon P, Gogolin-Ewens KJ, Hinds DA, Wapelhorst B, Morrison VA, Stirling B, Mitra M, et al (1998) A second-generation screen of the human genome for susceptibility to insulin-dependent diabetes mellitus. Nat Genet 19:292–296 [DOI] [PubMed] [Google Scholar]
- Copeman JB, Cucca F, Hearne CM, Cornall RJ, Reed PW, Ronnigen KS, Undlien DE, et al (1995) Linkage disequilibrium mapping of a type 1 diabetes suceptibility gene (IDDM7) to chromosome 2q31-q33. Nat Genet 9:80–85 [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Todd JA (1995) Multifactorial inheritance in type 1 diabetes. Trends Genet 11:499–504 [DOI] [PubMed] [Google Scholar]
- Cordell HJ, Todd JA, Lathrop GM (1998) Mapping multiple linked quantitative trait loci in non-obese diabetic mice using a stepwise regression strategy. Genet Res 71:51–64 [DOI] [PubMed] [Google Scholar]
- Cornall RJ, Prins J, Todd J, Pressey A, DeLarato NH, Wicker LS, Peterson LB (1991) Type 1 diabetes in mice is linked to the interleukin-1 receptor and Lsh/lty/Bcg genes on chromosome 1. Nature 353:262–265 [DOI] [PubMed] [Google Scholar]
- Danciger M, Kozak CA, Tsuda M, Shinohara T, Farber DB (1989) The gene for retinal S-antigen (48-kDa protein) maps to the centromeric portion of mouse chromosome 1 near Idh-1. Genomics 5:378–381 [DOI] [PubMed] [Google Scholar]
- Davies JL, Kawaguchi Y, Bennett ST, Copeman JB, Cordell HJ, Pritchard LE, Reed PW, et al (1994) A genome-wide search for human type 1 diabetes susceptibility genes. Nature 371:130–136 [DOI] [PubMed] [Google Scholar]
- Deng GY, Muir A, Maclaren NK, She JX (1995) Association of LMP2 and LMP7 genes within the major histocompatibility complex with insulin-dependent diabetes mellitus: population and family studies. Am J Hum Genet 56:528–534 [PMC free article] [PubMed] [Google Scholar]
- Erlich HA, Rotter JI, Chang JD, Shaw SJ, Raffel LJ, Klitz W, Bugawan TL, et al (1996) Association of HLA-DPB1*0301 with IDDM in Mexican-Americans. Diabetes 45:610–614 [DOI] [PubMed] [Google Scholar]
- Esposito L, Hill NJ, Pritchard LE, Cucca F, Muxworthy C, Merriman ME, Wilson A, et al (1998) Genetic analysis of chromosome 2 in type 1 diabetes: analysis of putative loci IDDM7,IDDM12, and IDDM13 and candidate genes NRAMP-1 and IA-2 and the interleukin-1 gene cluster. Diabetes 47:1797–1799 [DOI] [PubMed] [Google Scholar]
- Field LL, Tobias R, Magnus T (1994) A locus on chromosome 15q26 (IDDM3) produces susceptibility to insulin-dependent diabetes mellitus. Nat Genet 8:189–194 [DOI] [PubMed] [Google Scholar]
- Fox CJ, Danska JS (1997) Interleukin-4 expression at the onset of islet inflammation predicts non-destructive inflammation in NOD mice. J Immunol 158:2414–2424 [PubMed] [Google Scholar]
- ——— (1998) Independent genetic regulation of T cell and antigen presenting cell participation in autoimmune islet inflammation. Diabetes 47:331–338 [DOI] [PubMed] [Google Scholar]
- Frankel WN, Schork NJ (1996) Who's afraid of epistasis? Nat Genet 14:371–373 [DOI] [PubMed] [Google Scholar]
- Galli J, Li LS, Glaser A, Ostenson CG, Jiao H, Fakhrai-Rad H, Jacob HJ, et al (1996) Genetic analysis of non-insulin dependent diabetes mellitus in the GK rat. Nat Genet 12:31–37 [DOI] [PubMed] [Google Scholar]
- Garchon H, Bedossa P, Eloy L, Bach J-F (1991) Identification and mapping to chromosome 1 of a susceptibility locus for peri-insulitis in non-obese diabetic mice. Nature 353:260–262 [DOI] [PubMed] [Google Scholar]
- Garchon HJ, Luan JJ, Eloy L, Bedossa P, Bach JF (1994) Genetic analysis of immune dysfunction in non-obese diabetic (NOD) mice: mapping of a susceptibility locus close to the Bcl-2 gene correlates with increased reistance of NOD T cells to apoptosis induction. Eur J Immunol 24:380–384 [DOI] [PubMed] [Google Scholar]
- Gauguier D, Froguel P, Parent V, Bernard C, Bihoreau MT, Portha B, James MR, et al (1996) Chromosomal mapping of genetic loci associated with non-insulin dependent diabetes in the GK rat. Nature genet 12:38–43 [DOI] [PubMed] [Google Scholar]
- Ghosh S, Palmer S, Rodrigues N, Cordell H, Hearne C, Cornall R, Prins J-B, et al (1993) Polygenic control of autoimmune diabetes in non-obese diabetic mice. Nat Genet 4:404–409 [DOI] [PubMed] [Google Scholar]
- Govoni G, Gros P (1998) Macrophage NRAMP1 and its role in resistance to microbial infections. Inflamm Res 47:277–284 [DOI] [PubMed] [Google Scholar]
- Graff RJ, Hauptfeld V, Riordan K, Kurtz M (1994) Continued mapping of chromosome 2 genes. Immunogenetics 40:21–26 [DOI] [PubMed] [Google Scholar]
- Hanninen A, Jalkanen S, Salmi M, Toikkanen S, Nikolakaros G, Simell O (1992) Macrophages, T cell receptor usage, and endothelial cell activation in the pancreas at the onset of insulin-dependent diabetes mellitus. J Clin Invest 90:1901–1910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hattori M, Buse JB, Jackson RA, Glimcher L, Dorf ME, Minami M, Makino S, et al (1986) The NOD mouse: recessive diabetogenic gene in the major histocompatibility complex. Science 231:733–735 [DOI] [PubMed] [Google Scholar]
- Hattori M, Yamato E, Itoh N, Senpuku H, Fujisawa T, Yoshino M, Fukuda M, et al (1999) Cutting edge: homologous recombination of the MHC class I K region defines new MHC-linked diabetogenic susceptibility gene(s) in nonobese diabetic mice. J Immunol 163:1721–1724 [PubMed] [Google Scholar]
- Heidmann O, Buonanno A, Geoffroy B, Robert B, Guenet JL, Merlie JP, Changeux JP (1986) Chromosomal localization of muscle nicotinic acetylcholine receptor genes in the mouse. Science 234:866–888 [DOI] [PubMed] [Google Scholar]
- Ikegami H, Makino S, Yamoto E, Kawaguchi Y, Ueda H, Sakamoto T, Takekawa K, et al (1995) Identification of a new susceptibility locus for insulin-dependent diabetes mellitus by ancestral haplotype congenic mapping. J Clin Invest 96:1936–1942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh N, Hanafusa T, Miyazaki A, Miyagawa J, Yamagata K, Yamamote K, Waguri M, et al (1993) Mononuclear cell infiltration and its relation to the expression of major histocompatibility complex antigens and adhesion molecules in pancreas biopsy specimens from newly diagnosed insulin-dependent diabetes mellitus patients. J Clin Invest 92:2313–2322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen A, Homo-Delarche F, Hooijkaas H, Leenen PJ, Dardenne M, Drexhage HA (1994) Immunohistochemical characterization of monocytes-macrophages and dendritic cells involved in the initiation of insulitis and b-cell destruction in NOD mice. Diabetes 43:667–675 [DOI] [PubMed] [Google Scholar]
- Jarpe AM, Hickman J, Anderson J, Winter W, Peck A (1990–91) Flow cytometric enumeration of mononuclear cell populations infiltrating the islets of Langerhans in prediabetic NOD mice: development of a model of autoimmune insulitis for type I diabetes. Reg Immunol 3:305–317 [PubMed] [Google Scholar]
- Lander ES, Green P, Abrahamson J, Barlow A, Daly MJ, Lincoln SE, Newburg L (1987) MAPMAKER: an interactive computer package for constructing primary genetic linkage maps of experimetnal and natural populations. Genomics 1:174–181 [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L (1995) Genetic dissection of complex traits: guideline for interpreting and reporting linkage results. Nat Genet 11:241–247 [DOI] [PubMed] [Google Scholar]
- LeBoeuf RC, Xia YR, Oram JF, Lusis AJ (1994) Mapping of the gene for high-density lipoprotein binding protein (Hdlbp) to proximal mouse chromosome 1. Genomics 23:296–298 [DOI] [PubMed] [Google Scholar]
- Leiter EH, Serreze DV (1992) Antigen presenting cells and the immunogenetics of autoimmune diabetes in NOD mice. Reg Immunol 4:263–273 [PubMed] [Google Scholar]
- Leo O, Foo M, Sachs DH, Samelson LE, Bluestone JA (1987) Identification of a monoclonal antibody specific for a murine T3 polypeptide. Proc Natl Acad Sci USA 84:1374–378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liao X, Zhou R, Gilbert DJ, Copeland NG, Jenkins NA (1996) Receptor tyrosine kinase gene Tyro3 maps to mouse chromosome 2, closely linked to Ltk. Mamm Genome 7:395–396 [DOI] [PubMed] [Google Scholar]
- Makino S, Kunimoto K, Muraoka Y, Mizushima Y, Katagiri K, Tochino Y (1980) Breeding of a non-obese, diabetic strain of mice. Jikken Dobotsu 29:1–13 [DOI] [PubMed] [Google Scholar]
- Martin A-M, Blankenhorn EP, Maxson MN, Zhao M, Leif J, Mordes JP, Greiner DL (1999) Non–major histocompatibility complex–linked diabetes susceptibility loci on chromosomes 4 and 13 in a backcross of the DP-BB/Wor rat to the WF rat. Diabetes 48:50–58 [DOI] [PubMed] [Google Scholar]
- McAleer MA, Reifsnyder P, Palmer SM, Prochazka M, Love JM, Copeman JB, Powell EE, et al (1995) Crosses of NOD mice with the related NON strain: a polygenic model for IDDM. Diabetes 44:1186–1195 [DOI] [PubMed] [Google Scholar]
- Mein CA, Esposito L, Dunn MG, Johnson GCL, Timms AE, Goy JV, Smith AN, et al (1998) A search for type 1 diabetes susceptibility genes in families from the United Kingdom. Nat Genet 19:297–300 [DOI] [PubMed] [Google Scholar]
- Merriman TR, Eaves IA, Twells RC, Merriman ME, Danoy PA, Muxworthy CE, Hunter KM, et al (1998) Transmission of haplotypes of microsatellite markers rather than single marker alleles in the mapping of a putative type 1 diabetes susceptibility gene (IDDM6). Hum Mol Genet 7:517–524 [DOI] [PubMed] [Google Scholar]
- Merriman T, Twells R, Merriman M, Eaves I, Cox R, Cucca F, McKinney P, et al (1997) Evidence by allelic association-dependent methods for a type 1 diabetes polygene (IDDM6) on chromosome 18q21. Hum Mol Genet 6:1003–1010 [DOI] [PubMed] [Google Scholar]
- Michaelson J (1983) Genetics of beta-2 microglobulin in the mouse. Immunogenetics 17:219–260 [DOI] [PubMed] [Google Scholar]
- Mohan C, Morel L, Yang P, Wakeland EK (1997) Genetic dissection of systemic lupus erythematosus pathogensis. J Immunol 159:454–465 [PubMed] [Google Scholar]
- Morahan G, Huang D, Tait BD, Colman PG, Harrison LC (1996) Markers on distal chromosome 2q linked to insulin-dependent diabetes mellitus. Science 272:1811–1813 [DOI] [PubMed] [Google Scholar]
- Naggert JK, Mu ML, Frankel W, Bailey DW, Paigen B (1995) Genomic analysis of the C57BL/Ks mouse strain. Mamm Genome 6:131–133 [DOI] [PubMed] [Google Scholar]
- Nakamura YS, Hakeda Y, Takahura N, Kameda T, Hamaguchi I, Miyamoto T, Kakudo S, et al (1998) Tyro 3 receptor tyrosine kinase and its ligand, Gas6, stimulate function of osteoclasts. Stem Cells 16:229–238 [DOI] [PubMed] [Google Scholar]
- Noble JA, Cavalli AS, Erlich HA (1996a) DPB1*5901a: a novel HLA-DPB1 allele from a Caucasian family with insulin-dependent diabetes mellitus. Tissue Antigens 47:159–162 [DOI] [PubMed] [Google Scholar]
- Noble JA, Valdes AM, Cook M, Klitz W, Thomson G, Erlich HA (1996b) The role of HLA class II genes in insulin-dependent diabetes mellitus: molecular analysis of 180 Caucasian, multiplex families. Am J Hum Genet 59:1134–1148 [PMC free article] [PubMed] [Google Scholar]
- Noble JA, Valdes AM, Thomson G, Erlich HA (2000) The HLA class II locus DPB1 can influence susceptibility to type 1 diabetes. Diabetes 49:121–125 [DOI] [PubMed] [Google Scholar]
- Oi VT, Jones PP, Goding JW, Hertzenberg LA, Hertzenberg LA (1978) Properties of monoclonal antibodies to mouse Ig allotypes, H-2, and Ia antigens. Curr Top Microbiol Immunol 81:115–120 [DOI] [PubMed] [Google Scholar]
- Owerbach D, Gabbay KH (1995) The HOXD8 locus (2q31) is linked to type I diabetes: interaction with chromosome 6 and 11 disease susceptibility genes. Diabetes 44:132–136 [DOI] [PubMed] [Google Scholar]
- Podolin PL, Denny P, Armitage N, Lord CJ, Hill NJ, Levy HR, Peterson LB, et al (1998) Localization of two insulin-dependent diabetes (Idd) genes to the Idd10 region on mouse chromosome 3. Mamm Genome 9:283–286 [DOI] [PubMed] [Google Scholar]
- Podolin PL, Denny P, Lord CJ, Hill NJ, Todd JA, Peterson LB, Wicker LS, et al (1997) Congenic mapping of the insulin-dependent diabetes (Idd) gene, Idd10, localizes two genes mediating the Idd10 effect and eliminates the candidate Fcgr1. J Immunol 159:1835–1843 [PubMed] [Google Scholar]
- Pozzilli P, Signore A, Williams AJK, Beales PE (1993) NOD colonies around the world: recent facts and figures. Immunol Today 14:193–196 [DOI] [PubMed] [Google Scholar]
- Prochazka M, Serreze DV, Frankel WN, Leiter EH (1992) NOR/Lt Mice: MHC-matched diabetes resistant control strain for NOD mice. Diabetes 41:98–106 [DOI] [PubMed] [Google Scholar]
- Reifsnyder PC, Plynne JC, Gavin AL, Simone EA, Sharp JJ, Herberg L, Leiter EH (1999) Genotypic and phenotypic characterization of six new recombinant congenic strains derived from NOD/Shi and CBA/J genomes. Mamm Genome 10:161–167 [DOI] [PubMed] [Google Scholar]
- Roca AI, Cox MM (1997) RecA protein: structure, function, and role in recombinational DNA repair. Prog Nucleic Acid Res Mol Biol 56:129–133 [DOI] [PubMed] [Google Scholar]
- Schurr E, Skamene E, Morgan K, Chu ML, Gros P (1990) Mapping of Col3a1 and Col6a3 to proximal murine chromosome 1 identifies conserved linkage of structural protein genes between murine chromosome 1 and human chromosome 2q. Genomics 8:477–486 [DOI] [PubMed] [Google Scholar]
- Seldin MF (1996) Mouse chromosome 1. Mamm Genome 6:S28–S50 [PubMed] [Google Scholar]
- ——— (1997) Mouse chromosome 1. Mamm Genome 7:S3–S27 [DOI] [PubMed] [Google Scholar]
- Sempe P, Bedossa P, Richard M-F, Villa M-C, Bach J-F, Boitard C (1991) Anti-α/β T cell receptor monoclonal antibody provides an efficient therapy for autoimmune diabetes in nonobese (NOD) mice. Eur J Immunol 21:1163–1169 [DOI] [PubMed] [Google Scholar]
- Serreze DV, Bridgett M, Chapman HD, Chen E, Richard SD, Leiter EH (1998) Subcongenic analysis of the Idd13 locus in NOD/Lt Mice: evidence for several susceptibility genes including a possible diabetogenic role for b2-microglobulin. J Immunol 160:1472–1478 [PubMed] [Google Scholar]
- Serreze DV, Prochazka M, Reifsnyder PC, Bridgett MM, Leiter EH (1994) Use of recombinant congenic and congenic strains of NOD mice to identify a new insulin-dependent diabetes resistance gene. J Exp Med 180:1553–1558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehy MJ, Scharf SJ, Rowe JR, Neme de Gimenez MH, Meske LM, Erlich HA, Nepom BS (1989) A diabetes-susceptible HLA haplotype is best defined by a combination of HLA-DR and -DQ alleles. J Clin Invest 83:830–835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Signore A, Pozzilli P, Gale EA, Andreani D, Beverley PC (1989) The natural history of lymphocyte subsets infiltrating the pancreas of NOD mice. Diabetologia 32:282–289 [DOI] [PubMed] [Google Scholar]
- Signore A, Procaccini E, Toscano AM, Ferretti E, Williams AJK, Beales PE, Cugini P, et al (1994) Histological study of pancreatic beta-cell loss in relation to the insulitis process in the non-obese diabetic mouse. Histochemistry 101:263–269 [DOI] [PubMed] [Google Scholar]
- Siracusa LD, Abbott CM, Morgan JL, Zuberi AR, Pomp D, Peters J (1997) Mouse chromosome 2. Mamm Genome 7:S28–S44 [DOI] [PubMed] [Google Scholar]
- Siracusa L, Morgan JL, Fisher JK, Abbott CM, Peters J (1996) Mouse chromosome 2. Mamm Genome 6:S51–S63 [PubMed] [Google Scholar]
- Snijders AJ, Ho SC, Haase VH, Pillai S, Bernards A (1997) A lymphocyte-specific Ltk tyrosine kinase isoform is retained in the endoplasmic reticulum in association with calnexin. J Biol Chem 272:1297–1301 [DOI] [PubMed] [Google Scholar]
- Stein PH, Rees MA, Singer A (1992) Reconstitution of (BALB/c X B6)F1 normal mice with stem cells and thymus from non-obese diabetic mice results in autoimmune insulitis of the normal hosts' pancreases. Transplantation 53:1347–1352 [DOI] [PubMed] [Google Scholar]
- Takahasi E, Matsuda Y, Hori T, Yasuda N, Tsuji S, Mori M, Yoshimura Y, et al (1994) Chromosome mapping of the human (RECA) and mouse (Reca) homologs of the yeast RAD51 and Escherichia coli recA genes to human (15q15.1) and mouse (2F1) chromosomes by direct R-banding fluorescence in situ hybridization. Genomics 19:376–378 [DOI] [PubMed] [Google Scholar]
- Todd JA, Aitman TJ, Cornall RJ, Ghosh S, Hall JRS, Hearne CM, Knight AM, et al (1991) Genetic analysis of autoimmune type 1 diabetes mellitus in mice. Nature 351:542–547 [DOI] [PubMed] [Google Scholar]
- Todd JA, Bell JI, McDevitt HO (1987) HLA-DQβ gene contributes to susceptibility and resistance to insulin-dependent diabetes mellitus. Nature 329:599–604 [DOI] [PubMed] [Google Scholar]
- Toyoda H, Formby B (1998) Contribution of T cells to the development of autoimmune diabetes in the NOD mouse model. Bioessays 20:750–757 [DOI] [PubMed] [Google Scholar]
- Vafiadis P, Bennett ST, Todd JA, Nadeau J, Grabs R, Goodyer CG, Wickramasinghe S, et al (1997) Insulin expression in human thymus is modulated by INS VNTR allelels at the IDDM2 locus. Nat Genet 15:289–292 [DOI] [PubMed] [Google Scholar]
- Vyse TJ, Morel L, Tanner FJ, Wakeland E, Kotzin BL (1996) Backcross analysis of genes linked to antibody production in New Zealand white mice. J Immunol 157:2719–2727 [PubMed] [Google Scholar]
- Vyse TJ, Todd JA (1996) Genetic analysis of autoimmune disease. Cell 85:311–318 [DOI] [PubMed] [Google Scholar]
- Waters SH, O'Neill JJ, Melican DT, Appel MC (1992) Multiple TCR Vb usage by infiltrates of young NOD mouse islets of Langerhans. Diabetes 41:308–312 [DOI] [PubMed] [Google Scholar]
- Wicker LS, Appel MC, Dotta F, Pressey A, Miller BJ, DeLarato NH, Fisher PA, et al (1992) Autoimmune syndromes in major histocompatibility complex (MHC) congenic strains of non-obese diabetic (NOD) mice. The NOD MHC is dominant for insulitis and cyclophosphamide-induced diabetes. J Exp Med 176:67–77 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker LS, Miller BJ, Coker LZ, McNally SE, Scott S, Mullen Y, Appel MC (1987) Genetic control of diabetes and insulitis in the non-obese diabetic (NOD) mouse. J Exp Med 165:1639–1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wicker LS, Todd JA, Peterson LB (1995) Genetic control of autoimmune diabetes in the NOD mouse. Annu Rev Immunol 13:179–200 [DOI] [PubMed] [Google Scholar]
- Xia YR, Klisak I, Sparkes RS, Oram J, Lusis AJ (1993) Localization of the gene for high-density lipoprotein binding protein (HDLBP) to human chromosome 2q37. Genomics 16:524–525 [DOI] [PubMed] [Google Scholar]
- Yui MA, Muralidharan K, Moreno-Altamirano B, Perrin G, Chestnut K, Wakeland EK (1996) Production of congenic mouse strains carrying NOD-derived diabetogenic genetic intervals: an approach for the genetic dissections of complex traits. Mamm Genome 7:331–334 [DOI] [PubMed] [Google Scholar]
- Zamani M, Posciot F, Raeymaekers P, Nerup J, Cassiman JJ (1996) Linkage of type I diabetes to 15q26 (IDDM3) in the Danish population. Hum Genet 98:491–496 [DOI] [PubMed] [Google Scholar]