Abstract
A Klebsiella pneumoniae clinical isolate was resistant to cefoxitin, cefotaxime, ceftazidime, ceftazidime-clavulanate, piperacillin-tazobactam (MICs, >256 μg/ml in all cases), and meropenem (MIC, 16 μg/ml) and was intermediate to imipenem (MIC, 8 μg/ml). Decreased expression of the OmpK36 porin and expression of an SHV-2 β-lactamase contributed to the observed resistance to these β-lactam-containing agents.
Expanded-spectrum cephalosporins play an important role in the treatment of infections due to Klebsiella pneumoniae. However, the presence of a carbapenemase or the expression of AmpC or SHV-2 β-lactamases combined with decreased outer membrane permeability provide resistance to these agents (3, 7, 10).
In this report, we investigate the resistance mechanisms in a K. pneumoniae clinical isolate resistant to oxyiminocephalosporins, α-methoxycephalosporins, and carbapenems. K. pneumoniae 103624 was isolated from blood cultures of a 60-year-old male with pneumonia. He was neutropenic following chemotherapy for Hodgkin's lymphoma, developed pneumonia, and was treated empirically with ceftazidime. After receipt of antibiotic susceptibility test results, treatment was changed to a combination of imipenem, gentamicin, and ciprofloxacin. Following 2 weeks of treatment, all signs of pneumonia had resolved. The patient had received 7 days of treatment with piperacillin-tazobactam and ceftazidime 6 weeks prior to this episode. The susceptibility of K. pneumoniae 103624 to antimicrobials (Table 1) indicated resistance to α-methoxycephalosporins (cefoxitin); oxyiminocephalosporins (cefotaxime and ceftazidime); β-lactam-β-lactamase inhibitor combinations such as ceftazidime-clavulanate, piperacillin-tazobactam (MICs, >256 μg/ml in all cases), and amoxicillin-clavulanate (MICs, 64 and 32 μg/ml, respectively); and meropenem (MIC, 16 μg/ml), and intermediate status for imipenem (MIC, 8 μg/ml). However, the strain was susceptible to all of the non-β-lactam compounds tested, such as gentamicin, amikacin, tobramycin, ciprofloxacin, and tetracycline.
TABLE 1.
MICs of β-lactam antibiotics for strains of E. coli and K. pneumoniae
| Strain | Plasmid(s)a | Porinb | MIC (μg/ml)c
|
||||||
|---|---|---|---|---|---|---|---|---|---|
| FOX | CTX | CAZ | C-Cd | P-Te | MP | IP | |||
| E. coli K-12 14R525 | NDf | ND | ≤0.5 | ≤0.5 | ≤0.5 | ≤0.125 | 0.5 | ≤0.06 | ≤0.125 |
| E. coli K-12 14R525 | p90 | ND | ≤0.5 | 8 | 16 | 0.25 | 32 | ≤0.06 | ≤0.125 |
| K. pneumoniae 103624 | p3, p90 | OmpK36 | >256 | >256 | >256 | >256 | >256 | 16 | 8 |
| K. pneumoniae 103624 | pSHA25K | wtOmpK36 | 4 | 2 | 8 | 0.125 | 2 | 0.125 | 0.75 |
| K. pneumoniae 103624 | pQE26 | isOmpK36 | 4 | 2 | 8 | ND | ND | 0.125 | 0.75 |
| K. pneumoniae CSUB10R | ND | None | 128 | 512 | >2,048 | 4 | ND | 4 | 1 |
| K. pneumoniae CSUB10R | pSHA25K | wtOmpK36 | 2 | 16 | 512 | 0.25 | ND | 0.015 | 0.25 |
| K. pneumoniae CSUB10R | pQE26 | isOmpK36 | 2 | 16 | 512 | 0.25 | ND | 0.015 | 0.25 |
Indigenous plasmids carried by isolates 103624 (p3 and p90) and CSUB10R (not determined) are not indicated on their derivative strains for clarity. Plasmid p90 was transferred from isolate 103624 by conjugation. Porin-encoding plasmids were introduced by electroporation.
K. pneumoniae porins OmpK36 and OmpK35 were analyzed by SDS-PAGE. wtOmpK36 is the OmpK36 cloned from strain C3 (accession no. Z33506), and isOmpK36 is OmpK36 cloned from isolate 103624.
FOX, cefoxitin; CTX, cefotaxime; CAZ, ceftazidime; C-C, CAZ-clavulanate; P-T, piperacillin-tazobactam; MP, meropenem; IP, imipenem.
Clavulanate, 4 μg/ml.
Tazobactam, 4 μg/ml.
ND, not determined.
Isoelectric focusing analysis of K. pneumoniae 103624 was performed as previously described (13) and yielded one single β-lactamase band with pI 7.6 that was inhibited by clavulanate but not by EDTA. Plasmid analysis of K. pneumoniae 103624 showed that the organism carried two plasmids of 3 and 90 kb designated p3 and p90, respectively. The plasmid carrying the β-lactamase gene (p90) was identified after conjugation-mediated transfer to Escherichia coli K-12 14R525 and selection on amoxicillin (50 μg/ml) and nalidixic acid (30 μg/ml). It was associated with resistance to ceftazidime and cefotaxime and did not confer resistance to other antibiotics, including cefoxitin, imipenem, and meropenem (Table 1). Transconjugants were more resistant to ceftazidime-clavulanate and piperacillin-tazobactam but remained susceptible to gentamicin, amikacin, ciprofloxacin, and tetracycline (MICs of 0.125, 1.0, 0.125, and 0.5 μg/ml, respectively). Isoelectric focusing analysis also revealed a single β-lactamase band with pI 7.6 that was inhibited by clavulanate. PCR amplification using the SHV primers GCCCGGGTTATTCTTATTTGTCGC and TCTTTCCGATGCCGCCGCCAGTCA (5′-to-3′ sequences) (11) and restriction fragment length polymorphism (RFLP) analysis of the PCR amplicon from isolate 103624 with NheI produced two fragments of 770 and 247 bp, confirming the presence of the SHV-2 gene, and an unrestricted product of 1,017 bp, which indicated the additional presence of SHV-1. The same PCR-RFLP analysis applied to the transconjugant produced only the 770- and 247-bp fragments, corresponding to the presence of SHV-2 alone. This was further confirmed by DNA sequencing (Applied Biosystems sequencer and dye-terminator chemistry). These data indicated that K. pneumoniae 103624 expresses a plasmid-borne extended-spectrum β-lactamase, SHV-2, and implied that another nontransferable mechanism should be responsible for the observed resistance to expanded-spectrum cephalosporins, cefoxitin, and carbapenems. Since porin deficiency is a well-established mechanism of resistance in K. pneumoniae and other species, the outer membrane of the isolate was characterized.
Outer membrane proteins (OMPs) and porins were isolated by differential solubilization in sodium lauryl sarkosinate and by a combination of methods, respectively, as described before (1). Electrophoretic analysis was performed in sodium dodecyl sulfate (SDS)-11% polyacrylamide gels as described previously (6).
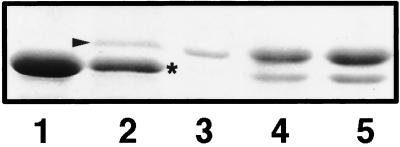
When grown in the low-osmolarity medium nutrient broth (71 mosmol/kg), K. pneumoniae 103624 showed a single band migrating above the constitutively expressed OmpA (Fig. 1, lane 2) which was not expressed in the high-osmolarity (1,511 mosmol/kg) Luria-Bertani broth (Fig. 1, lane 1). The apparent molecular mass of the osmolarity-regulated protein supported the conclusion that it was a porin, and this was confirmed because the protein could be isolated by using porin isolation methods (Fig. 1, lane 3) based on trypsin resistance and differential solubilization as previously described (9). Its regulation by osmolarity is similar to that of the OmpF-type porins, like the K. pneumoniae porin OmpK35, and differs from that of the OmpC-type porins, like the K. pneumoniae porin OmpK36 (6). Expression of porin OmpK37 was not detected by Western blotting experiments (data not shown) performed as previously described (4). This is consistent with the observed resistance of the isolate to imipenem and meropenem, since OmpK37 confers sensitivity to them while being less permeable to cephalosporins (4). However, the expression of OmpK35 by isolate 103624, as deduced from the SDS-polyacrylamide gel electrophoresis (PAGE) experiments, contradicts our previous observation that most K. pneumoniae clinical isolates expressing extended-spectrum β-lactamases express the OmpK36 porin but do not express, or express reduced amounts of, OmpK35.
FIG. 1.
SDS-PAGE analysis of OMPs from isolate 103624. Shown are OMPs from isolate grown in high- (lane 1) and low-osmolarity media (lane 2), porin from isolate 103624 (lane 3), and OMPs from isolate 103624 expressing the wild-type OmpK36 porin cloned from strain C3 (lane 4) and from isolate 103624 (lane 5). OMPs (100 μg) or isolated porin (5 μg) were boiled for 5 min in electrophoresis sample buffer before analysis. Porins and putative OmpA are indicated by the arrowhead and asterisk, respectively.
Thus, we performed three types of concurrent experiments to clarify the contribution of porins to the observed resistance. First, we PCR amplified and sequenced the ompK36 and ompK35 porin genes of the isolate. Second, we analyzed the expression of these two genes by RNA isolation and reverse transcriptase PCR (RT-PCR)-specific amplification. Third, we isolated and sequenced the N terminus of the porin.
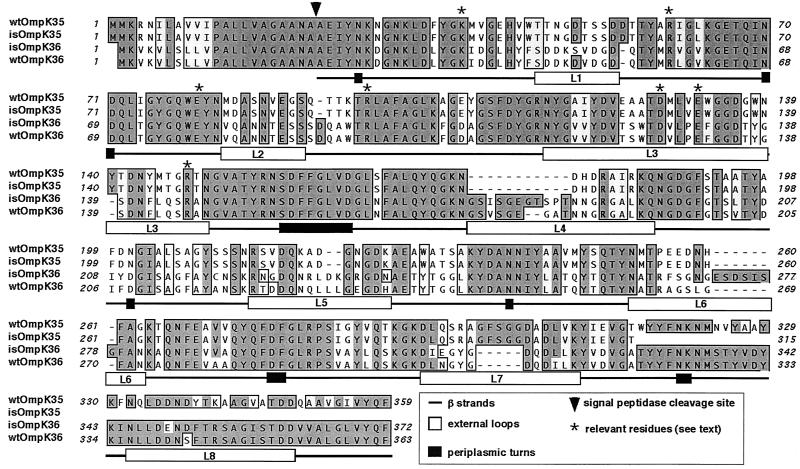
Compared to the wild type (Fig. 2) (unpublished data), the deduced OmpK35 sequence from isolate 103624 contained three silent changes at positions 474 (T to C, Ser), 648 (C to G, Val), and 786 (C to T, Ala), with the A of the ATG start codon of the wild-type ompK35 gene considered position 1. Most importantly, one change was found in position 847, where codon TGG changed to the stop codon TAG. This should result (Fig. 2) in a truncated porin lacking the last 44 amino acids and without expression in the outer membrane, since the terminal F residue is necessary for such expression (12). Comparison of the sequence of OmpK36 from the isolate with that of wild-type OmpK36 revealed that most differences were found in the predicted external loops (Fig. 2). These included amino acid replacements (17 changes) and insertions (nine amino acids), whereas the predicted β strands, which constitute most of the secondary structure, were quite conserved (five replacements).
FIG. 2.
Comparison by alignment of the deduced OmpK35 and OmpK36 sequences from isolate 103624 (isOmpK35 and isOmpK36, respectively) and the same porin sequences from the wild-type porins (wtOmpK36 and wtOmpK35) available in GenBank, EMBL, and DDBJ. Secondary structural motifs are described based on the crystal structure of wtOmpK36 (5). The numbering is based on the mature wild-type OmpK36.
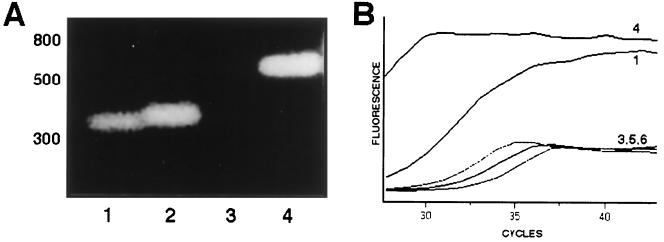
Expression of porin genes in the isolate was studied by RT-PCR (Fig. 3). RNA was extracted with an RNeasy mini kit (Qiagen), and chromosomal DNA was extracted using cetyltrimethylammonium bromide. These were amplified with primers U681 and L1316, which anneal to both porin genes and have been described before (4). Primers UE36 and LE35, GCAGCGTCAGCGGCGAAGG and GGAGAAGCCAGCACGCGAC (5′ to 3′), respectively, are specific for the ompK36 and ompK35 genes. RT-PCR amplifications with the U681-LE35 and UE36-L1316 primer pairs were performed using a Promega Access RT-PCR System kit, and amplicons were detected by agarose electrophoresis. Real-time RT-PCR was also performed using Light Cycler (Roche) and the Light Cycler RNA Master SYBR Green I kit and 3 mM MnCl2. The results of both the RT-PCR (Fig. 3A) and real-time RT-PCR (Fig. 3B) experiments confirmed that it was the ompK36 gene, not the ompK35 gene, that was being expressed by isolate 103624.
FIG. 3.
Analysis of porin gene expression from isolate 103624 by RT-PCR and agarose electrophoresis of the amplicons (A) or by real-time RT-PCR (B). In panel A, ompK36 amplicons are shown in lanes 1 and 2 and ompK35 amplicons are shown in lanes 3 and 4. Odd lanes show amplicons from RNA, and even lanes show amplicons from DNA. The numbers in panel B correspond to the lane numbers in panel A, except that lane 2 was not included and additional lanes 5 and 6 correspond to negative controls (water instead of RNA or DNA).
Finally, determination of the N-terminal sequence of the porin expressed by isolate 103624 confirmed that the porin being expressed is the OmpK36 homologue in this strain and not OmpK35: the sequence AEIYNKDGNKLD was determined, which coincides with that of wild-type OmpK36 (1), with the seventh residue being an N in OmpK35 (unpublished data; see also the data listed in GenBank under accession no. AJ011501) (Fig. 2).
It is difficult to reconcile OmpK36 expression with the observed MICs because expression of wild-type OmpK36 should cause sensitivity to, e.g., cefoxitin (2, 9) unless the porin being expressed has some structural changes making it less permeable to antimicrobials. Sequence inspection showed minor changes and the presence of all of the amino acids that are important for OmpK36 pore function (Fig. 2), including the arginine cluster R37-R75-R125 (numbering based on the mature wild-type OmpK36) and K16 that in the tertiary structure are found opposed to D106 and E110 across the pore and that play major roles in charge segregation (5). These sequence changes do not contribute to the observed MICs, as demonstrated by liposome experiments performed as previously described (4, 5) with the OmpK36 porins isolated from strain 103624 and from wild-type strain C3 (1). In these experiments, liposomes containing the OmpK36 porin from the isolate were in fact slightly more permeable than those with the wild-type OmpK36, which therefore does not explain the MICs (Table 2).
TABLE 2.
Sugar permeation through OmpK36 porins extracted from wild-type strain C3 (wtOmpK36) and isolate 103624 (isOmpK36)
| Sugar | Mol wt | Sugar permeation (mean rate ± SD)a through liposomes reconstituted with:
|
|
|---|---|---|---|
| wtOmpK36 | isOmpK36 | ||
| Arabinose | 150.1 | 100 | 100 |
| Galactose | 180.2 | 85.3 ± 6.5 | 87.4 ± 4.1 |
| Mannose | 180.2 | 74.9 ± 14.0 | 81.0 ± 1.5 |
| NAcGlu | 221.1 | 49.8 ± 10.6 | 60.7 ± 3.0 |
| Lactose | 360.3 | 4.4 ± 4.1 | 6.5 ± 1.8 |
| Trehalose | 378.3 | ND | ND |
Means of three independent experiments expressed as the percentage of the value for arabinose. ND, not detected.
The contribution of reduced OmpK36 expression was studied by expressing in the isolate both OmpK36 cloned from the isolate and the wild-type porin from strain C3. The ompK36 gene was amplified by PCR using primers ompk36-0 (AAGCTTGTTGGATTATTCTGC) and ompk36-end (CAAGCTTAGAACTGGTAAACC) (5′-to-3′ sequences), which in the wild-type OmpK36 (GenBank accession no. Z33506) anneal 95 and 1,098 nucleotides upstream and downstream of the ATG start codon, respectively, thus amplifying the gene and promoter, and was cloned in vector pCR2.1 (Invitrogen). Cloned products were introduced in the isolate by electroporation and selection with kanamycin, and porin expressions were confirmed by SDS-PAGE analysis of the OMPs. The increased OmpK36 expression resulting from the cloning of ompK36 from wild-type strain C3 and from isolate 103624 (Fig. 1, lanes 4 and 5) produced a reduction in the MICs of all tested antimicrobials. These reductions were the same for both porins, thus confirming that reduced porin expression in the isolate, not alterations in the porin sequence, contributed to the observed MICs. This was further confirmed by expression of OmpK36 cloned from both wild-type strain C3 and from isolate 103624 in the porinless isolate CSUB10R (2). Expression of either porin caused the same reduction in the MICs of all antimicrobials tested.
The expression of AmpC-type β-lactamases and the loss of porin expression as a cause of cephalosporin and carbapenem resistance in K. pneumoniae (2, 3, 10) and in other species has been described previously. Isolate 103624 does not exhibit the above-mentioned mechanisms, because it expresses an SHV-2 enzyme rather than an AmpC-type enzyme and because it is porin sufficient, since it expresses a porin. A carbapenem-resistant K. pneumoniae isolate has also been isolated in the United Kingdom, but its resistance mechanism, increased expression of an SHV-2 β-lactamase and loss of an OMP, differs from that of our isolate (8). Carbapenem resistance is still rare in K. pneumoniae, but the results with our isolate illustrate that it is not always sufficient to analyze the expression of porins by SDS-PAGE when studying their possible contribution to antimicrobial resistance. Genetic studies demonstrated that the decreased amount of OmpK36 porin expressed by the isolate, together with the expression of SHV-1 and SHV-2, contributed to the observed resistance to β-lactam-containing agents.
Nucleotide sequence accession numbers.
The DNA sequences of the ompK35 and ompK36 genes from isolate 103624 have been deposited in GenBank and EMBL under accession no. AJ303057 and AJ344089, respectively.
Acknowledgments
We thank L. Martínez-Martínez (University of Seville) for helpful discussions throughout this work and A. Johnson (PHLS Colindale, London, England) for help with MIC determinations.
This work was partially supported by grants from the Spanish Government to V.J.B. A.D.-S. was supported by a predoctoral fellowship from Ministerio de Educación y Cultura of the Spanish Government.
REFERENCES
- 1.Albertí, S., F. Rodríguez-Quiñones, T. Schirmer, G. Rummel, J. M. Tomás, J. P. Rosenbusch, and V. J. Benedí. 1995. A porin from Klebsiella pneumoniae: sequence homology, three-dimensional structure, and complement binding. Infect. Immun. 63:903-910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ardanuy, C., J. Liñares, M. A. Domínguez, S. Hernández-Allés, V. J. Benedí, and L. Martínez-Martínez. 1998. Outer membrane profiles of clonally related Klebsiella pneumoniae isolated from clinical samples and activity of cephalosporins and carbapenems. Antimicrob. Agents Chemother. 42:1636-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bradford, P., C. Urban, N. Mariano, S. Projan, J. Rahal, and K. Bush. 1997. Imipenem resistance in Klebsiella pneumoniae is associated with the combination of ACT-1, a plasmid-mediated AmpC β-lactamase, and the loss of an outer membrane protein. Antimicrob. Agents Chemother. 41:563-569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Doménech-Sánchez, A., S. Hernández-Allés, L. Martínez-Martínez, V. J. Benedí, and S. Albertí. 1999. Identification and characterization of a new porin gene of Klebsiella pneumoniae: its role in β-lactam antibiotic resistance. J. Bacteriol. 181:2726-2732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dutzler, R., G. Rummel, S. Albertí, S. Hernández-Allés, P. S. Phale, J. P. Rosenbusch, V. J. Benedí, and T. Schirmer. 1999. Crystal structure and functional characterization of OmpK36, the osmoporin from Klebsiella pneumoniae. Structure 7:425-434. [DOI] [PubMed] [Google Scholar]
- 6.Hernández-Allés, S., S. Albertí, D. Álvarez, A. Doménech-Sánchez, L. Martínez-Martínez, J. Gil, J. M. Tomás, and V. J. Benedí. 1999. Porin expression in clinical isolates of Klebsiella pneumoniae. Microbiology 145:673-679. [DOI] [PubMed] [Google Scholar]
- 7.Livermore, D. M. 1997. Acquired carbapenemases. J. Antimicrob. Chemother. 39:673-676. [DOI] [PubMed] [Google Scholar]
- 8.MacKenzie, F. M., K. J. Forbes, T. Dorai-John, S. G. Amyes, and I. M. Gould. 1997. Emergence of a carbapenem-resistant Klebsiella pneumoniae. Lancet 350:783.. [DOI] [PubMed] [Google Scholar]
- 9.Martínez-Martínez, L., S. Hernández-Allés, S. Albertí, J. M. Tomás, V. J. Benedí, and G. A. Jacoby. 1996. In vivo selection of porin-deficient mutants of Klebsiella pneumoniae with increased resistance to cefoxitin and expanded-spectrum cephalosporins. Antimicrob. Agents Chemother. 40:342-348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Martínez-Martínez, L., A. Pascual, S. Hernández-Allés, D. Álvarez-Díaz, A. I. Suárez, J. Tran, V. J. Benedí, and G. A. Jacoby. 1999. Role of β-lactamases and porins in the activity of carbapenems and cephalosporins against Klebsiella pneumoniae. Antimicrob. Agents Chemother. 43:1669-1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nuesch-Inderbinen, M. T., H. Hachler, and F. H. Kayser. 1996. Detection of genes coding for extended-spectrum SHV beta-lactamases in clinical isolates by a molecular genetic method, and comparison with the E test. Eur. J. Clin. Microbiol. Infect. Dis. 15:398-402. [DOI] [PubMed] [Google Scholar]
- 12.Struyvé, M., M. Moons, and J. Tommassen. 1991. Carboxy-terminal phenylalanine is essential for the correct assembly of a bacterial outer membrane protein. J. Mol. Biol. 218:141-148. [DOI] [PubMed] [Google Scholar]
- 13.Thomson, K. S., C. C. Sanders, and J. A. Washington. 1991. High-level resistance to cefotaxime and ceftazidime in Klebsiella pneumoniae isolates from Cleveland, Ohio. Antimicrob. Agents Chemother. 35:1001-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]