Abstract
The expression of chromosomal AmpC β-lactamase in Pseudomonas aeruginosa is negatively regulated by the activity of an amidase, AmpD. In the present study we examined resistant clinical P. aeruginosa strains and several resistant variants isolated from in vivo and in vitro biofilms for mutations in ampD to find evidence for the genetic changes leading to high-level expression of chromosomal β-lactamase. A new insertion sequence, IS1669, was found located in the ampD genes of two clinical P. aeruginosa isolates and several biofilm-isolated variants. The presence of IS1669 in ampD resulted in the expression of high levels of AmpC β-lactamase. Complementation of these isolates with ampD from the reference P. aeruginosa strain PAO1 caused a dramatic decrease in the expression of AmpC β-lactamase and a parallel decrease of the MIC of ceftazidime to a level comparable to that of PAO1. One highly resistant, constitutive β-lactamase-producing variant contained no mutations in ampD, but a point mutation was observed in ampR, resulting in an Asp-135→Asn change. An identical mutation of AmpR in Enterobacter cloacae has been reported to cause a 450-fold higher AmpC expression. However, in many of the isolates expressing high levels of chromosomal β-lactamase, no changes were found in either ampD, ampR, or in the promoter region of ampD, ampR, or ampC. Our results suggest that multiple pathways may exist leading to increased antimicrobial resistance due to chromosomal β-lactamase.
Many bacterial species express a chromosomally encoded class I β-lactamase, AmpC (2). In the absence of β-lactam antibiotics AmpC is normally produced at low levels, but in the presence of inducing β-lactams the expression may increase 100- to 1,000-fold (28). Resistance to β-lactam antibiotics is a serious problem in chronic Pseudomonas aeruginosa lung infections in cystic fibrosis (CF) patients (5, 10, 30). The resistance to β-lactam antibiotics is frequently a consequence of constitutive hyperproduction of AmpC β-lactamase (stable derepression) (28, 29).
The regulatory mechanisms that underlie the AmpC expression have been studied in detail in, for example, Enterobacter cloacae. Similar regulatory mechanisms seem to regulate β-lactamase expression in P. aeruginosa (21, 22, 26-28). Increased ampC transcription is dependent on AmpR, a positive transcriptional regulator of the LysR family (26, 28). The transcriptional regulatory activity of AmpR is linked to peptidoglycan recycling (7, 14, 21, 22). Anhydromuropeptides released from the bacterial peptidoglycan are transported to the cytoplasm via the transmembrane permease AmpG (6, 28, 31). In the cytoplasm, muropeptides act as signal molecules modulating the regulatory activity of AmpR, leading to upregulation of ampC transcription (25). The cytosolic amidase AmpD is essential in the turnover and recycling of muropeptides. Thus, AmpD has been shown to be a negative regulator of AmpC β-lactamase expression (21). However, studies of P. aeruginosa ampD knockout mutants have only partially elucidated changes leading from low basal level to high basal level and constitutive β-lactamase expression (22). Langaee et al. showed that inactivation of ampD in PAO1 resulted in partially derepressed AmpC expression and suggested that further genetic changes at another bla locus may be required to achieve full AmpC derepression (22).
In the present study we report the presence of a new 1,669-bp insertion sequence (IS1669) located in the ampD gene of several resistant clinical P. aeruginosa isolates with stable derepressed production of chromosomal β-lactamase. To study these AmpD-defective isolates, we performed ampD complementation analyses and phenotypically characterization of the isolates.
To screen for mutations affecting the expression of chromosomal β-lactamase in P. aeruginosa other than the presence of IS1669, we sequenced ampD, including the promoter region of several clinical isolates and resistant variants producing high levels of chromosomal β-lactamase but with no IS1669 inserted in ampD. In addition, ampR and the promoter region of ampR and ampC were sequenced.
MATERIALS AND METHODS
Bacterial isolates.
The clinical strains of P. aeruginosa PA258, 17107B, and 19676A included in the present study were isolated from the sputum of different CF patients attending the Danish CF Center every third month, Rigshospitalet, Copenhagen. J. R. W. Govan, Edinburgh, United Kingdom, kindly provided strain PAO579. We have previously used these clinical isolates for in vivo (rat model of chronic lung infection [15, 34]) and in vitro (modified Robbins device [8, 16]) biofilm models to examine the development of resistance during intensive exposure to β-lactams (ceftazidime) (1). From these experiments resistant variants of 17107B, 19676A, and PAO579 were isolated (1). Pulsed field gel electrophoresis confirmed the variants to be progeny of the clinical strains, expressing β-lactamases showing identical pI values and inhibition patterns (1). The resistance of the variants to ceftazidime was found to be due to increased expression of chromosomal β-lactamase compared to the clinical isolates (Table 1). However, the genetic regulatory mechanisms leading to increased expression of chromosomal β-lactamase in these isolates have not been demonstrated previously. All isolates included in the present study are shown in Table 1. As a reference strain, we used PAO1. The bacteria were grown in Luria-Bertani (LB) broth or LB agar containing the appropriate antibiotics.
TABLE 1.
β-Lactamase expression and MICs of ceftazidime against selected P. aeruginosa isolates before and after complementation with ampD
| Isolate | Sourcea | ampD genotypeb | Ceftazidime MIC (μg/ml) before/aftercampD complementation | β-Lactamase activity (mU) before/aftercampD complementation
|
|
|---|---|---|---|---|---|
| Basal | Induced | ||||
| PAO1 | Reference | + | 0.064/0.064 | 24/22 | 218/152 |
| PA258 | Clinical | − | 64/1.00 | 10,100/27 | 12,000/1,500 |
| 17107B | Clinical | − | 100/0.25 | 504/10 | 947/345 |
| 17107B-1 | Variant, in vivo | − | >256/0.75 | 1,090/14 | 4,120/521 |
| 17107B-2 | Variant, in vivo | − | >256/0.38 | 1,560/2.4 | 2,260/223 |
| 17107B-3 | Variant, in vivo | − | >256/8 | 9,160/34 | 7,950/177 |
| 17107B-4 | Variant, in vivo | − | >256/1.00 | 12,000/9 | 12,600/288 |
| 19676A | Clinical | + | 50/− | 38/− | 957/− |
| 19676A-1 | Variant, in vitro | + | 200/− | 687/− | 2,200/− |
| 19676A-2 | Variant, in vivo | + | 96/96 | 11,000/7,810 | 12,700/11,400 |
| 19676A-3 | Variant, in vivo | + | >256/>256 | 17,300/13,500 | 16,700/16,000 |
| PAO579 | Clinical | + | 0.8/− | 19/− | 550/− |
| PAO579-1 | Variant, in vivo | + | 12.5/− | 114/− | 275/− |
| PAO579-2 | Variant, in vitro | + | 200/− | 984/− | 627/− |
| PAO579-3 | Variant, in vitro | + | 200/− | 1,130/− | 647/− |
Variants from in vivo and in vitro biofilm experiments have previously been published (1).
+, ampD gene present; −, ampD gene interrupted by insertion of IS1669.
Before, isolates transformed with pNB100; after, isolates transformed with pNB101. A “−” sign indicates no transformation was performed with pNB101. One milliunit of β-lactamase is defined as 1 nmol of nitrocefin hydrolyzed per min per mg of protein.
Plasmids.
For the complementation experiments with ampD, the shuttle plasmids pNB100 and pNB101 were constructed. Plasmid pNB101 was constructed by blunt-end cloning of a 1,088-bp PCR product, including gene ampD into the SmaI site of pNB100. pNB100 was derived from pUCP22 after digestion with ScaI-XmnI, followed by religation. Excluding this 119-bp ScaI-XmnI fragment resulted in knockout of the bla gene. A 1,088-bp ampD-coding region of PAO1 was amplified by PCR with the primers D1 (5′-GCTGCGTGTCGGCAGTTGGGTGGAGATCCAG-3′) and D2 (5′-GAGGGCAGATCCTCGACCAGGCTCAGCCAG-3′), which annealed 406 bp upstream and 115 bp downstream of ampD, respectively. The promoter region was included in the upstream sequence (21, 22). Genomic DNA was extracted by standard procedures and used as a template in the PCR amplification (36). PCR was performed in a final volume of 100 μl containing 1.5 mM MgCl2, 10 mM Tris-HCl (pH 8.8), 150 mM KCl, 0.1% Triton X-100, a 250 μM concentration of deoxynucleoside triphosphate, 0.4 μM concentrations of each primer, ca. 500 ng of genomic DNA, 3% dimethyl sulfoxide, and 2.5 U of proofreading Pwo DNA polymerase (Roche). After an initial incubation of 97°C for 10 min, amplification was performed in 35 cycles of 94°C for 1.5 min, 66°C for 2 min, and 72°C for 2.5 min. The third step in the cycle (72°C, 2.5 min) was extended 5 s in each cycle. The samples were subsequently stored at 4°C. PCR products were blunt ended with the Klenow enzyme, phosphorylated by using T4 polynucleotide kinase, Microspin column purified, and finally ligated into pNB100 according to the instructions of the SureClone Ligation Kit (Amersham Pharmacia Biotech). The orientation of the insert was controlled by restriction fragment length polymorphism after digestion with both EcoRI and SmaI. The transcriptional orientation of ampD in pNB101 was opposite to the transcriptional orientation of the gentamicin marker and LacZα. For control experiments without ampD complementation, pNB100 was used. The Escherichia coli strain used for cloning was JM105 [thi rpsL (Strr) end sbcB15 sbcC hsdR4 (rK− mK−) Δ(lac-proAB)123 F′ trad36 lacIq Δ(lacZ)M15 proA+B+ 125]. Plasmid isolation was performed by using QIAprep spin Miniprep kits (Qiagen).
Transformation.
Transformations of E. coli JM105 with shuttle plasmids (pUCP22, pNB100, or pNB101) were performed as described by Sambrook et al., although slightly modified (36). Competent E. coli JM105 were prepared by harvesting 60 ml of culture (in LB medium and 10 mM MgCl2) in late logarithmic phase (optical density at 550 nm [OD550] = 0.5 to 0.6) by centrifugation (750 × g for 4 min, 4°C), followed by two successive washes with 8 ml of ice-cold FB buffer (10% [wt/vol] glycerin, 10 mM potassium acetate [pH = 7.5], 0.1 M KCl, 50 mM CaCl2). After the final wash, the bacteria were resuspended in 2 ml of FB buffer, divided into aliquots, and frozen at −80°C. A 100-μl aliquot of thawed competent bacteria was mixed with ca. 1 μg of plasmid DNA, loaded into a microcentrifuge tube, and placed on ice for 10 min. The cells were heated for 45 s at 42°C and subsequently placed on ice for 5 min. To each tube was added 900 μl of SOC (2.5 mM KCl, 10 mM MgCl2, 10 mM MgSO4, and 20 mM glucose in LB medium), and the bacteria were recovered after 1 h of gentle shaking at 37°C. A total of 300 μl was inoculated onto LB agar containing 25 μg of gentamicin/ml, followed by incubation at 37°C for 16 h.
Transformation of P. aeruginosa with pNB100 or pNB101 was performed by electroporation. Electroporation-competent P. aeruginosa isolates were prepared by pelleting 250 ml of culture in late logarithmic phase (OD600 = 0.5 to 1.0) by centrifugation (6,000 × g for 5 min at 4°C). The pellet was washed three times with 20 ml of ice-cold SMEB (1 mM HEPES [pH 7.0], 300 mM sucrose, 1 mM MgCl2). The bacteria were then resuspended in 2 ml of SMEB, and aliquots frozen at −80°C. Then, 100 μl of thawed competent bacteria were mixed with 1 μg of plasmid DNA, loaded into prechilled 2-mm cuvettes, and placed on ice for 1 min. The bacteria were electroporated at 12.5 kV/cm, 200 Ω, and a capacitance at 25 μF for ca. 5 ms. Subsequently, 900 μl of SOC was added, and the suspension was transferred to a microcentrifuge tube and placed on ice for 30 min. Subsequently, the bacteria were recovered during 1.5 h with gentle shaking at 37°C. Next, 300 μl was inoculated on LB agar containing 400 μg of gentamicin per ml to select for P. aeruginosa transformants with pNB100 or pNB101, followed by incubation at 37°C for 16 h.
Susceptibility test.
The MIC was determined by using the E-test system (AB Biodisk, Sweden) according to the instructions of the manufacturer.
β-Lactamase assay.
The basal β-lactamase level, as well as the induced β-lactamase levels after 2.5 h of induction with benzylpenicillin (500 μg/ml), were measured spectrophotometrically with nitrocefin (51.6 μg/ml) as the substrate as previously described (10, 32).
PCR amplification and sequencing of amp genes.
PCR amplification of ampDE, ampR, and the promoter region of ampC and ampR were performed as described above for ampD. Modified annealing temperatures were, however, used for the specific primers for ampDE, ampR, and the promoter region of ampC and ampR. ampDE (including the promoter region and the complete genes of ampD and ampE) was amplified by using the primers DE1 (5′-GTACGCCTGCTGGACGATGCCTTGCTGTTCGAC-3′) and DE2 (5′-GGAGTCCGCTCGGTAGTGGCCTTTCGAAATCATGC-3′), which annealed 234 bp upstream of ampD and 91 bp downstream of ampE. The applied annealing temperature was 65°C. Amplification of ampR (the complete gene), including the promoter regions of both ampR and ampC and a 117-bp sequence of ampC (117 bp downstream of the translational start codon) were performed by using the primers RC1 (GTCGACCAGTGCCTTCAGGCGATCC) and RC2 (CTCGAGAGCGAGATCGTTGCGGCACG) and an annealing temperature of 63°C.
Sequencing was performed on the ABI Prism 377 DNA Sequencer (Perkin-Elmer Applied Biosystems) by using the ABI Prism BigDye terminator cycle sequencing ready reaction kit as outlined by the manufacturer.
RESULTS
Identification of a new insertion sequence (IS1669).
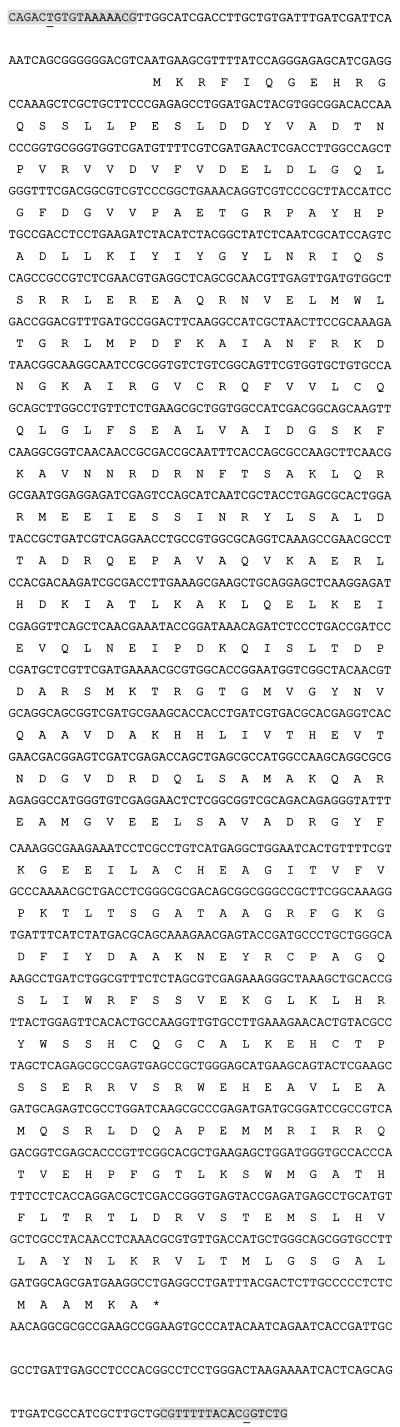
Sequencing of the ampD gene in the two clinical strains PA258 and 17107B and in the four independent resistant biofilm-variants 17107B-1, 17107B-2, 17107B-3, and 17107B-4 showed the presence of a new 1,669-bp insertion sequence (Fig. 1). In all isolates, the nucleotide sequence indicated insertion at the same position in ampD, 74 bp downstream of the start codon. The insertion sequence was not present in ampD in the reference strain PAO1. The insertion sequence located in ampD was found to be 100% identical in the two clinical strains and in the four resistant biofilm variants. The new insertion sequence was named IS1669 due to the size of the IS element. IS1669 was found to contain 17-bp inverted terminal repeats, although imperfect with a single mismatch at the terminal 6 position. In connection to the insertion sequence a 4-bp target DNA duplication was found, although imperfect as GTCC was duplicated as GGCC. The GC content of IS1669 was 58.1% compared to an overall GC content of 66.6% in the complete sequence of PAO1 (http://www.pseudomonas.com). An open reading frame in IS1669 from nucleotides 71 to 1,501 was found to encode a 476-amino-acid protein, probably a transposase, having 46% similarity to the transposase of the insertion sequence IS1194 from Streptococcus thermophilus (GenBank accession no. Y13626). IS1669 was found to show 77% identity to a genomic sequence in the published PAO1 (http://www.pseudomonas.com, PA sequence range from 1482826 to 1481163 of the updated sequence database, PAO1-2001-09). This sequence includes a hypothetical, unclassified, unknown protein (confidence level 4) annotated PA1368. BLAST analysis of PA1368 to the 1,431-bp open reading frame of IS1669 showed 80% identity.
FIG. 1.
Total sequence of the new insertion sequence, IS1669. The shaded 17-bp sequences in the upstream and downstream parts of IS1669 represent the “left inverted repeated sequence” and the “right inverted repeated sequence,” respectively. The inverted repeated sequences were imperfect in one nucleotide position (underlined). The amino acid composition of a probably 476-amino-acid transposase is shown.
IS1669 was not found in ampD of 19676A, in PAO579, or in the ampD gene of any of the resistant 19676 and PAO579 variants (Table 1). The nucleotide sequences of ampD and the 201-bp upstream regions of the 19676A variants and the PAO579 variants were identical to those of the mother strains. The ampD sequence of PAO579 and the variants were identical to the published sequence of PAO1, whereas ampD of 19676A and its three variants contained several base substitutions. The nucleotide sequence relative to the start codon of ampD differed in position 32 (G-to-T substitution), position 102 (T-to-C substitution), position 144 (T-to-C substitution), position 443 (G-to-C substitution), and position 547 (G-to-T substitution). These nucleotide substitutions caused amino acid substitutions in position 11 (Arg to Leu), position 148 (Gly to Ala), and position 183 (Asp to Tyr).
The sequence of ampD in our PAO1 strain was identical to the gene in the published sequenced PAO1 genome.
Chromosomal β-lactamase activity.
The isolates containing IS1669 in ampD were all producing either moderate to high basal levels that could be further induced (17107B, 17107B-1, and 17107B-2) or stable derepressed levels (PA258, 17107B-3, and 17107B-4) (Table 1). Thus, the lack of AmpD activity correlated with high levels of chromosomal β-lactamase activity of the isolates. However, although the clinical strain 17107B expressed no functional AmpD, its variants could increase their production of chromosomal β-lactamase to a stable derepressed level exceeding 10,000 mU (17107B-4).
Basal and induced β-lactamase production of the 19676 variants and the PAO579 variants were increased compared to the mother strains. 19676A-2 and 19676A-3 produced high derepressed levels, higher levels compared to the other 19676 and PAO579 variants, which expressed moderate basal inducible levels. The mother strains produced low basal levels, which could be induced to higher levels. The basal level of β-lactamase expression in PAO1 was low and could be induced ninefold to 218 U (Table 1).
Complementation with wild-type ampD.
The pNB101 plasmid (carrying the wild-type ampD from P. aeruginosa PAO1) was transformed into the control strain PAO1, the clinical isolates PA258 and 17107B, and the resistant 17107 variants (Table 1). As a control of no ampD complementation, all isolates were transformed with the plasmid pNB100. Experiments with or without ampD complementation in the control strain PAO1 resulted in similarly low levels of basal and induced specific β-lactamase activity, as well as a low ceftazidime MIC (Table 1 and Fig. 2). However, in all isolates with a defective ampD gene caused by IS1669, the complementation with ampD resulted in low levels of both basal and induced specific β-lactamase activity that were comparable to the level of PAO1. To examine the effect of the three amino acid substitutions identified in ampD gene of the 19676 isolates, the two variants 19676A-2 and 19676A-3 were complemented with ampD from PAO1. However, no decreases in specific β-lactamase activity were observed.
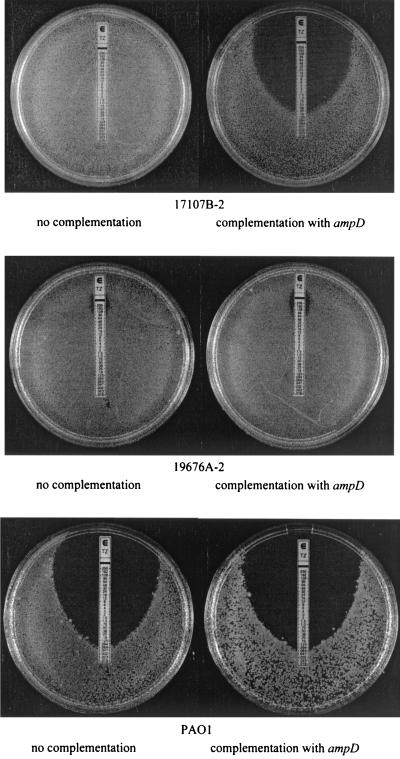
FIG. 2.
Results of E-test measurements for the MIC of ceftazidime for three P. aeruginosa isolates with or without ampD complementation. The isolate 17107B-2 was AmpD defective since IS1669 was found in ampD. This was not the case for strains 19676A-2 or PAO1. ampD from PAO1 was used for complementation on the plasmid pNB101. Control (no complementation) was performed by transforming the isolates with pNB100 containing no ampD.
The phenotypic effect of the ampD complementation on 17107B-2, 19676A-2, and PAO1 is illustrated in Fig. 2, showing the ceftazidime MIC as determined by the E-test system. As seen by the inhibition zones, the complementation only affected the MIC of ceftazidime against the AmpD-defective isolate 17107B-2. The MICs for 19676A-2 and PAO1 were unaffected. As expected, ampD complementation of strains PA258 and 17107B and the resistant 17107 variants resulted in changes in the MIC (Table 1). The results indicated a pronounced effect of the ampD complementation on the isolates, which harbored a nonfunctional ampD gene due to the insertion of IS1669.
Nucleotide sequencing of ampR and the ampR-ampC intergenic region.
To examine whether high-level β-lactamase expression could be due to mutations in the transcriptional regulator protein AmpR, the entire ampR gene was sequenced. The clustered ampC and ampR genes are divergently transcriped and, hence, the 148-bp intergenic region contains both the ampR and the ampC promoters (3, 26, 27). The ampR-ampC intergenic region of strain PA258 has been sequenced previously, and no nucleotide changes were observed (3). In the present study, the ampR-ampC intergenic region, ampR, and the 5′ part of ampC (117 nucleotides) of all of the isolates presented in Table 1, except for strains PA258 and 17107B-1, were sequenced and compared to the PAO1 genome sequence. All of the isolates, except 19676A-3, showed 100% identity in the ampC-ampR intergenic region, in ampR, and in the 177-bp 5′ region of ampC. In 19676A-3 one mutation was found in ampR since the guanine in position 402 (relative to the ampR start codon) was changed to an adenine, causing an amino acid substitution in position 135, as aspartic acid was changed to asparagine. This point mutation was confirmed by a second sequencing of a new PCR product of ampR. In addition, 19676A-3 was found to be the most resistant variant, producing the highest level of chromosomal β-lactamase.
DISCUSSION
The location of the new insertion sequence IS1669 in ampD caused disruption of the reading frame of ampD and production of a nonfunctional AmpD protein. All isolates containing IS1669 in ampD produced high levels of chromosomal β-lactamase. In accordance with this, it has previously been reported that AmpD defects cause increased expression of chromosomal β-lactamase from P. aeruginosa, E. cloacae, Citrobacter freundii, and some other species (7, 17, 19, 22, 23). The present study emphasized the importance of AmpD, since ampD complementation caused all of the Amp-defective isolates to express the same low basal levels of chromosomal β-lactamase activity, as well as induced levels of β-lactamase comparable to the levels found for the reference strain PAO1. The AmpD-defective 17107 variants were more resistant and expressed increased levels of chromosomal β-lactamase activity compared to the isogenic (as determined by pulsed-field gel electrophoresis) mother strain 17107B. This must be due to unidentified changes at genetic loci other than ampD. Therefore, it was remarkable that the ampD-complemented isolates all produced the same low level of chromosomal β-lactamase activity. Whatever the genetic changes in addition to the AmpD defect that caused the high-level expression of chromosomal β-lactamase, it was overruled when complemented with ampD.
Since no changes were found in ampD, including the promoter region of 19676A, in PAO579, or in any of the variants derived from these strains, the increased β-lactamase activity of these variants must be due to unidentified changes at genetic loci other than ampD. The identified amino acid changes of ampD from the 19676 isolates compared to the ampD gene from PAO1 did not have any effect on the β-lactamase activity, as shown by complementation. In addition, the results indicated that the increased copy number of ampD, corresponding to the copy number of the vector (pNB101), did not affect the expression of chromosomal β-lactamase. Thus, our results showed that AmpD defects are not essential for the development of resistance due to chromosomal β-lactamase activity, since the observed resistance development of all of the variants compared to the mother strains could not be explained by changes in ampD. In E. cloacae, on the other hand, it has been found that altered β-lactamase expression is commonly associated with mutations in ampD (9, 17, 24). However, in the present study it was obvious to look for changes in other amp genes, since this may explain the observed high-level expression of β-lactamase.
Our results demonstrated that the ampC-ampR intergenic region, as well as the 177-bp 5′ region of ampC, was conserved in all of the isolates, indicating that mutations in these regions are probably not typical causes of high-level expression of chromosomal β-lactamase in P. aeruginosa. In accordance with this, Campbell et al. screened clinical isolates of P. aeruginosa from CF patients for changes in ampC and in the ampC-ampR intergenic region but screened as well for the presence of duplications of ampC in the genome (3). No ampC duplications were detected, no indications of changes in ampC were found, and no mutations in the ampC-ampR intergenic region could account for high-level AmpC expression (3). The results for P. aeruginosa thus contradict the findings from E. coli, since mutations in the ampC promoter region are numerous and have been shown to cause >100-fold changes in the AmpC expression (4).
Since we did not find mutations in the ampC promoter region or in ampC in the present study, it was of interest to examine whether the high level of β-lactamase expression was caused by changes in AmpR. Our results showed that the sequence of ampR was well conserved in almost all of the isolates. Only one isolate, 19676A-3, was found to have a single point mutation in ampR. It was remarkable that 19676A-3 expressed a constitutively very high level of chromosomal β-lactamase, a level considerably higher than with the other isolates examined here. It has not been demonstrated previously that mutations in ampR of P. aeruginosa may be linked to high-level expression of chromosomal β-lactamase. However, in E. cloacae specific mutations in AmpR have been shown to cause considerably increase in chromosomal β-lactamase activity (20). The AmpR amino acid sequences of E. cloacae and P. aeruginosa are very similar, and the amino acid identity is more than 60% (26). The amino acid identity of residues 127 to 157 is >90% (26). Amino acids 128 to 135 in AmpR from P. aeruginosa, E. cloacae, C. freundii, and Yersinia enterolitica are 100% identical (26). Kuga et al. found an aspartic acid-to-asparagine substitution in position 135 that caused a 450-fold increase in class C β-lactamase activity of E. cloacae (20). Our finding of the same amino acid substitution in position 135 of AmpR from strain 19676A-3 supports the possibility that this point mutation could be responsible for the high level of β-lactamase activity.
For E. cloacae it has been reported that specific mutations in ampR seem to be required to improve the effect of AmpR as a positve regulator on AmpC (20). However, it seems more likely that mutations in AmpD cause high-level expression of chromosomal β-lactamase, since specific mutations are not needed to knock out the function of AmpD. Thus, numerous studies have shown the impact of AmpD knockout on β-lactamase production, and this was confirmed in our study as well (11, 12, 19, 21, 23, 35, 37). However, the present results have also pointed to other mechanisms, other than AmpD defects, that are involved in the increased expression of chromosomal β-lactamase, as shown in the resistant variants of clinical strains.
Little is known about the transcriptional regulation of ampD or the permease gene ampG, both of which are essential for the recycling of muropeptides (18, 31, 33). Changes in the transcriptional regulation of these genes may affect the concentrations of the muropeptides, which affect the activity of AmpR. Thus, decreased expression of AmpD or increased expression of AmpG may affect the concentration of the muropeptides (primary anhMurNac-tripeptides) that induces AmpR to an activator of ampC expression (13). Experiments involving DNA arrays are now in progress to study AmpC regulating genes. We are also in the process of screening PA-isolates from CF patients for the presence of IS1669, since a mobile insertion sequence could be important for the ability of P. aeruginosa to adapt in the complex lung environment of CF patients.
Acknowledgments
We thank Solveig Kreisby for technical assistance. We thank H. P. Schweizer for the pUCP22 shuttle plasmid.
The Danish Health Medical Research Council supported this study.
REFERENCES
- 1.Bagge, N., O. Ciofu, L. T. Skovgaard, and N. Høiby. 2000. Rapid development in vitro and in vivo of resistance to ceftazidime in biofilm-growing Pseudomonas aeruginosa due to chromosomal β-lactamase. APMIS 108:589-600. [DOI] [PubMed] [Google Scholar]
- 2.Bush, K., G. A. Jacoby, and A. A. Medeiros. 1995. A functional classification scheme for β-lactamases and its correlation with molecular structure. Antimicrob. Agents Chemother. 39:1211-1233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell, J. I. A., O. Ciofu, and N. Høiby. 1997. Pseudomonas aeruginosa isolates from patients with cystic fibrosis have different β-lactamase expression phenotypes but are homogeneous in the ampC-ampR genetic region. Antimicrob. Agents Chemother. 41:1380-1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caroff, N., E. Espaze, D. Gautreau, H. Richet, and A. Reynaud. 2000. Analysis of the effects of −42 and −32 ampC promoter mutations in clinical isolates of Escherichia coli hyperproducing AmpC. J. Antimicrob. Chemother. 45:783-788. [DOI] [PubMed] [Google Scholar]
- 5.Ciofu, O., B. Giwercman, S. S. Pedersen, and N. Høiby. 1994. Development of antibiotic resistance in Pseudomonas aeruginosa during two decades of antipseudomonal treatment at the Danish CF Center. APMIS 102:674-680. [PubMed] [Google Scholar]
- 6.Dietz, H., D. Pfeifle, and B. Wiedemann. 1996. Location of N-acetylmuramyl-l-alanyl-d-glutamylmesodiaminopimelic acid, presumed signal molecule for β-lactamase induction, in the bacterial cell. Antimicrob. Agents Chemother. 40:2173-2177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietz, H., D. Pfeifle, and B. Wiedemann. 1997. The signal molecule for β-lactamase induction in Enterobacter cloacae is the anhydromuramyl-pentapeptide. Antimicrob. Agents Chemother. 41:2113-2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Domingue, G., B. Ellis, M. Dasgupta, and J. W. Costerton. 1994. Testing antimicrobial susceptibilities of the adherent bacteria by a method that incorporates guidelines of the national committee for clinical laboratory standards. J. Clin. Microbiol. 32:2564-2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ehrhardt, A. F., C. C. Sanders, J. R. Romero, and J. S. Leser. 1996. Sequencing and analysis of four new Enterobacter ampD alleles. Antimicrob. Agents Chemother. 40:1953-1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Giwercman, B., P. A. Lambert, V. T. Rosdahl, G. H. Shand, and N. Høiby. 1990. Rapid emergence of resistance in Pseudomonas aeruginosa in cystic fibrosis patients due to in vivo selection of stable partially derepressed β-lactamase producing strains. J. Antimicrob. Chemother. 26:247-259. [DOI] [PubMed] [Google Scholar]
- 11.Höltje, J. V., U. Kopp, A. Ursinus, and B. Wiedemann. 1994. The negative regulator of β-lactamase induction AmpD is a N-acetyl-anhydromuramyl-l-alanine amidase. FEMS Microbiol. Lett. 122:159-164. [DOI] [PubMed] [Google Scholar]
- 12.Jacobs, C., B. Joris, M. Jamin, K. Klarsov, J. Van Beeumen, D. Mengin-Lecreulx, J. Van Heijenoort, J. T. Park, S. Normark, and J. M. Frère. 1995. AmpD, essential for both β-lactamase regulation and cell wall recycling, is a novel cytosolic N-acetylmuramyl-l-alanine amidase. Mol. Microbiol. 15:553-559. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs, C., J. M. Frère, and S. Normark. 1997. Cytosolic intermediates for cell wall biosynthesis and degradation control inducible β-lactam resistance in gram-negative bacteria. Cell 88:823-832. [DOI] [PubMed] [Google Scholar]
- 14.Jacobs, C., L. J. Huang, E. Bartowsky, S. Normark, and J. T. Park. 1994. Bacterial cell wall recycling provides cytosolic muropeptides as effectors for β-lactamase induction. EMBO J. 13:4684-4694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Johansen, H. K., F. Espersen, S. S. Pedersen, H. P. Hougen, J. Rygaard, and N. Høiby. 1993. Chronic Pseudomonas aeruginosa lung infection in normal and athymic rats. APMIS 101:207-225. [PubMed] [Google Scholar]
- 16.Kharazmi, A., B. Giwercman, and N. Høiby. 1999. Robbins device in biofilm research. Methods Enzymol. 310:207-215. [DOI] [PubMed] [Google Scholar]
- 17.Kopp, U., B. Wiedemann, S. Lindquist, and S. Normark. 1993. Sequences of wild-type and mutant ampD genes of Citrobacter freundii and Enterobacter cloacae. Antimicrob. Agents Chemother. 37:224-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korfmann, G., and C. C. Sanders. 1989. ampG is essential for high-level expression of AmpC β-lactamase in Enterobacter cloacae. Antimicrob. Agents Chemother. 33:1946-1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Korfmann, G., C. C. Sanders, and E. S. Moland. 1991. Altered phenotypes associated with ampD mutations in Enterobacter cloacae. Antimicrob. Agents Chemother. 35:358-364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kuga, A., R. Okamoto, and M. Inoue. 2000. ampR gene mutations that greatly increase class C β-lactamase activity in Enterobacter cloacae. Antimicrob. Agents Chemother. 44:561-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Langaee, T. Y., M. Dargis, and A. Huletsky. 1998. An ampD gene in Pseudomonas aeruginosa encodes a negative regulator of AmpC β-lactamase expression. Antimicrob. Agents Chemother. 42:3296-3300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Langaee, T. Y., L. Gagnon, and A. Huletsky. 2000. Inactivation of the ampD gene in Pseudomonas aeruginosa leads to moderate-basal-level and hyperinducible AmpC β-lactamase expression. Antimicrob. Agents Chemother. 44:583-589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lindberg, F., S. Lindquist, and S. Normark. 1987. Inactivation of the ampD gene causes semiconstitutive overproduction of the inducible Citrobacter freundii β-lactamase. J. Bacteriol. 169:1923-1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lindquist, S., M. Galleni, F. Lindberg, and S. Normark. 1989. Signalling proteins in enterobacterial AmpC β-lactamase regulation. Mol. Microbiol. 3:1091-1102. [DOI] [PubMed] [Google Scholar]
- 25.Lindquist, S., F. Lindberg, and S. Normark. 1989. Binding of the Citrobacter freundii AmpR regulator to a single DNA site provides both autoregulation and activation of the inducible ampC β-lactamase gene. J. Bacteriol. 171:3746-3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lodge, J., S. Busby, and L. Piddock. 1993. Investigation of the Pseudomonas aeruginosa ampR gene and its role at the chromosomal ampC β-lactamase promoter. FEMS Microbiol. Lett. 111:315-320. [DOI] [PubMed] [Google Scholar]
- 27.Lodge, J. M., S. D. Minchin, L. J. V. Piddock, and S. J. W. Busby. 1990. Cloning, sequencing, and analysis of the structural gene and regulatory region of the Pseudomonas aeruginosa chromosomal ampC β-lactamase. Biochem. J. 272:627-631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lodge, J. M., and L. J. V. Piddock. 1991. The control of class I β-lactamase expression in Enterobacteriaceae and Pseudomonas aeruginosa. J. Antimicrob. Chemother. 28:167-172. [DOI] [PubMed] [Google Scholar]
- 29.Medeiros, A. A. 1997. Evolution and dissemination of β-lactamases accelerated by generations of β-lactam antibiotics. Clin. Infect. Dis. 24:S19-S45. [DOI] [PubMed] [Google Scholar]
- 30.Mouton, J. W., J. G. den Hollander, and A. M. Horrevorts. 1993. Emergence of antibiotic resistance amongst Pseudomonas aeruginosa isolates from patients with cystic fibrosis. J. Antimicrob. Chemother. 31:919-926. [DOI] [PubMed] [Google Scholar]
- 31.Normark, S. 1995. β-Lactamase induction in gram-negative bacteria is intimately linked to peptidoglycan recycling. Microb. Drug Resist. 1:111-114. [DOI] [PubMed] [Google Scholar]
- 32.O'Callaghan, C. H., A. Morris, S. M. Kirby, and A. H. Shingler. 1972. Novel method for detection of β-lactamases by using a chromogenic cephalosporin substrate. Antimicrob. Agents Chemother. 1:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Park, J. T. 1996. The convergence of murein recycling research with β-lactamase research. Microb. Drug Resist. 2:105-112. [DOI] [PubMed] [Google Scholar]
- 34.Pedersen, S. S., G. H. Shand, B. L. Hansen, and G. N. Hansen. 1990. Induction of experimental chronic Pseudomonas aeruginosa lung infection with P. aeruginosa entrapped in alginate microspheres. APMIS 98:203-211. [PubMed] [Google Scholar]
- 35.Peter, K., G. Korfmann, and B. Wiedemann. 1988. Impact of the ampD gene and its product on β-lactamase production in Enterobacter cloacae. Rev. Infect. Dis. 10:800-805. [DOI] [PubMed] [Google Scholar]
- 36.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 37.Stapleton, P., K. Shannon, and I. Phillips. 1995. DNA sequence differences of ampD mutants of Citrobacter freundii. Antimicrob. Agents Chemother. 39:2494-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]