Abstract
Hereditary lymphedema is a chronic swelling of limbs due to dysfunction of lymphatic vessels. An autosomal dominant, congenital form of the disease, also known as “Milroy disease,” has been mapped to the telomeric part of chromosome 5q, in the region 5q34-q35. This region contains a good candidate gene for the disease, VEGFR3 (FLT4), that encodes a receptor tyrosine kinase specific for lymphatic vessels. To clarify the role of VEGFR3 in the etiology of the disease, we have analyzed a family with hereditary lymphedema. We show linkage of the disease with markers in 5q34-q35, including a VEGFR3 intragenic polymorphism, and we describe an A→G transition that cosegregates with the disease, corresponding to a histidine-to-arginine substitution in the catalytic loop of the protein. In addition, we show, by in vitro expression, that this mutation inhibits the autophosphorylation of the receptor. Thus, defective VEGFR3 signaling seems to be the cause of congenital hereditary lymphedema linked to 5q34-q35.
Introduction
Lymphedema is caused by a dysfunction of the lymphatic system. Lymphangiography demonstrates hypoplasia or aplasia of lymphatic channels (Kinmonth 1982). It provokes disfiguration and disabling swelling, mostly localized at the extremities (Esterly 1965). Familial lymphedema, usually segregating as an autosomal dominant trait, can be classified according to the age at onset, as early-onset lymphedema (primary congenital lymphedema or Milroy disease [MIM 153100]) or late-onset lymphedema (lymphedema praecox or Meige lymphedema [MIM 153200]). Lymphedema can also occur in association with other clinical features (e.g., autosomal dominant lymphedema with distichiasis that has been linked to 16q24.3 [MIM 153400]), or as a component of a syndrome (e.g., Turner or Noonan syndrome [MIM 163950]).
Linkage between early-onset lymphedema and markers in the telomeric part of chromosome 5q (5q34-q35) has been demonstrated for four families in one study (Ferrell et al. 1998). In another study, linkage with 5q35.3 has been demonstrated in five families, including one large four-generation family (Evans et al. 1999). The vascular endothelial growth factor receptor-3 (VEGFR3, FLT4) gene, located in this region, is a strong candidate for being causative because of its importance in lymphatic vessels. The gene is expressed in all vessels early during embryogenesis, and, thereafter, expression is limited to the developing and adult lymphatic vessels (Kaipainen et al. 1995). Disruption of Vegfr3 in mice leads to embryonic death at day 9.5 due to defective development of large blood vessels and cardiovascular failure (Dumont et al. 1998). Moreover, cutaneous overexpression of its ligand, vascular endothelial growth factor C (VEGFC), induces selective hyperplasia of the lymphatic vasculature (Jeltsch et al. 1997). A leucine for proline substitution (P1114L) has been detected in the VEGFR3 protein in one family with lymphedema (Ferrell et al. 1998). However, this family was too small to be informative for linkage, and no other putative mutation was detected in the four families sharing linkage to this region.
We describe a two-generation family with five individuals affected by congenital hereditary lymphedema, show linkage between the phenotype and markers in the 5q34-q35 region, and describe an H1035R substitution in a highly conserved residue in the catalytic loop of the VEGFR3 receptor. In addition, we show by in vitro expression studies that this amino acid substitution causes loss of VEGFR3 tyrosine kinase activity. Thus, defective VEGFR3 signaling is responsible for 5q34-q35–linked lymphedema.
Subjects and Methods
Family
Congenital lymphedema had been identified in all five affected family members (fig. 1A), and thus the family had been referred to a genetic counselor (K.D.). The father, individual I.1, had bilateral congenital lymphedema and frequent erysipelas. Individual II.2 had bilateral congenital lymphedema of the feet, more pronounced on the right side and not present above the ankles. Prominent veins were observed on the lower limbs. Individual II.3 had congenital lymphedema, limited to the dorsum of the right foot, and prominent left pretibial vein. Individual II.5 had bilateral congenital lymphedema involving the lower limbs below the knee. Individual II.7 had bilateral congenital lymphedema of the feet, extending to the lower part of the lower limbs. She was born at 32 weeks, with premature rupture of the membranes; hyalinic membrane disease, grade II; hyperbilirubinism; and transient hyperphenylalaninemia, probably unrelated to the disorder. In addition, the affected children (subjects II.2, II.3, II.5, and II.7) had flat, broad nasal bridges and epicanthic folds. This was not obvious in their affected father. Venous blood samples were drawn after informed consent was obtained from all family members, as approved by the ethics committee of Katholieke Universiteit Leuven.
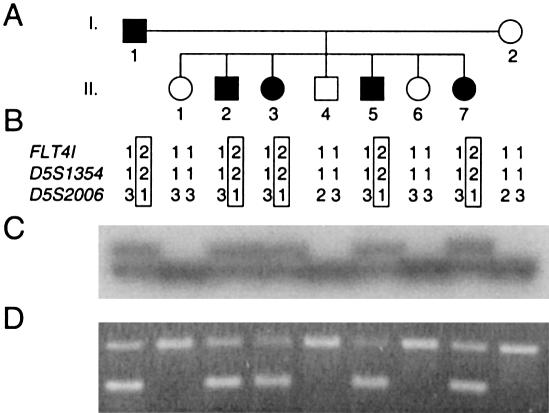
Figure 1.
A, Pedigree of the family with inherited congenital lymphedema. Blackened symbols represent affected individuals. B, Deduced haplotypes for three markers in 5q34-35. FLT4I, VEGFR3 intragenic polymorphism. Haplotype transmitted with disorder is boxed. C, Autoradiogram of the intragenic FLT4I marker. D, Allele-specific PCR for the A3123G mutation. Lower band, mutated allele. Upper band, internal control for PCR.
Linkage Analysis
Genomic DNA was extracted from buffy coats (QIAamp DNA Blood Mini Kit [Qiagen]). Individuals were genotyped as described elsewhere (Boon et al. 1994). For linkage analyses, we used microsatellite markers D5S1354 and D5S2006, located in the VEGFR3 region (Ferrell et al. 1998), as well as an intragenic polymorphism of the VEGFR3 gene (detectable by primers FLT4-CA-F: 5′-CCTTGGGCAAGTCGTGGC-3′ and FLT4-CA-R: 5′-GAGAGAGACTCCATCAGG-3′ (Ferrell et al. 1998). Linkage analyses were performed with autosomal dominant mode of inheritance, either 100% or 90% penetrance, a disease allele frequency of 1/10,000, and equal allele frequencies for markers. Calculations were performed with Linkage Package version 5.1 (Lathrop et al. 1984).
VEGFR3 Gene Analysis
To screen part of the VEGFR3 gene encoding the intracellular kinase domain for mutations by SSCP and heteroduplex analyses, primers (synthesized by Gibco BRL) were designed on the basis of intronic sequences (K. Iljin, M. J. Karkkainen, E. C. Lawrence, M. A. Kimak, M. Uutela, J. Taipale, R. E. Ferrell, and K. Alitalo, unpublished data). The primer sequences (from 5′ to 3′) are as follows: FLT4I-1: CAACCCAGTCAGCTCCTTC, FLT4I-2: ACCTGTCTCCACGCTCACC, FLT4I-3: CTGCTCCTCACCAGCTAGG, FLT4I-4: GAACGGGGACCTGCCAGG, FLT4I-5: GGGGTCTCGCCGTCCCAG, FLT4I-6: CGGGTGCAAACGCGGAGC, FLT4I-7: GCAGAGGCGCCTCCATTCC, FLT4I-8: TCTGCGGCGGACGACTGG, FLT4I-9: GGCCCGTCAGGCACTTGG, FLT4I-10: CCTGGGGCTGGGTAGATGG, FLT4I-11: GTGGGGATGCACCCTTTTCC, FLT4I-12: TATTACCCCAGGGGCCTGC, FLT4I-13: TCACCTGTTCCGCCCCACG, FLT4I-14: TCCCTCCGTCTCCCCATCC, FLT4I-15: CGCTTGCTGTCCCCAAAACC, FLT4I-16: ACGGGGGTGGGGTGCATG, FLT4I-17: CAGGGGCCAAAGGCCATAG, FLT4I-18: CAGTGGCTGGTGGTTTCTGG, FLT4I-19: GCTTTCTCCCACCCTACTCC, FLT4I-20: GCTGCAGCGCGTTCCTCAG, FLT4I-21: CAGCCCTGAGCCGAGAGC, FLT4I-22: TGCAGGAGGGCCTCAGGC, FLT4I-23: AGGGCAACATCGATACCTGC, FLT4I-24: CTGGCCACGTGGGCGCTG. Primers were end-labeled with 32P, using polynucleotide kinase (Takara) and γ[32P]-ATP (Amersham Pharmacia Biotech), and were used to amplify exons 16–27 (according to the numbering of Iljin et al., unpublished data). These exons encode the intracellular part of the receptor. Only genomic DNA was available from this family, and thus only the intracellular part of VEGFR3, for which the exon-intron structure had been identified, was examined. The PCRs were performed in a volume of 20 μl as described by Boon et al. (1999), with the following cycling conditions: 95°C for 5 min; 32 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 30 s; and 72°C for 10 min. The size of the amplification products varied between 184 and 241 bp. The PCR reactions were divided into two aliquots before loading onto nondenaturing polyacrylamide gels (MDE Mutation Detection gel solution, FMC BioProducts). EDTA (final concentration, 5 mM) and nondenaturing loading buffer (according to FMC) were added to the reactions for heteroduplex analysis, whereas a denaturing loading buffer (according to FMC) was added to the SSCP samples. After heat denaturation, the samples for SSCP analysis were immediately loaded onto SSCP gels. The samples for heteroduplex analysis were first cooled from 95°C to 37°C at 1°C/min, to increase the formation of heteroduplexes. Both gels were run for 14–16 h, SSCP gels at constant power (6–8 W) and heteroduplex gels at constant voltage (700 V). Gels were vacuum dried and exposed for 12–24 h to a Kodak X-Omat film (Amersham Pharmacia Biotech). PCR fragments showing abnormal migration were cloned. First, 3′-A overhangs were further added to the PCR products, using Biotools Tth DNA polymerase (B&M Labs) and dATP (Gibco BRL). The PCR products were then ligated in a pBluescript-SK+ (Stratagene) T vector prepared in the laboratory according to the protocol described elsewhere (Zhou et al. 1995). After transformation and overnight liquid culture, plasmid DNA was purified (Bio Rad Quantum Prep Kit). The inserts were cycle sequenced (Gibco BRL dsDNA Cycle Sequencing System) with 33P end-labeled M13-forward and M13-reverse primers, and the reactions were run on a denaturing polyacrylamide sequencing gel. After vacuum drying, the gels were exposed for 12–24 h to Kodak BioMax MR film (Amersham Pharmacia Biotech).
Allele-Specific PCR
To detect the A3123G transition in genomic DNA, an allele-specific primer, FLT4I-3123G (5′-CGCACCCCAGTGCATCCG-3′), was used with primer FLT4I-13. This primer pair produces a specific band of 180 bp only for the allele with a G in position 3123 of the cDNA. Primer FLT4I-12 was also added to the PCR mix, to produce a constant control band of 224 bp with the primer FLT4I-13. The concentration of each primer was 1 μM and that of dNTP was 50 μM, and we used 0.5 U Biotools Tth polymerase (B&M Labs) in a final volume of 50 μl. The cycling conditions were as follows: 95°C for 5 min; 35 cycles of 94°C for 30 s, 64°C for 30 s, and 72° for 30 s; and 72°C for 10 min. PCR fragments were resolved on a 3% Metaphor agarose gel (FMC Bioproducts).
Phosphorylation Analysis of Wild Type (WT) and H1035R Mutant VEGFR3
To analyze the phosphorylation of the WT and mutant VEGFR3 receptors, we used the long form of human VEGFR3 cDNA cloned into pcDNA3.1/Z(+) vector backbone (Invitrogen) (Korpelainen et al. 1999). For H1035R mutant receptor expression vector, the corresponding A→G mutation at nucleotide 3123 of cDNA was generated by the GeneEditor in vitro Site-Directed Mutagenesis System (Promega), using the oligonucleotide 5′-AAGTGCATCCGCAGAGACCTG-3′.
To study phosphorylation of the corresponding mutant VEGFR3, the plasmids encoding WT and mutant VEGFR3 were transfected into subconfluent 293T cells, using the calcium phosphate method and 1:0, 3:1, 1:1, 1:3 and 0:1 ratios of WT to mutant plasmid. The cells were grown in Dulbecco's modified Eagle's medium supplemented with 10% FCS (Gibco BRL), glutamine, and antibiotics. An empty vector was used for mock transfections. After 36 h, the cell monolayers were washed twice with cold PBS containing 2 mM vanadate and 2 mM phenylmethylsulfonyl fluoride (PMSF) and were lysed into PLCLB buffer (150 mM NaCl, 5% glycerol, 1% Triton X-100, 1.5 M MgCl2, and 50 mM Hepes, pH 7.5) containing 2 mM Vanadate, 2 mM PMSF, and 0.07 U/ml Aprotinin. The lysates were centrifuged for 10 min at 19,000 g, and the supernatants were incubated for 2 h on ice with 2 μg/ml of monoclonal anti-VEGFR3 antibodies (9D9f9) (Jussila et al. 1998). The immunocomplexes were incubated with protein A sepharose (Pharmacia) for 45 min with rotation at 4°C. The sepharose beads were washed three times with cold PLCLB buffer (containing 2 mM vanadate and 2 mM PMSF), analyzed by 7.5 % SDS-PAGE, and transferred to a nitrocellulose filter (Protran Nitrocellulose, Schleicher & Schuell), using semidry transfer apparatus. After blocking with 5% BSA in TBS-T buffer (10 mM Tris pH 7.5, 150 mM NaCl, and 0.1 % Tween 20), the filters were incubated with the phosphotyrosine-specific primary antibodies (Upstate Biotechnology), followed by biotinylated goat anti-mouse immunoglobulins (Dako) and biotin-streptavidin horseradish peroxidase (HRP) complex (Amersham). The bands were visualized by the enhanced chemiluminescence method. The filters were stripped with occasional agitation in 100 mM 2-mercaptoethanol, 2% SDS, and 62.5 mM Tris-HCl, pH 6.7, for 30 min at 55°C, washed with TBS-T, blocked, and analyzed for the presence of VEGFR3, using the 9D9f9 antibodies and HRP-conjugated rabbit anti-mouse immunoglobulins (Dako).
Results
The family identified showed autosomal dominant inheritance with high penetrance (fig. 1A). Linkage analysis was performed with microsatellite markers located around the VEGFR3 gene, in the 5q34-q35 region. The marker results for D5S1354 and D5S2006 were fully informative for linkage, as well as for the VEGFR3 intragenic polymorphism (fig. 1B and 1C). For these three markers, the LOD scores were 1.8 at recombination fraction (θ) 0 with 100% penetrance and 1.68 at θ=0 with 90% penetrance. These are the maximum LOD scores for this family.
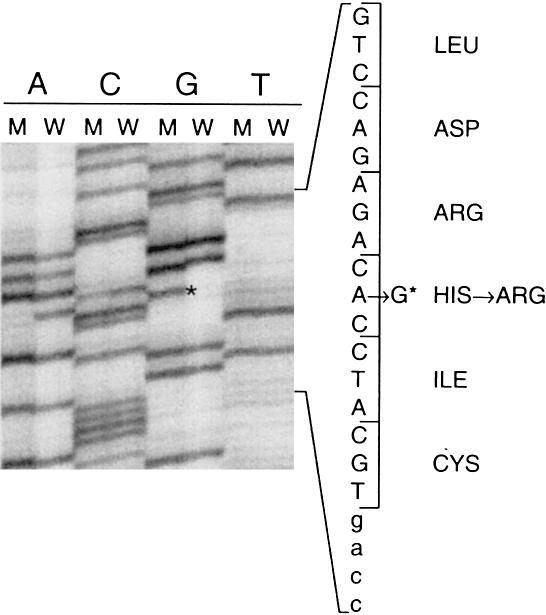
The exons corresponding to the intracellular part of the protein were screened for mutations with SSCP and heteroduplex analysis. A shift was observed in affected individuals for the PCR fragment corresponding to exon 21. This fragment was reamplified from genomic DNA of individual I.1 and cloned into a T vector. Six cloned fragments were sequenced, revealing the presence of an A→G transition, visible on both strands in two clones at nucleotide 3123 of the cDNA (Genbank accession number X68203) (fig. 2). This nucleotide change corresponds to a histidine-to-arginine substitution in the catalytic loop of the VEGFR3 receptor tyrosine kinase (residue 1035 of the protein, Swissprot accession number P35916). With an allele-specific PCR, we checked the cosegregation of this substitution with the disease in the family (fig. 1D). Similarly, we checked that this substitution was not present in 92 control individuals (184 alleles) of mixed European origin (data not shown).
Figure 2.
Autoradiogram showing sequences of two clones corresponding to wild-type (W) and mutant (M) alleles in individual I.1. Small letters, intronic sequence; capital letters, exonic sequence. * = mutation.
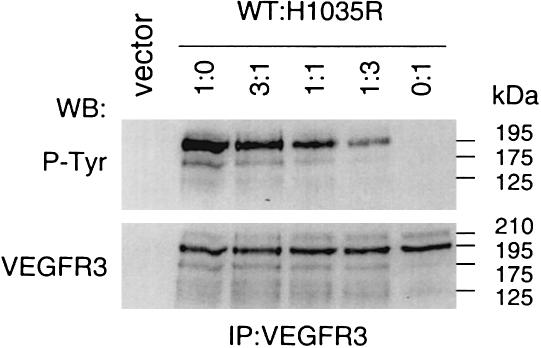
To test whether the single amino acid substitution identified in the family with lymphedema alters VEGFR3 function, the corresponding mutant receptor cDNA was generated in VEGFR3 expression vector by site-directed mutagenesis and analyzed by transient transfection. Under these conditions, overexpression results in ligand-independent dimerization and activation of the receptor tyrosine kinase, thus allowing its transphosphorylation to be studied. Transfections were made using 1:0, 3:1, 1:1, 1:3, and 0:1 ratios of WT and H1035R mutant VEGFR3 plasmids (fig. 3). The effect of the mutation on VEGFR3 tyrosyl phosphorylation was analyzed by immunoprecipitation with anti-VEGFR3 monoclonal antibodies followed by western blotting with antiphosphotyrosine antibodies (P-Tyr). In this analysis, the mature full-length VEGFR3 shows as a 195-kD polypeptide band, and smaller amounts of the VEGFR3 polypeptides of 175 kD and 125 kD represent the partially glycosylated precursor and the proteolytically processed receptor subunit chain containing the tyrosine kinase domain (Pajusola et al. 1994). In addition, minor amounts of a 210-kD form of VEGFR3 were observed in all samples. This form may be generated by additional glycosylation of the mature protein. No tyrosyl phosphorylation of the H1035R VEGFR3 was detected, whereas the WT receptor was strongly phosphorylated (fig. 3). Moreover, phosphorylation of the total receptor protein decreased with increasing amount of mutant vector transfected. Expression of equal amounts of the VEGFR3 protein was confirmed by immunoblotting with antibodies directed toward the extracellular domain of VEGFR3.
Figure 3.
Effect of H1035R mutation on tyrosyl autophosphorylation of VEGFR3. Expression plasmids for WT and mutant VEGFR3 were transfected into 293T human embryonic kidney cells (lanes 1:0 and 0:1, respectively) or cotransfected using 3:1, 1:1, and 1:3 ratios of WT to mutant plasmid. Receptor phosphorylation was analyzed from VEGFR3 immunoprecipitates by western blotting using antiphosphotyrosine antibodies (P-Tyr, upper part). Total amount of receptor protein is shown in VEGFR3 western blotting (lower part). Sizes on the right correspond to four different forms of VEGFR3 (see text).
Discussion
Using four multigeneration families with inherited congenital lymphedema, Ferrell et al. (1998) obtained a peak multipoint LOD score of 10 at marker D5S1354. They analyzed the cDNA sequence of the vascular endothelial growth factor receptor-3 gene (VEGFR3, FLT4) and found a P1114L substitution in one of the families. This family, however, was not big enough to be informative for linkage, and no putative mutations were found in the families with linkage for the parts of the gene analyzed. One family was recombinant for 5q34-q35, thus suggesting the existence of another gene responsible for early-onset lymphedema. More recently, another study has been published establishing a genetic linkage of congenital lymphedema with 5q35.3, with a combined multipoint LOD score of 16.55 (Evans et al. 1999). Thus, a gene causing congenital hereditary lymphedema maps to the distal part of chromosome 5q and could be VEGFR3 (FLT4).
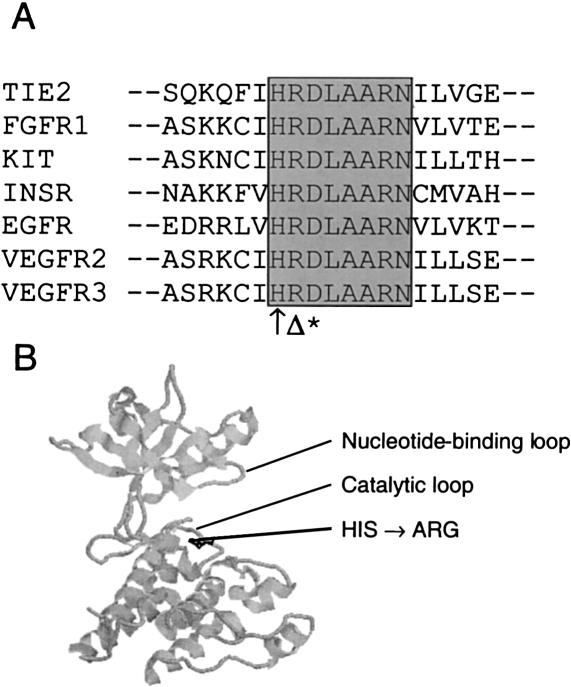
We describe the identification of a two-generation family with five individuals affected by congenital lymphedema. This family also showed linkage to 5q34-q35, and an A3123G transition corresponding to an H1035R substitution was identified in the affected family members. This transition was not seen in the genomic DNA of 92 control individuals. The substituted histidine is conserved in protein tyrosine kinases and in many serine/threonine kinases (fig. 4A; see Hanks and Hunter [1995] for a review on protein kinases), suggesting a very important structural and/or functional role. It is the first residue of the consensus sequence −HRDLAARN−, the central part of the catalytic loop of VEGFR3, with the aspartate residue considered to be the key catalytic residue. The position of the mutated residue is shown in the three-dimensional structure of the kinase domain of VEGFR2 homologous receptor (fig. 4B) (McTigue et al. 1999). For the part of the protein modeled, this receptor presents 77% identity with VEGFR3 at the amino acid level, with no insertions or deletions, and is thus likely to adopt a very similar structure. Of interest, mutation of the adjacent arginine to a glycine in the KIT (mast/stem cell growth factor receptor [MIM 172800]) (Spritz et al. 1993). For all these reasons, we thought that the H1035R mutation was likely to cause an impairment of VEGFR3 function.
Figure 4.
A, Amino acid sequence alignment for seven different human tyrosine-kinase receptors. Conserved catalytic loop residues are highlighted with gray background. ↑ = mutated histidine (H1035R). Δ = residue mutated in KIT causing piebaldism (R→G). * = catalytic aspartate. B, Crystal structure of VEGFR2, a tyrosine kinase receptor homologous to VEGFR3. Histidine residue mutated in VEGFR3 receptor of patients with lymphedema shown in black. The kinase activation loop was disordered in the crystal and is not shown.
In fact, our in vitro expression studies showed that the H1035R VEGFR3 has lost its autophosphorylation capacity. The H1035R mutation is located in the catalytic domain of VEGFR3, not in a region of tyrosyl autophosphorylation sites. Thus, the loss of autophosphorylation capacity by H1035R indicates loss of tyrosine kinase activity, rather than lack of kinase site recognition, and lymphedema is caused by lack of sufficient VEGFR3 signaling. Since the disorder is inherited as an autosomal dominant trait with high penetrance, it seems to be enough to lose the tyrosine kinase activity of one of the copies of the receptor. However, it is possible that, in the affected endothelia, additional alteration in VEGFR3 signaling occurs. This will be interesting to test when affected tissue becomes available from a patient with a verified VEGFR3 mutation.
An interesting aspect of this family is the appearance of mild specific facial features in the four affected members of the youngest generation. Each of the four had a flat, broad nasal bridge and epicanthic folds. This has not been reported for any of the other families with linkage to 5q34-q35 and thus could be specific for this family, and perhaps for this mutation. However, these facial features are seen in several other syndromes with edema (Opitz 1986).
VEGFR3 is not the only mutated gene causing lymphedema. Since one family with early-onset lymphedema has been shown to be recombinant in 5q34-q35 (Ferrell et al. 1998), a second gene must exist. Moreover, lymphedema with distichiasis—abnormal hairs arising from meibomian glands in the inner eyelid—has been mapped to chromosome 16q24.3 (Mangion et al. 1999). However, the linked interval of 16 cM does not contain any obvious candidate gene. Whether the products of these other genes function with VEGFR3 or in another pathway is not known. In addition, the cause of lymphedema in syndromes such as Turner or Noonan still needs to be discovered.
Finally, identification of VEGFR3 as the causative gene for hereditary congenital lymphedema might lead to some advances in the treatment of the disorder. Lymphedema is characterized by hypoplastic or aplastic lymphatic channels. Thus, loss of adequate VEGFR3 signaling seems to cause diminished or delayed development of lymphatic channels. On the contrary, overexpression of the ligand VEGFC in the skin induces hyperplasia of the lymphatic channels in transgenic mice. Although mice and humans differ (e.g. mice with loss of one Vegfr3 allele do not develop apparent peripheral lymphedema), local VEGFC supplementation might ameliorate the condition.
Acknowledgments
The authors thank the family members for their invaluable participation. Studies from the authors' laboratories were supported by grants from the Finnish Cancer Organization, the Finnish Cultural Foundation, the Ida Montini Foundation, the Emil Aaltonen Foundation, the Finnish Academy and the European Union (Biomed grant PL 963380 [to M.J.K. and K.A.]), and the Fonds de Développement Scientifique–Université catholique de Louvain, the Belgian Federal Service for Scientific, Technical and Cultural Affairs and European Commission (grant ERB4001GT963858 [to M.V.]). K.D. is a Senior Clinical Investigator of the Fund for Scientific Research–Flanders (Belgium) (F.W.O.–Vlaanderen). A.I. is supported by a fellowship from Fonds pour la formation à la recherche dans l’industrie et dans l’agriculture (F.R.I.A.).
Note added in proof.—While this article was in press, similar results were published by Karkkainen et al. (2000 [Nat Genet 25:153–159]).
Electronic-Database Information
Accession numbers and URLs for data in this article are as follows:
- GenBank, http://www.ncbi.nlm.nih.gov/Genbank/index.html (for accession numbers X68203 for VEGFR3 short form and S66407 for c-terminal tail of VEGFR3 long form)
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim (for early-onset lymphedema [MIM 153100], late-onset lymphedema [MIM 153200], lymphedema with distichiasis [MIM 153400], piebaldism [MIM 172800], and Noonan syndrome [MIM 163950])
- SwissProt, http://www.expasy.ch/sprot/ (for VEGFR3, accession number P35916)
References
- Boon LM, Brouillard P, Irrthum A, Karttunen L, Warman ML, Rudolph R, Mulliken JB, et al (1999) A gene for inherited cutaneous venous anomalies (“glomangiomas”) localizes to chromosome 1p21-22. Am J Hum Genet 65:125–133 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boon LM, Mulliken JB, Vikkula M, Watkins H, Seidman J, Olsen BR, Warman ML (1994) Assignment of a locus for dominantly inherited venous malformations to chromosome 9p. Hum Mol Genet 3:1583–1587 [DOI] [PubMed] [Google Scholar]
- Dumont DJ, Jussila L, Taipale J, Lymboussaki A, Mustonen T, Pajusola K, Breitman M, et al (1998) Cardiovascular failure in mouse embryos deficient in VEGF receptor-3. Science 282:946–949 [DOI] [PubMed] [Google Scholar]
- Esterly JR (1965) Congenital hereditary lymphoedema. J Med Genet 2:93–98 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans AL, Brice G, Sotirova V, Mortimer P, Beninson J, Burnand K, Rosbotham J, et al (1999) Mapping of primary congenital lymphedema to the 5q35.3 region. Am J Hum Genet 64:547–555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrell RE, Levinson KL, Esman JH, Kimak MA, Lawrence EC, Barmada MM, Finegold DN (1998) Hereditary lymphedema: evidence for linkage and genetic heterogeneity. Hum Mol Genet 7:2073–2078 [DOI] [PubMed] [Google Scholar]
- Hanks SK, Hunter T (1995) Protein kinases 6: the eukaryotic protein kinase superfamily: kinase (catalytic) domain structure and classification. FASEB J 9:576–596 [PubMed] [Google Scholar]
- Jeltsch M, Kaipainen A, Joukov V, Meng X, Lakso M, Rauvala H, Swartz M, et al (1997) Hyperplasia of lymphatic vessels in VEGF-C transgenic mice. Science 276:1423–1425 [DOI] [PubMed] [Google Scholar]
- Jussila L, Valtola R, Partanen TA, Salven P, Heikkila P, Matikainen MT, Renkonen R, et al (1998) Lymphatic endothelium and Kaposi's sarcoma spindle cells detected by antibodies against the vascular endothelial growth factor receptor-3. Cancer Res 58:1599–1604 [PubMed] [Google Scholar]
- Kaipainen A, Korhonen J, Mustonen T, van Hinsbergh VW, Fang GH, Dumont D, Breitman M, et al (1995) Expression of the fms-like tyrosine kinase 4 gene becomes restricted to lymphatic endothelium during development. Proc Natl Acad Sci USA 92:3566–3570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinmonth JB (1982) Methods of lymphography. In: Kinmonth JB (ed) The lymphatics: diseases, investigation and treatment. Arnold, London, pp 1–17 [Google Scholar]
- Korpelainen EI, Karkkainen M, Gunji Y, Vikkula M, Alitalo K (1999) Endothelial receptor tyrosine kinases activate the STAT signaling pathway: mutant TIE-2 causing venous malformations signals a distinct STAT activation response. Oncogene 18:1–8 [DOI] [PubMed] [Google Scholar]
- Lathrop GM, Lalouel JM, Julier C, Ott J (1984) Strategies for multilocus linkage analysis in humans. Proc Natl Acad Sci USA 81:3443–3446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangion J, Rahman N, Mansour S, Brice G, Rosbotham J, Child AH, Murday VA, et al (1999) A gene for lymphedema-distichiasis maps to 16q24.3. Am J Hum Genet 65:427–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McTigue MA, Wickersham JA, Pinko C, Showalter RE, Parast CV, Tempczyk-Russell A, Gehring MR, et al (1999) Crystal structure of the kinase domain of human vascular endothelial growth factor receptor 2: a key enzyme in angiogenesis. Structure Fold Des 7:319–330 [DOI] [PubMed] [Google Scholar]
- Opitz JM (1986) On congenital lymphedema. Am J Med Genet 24:127–129 [DOI] [PubMed] [Google Scholar]
- Pajusola K, Aprelikova O, Pelicci G, Weich H, Claesson-Welsh L, Alitalo K (1994) Signalling properties of FLT4, a proteolytically processed receptor tyrosine kinase related to two VEGF receptors. Oncogene 9:3545–3555 [PubMed] [Google Scholar]
- Spritz RA, Holmes SA, Itin P, Kuster W (1993) Novel mutations of the KIT (mast/stem cell growth factor receptor) proto-oncogene in human piebaldism. J Invest Dermatol 101:22–25 [DOI] [PubMed] [Google Scholar]
- Zhou MY, Clark SE, Gomez-Sanchez CE (1995) Universal cloning method by TA strategy. Biotechniques 19:34–35 [PubMed] [Google Scholar]