Abstract
Unequal crossover has long been suspected to play a role in the germline-specific instability of tandem-repeat DNA, but little information exists on the dynamics and processes of unequal exchange. We have therefore characterized new length alleles associated with flanking-marker exchange at the highly unstable human minisatellite CEB1, which mutates in the male germline by a complex process often resulting in the gene conversion–like transfer of repeats between alleles. DNA flanking CEB1 is rich in single-nucleotide polymorphisms (SNPs) and shows extensive haplotype diversity, consistent with elevated recombinational activity near the minisatellite. These SNPs were used to recover mutant CEB1 molecules associated with flanking-marker exchange, directly from sperm DNA. Mutants with both proximal and distal flanking-marker exchange were shown to contribute significantly to CEB1 turnover and suggest that the 5′ end of the array is very active in meiotic unequal crossover. Coconversions involving the interallelic transfer of repeats plus immediate flanking DNA were also common, were also polarized at the 5′ end of CEB1, and appeared to define a conversion gradient extending from the repeat array into adjacent DNA. Whereas many mutants associated with complete exchange resulted in simple recombinant-repeat arrays that show reciprocity, coconversions were highly gain-biassed and were, on average, more complex, with allele rearrangements similar to those seen in the bulk of sperm mutants. This suggests distinct recombination-processing pathways producing, on the one hand, simple crossovers in CEB1 and, on the other hand, complex conversions that sometimes extend into flanking DNA.
Introduction
Higher-eukaryotic genomes contain a wide variety of tandem-repeat DNA sequences differing in repeat size, array length, chromosomal distribution, and the processes of mutation that generate allele length variation. In humans, GC-rich minisatellites are preferentially found clustered in the recombination-proficient subtelomeric regions of chromosomes (Royle et al. 1988; Amarger et al. 1998) and sometimes can show substantial germline-specific instability detectable both in families (Jeffreys et al. 1988; Vergnaud et al. 1991) and in gametic (sperm) DNA (Jeffreys et al. 1994). Some unstable minisatellites, including human minisatellites MS32 (D1S8) and MS31A (D7S21), show significant similarity between their repeat sequence and the Chi sequence (Jeffreys et al. 1985; Wong et al. 1987), an element that promotes recombination in Escherichia coli, a finding that led to early suggestions that unequal crossover between misaligned alleles might be involved in generation of allele diversity (Jeffreys et al. 1985). Although flanking-marker analysis of new mutant alleles has ruled out unequal crossover as a major mechanism (Wolff et al. 1988, 1989; Vergnaud et al. 1991), detailed analysis of the structural rearrangements occurring within alleles during germline mutation has revealed a recombination-based mutation process most probably occurring at meiosis. These rearrangements, seen at all minisatellites studied (Buard and Vergnaud 1994; Jeffreys et al. 1994; May et al. 1996; Andreassen and Olaisen 1998; Buard et al. 1998; Tamaki et al. 1999), are often complex and can include duplications, deletions, and gene conversion–like transfers of blocks of repeats between alleles. Occasionally, however, minisatellite length changes can be associated with exchange of DNA markers flanking the repeat array, as seen, in families, at minisatellite MS31A and, in sperm, at minisatellite MS32 (Jeffreys et al. 1998b). In most cases, these exchanges appear to arise by classic unequal crossover between alleles, resulting in simple recombinant repeat arrays and exchange of all flanking markers; such mutants establish that unequal crossover does indeed play a role in minisatellite mutation. In a few cases, however, exchange is incomplete, indicating that mutation can sometimes involve the conversional transfer, between alleles, not only of repeat DNA but also of proximal, but not distal, flanking markers. We refer to such conversion events involving the minisatellite plus adjacent flanking DNA as “coconversions.”
Our understanding of unequal crossover and coconversion processes at human minisatellites is far from complete, given both the technical difficulty of analysis of reciprocal unequal crossover events at MS32 in sperm and the rarity of coconversion events at this locus (Jeffreys et al. 1998b). We have therefore extended these analyses to minisatellite CEB1 (D2S90), a highly unstable locus that, for some alleles, has male-specific mutation rates as high as 20%/sperm (Buard et al. 1998). Although the 40-bp CEB1 repeat unit shows no significant similarity to the Chi element, mutation at this locus is complex, germline-specific, and recombinational in nature, although most mutations involve intra-allelic rearrangements rather than conversional transfers of repeats between alleles (Buard et al. 1998). It has been proposed that repair, by either strand invasion of the allelic partner or single-strand annealing, of a meiosis-specific double-strand break (DSB) initiated by staggered nicks in the tandem array could account for the diversity and complexity of minisatellite rearrangements observed in sperm (Buard and Vergnaud 1994; Buard and Jeffreys 1997). Both the small size (<3 kb; Vergnaud et al. 1991) of most CEB1 alleles and their extreme germline instability should facilitate crossover and coconversion analysis. We have therefore identified single-nucleotide polymorphisms (SNPs) near CEB1 and have used these to recover and characterize flanking marker-exchange events in sperm DNA.
Material and Methods
Detection of SNPs
SNPs were identified by resequencing of 2.7 kb of 5′ flanking DNA and 2.5 kb of 3′ flanking DNA around CEB1 (GenBank accession number AF048727), in four Europeans and four Africans (Zimbabweans). Sequences of overlapping amplicons that were, on average, 700 bp long, were determined by Big Dye chemistry (PE Biosystems), assembled by AutoAssembler software (PE Biosystems), and screened visually for variants. Each potential base-substitutional polymorphism was validated either by PCR-RFLP or by allele-specific PCR (Newton et al. 1989).
Allele-Specific Primers and Universal Primers
Sequences of the CEB1 minisatellite variant repeat (MVR)–specific primers used for allele-structural analysis and of the allele-specific primers −4A, −4C, −72A, −72G, +256A, +256G, +384A, and +384G have been reported elsewhere (Buard and Vergnaud 1994; Buard et al. 1998). Primers are named according to whether they are located 5′ (−) or 3′ (+) to the repeat array, with the number corresponding to the distance (in base pairs) between the 3′ base of the oligonucleotide and the repeat array. Allele-specific primers are further discriminated by their 3′ nucleotide. All primers were orientated toward the repeat array, except for −183 and +39. The universal (not allele-specific) primers used were −3556 (cccttgctgaaggctgcgtgtg), −196 (gaggctgagaccccagcagtg), −183 (aagcgtggacacacctagacctg), +39 (tcctgccaggtaaagggaaagtg), +782 (gggtaactggatgctaaaac), and +2496 (gcccaggccagaatctcagagg). The allele-specific primers were −631G (tcccgaacagctccacag), −631T (tcccgaacagctccacat), −934C (cagccacccgccccccc), −934T (cagccacccgcccccct), −1088G (gtcaccgggaggccgg), −1088A (gtcaccggggaggccga), −2085G (ctcggcagatgtgaggag), −2085T (ctcggcagatgtgaggat), −2690G (ctgtggggtgggggctgcg), −2690T (ctgtggggtgggggctgct), +774G (ggatgctgaaaactgtgagg), +774T (ggatgctgaaaactgtgagt), +1017A (accgcagcggccactga), +1017C (accgcagcggccactgc), +1246G (cttctcaatgcacagcccg), +1246A (cttctcaatgcacagccca), +1529G (agggtatatgacagccacg), +1529A (agggtatatgacagccaca), +1713G (cagagctggggtggggg), +1713A (cagagctggggtgggga), +2248G (agctgggaggggggcgg), +2248A (agctgggaggggggcga), +2476T (gccacaggtscccggtat), and +2476A (gccacaggtscccggtaa).
SNP Typing
The 5′ and 3′ regions of CEB1 were amplified from 15 ng of genomic DNA/individual, in 10-μl PCR reactions, at 96°C for 45 s, followed by 30 cycles of treatment at 96°C for 30 s, 68°C for 30 s, and 70°C for 3 min, with PCR buffer described elsewhere (Jeffreys et al. 1990), plus 1 μM primers −3556/−183 or +39/+2496, respectively. Genotypes for most SNPs were established by multiplex allele-specific PCR using these amplicons. One nanogram of PCR product was reamplified at 96°C for 45 s, followed by 11 cycles of treatment at 96°C for 30 s, 59°C for 20 s, and 70°C for 2.5 min, in three reactions each containing 1 μM universal primer (−183 or +36) plus a subset of allele-specific primers at 0.08–0.2 μM each and chosen to minimize the risk of primer incompatibility. The three sets of PCR products were compared by electrophoresis on a 1% Seakem LE agarose gel (FMC BioProducts) and by visualization based on staining with ethidium bromide, to determine, for each SNP site, which allele-specific primer(s) successfully produced the appropriate-sized PCR product. 5′ and 3′ Haplotypes were similarly established by allele-specific PCR, by use of the most distal heterozygous SNP per individual, to create two amplicons corresponding to the two haplotypes, followed by multiplex allele-specific PCR as described above, to type the status of more-proximal heterozygous SNPs. The +129C/T SNP creates a Psp1406I+/− restriction-site polymorphism and, instead, was typed by PCR-RFLP analysis.
Selection of Sperm Donors for Detection of CEB1 Recombinants
Two sperm donors (individuals A and B) were chosen for detection of CEB1 rearrangements associated with flanking-marker exchange. Individual A was heterozygous for alleles AU (41 repeats, linked to −72A, −4C, and +384G) and AL (18 repeats, linked to −72G, −4A, and +384A). Allele AU showed a 4.7% sperm-mutation rate, with ∼70% of mutations involving gains of repeats. Allele AL had a 1.4% mutation rate, with 90% “gain” mutants divided approximately equally between intra-allelic duplications and interallelic transfers of repeats. Individual B showed a typical, 12.8% mutation rate at CEB1, with alleles BU (44 repeats, 65% gain mutants and linked to −4A and +256A) and BL (29 repeats, 70% gain mutants and linked to −4C and +256G) and with 10%–20% of gain mutants showing interallelic transfers of repeats, for both alleles. Alleles AL, BU, and BL are alleles C, E, and F, respectively, in the study by Buard et al. (1998).
Detection of Recombinants in Fractionated Sperm DNA
The preparation of sperm DNA and all subsequent manipulations were carried out under conditions designed to minimize the risk of contamination (Jeffreys et al. 1990, 1994). Size enrichment of CEB1 mutant molecules from 10 μg of sperm DNA was performed as described elsewhere (Jeffreys and Neumann 1997; Buard et al. 1998), following digestion with restriction enzyme BglI, which releases CEB1 plus 843 bp of upstream and 950 bp of downstream flanking sequence. The number of amplifiable mutant molecules contained in each size fraction was estimated by long PCR (Cheng et al. 1994) with 1 μM universal primers −196 and +782, in the presence of 5 μg of herring-sperm DNA/ml as carrier, plus Taq and Pfu DNA polymerases (20:1 ratio, 0.05 U/μl), 50 mM Tris base, and 9% (w/v) glycerol. Different fractions were subsequently pooled to generate a relatively homogeneous distribution of mutant molecules across the size window tested. Multiple aliquots of this pool were preamplified by long PCR with primers −196 and +782, as described above, with cycling at 96°C for 45 s, followed by 18 cycles of treatment at 96°C for 30 s, 66°C for 20 s, and 70°C for 6 min. PCR products were diluted 100-fold into 5 mM Tris-HCl (pH 7.5) and 5 μg of herring-sperm DNA/ml, and 1.5 μl of this dilution was used to seed a new, 7-μl PCR reaction (without addition of Pfu polymerase) containing combinations of 5′ and 3′ allele-specific primers for detection of potential recombinants. Touchdown-PCR cycling conditions were 96°C for 45 s, followed by 10 cycles of treatment at 96°C for 30 s, A1°C for 20 s, and 70°C for 4 min and then 12 cycles of treatment at 96°C for 30 s, A2°C for 20 s, and 70°C for 4 min. Annealing temperatures A1°C and A2°C were optimized for each primer combination, as follows: −72G/+384G, 67°C and 65°C; −4A/+384G, 69°C and 67°C; −72A/+384A, 66°C and 64°C; −4C/+384A, 70°C and 68°C; −4C/+256A, 69°C and 67°C; and −4A/+256G, 67°C and 65°C. PCR products were analyzed by electrophoresis in a 40-cm-long 1% Seakem HGT agarose gel, followed by Southern blot hybridization with a [32P]-labeled CEB1 probe, as described elsewhere (Buard and Vergnaud 1994). Mutants showing flanking exchange were reamplified for 15 cycles and were purified after agarose-gel electrophoresis and visualization based on staining with ethidium bromide. The structure of mutant CEB1 repeat arrays was determined by MVR-PCR, as described elsewhere (Buard and Vergnaud 1994; Buard et al. 1998).
Detection of Recombinants in Unfractionated Sperm DNA
Multiple 3-ng aliquots of HincII-digested sperm DNA, each containing ∼800 amplifiable CEB1 molecules, were preamplified in 10 μl of long-PCR reactions, with 1 μM each of distal primers −3556 and +2496, for 96°C for 45 s, followed by 19 cycles of treatment at 96°C for 30 s, 66°C for 20 s, and 70°C for 10 min. These batches of preamplified CEB1 molecules, including progenitor alleles and mutants, were diluted 100- fold, as described above, and 1.5 μl of each dilution was used to seed secondary normal PCR reactions containing 0.2 μM of each 5′ and 3′ allele-specific primer. Cycling was at 96°C for 45 s, followed by C1 cycles of treatment at 96°C for 30 s, A1°C for 20 s, and 70°C for 4 min and by C2 cycles of treatment at 96°C for 30 s, A2°C for 20 s, and 70°C for 4 min. Optimal annealing temperatures/cycles A1°C/C1 and A2°C/C2 were −4A/+256G, 70°C/10 and 69°C/10; −4A/+1529G, 64°C/10 and 62°C/15; and −1088C/+256G, 66°C/11 and 64°C/12. PCR products were analyzed by gel electrophoresis and Southern blot hybridization, as described above.
Results
SNPs in DNA Flanking Minisatellite CEB1
Sequence analysis of 16 chromosomes from four Europeans and four Africans revealed 10 SNPs in 2.7 kb of 5′ flanking region and 21 SNPs in 2.5 kb of 3′ flanking. These 31 base-substitutional polymorphisms consisted of 15 transitions, 13 transversions, and 3 insertion/deletions. Eighteen of these SNPs were shared by the sequenced European and African individuals.The overall nucleotide diversity in Europeans and Africans, estimated from the normalized number of variant sites by Watterson's (1975) statistic, is 2.7×10-3 and 2.4×10-3 respectively, compared with 5.4×10-4 and 6.8×10-4 estimated for these two populations in large-scale SNP surveys of noncoding sequences of the human genome (Halushka et al. 1999). SNPs therefore are unusually abundant in DNA flanking CEB1.
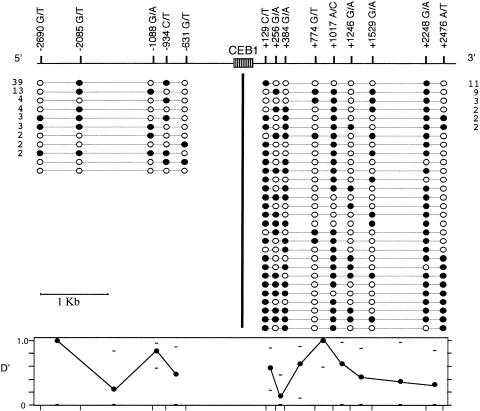
To investigate patterns of haplotype diversity around CEB1, we used multiplex allele-specific PCR (see the Material and Methods section) to establish complete 5′ and 3′ haplotypes from 37 and 26 different Europeans, respectively (fig. 1). Haplotype diversity was lower upstream of CEB1, in part because of the relative paucity of SNPs in this region and in part because of the generally low heterozygosities of these SNPs. In contrast, 9 SNPs 3′ to CEB1 defined 29 different haplotypes among 52 chromosomes tested, among which 23 haplotypes were observed only once. Analysis of adjacent SNPs revealed all four possible haplotypes for most intervals, both upstream and downstream of CEB1, establishing that recombination as well as base substitution has actively contributed to haplotype diversification. Corresponding linkage-disequilibrium analysis showed essentially free association between most pairs of adjacent markers, even those as close as 128 bp. Taken together, these results suggest that CEB1 is located within a recombinationally active region of the human genome.
Figure 1 .
Haplotype diversity around minisatellite CEB1. European SNP haplotypes upstream and downstream of CEB1 (74 and 52 haplotypes, respectively) are shown, with the status of each locus indicated by a blackened or unblackened circle, respectively, for the two alleles shown (e.g., −2690G [blackened] and −2690T [unblackened]). The number of occurrences of each haplotype found more than once is shown. 5′ and 3′ Haplotypes were not linked into single haplotypes spanning CEB1. The normalized disequilibrium measure D′ of Lewontin (Hedrick 1988) was determined for each adjacent pair of markers, together with the upper and lower 95% confidence limits.
Detection of Sperm Mutants Showing Exchange of Flanking Markers
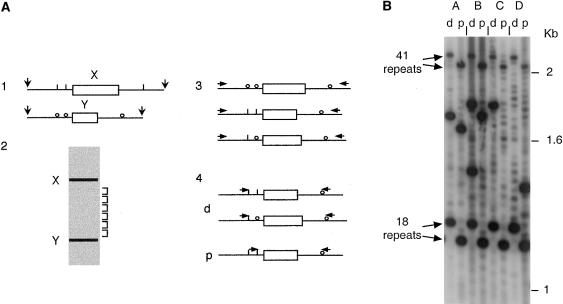
To determine whether CEB1 mutants are ever associated with flanking-marker exchange, we isolated recombinant CEB1 molecules directly from sperm DNA, using methods developed previously for crossover analysis at minisatellite MS32 (Jeffreys et al. 1998b). Two African sperm donors (individuals A and B; for genotypes and mutation rates, see the Material and Methods section) were chosen who were heterozygous at several flanking sites on each side of the tandem array and who had CEB1 alleles with a size range of 18–44 repeats (0.72–1.76 kb). Mutant CEB1 molecules were enriched from sperm DNA by size fractionation (fig. 2A). Batches of enriched mutant molecules were then preamplified by long-PCR using universal primers distal to flanking SNP sites, ensuring that all molecules, whether recombinant or not, were amplified with equal efficiency. These primary PCR products were then surveyed by PCR using allele-specific primers on each side of the minisatellite, in repulsion phase, to selectively amplify any molecules showing exchange between markers 5′ and 3′ to the repeat array.
Figure 2.
Detection of CEB1 sperm mutants associated with flanking-marker exchange. A, Schematic representation of the size-enrichment and small-pool PCR strategy. 1, Sperm DNA from a man multiply heterozygous for SNPs flanking CEB1, digested with restriction enzymes releasing fragments containing both the SNPs and the CEB1 minisatellite. 2, Size fractionation of digested DNA. The DNA was electrophoresed; and fractions were collected depleted in progenitor alleles X and Y and, therefore, enriched in CEB1 mutant molecules with abnormal array length. 3, Pooled fractions containing mutant molecules, whether recombinant or not, amplified with distal universal primers. 4, Preamplified mutants subjected to allele-specific PCR, in repulsion phase with various combinations of allele specific-primers, to selectively detect any mutants associated with distal (d) or proximal (p) marker exchange. B, Example of detection of sperm mutants associated with flanking-marker exchange. Batches (A–D) of preamplified sperm mutants with sizes intermediate between the 18- and 41-repeat progenitor alleles of individual A were screened by allele-specific PCR for distal (−72G/+384G; lanes d) and more proximal (−4A/+384G; lanes p) flanking exchanges. Low-level mispriming from the wrong allele allows the progenitor alleles and nonexchanged mutants in each batch to be detected. True exchange mutants show much stronger hybridization signals. Two mutants show exchange of both 5′ markers (batch A and the larger mutant in batch B), two show distal but not proximal exchange (batch C and the smaller mutant in batch B), and one shows proximal but not distal exchange (batch D).
CEB1 sperm mutants from individual A that were 19–39 repeats long, selected because they were intermediate in size between the two progenitor alleles AL (18 repeats) and AU (41 repeats), were screened in batches of, on average, 20 mutants (96 batches in total), for exchanges between the −72A/G and −4A/C heterozygous SNPs, located, respectively, 72 bp and 4 bp upstream of CEB1, and the +384A/G SNP, located 384 bp downstream of the minisatellite. Figure 2B shows an example of such screening. None of the recombinant mutants detected fell outside the selected size window, and all show a much stronger and “clonal” signal intensity relative to the continuous background ladder, strongly suggesting that these are authentic sperm mutants rather than template-switching or base-misincorporation PCR artifacts.
Of the 1,920 mutants screened, derived from ∼48,000 amplifiable molecules, 44 showed association between both 5′ markers from allele AL and the 3′ marker from allele AU, and 31 showed the reciprocal combination, of 5′ AU/3′ AL markers (table 1). These similar numbers are consistent with reciprocal unequal crossover and suggest an unequal crossover rate of 1.6×10-3/sperm (i.e., 75 crossovers in 48,000 sperm analyzed). This rate will be an underestimate, since any recombinant molecules lying outside the selected size window of 19–39 repeats will have been lost during size enrichment. There also were four examples of exchange of distal (but not of proximal) markers, yielding recombinants with the −72 marker from AU linked to the −4 and +384 markers from AL and indicating an extremely high rate of exchange within the very small (67 bp) physical interval between −72 and −4.
Table 1.
CEB1 Molecules in Sperm DNA That Are Associated with Different Combinations of Flanking Markers
| No. of CEB1 Repeats | Flanking Markers | No. of Molecules | ||||
| A. Individual Aa |
||||||
| −72 |
−4 |
CEB1 |
+384 |
|||
| Progenitor AU: | ||||||
| 41 | A | C | –––– | G | ||
| Progenitor AL: | ||||||
| 18 | g | a | –––– | a | ||
| Total | ∼46,000 | |||||
| No exchange: | ||||||
| 19–39 | A | C | –––– | G | ∼700 | |
| g | a | –––– | a | ∼1,000 | ||
| Exchanges: | ||||||
| 19–39 | A | C | –––– | a | 44 | |
| g | a | –––– | G | 31 | ||
| 19–39 | g | C | –––– | a | 91 | |
| A | a | –––– | G | 9 | ||
| 19–39 | g | C | –––– | G | 4 | |
| A |
a |
–––– |
a |
0 | ||
| B. Individual B—Two Size Ranges of CEB1 Mutantsb |
||||||
| −4 |
CEB1 |
+256 |
||||
| Progenitor BU: | ||||||
| 44 | A | –––– | A | |||
| Progenitor BL | ||||||
| 29 | c | –––– | g | |||
| Total | ∼14,500 | |||||
| No exchange: | ||||||
| 31–42 | A | –––– | A | ∼500 | ||
| c | –––– | g | ∼600 | |||
| Exchanges: | ||||||
| 31–42 | A | –––– | g | 15 | ||
| c | –––– | A | 5 | |||
| No exchange: | ||||||
| 46–75 | A | –––– | A | ∼950 | ||
| c | –––– | g | ∼50 | |||
| Exchanges: | ||||||
| 46–75 | A | –––– | g | 15 | ||
| c |
–––– |
A |
9 |
|||
| C. Individual B: Unfractionated Sperm DNAc |
||||||
| −1088 |
−4 |
CEB1 |
+256 |
+1529 |
No. of Sperm |
|
| Progenitor BU: | ||||||
| 44 | C | A | –––– | A | A | |
| Progenitor BL: | ||||||
| 29 | t | c | –––– | g | g | |
| Total | ∼17,400 | |||||
| No exchange: | ||||||
| 5–100 | C | A | –––– | A | A | ∼1,400 |
| t | c | –––– | g | g | ∼1,200 | |
| Exchanges: | ||||||
| 5–100 | C | A | –––– | g | g | 40 |
| t | A | –––– | g | g | 4 | |
| C | A | –––– | g | A | 1 | |
One size range (19–39 repeats) of CEB1 mutants was screened, after size fractionation of sperm DNA, for exchanges between two markers 5′ and one marker 3′ of the tandem array. Given the respective mutation rates and gain biases for allele AL and AU, the number of mutants showing losses of repeats of AU or gains in AL (including ∼55% intra-allelic duplications and ∼45% interallelic transfers without exchange) could be estimated as being 700 and 1,000, respectively, within the size window screened.
Two size ranges (31–42 repeats and 46–75 repeats) of CEB1 mutants were screened for exchange between one proximal marker 5′ and one proximal marker 3′ of CEB1 after fractionation.
Unfractionated sperm DNA was screened for exchanges involving one distal and one proximal marker on each side of the tandem array. Other combinations involving exchanges of distal and proximal markers were not tested.
This survey also revealed many cases of coconversion events (fig. 2B and table 1A). The most common class, seen 91 times in the 1,920 mutants screened, showed association between the proximal 5′ −4 marker from allele AU and the distal 5′ −72 and 3′ +384 markers from allele AL. The reciprocal combination—the combination of the proximal 5′ marker from AL and the distal markers from AU—was 10 times less frequent (frequency 9/1,920). This apparent asymmetry of coconversion events probably results from the size range of mutants scored, given the strong tendency of conversion events within the repeat array itself to be associated with gains of repeat units; the size window analyzed therefore will contain conversional transfers from allele AU into AL, but not vice versa. The specific gain bias of events with proximal marker exchange alone is particularly striking if one compares the 10:1 disparity of coconversion events (gC––a vs. Aa––G; see table 1A) with the 3:2 ratio between events with exchange of both proximal and distal markers (ga––G vs. AC––a).
Flanking-marker–exchange analysis was extended to sperm DNA from a second source, individual B, with 44- and 29-repeat CEB1 alleles (alleles BU and BL, respectively). Mutant molecules intermediate in size between the two alleles (31–42 repeats) and larger than allele BU (46–75 repeats) were screened for exchanges of single 5′ and 3′ SNP markers at −4 and +256. Again, exchanges were seen in both size ranges, at an overall frequency of 2.7×10-3/sperm (i.e., 44 exchanges/16,500 sperm analyzed; see table 1B). Furthermore, there was, as before, evidence of exchange asymmetry, particularly in the intermediate-size mutants (15 A–––g mutants vs. 5 c–––A mutants), suggesting that some of these exchanges may be the result of coconversion events, and not of crossover.
Both to investigate marker exchange farther away from the CEB1 minisatellite and to prevent biases arising from size selection (see the Material and Methods section), unfractionated sperm DNA from individual B was analyzed by including more-distal 5′ and 3′ markers, located, respectively, 1,088 and 1,529 bp away from the array. Screening of 108 ng of DNA (equivalent to ∼20,000 sperm analyzed) yielded 45 exchange mutants with 5′ markers from allele BU that were associated with 3′ markers from allele BL (because of poor specificity of the corresponding allele-specific primers, the reciprocal combination of markers could not be tested) (table 1C). Thirty-six of these mutants lay within the size window previously screened for exchanges after DNA fractionation, giving an exchange frequency slightly lower, albeit not significantly different from, that previously established (χ2=0.42, 1 df; P>.7) and suggesting that interallelic-jumping PCR artifacts do not contribute significantly to the rearrangements detected in unfractionated DNA. Of the 45 mutants analyzed, 40 showed complete exchange of 5′ and 3′ markers, consistent either with unequal crossover or with conversion domains >1 kb. If they are true crossovers, this gives an unequal-crossover rate of 2×10-3 across CEB1, similar to other estimates for individuals A and B. Of the remaining exchanges, four showed conversion of the proximal −4 SNP site alone, suggesting that the immediate 5′ flanking DNA also is prone to conversions in individual B, although not at the frequency seen in individual A; and one mutant showed conversion of the proximal +256 site.
Minisatellite Rearrangements Associated with Flanking Exchange
To investigate the nature of CEB1 rearrangements associated with flanking exchange, 77 mutants recovered by size fractionation from individuals A and B and showing complete exchange of 5′ and 3′ markers were analyzed by MVR-PCR, to determine internal allele structure (fig. 3). Although only single 5′ and 3′ markers were used in the case of individual B, analysis of unfractionated sperm DNA showed that ∼90% of these mutants will show complete exchange of proximal and distal markers (see above). All mutants were different, as expected for products of meiotic recombination. Most of (65/77) these mutants showed recombinant repeat arrays. Of these, ∼50% (32/65), including mutants with junctional repeats that can come from one or the other progenitor allele (e.g., A4), showed a very simple structure consisting of a perfect fusion between the beginning of one allele and the end of the other allele.
Figure 3.
Structures of CEB1 sperm mutants associated with flanking-marker exchange. Mutants from individual A, showing partial or complete exchange of flanking markers −72, −4 and +384, and from individual B, with exchanges between markers −4 and +256, were subjected to MVR-PCR analysis (Jeffreys et al. 1991; Buard and Vergnaud 1994), and allele structures were compared with the larger and smaller progenitor alleles. Each letter within the MVR maps represents a different CEB1 variant repeat encoded as described elsewhere (Buard et al. 1998). The numbers of repeats and the flanking SNP status of each mutant also are shown, together with the origin of repeats (larger allele [green] and smaller allele [red]). Deletions are denoted by dashes, duplications by underlining of the 5′ duplication, the 3′ duplicate by boldface. Also shown are repeats of uncertain origin (black), junctional repeats that could be derived from either allele (pink), and repeats, at crossover junctions, that are interpreted as hybrid recombinant repeats (blue). Repeats showing slight sequence variations compared with the repeat originally found at the same position in the progenitor allele (i.e., one site change among the three sites tested by MVR-PCR) are shown in small caps ; these are rare and may have arisen by base misincorporation during PCR.
Furthermore, these simple fusion events appear to be completely reciprocal. In particular, in individual A there were eight mutants (mutants A4, A6–A8, A10, A13, A15, and A17) showing a simple fusion of the 5′ end of allele AL with a 3′ end of AU, together with eight reciprocal AU-AL recombinants (mutants A21, A23, A24, A27, A29, A31, A33, and A34). The AL-AU recombinants were, on average, 14.8 repeats shorter than allele AU, and, similarly, the AU-AL recombinants were 13.5 repeats larger than allele AL. More-detailed analysis of these size changes for the two classes of recombinants showed that they were undistinguishable (Student's t-test: t=.899, P=.384), strongly suggesting that this simple recombination process within the repeat array is completely reciprocal and does not result in the net gain or loss of repeats from the two interacting alleles. Similarly, for individual B, it is worth noting that the 8 “A–g” simple exchange mutations with size intermediate between the two progenitor alleles are, on average, six repeats shorter than allele BU and that the simple fusion mutant with a reciprocal combination of flanking markers within this size window (mutant B40) is six repeats larger than allele BL.
The remainder of the exchange mutants were associated with various rearrangements, including duplications of repeat motifs at the site of crossover (e.g., mutants A1 and A3), slight alteration of sequence, at the subrepeat level, close to the site of crossover (e.g., mutants A5 and B14), more-complex and uninterpretable rearrangements at the crossover junction (e.g., mutant B9), and duplication/deletion rearrangement distal to the site of crossover (e.g., mutants A10, B6, and B8). The remaining 12 putative exchange mutants either showed no evidence of recombinant arrays but, instead, rearrangements occurring within a single allele (e.g., mutants A19 and A20) or, alternatively, conversional transfer, without creation of a true recombinant array, of a repeat segment from one allele to the other (e.g., mutant B11).
MVR analysis of four mutants from individual A that showed CEB1 mutation accompanied by exchange in the interval between the 5′ markers 72 and 4 bp upstream of the minisatellite (mutants A49-A52; see fig. 3) revealed only intra-allelic deletions, with no evidence for recombinant repeat arrays. This raises the possibility that these mutants have arisen by two separate events—namely, an intra-allelic deletion and a crossover outside the repeat array.
Finally, MVR analysis was extended to 12 mutants from individual A that showed exchange limited only to the proximal 5′ marker at position −4, and not including the more distal −72 marker (mutants A37–A48). Nine of these mutants showed clear evidence of a recombinant repeat array, consistent with a single coconversion event transferring the flanking marker plus the beginning of the repeat array from one allele to the other. Only one of these mutants (mutant A47) contained a simple recombinant array; the remainder showed additional rearrangements, sometimes complex, within or adjacent to the conversion domain. Only two mutants (mutants A44 and A45) showed no evidence of interallelic transfers between repeat arrays; both instead showed association between a deletion within the beginning of the repeat array in the recipient allele and transfer of the flanking SNP site.
Discussion
A Gradient of Conversion Events at CEB1
Conversion-like events involving the transfer of minisatellite repeats between alleles are well documented for several unstable minisatellites (Jeffreys et al. 1994; Buard et al. 1998; Tamaki et al. 1999) and almost exclusively result in a gain of repeats. At CEB1, these rearrangements show polarity toward the 5′ end of the tandem array, although less strongly than for minisatellite MS32 (Buard et al. 1998). The present study shows that these polarized conversion events at CEB1 sometimes can extend into the proximal 5′ flanking DNA, resulting in coconversion of repeat DNA and a flanking SNP. These coconversions contribute significantly to minisatellite instability and show a gain bias similar to that observed for the bulk of interallelic transfers at CEB1. It is possible that biased repair of heterologies at flanking SNPs contained in putative long stretches of heteroduplex DNA also contribute to the apparent asymmetry of coconversion events. Elsewhere we have shown that approximately half of the gain mutants of allele AL are interallelic transfers without disjunction of the 5′ marker (7/16 in Buard et al. 1998), representing ∼440 mutants among the mutants screened in the present study (table 1A). Thus, interallelic events at allele AL can be subdivided roughly as follows: 75% of events with no exchange of flanking markers, 16% accompanied by coconversion of the immediate 5′ flanking marker 4 bp upstream of the repeat array (91 “gC––a” events in table 1A), and 8% with exchange of both −72 and −4 5′ markers (44 “AC––a” events). Even if the latter events were due to coconversions extending over both 5′ flanking markers, rather than unequal crossovers, this still implies a gradient of conversion events at CEB1, with maximum activity occurring within the beginning of the tandem array and declining upstream into 5′ flanking DNA. Such gradients of conversion have been observed in the yeast Saccharomyces cerevisiae, in which they are associated with meiotic recombination hotspots (Schultes and Szostak 1990). Curiously, individual B shows a markedly different contribution of coconversion to CEB1 instability, at least for the single exchange combination tested (table 1C). Only 10% of 5′ exchanges involve the immediate 5′ marker at position −4 alone, compared with 60% in individual A; the remaining 90% of recombinants in individual B show complete exchange for both the proximal and distal (−1088) markers, consistent with unequal crossover. Furthermore, the 5′ coconvertants account for only ∼2%–4% of interallelic mutants, compared with 16% in individual A. The reason for such conversion-efficiency variability between flanking sequences of different CEB1 alleles is unclear.
Unequal Crossovers and Coconversion Products: Reciprocity and Complexity
In contrast to the complexity of most interallelic events occurring at CEB1 (Buard and Vergnaud 1994; Buard et al. 1998), a substantial proportion of mutants with complete flanking-marker exchange show simple recombinant arrays. Although some of these apparent unequal crossovers may be coconvertants involving all markers tested, the simplicity of the process is very similar to that observed for unequal crossovers at minisatellites MS32 and MS31A (Jeffreys et al. 1998b). In contrast, coconversion events are associated more often with complex rearrangements in the tandem array, a result that is reminiscent of the complexity observed for the bulk of CEB1 interallelic events in sperm (Buard et al. 1998). Thus for individual A, 9/10 coconversions show complex rearrangements, compared with only 14/30 mutants with proximal and distal marker exchange (fig. 3; Fisher's exact test, two-tailed, P=.018).
Furthermore, the subset of simple fusion events among “complete”-exchange recombinants appears to be completely reciprocal, with the number of repeats gained for one combination of marker exchange being identical to the number of repeats lost for the reciprocal combination. In contrast, coconversion events are heavily biased toward expansions, a result reminiscent of the gain bias observed for interallelic transfers without involvement of flanking DNA. A nonconservative process for coconversion events is further substantiated by tetrad analysis of mutation events for human minisatellite MS1 integrated into the yeast genome, where most interallelic events are complex coconversion events occurring as single mutants in only one of the spores in a tetrad (Berg et al. 2000).
Altogether, these data strongly suggest that, at CEB1, simple fusion mutants are processed differently from conversion events and that these latter events, regardless of whether involving conversion of flanking DNA, are processed through a common pathway. On the basis of these striking differences, we suggest that sperm mutants consisting of simple fusion events and associated with complete flanking-marker exchange are genuine crossover products.
Unstable Minisatellites: Activity in Meiotic Recombination
To date, the best-characterized human minisatellites are MS32 and MS31A, both of which mutate, in the male germline, mainly by gene-conversion repeat-segment transfers between alleles that rarely extend into flanking DNA and, occasionally, by unequal crossover (Jeffreys et al. 1994, 1998b). The present study indicates that minisatellite CEB1, which shows no similarity to the Chi sequence and mutates mainly by intra-allelic rearrangement, also is proficient in meiotic crossover. Although only 1%–2% of CEB1 mutants show complete flanking-marker exchange associated with simple junctions, the absolute frequency of these events is extremely high for such a small physical interval (1.1–2.0 kb separating −4A/C and +384A/G). This indicates that meiotic unequal crossover occurs in CEB1 at a frequency ⩾70-fold higher than what would be expected on the basis of the mean rate of crossover at male meiosis (0.89 cM/Mb ; Weissenbach et al. 1992; Gyapay et al. 1994). CEB1 therefore constitutes a recombination hotspot, strengthening the evidence that meiotic recombination drives instability at GC-rich minisatellites.
As in the case of MS32 (Jeffreys et al. 1998a), crossover activity at CEB1 may not be limited to the repeat array itself but may also extend into flanking DNA, as shown by the breakdown of linkage disequilibrium near CEB1, even between very closely linked markers. Furthermore, there is more-direct evidence for potentially intense crossover activity in flanking DNA; for example, the interval 72–4 bp upstream of the array shows exchanges occurring at a frequency of 8×10-5/sperm, among CEB1 mutants (4 exchanges in 48,000 sperm; table 1A). This recombination activity in flanking DNA appears to be less intense, per unit of physical distance, on the other side of the repeat array, given the fact that only two exchange mutants have recombined in the interval between the end of CEB1 and either the +256 or the +384 3′ flanking marker (e.g., mutants A36 and B3; see fig. 3).
The recombination hotspot at CEB1 therefore could extend from the 5′ end of the minisatellite, for an as-yet-unknown distance, into the adjacent flanking DNA. If this is the case, it would be reminiscent of the localized crossover hotspot identified just upstream of minisatellite MS32 (Jeffreys et al. 1998a); however, direct analysis of the recombinational behavior of DNA flanking CEB1 will require the development of sperm DNA–typing systems capable of detecting all crossovers near CEB1, regardless of whether they are associated with minisatellite mutation (Jeffreys et al. 1998a).
Recombination Models for Minisatellite Instability: Human and Yeast
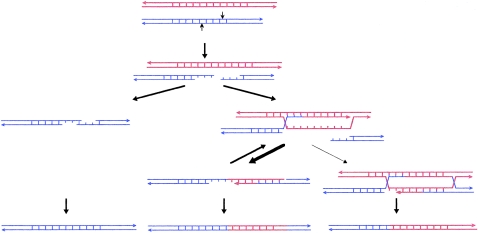
We propose an extension of the previous model for minisatellite instability, a model that involves staggered nicks and DSB repair (Buard and Vergnaud 1994; Buard and Jeffreys 1997). In this synthesis-dependent strand-annealing model (fig. 4), DNA breaks are most often formed within the tandem array near the 5′ end of the minisatellite. The resulting protruding single-stranded ends then invade the allelic partner or sister chromatid, allowing templated synthesis and extension of the ends. Most strand-invasion events are aborted after a limited extension of the broken single strands, perhaps as a result of mismatch-repair systems (MRS) detecting heterologies between interacting repeats of the two alleles. Both extended single strands then anneal together. Heteroduplexed regions detected by the MRS might result in separation of the two extended strands, initiating a new round of invasion/synthesis and single-strand annealing. This “flapping” and extension process of single strands (Pâques and Haber 1999) could readily account both for the observed complexity of minisatellite sperm rearrangements and, in particular, for highly complex, patchwork interallelic transfers (Tamaki et al. 1999). Ultimately, single-strand gaps are filled in, and heteroduplexed regions are repaired, producing interallelic insertions with no involvement of flanking DNA. In a few cases, DNA synthesis using the allelic partner as template would extend into the nonrepeated, flanking DNA (the case depicted in fig. 4). Again, some of these larger extension events would then be aborted, and single-strand annealing would take place between the extended strands. Heteroduplexed regions, extending outside the tandem array, would be repaired, sometimes by alternate use of the two strands to generate patchwork conversion events that involve flanking DNA. In a minority of cases, the process would not be aborted, and classical Holliday junctions would be formed and further resolved either as crossover products (the case depicted in fig. 4) or as simple conversion events.
Figure 4.
DSB repair and synthesis-dependent strand-annealing recombination model for minisatellite instability. Protruding single-strand ends formed by staggered nicks in a minisatellite allele can either anneal together out of register, producing an intra-allelic duplication after synthesis and ligation, or can invade the allelic partner. Most strand-invasion events are aborted, either before or, in a minority of cases, after synthesis extends into the minisatellite flanking DNA (depicted here). Reinvasion and synthesis at another location in the template allele frequently occur. Ultimately, extended strands anneal together out of register, single-strand gaps are filled in, and heteroduplexed regions are repaired. In a minority of cases, the extension process is not aborted, and Holliday junctions are formed and further resolved as either crossover products (depicted here) or simple conversion events.
A similar synthesis-dependent strand-annealing model, but one that includes DSBs formed outside the tandem array, has been proposed to account for meiotic rearrangements of CEB1 integrated into the yeast genome (Debrauwere et al. 1999). However, the few CEB1 interallelic events characterized in wild-type strains of S. cerevisiae do not show the complex rearrangements that are observed in the vast majority of these events in human sperm. In contrast, they are all very similar, showing simple fusion events associated with “complete”’ marker exchange, as described in the present study. Thus, DSBs formed outside the tandem array could occur in human sperm, and their repair would result in simple recombinant arrays, perhaps because of an initial interaction between the extending strand and the invaded strand, over almost identical flanking DNA sequences, that is more stable than the multiple mismatches arising from annealing of heterogeneous variant repeats in the minisatellite itself. Conversely, this predicts that a modified CEB1 yeast experimental system with DSBs occurring within the tandem array should produce complex interallelic transfers without flanking-sequence exchange. Further analyses of the dynamics of minisatellite mutation in human spermatogenic cells are necessary in order to allow us to characterize directly some of the recombination intermediates predicted by these models and, more generally, to dissect the mechanisms, at the molecular level, of meiotic recombination in man.
Acknowledgments
We thank Jane Blower for supplying semen samples from anonymous donors, and we thank Kathryn Lilley and Stuart Bayliss for oligonucleotide synthesis and assistance with automated sequencing. We are grateful to Yuri Dubrova, for statistical analyses; to colleagues, for helpful discussions; and to the anonymous reviewers of the manuscript, for their insightful comments. This work was supported in part by an International Research Scholars Award from the Howard Hughes Medical Institute and in part by grants from the Wellcome Trust, the Medical Research Council, and the Royal Society (all to A.J.J.). J.B. is a Medical Research Council fellow.
References
- Amarger V, Gauguier D, Yerle M, Apiou F, Pinton P, Giraudeau F, Montfouilloux S, et al (1998) Analysis of distribution in the human, pig, and rat genomes points toward a general sub-telomeric origin of minisatellite structures. Genomics 52:62–71 [DOI] [PubMed] [Google Scholar]
- Andreassen R, Olaisen B (1998) De novo mutations and allelic diversity at minisatellite locus D7S22 investigated by allele-specific four-state MVR-PCR analysis. Hum Mol Genet 7:2113–2120 [DOI] [PubMed] [Google Scholar]
- Berg I, Cederberg H, Rannug U (2000) Tetrad analysis shows that gene conversion is the major mechanism involved in mutation at the human minisatellite MS1 integrated in Saccharomyces cerevisiae. Genet Res 75:1–12 [DOI] [PubMed] [Google Scholar]
- Buard J, Bourdet A, Yardley J, Dubrova Y, Jeffreys AJ (1998) Influences of array size and homogeneity on minisatellite mutation. EMBO J 17:3495–3502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buard J, Jeffreys AJ (1997) Big, bad minisatellites. Nat Genet 15:327–328 [DOI] [PubMed] [Google Scholar]
- Buard J, Vergnaud G (1994) Complex recombination events at the hypermutable minisatellite CEB1 (D2S90). EMBO J 13:3203–3210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng S, Fockler C, Barnes WM, Higuchi R (1994) Effective amplification of long targets from cloned inserts and human genomic DNA. Proc Natl Acad Sci USA 91:5695–5699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Debrauwere H, Buard J, Tessier J, Aubert D, Vergnaud G, Nicolas A (1999) Meiotic instability of the human minisatellite CEB1 in yeast requires DNA double-strand breaks. Nat Genet 23:367–371 [DOI] [PubMed] [Google Scholar]
- Gyapay G, Morisette J, Vignal A, Dib C, Fizames C, Milasseau P, Marc S, et al (1994) The 1993–94 Généthon human genetic linage map. Nat Genet 7:246–339 [DOI] [PubMed] [Google Scholar]
- Halushka MK, Fan JB, Bentley K, Hsie L, Shen N, Weder A, Cooper R, et al (1999) Patterns of single-nucleotide polymorphisms in candidate genes for blood-pressure homeostasis. Nat Genet 22:239–247 [DOI] [PubMed] [Google Scholar]
- Hedrick PW (1988) Inference of recombinational hotspots using gametic disequilibrium values. Heredity 60:435–438 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, MacLeod A, Tamaki K, Neil DL, Monckton DG (1991) Minisatellite repeat coding as a digital approach to DNA typing. Nature 354:204–209 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Murray J, Neuman R (1998a) High resolution mapping of cross-overs in human sperm defines a minisatellite-associated recombination hotspot. Mol Cell 2:267–273 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Neil DL, Neumann R (1998b) Repeat instability at human minisatellites arising from meiotic recombination. EMBO J 17:4171–4157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffreys AJ, Neumann R (1997) Somatic mutation processes at a human minisatellite. Hum Mol Genet 6:129–136 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Neumann R, Wilson V (1990) Repeat unit sequence variation in minisatellites: a novel source of DNA polymorphism for studying variation and mutation by single molecule analysis. Cell 60:473–485 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Royle NJ, Wilson V, Wong Z (1988) Spontaneous mutation rates to new length alleles at tandem-repetitive hypervariable loci in human DNA. Nature 332:278–281 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Tamaki K, MacLeod A, Monckton DG, Neil DL, Armour JAL (1994) Complex gene conversion events in germline mutation at human minisatellites. Nat Genet 6:136–145 [DOI] [PubMed] [Google Scholar]
- Jeffreys AJ, Wilson V, Thein SL (1985) Hypervariable “minisatellite” regions in human DNA. Nature 314:67–73 [DOI] [PubMed] [Google Scholar]
- May CA, Jeffreys AJ, Armour JAL (1996) Mutation rate heterogeneity and the generation of allele diversity at the human minisatellite MS205 (D16S309). Hum Mol Genet 5:1823–1833 [DOI] [PubMed] [Google Scholar]
- Newton CR, Graham A, Heptinstall LE, Powell SJ, Summers C, Kalsheker N, Smith JC, et al (1989) Analysis of any point mutation in DNA: the amplification refractory mutation system (ARMS). Nucleic Acids Res 17:2503–2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pâques F, Haber JE (1999) Multiple pathways of double strand break-induced recombination in Saccharomyces cerevisiae. Microbiol Mol Biol Rev 63:349–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Royle NJ, Clarkson RE, Wong Z, Jeffreys AJ (1988) Clustering of hypervariable minisatellites in the proterminal regions of human autosomes. Genomics 3:352–360 [DOI] [PubMed] [Google Scholar]
- Schultes NP, Szostak JW (1990) Decreasing gradients of gene conversion on both sides of the initiation site for meiotic recombination at the ARG4 locus in yeast. Genetics 126:813–822 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaki K, May CA, Dubrova YE, Jeffreys AJ (1999) Extremely complex repeat shuffling during germline mutation at human minisatellite B6.7. Hum Mol Genet 8:879–888 [DOI] [PubMed] [Google Scholar]
- Vergnaud G, Mariat D, Apiou F, Aurias A, Lathrop M, Lauthier V (1991) The use of synthetic tandem repeats to isolate new VNTR loci: cloning of a human hypermutable sequence. Genomics 11:135–144 [DOI] [PubMed] [Google Scholar]
- Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Popul Biol 7:256–276 [DOI] [PubMed] [Google Scholar]
- Weissenbach J, Gyapay G, Dib C, Vignal A, Morissette J, Millasseau P, Vaysseix G, et al (1992) A second-generation linkage map of the human genome. Nature 359:794–801 [DOI] [PubMed] [Google Scholar]
- Wolff RK, Nakamura Y, White R (1988) Molecular characterization of a spontaneously generated new allele at a VNTR locus: no exchange of flanking DNA sequence. Genomics 3:347–351 [DOI] [PubMed] [Google Scholar]
- Wolff RK, Plaetke R, Jeffreys AJ, White R (1989) Unequal crossingover between homologous chromosomes is not the major mechanism involved in the generation of new alleles at VNTR loci. Genomics 5:382–384 [DOI] [PubMed] [Google Scholar]
- Wong Z, Wilson V, Patel I, Povey S, Jeffreys AJ (1987) Characterization of a panel of highly variable minisatellites cloned from human DNA. Ann Hum Genet 51:269–288 [DOI] [PubMed] [Google Scholar]