Abstract
A population study of heteroplasmy in the hypervariable region 1 (HV1) portion of the human mtDNA control region was performed. Blood samples from 253 randomly chosen individuals were examined using a sensitive denaturing gradient-gel electrophoresis (DGGE) system. This method is capable of detecting heteroplasmic proportions as low as 1% and virtually all heteroplasmy where the minor component is ⩾5%. Heteroplasmy was observed in 35 individuals (13.8%; 95% confidence interval [CI] 9.6–18.0). Of these individuals, 33 were heteroplasmic at one nucleotide position, whereas 2 were heteroplasmic at two different positions (a condition known as “triplasmy”). Although heteroplasmy occurred at a total of 16 different positions throughout HV1, it was most frequently observed at positions 16093 (n=13) and 16129 (n=6). In addition, the majority of heteroplasmic variants occurred at low proportions and could not be detected by direct sequencing of PCR products. This study indicates that low-level heteroplasmy in HV1 is relatively common and that it occurs at a broad spectrum of sites. Our results corroborate those of other recent reports indicating that heteroplasmy in the control region is more common than was previously believed—a finding that is of potential importance to evolutionary studies and forensic applications that are based on mtDNA variation.
Introduction
The control region (CR) of human mtDNA, which is the only significant noncoding portion of the mtDNA genome (Clayton 1992), contains two generally recognized hypervariable regions—hypervariable region 1 (HV1) and hypervariable region 2 (HV2) (Greenberg et al. 1983; Horai and Hayasaka 1990). Sequence variation in HV1 (length ∼350 bp) within human populations has been intensively studied, making it the single most important genetic locus for our understanding of human evolution and population genetics history (e.g., Vigilant et al. 1989, 1991; Richards et al. 1996). In addition, the level of polymorphism in HV1 is high enough to permit its use as an important tool in human identity testing (reviewed in Holland and Parsons 1999). This high level of polymorphism is thought to reflect an extremely high and variable rate of mutation in the CR (Parsons et al. 1997; Jazin et al. 1998; Parsons and Holland 1998; Excoffier and Yang 1999; Meyer et al. 1999). Numerous studies have resulted in the determination of thousands of individual HV1 sequences (Handt et al. 1998), specifying, within an individual, a single identifiable sequence type that very often can be distinguished from that of individuals from other mtDNA lineages. The presence of many hundreds of copies of mtDNA per cell, together with a high mutation rate, creates the potential for widespread heteroplasmy—that is, the presence of multiple sequence types within a cell, a tissue, or an individual. However, the observation of heteroplasmy in routine sequencing studies is the exception rather than the rule. This is thought to be due to a bottleneck in the transmission of a woman's mtDNA population to her offspring that greatly restricts the amount of mtDNA variation that is passed between generations (Hauswirth and Laipis 1982; Olivo et al. 1983; Koehler et al. 1991; Jenuth et al. 1996).
Heteroplasmy in human mtDNA was investigated in early cloning and sequencing studies done in the 1980s, with little sequence heterogeneity observed among tissues from normal individuals (Monnat and Loeb 1985; Monnat and Reay 1986; Bodenteich 1991). Heteroplasmy has been widely reported in association with mitochondrial disorders, where affected individuals harbor both mutant and wild-type mtDNA alleles (Wallace 1995); however, the pathogenic nature of these mutations ensures their detection even if they are rare. For a long time, on the basis of results of population sequencing studies, heteroplasmy in the CR was not considered to be a significant factor in CR sequence variation (Stoneking 1996). The first report of heteroplasmy in the CR was in the forensic identity case of Russian Tsar Nicholas II (Gill et al. 1994; Ivanov et al. 1996). Subsequently, there have been a number of reports of CR point-mutation heteroplasmy (Comas et al. 1995; Bendall et al. 1996; Howell et al. 1996; Jazin et al. 1996; Sullivan et al. 1996; Bendall et al. 1997; Parsons et al. 1997; Wilson et al. 1997). In addition, it is well documented that length heteroplasmy is commonly associated with polycytosine stretches within both HV1 (Bendall and Sykes 1995; Levin et al. 1996) and HV2 (Marchington et al. 1997; Parsons et al. 1997), apparently occurring quite readily in sequences that have a run of ⩾8 cytosines.
The frequency of CR point-mutation heteroplasmy is a question of importance, since it has a central relationship to the intrinsic rate of base misincorporation in mtDNA, the size of the intergenerational bottleneck, and the rate of sequence substitutions within lineages. The frequency of heteroplasmy is also of practical interest in forensic identity testing (Holland and Parsons 1999), where mtDNA analysis serves as a valuable and increasingly used tool for identification of old or degraded biological evidentiary samples. Although it is now clear that CR heteroplasmy occurs at an appreciable frequency, there has been no reliable quantitative estimate of the population incidence. (However, given the untold billions of mtDNAs in an individual, we are all certainly heteroplasmic at some minuscule level.) Direct sequence data have not provided a very useful estimate because of the difficulties in distinguishing heteroplasmy from various types of sequence background signal, although recent advances in automated fluorescent sequencing chemistry have reduced this problem. However, in some instances, sequence context–dependent signal-intensity artifacts (Parker et al. 1995, 1996) can also complicate the characterization of heteroplasmy when fluorescent terminator-sequencing chemistry is used (see the Results section).
We have implemented a denaturing gradient-gel electrophoresis (DGGE) assay, to perform a population survey of heteroplasmy in the HV1 portion of the mtDNA CR. DGGE has been successfully used to identify mutant sequences in nuclear genes (Traystman et al. 1990; Laubscher et al. 1994) as well as heteroplasmy in the mitochondrial genome (Yoon et al. 1991; Lombes et al. 1992; Hanekamp et al. 1996; Jazin et al. 1996; Michikawa et al. 1997; Danan et al. 1999; Michikawa et al. 1999). In the present study, we report both the frequency of HV1 heteroplasmy in the population, on the basis of a survey of blood samples from 253 individuals, and estimates of the heteroplasmic ratios present in those individuals determined to be heteroplasmic.
Material and Methods
Blood samples, either whole blood or blood spotted onto filter paper and air-dried, were collected from 253 randomly chosen individuals, none of whom were known or suspected to be maternally related. All participants provided appropriate informed consent. Genomic DNA was isolated using either the Puregene DNA Isolation Kit (for whole blood; Gentra Systems), according to the manufacturer's protocol, or Chelex 100 chelating resin (for blood spots; Biorad) (Walsh et al. 1991; Holland et al. 1993).
HV1 was analyzed as two overlapping fragments, by use of PCR primer pairs F15989 (5′-CCC AAA GCT AAG ATT CTA AT-3′)/R16258GC (5′-GC TGG CTT TGG AGT TGC AGT TG-3′) and F16144GC (5′-GC TGA CCA CCT GTA GTA CAT AA-3′)/R16410 (5′-GAG GAT GGT GGT CAA GGG AC-3′) (numbering system of Anderson et al. [1981]). These primer pairs were designed on the basis of the melt profiles generated by MacMelt Software (DNA Melt Profile Macintosh Software for the D Gene System, version 1.0) for the HV1 region (Steighner et al. 1999). The DNA from individuals with a C→T transition at position 16189 was reanalyzed using primer set F15989GC (5′-GC CCC AAA GCT AAG ATT CTA AT-3′)/R16175 (5′-TGG ATT GGG TTT TTA TGT A-3′). A GC-clamp (5′-CGC CCG CCG CGC CCC GCG CCC GTC CCG CCG CCC CCG CCC G-3′), which was added to the 5′ end of one primer in each pair (GC), made the resulting PCR products suitable for DGGE analysis (Myers et al. 1985; Sheffield et al. 1989; Abrams et al. 1990). PCR amplification was performed by combining 10–50 ng DNA template with 1 × Pfu reaction buffer (20 mM Tris-HCl pH 8.8, 2 mM MgSO4, 10 mM KCl, 10 mM (NH4)2SO4, 0.1% Triton X-100, 0.1 mg nuclease-free BSA per ml), 200 μM each dNTP (Boehringer Mannheim), 0.2 μM each primer (Synthetic Genetics), 2 μg BSA (Sigma Chemical), 2.5 U Pfu polymerase, and sufficient sterile distilled water (Gibco BRL Life Technologies) to bring the final volume to 50 μl. (The accepted SI unit of concentration, mol/L, has been represented by the symbol “M,” in order to conform to the conventions of this journal.) PCR cycling consisted of initial denaturation at 95°C for 30 s, followed by 35 cycles consisting of denaturation at 95°C for 30 s, primer annealing at 50°C (F15989/R16258GC) or 60°C (F16144GC/R16410) for 30 s, and extension at 72°C for 30 s. Prior to DGGE analysis, the PCR products were heated to 95°C to denature the DNA and then were slowly cooled to room temperature to allow for reannealing of the DNA strands. This heating-and-cooling process resulted in the formation of a single homoduplex fragment in homoplasmic individuals and a series of homoduplex and heteroduplex fragments in heteroplasmic individuals.
DGGE was performed using a 6.5% polyacrylamide gel (37.5:1 acrylamide/bisacrylamide [Biorad]) with a 40%–50% chemical (i.e., urea/formamide) denaturing gradient for 15 h at 100 volts and 60°C. The gel was then stained with 1 × SYBR Green (Molecular Probes) for 20 min and was visualized using an ultraviolet-wavelength transilluminator. DGGE separation of homoduplex and heteroduplex fragments, followed by elution, reamplification, and sequencing of the DNA from DGGE bands, allowed for identification of the heteroplasmic positions. The public domain Image program (developed at the United States National Institutes of Health) was used to estimate the relative heteroplasmic proportions (Steighner et al. 1999). Confidence limits were calculated for the observed population frequency of heteroplasmy, by use of a standard equation for true proportions (Colton 1974).
Sequencing analyses were performed using the ABI PRISM Dye Terminator Cycle Sequencing Kit Taq FS and the ABI 310 Capillary Electrophoresis Analyzer (PE Biosystems). Heteroplasmy suspected at position 16189 was evaluated by ligation of the reamplified heteroduplex fragment into the pCR-Script Amp SK+ cloning vector (Stratagene), followed by transformation into Epicurian Coli XLJ-Blue supercompetent cells, according to the manufacturer's protocol. After plasmid purification (Wizard Plus Minipreps DNA Purification System [Promega]), 10 of the clones were sequenced.
Results
The use of DGGE for mutation or heteroplasmy detection in HV1 was characterized in a previous study (Steighner et al. 1999), which demonstrated that this strategy reliably resolved single-base-pair differences at >40 sites spread evenly throughout HV1. In the present study, the detection limit of the DGGE assay was investigated at four positions—16126, 16220, 16298, and 16343—by means of a series of dilution-mixing experiments down to a minority variant proportion of 1:100 (fig. 1). At positions 16126 and 16298, detection of the minor variant was reliable at the 1% level, whereas, at 16343, the lowest detection was at the 1.3% level (1:75). Position 16220 was reliably detected at the 5% level but not below that level. In the course of our population survey, heteroplasmy was quantified at levels <5% at six additional scattered sites, three of which were at or below the 1% level. We therefore consider the sensitivity of the assay for heteroplasmy detection at any site within HV1 to be 5% or better. Quantitation of heteroplasmic ratios was performed by densitometric comparison of heteroduplex and homoduplex band intensity on polyacrylamide-gel images. The results of experiments with mixtures of known quantities showed that this provided a reasonable approximation of the mixture ratios, with some tendency toward overestimation of the minority component as a result of saturation of brighter bands (fig. 2).
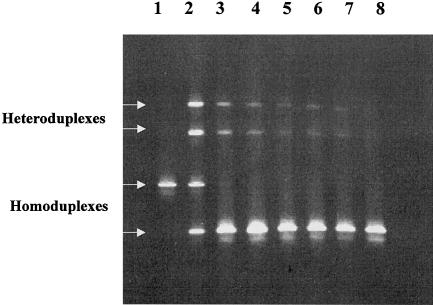
Figure 1.
DGGE gel from sensitivity experiments performed to determine the detection limit of the DGGE system. DNA from an individual with the standard reference sequence was combined with DNA from an individual with a T→C polymorphism at position 16126, prior to PCR and testing by DGGE. The concentration of the minority species could thus be intentionally varied to include 50% (lane 2), 10% (lane 3), 5% (lane 4), 2% (lane 5), 1.3% (lane 6), and 1% (lane 7). Lanes 1 and 8 represent single PCR products from the reference sequence and the 16126C polymorphism, respectively. Although the homoduplex band representing the reference sequence was not visible at a level ⩽10%, the heteroduplexes were clearly seen at levels as low as 1%. (Reprinted with permission from the American Society for Testing and Materials.)
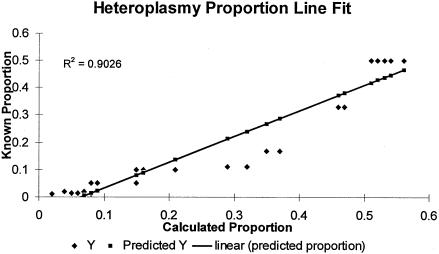
Figure 2.
Least-squares line fit for known and calculated heteroplasmic proportions. A series of simulated heteroplasmic samples with various proportions of the minor species were analyzed by DGGE. The known heteroplasmic proportions were plotted against the calculated proportions after densitometric analysis of DGGE gel photographs (coefficient of correlation .9026).
We performed the DGGE heteroplasmy assay on blood samples from a study population of 253 randomly selected individuals with the following ethnic distribution: 205 whites, 26 African Americans, 12 Hispanic Americans, and 10 Asians. Samples that appeared to be heteroplasmic were analyzed a second time, beginning with new DNA extracts, to confirm that heteroplasmy was present. Overall, point heteroplasmy was reproducibly detected in 35/253 individuals in the study population, resulting in an overall incidence of 13.8% (95% CI 9.6–18.0). In two individuals, two sets of heteroduplex bands were observed, indicating the presence of heteroplasmy at two different positions—a condition known as “triplasmy.” Although the majority of individuals in the study population were white, heteroplasmy was detected in all four racial groups studied, with no statistically significant differences in the frequencies observed (table 1). Length heteroplasmy associated with the HV1 polycytosine stretch produced a complex DGGE pattern and was detected in 30 additional individuals. This C-stretch length heteroplasmy generally results from a T→C transition at position 16189 that produces an uninterrupted string of 10 C's that is copied with low fidelity during mtDNA replication (Bendall and Sykes 1995). This was the case in each of the 30 examined individuals with C-stretch length heteroplasmy.
Table 1.
Frequency of HV1 mtDNA Heteroplasmy, as Determined by DGGE, in a Study Population of 253 Individuals from Four Ethnic Groups
|
No. Individuals |
|||
| Ethnic Group | Tested | Heteroplasmic | Frequency (95% CI) |
| White | 205 | 30 | 14.6% (9.8–19.4) |
| African American | 26 | 3 | 11.5% (0–23.8) |
| Hispanic | 12 | 1 | 8.3% (0–23.9) |
| Asian | 10 | 1 | 10.0% (0–28.6) |
The position at which heteroplasmy occurred in a sample was determined by excising both homoduplex bands from the DGGE gel and by eluting, reamplifying, and sequencing the DNA (fig. 3). When the proportion of the minor heteroplasmic species was too low to produce a visible homoduplex band, the single major homoduplex band (resulting from the predominant heteroplasmic species) and one or both heteroduplexes were excised from the DGGE gel and were eluted, reamplified, and sequenced. Since reamplification of either heteroduplex results in a PCR product containing approximately equal proportions of both heteroplasmic variants, comparison of the DNA sequences from the homoduplex and heteroduplex bands permitted identification of the heteroplasmic site (fig. 4). In some instances, there were pronounced differences in the intensity of fluorescent sequence signal for different variants at the position of interest, on the basis of sequence-specific-strength artifacts. This made it difficult to identify the heteroplasmic position by sequencing of the excised heteroduplex band (fig. 5). In those cases, the excised heteroduplex band was reamplified to produce equal proportions of the two sequence types, and subsequent DGGE gave two strong homoduplex bands. Comparison of sequences from the two homoduplexes then confirmed the heteroplasmic position (fig. 6).
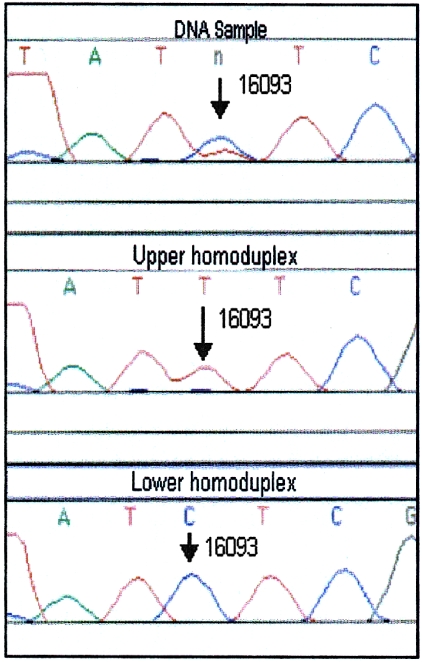
Figure 3.
mtDNA heteroplasmy at position 16093. The original PCR product (top) appears to be heteroplasmic (C/T) at position 16093. The sequence data from both homoduplexes (middle and bottom) confirm the heteroplasmic position and sequence.
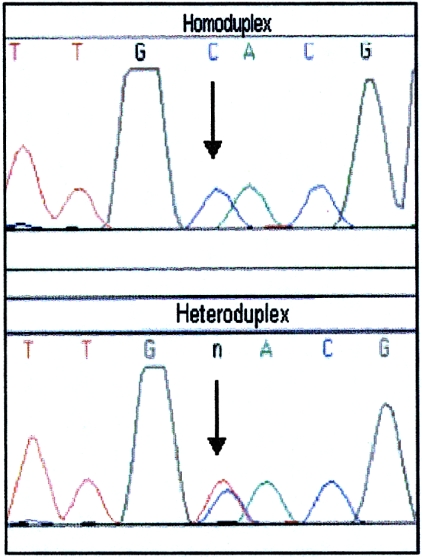
Figure 4.
Homoduplex and heteroduplex DNA sequences. Sequence electropherograms of reamplified homoduplex (top) and heteroduplex (bottom) DNA from a simulated heteroplasmic sample (prepared by combining two homoplasmic samples differing at a single position) are shown. Comparison of the sequence data allows for identification of the heteroplasmic position.
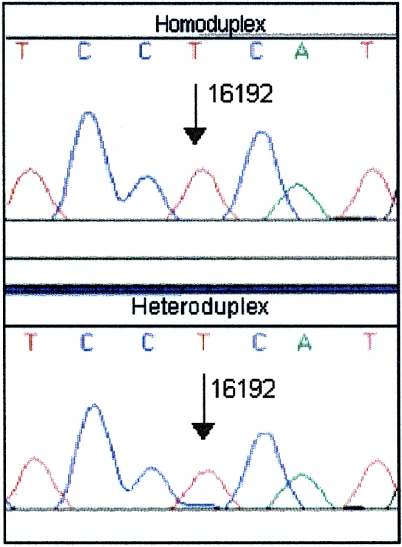
Figure 5.
Potential heteroplasmy at position 16192. The sequence electropherograms of reamplified homoduplex (top) and heteroduplex (bottom) DNA are shown. A very low level of a second sequence (C) is seen at position 16192 in the heteroduplex. However, this is insufficient for identification of heteroplasmy.
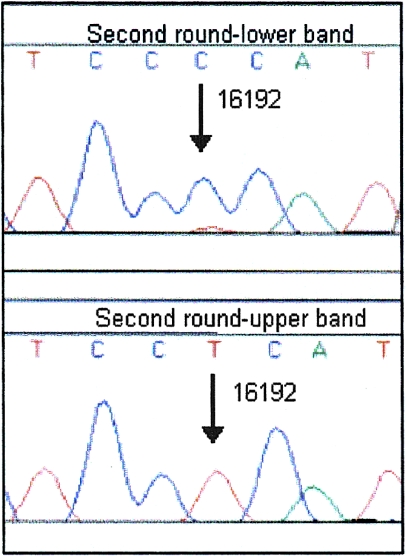
Figure 6.
Sequence electropherograms after analysis of heteroduplex DNA by second-round DGGE. The sequences of the lower band (top) and upper band (bottom) confirmed 16192 as the heteroplasmic position.
Among the 35 heteroplasmic individuals, heteroplasmy was found at 16 different positions and at a wide range of heteroplasmic ratios (table 2). Six positions showed heteroplasmy in more than one individual. There were two observations of heteroplasmy at positions 16104, 16192, 16362, and 16390. Of note were two positions that showed an extreme propensity for heteroplasmy: position 16129 (A/G) was heteroplasmic in six individuals, and position 16093 (C/T) was heteroplasmic in 13 individuals. In one individual, C/T heteroplasmy was observed at position 16189; this was also associated with length heteroplasmy in the HV1 C-stretch within the population of molecules that had C at position 16189.
Table 2.
HV1 Heteroplasmy Data from a Population Survey of 253 Unrelated Individuals
| Relative Rate of Evolutionary Substitutions Per Site, as Estimated by |
|||||||
| Position (Type)a | No. of Observations | Reference Sequencea | Estimated Proportion of Minority Species in Heteroplasmic Individuals b | Hasegawa et al. (1993) | Excoffier and Yang (1999) | Meyer et al. (1999) | Population Frequency of Minority Variantc |
| 16093 (C/T) | 13 | T | .48C, .41T, .26T, .19T, .16T, .13T, .09T, .08T, .07T, .05T*, .05T, .04T, .03C | 8 | 4.4 | 3.0 | .06 |
| 16129 (A/G) | 6 | G | .11A, .09G, .07A, .05A, (BQT)A, (BQT)A | 15 | 4.4 | 4.8 | .12 |
| 16104 (C/T) | 2 | C | .10C, .06C | 0 | .6 | .5 | .003 |
| 16192 (C/T) | 2 | C | .06C, .03C | 4 | 4.4 | 3.6 | .04 |
| 16362 (C/T) | 2 | T | .05C, .03C† | 13 | 4.4 | 4.7 | .17 |
| 16390 (A/G) | 2 | G | .06A†, .04A | 9 | … | … | … |
| 16124 (C/T) | 1 | T | .10C* | 4 | 1.1 | .8 | .03 |
| 16172 (C/T) | 1 | T | .12T | 8 | 4.4 | 3.8 | .07 |
| 16185 (C/T) | 1 | C | .23C | 3 | 1.0 | .6 | .01 |
| 16189 (C/T) | 1 | T | (ND)C | 15 | 4.4 | 4.8 | .24 |
| 16234 (C/T) | 1 | C | .22T | 4 | 2.2 | .8 | .01 |
| 16256 (C/T) | 1 | C | .16T | 5 | 2.6 | 2.3 | .04 |
| 16293 (A/G) | 1 | A | .16A | 8 | 4.4 | 3.9 | .04 |
| 16295 (C/T) | 1 | C | (BQT)C | 2 | .8 | .7 | .02 |
| 16301 (C/T) | 1 | C | .27T | 2 | 1.2 | .3 | .004 |
| 16309 (A/G) | 1 | A | .25A | 3 | 4.3 | 4.0 | .05 |
Positions where heteroplasmy was observed. Numbering and sequence are according to Anderson et al. (1981).
Underlined type denotes heteroplasmy that was distinguishable by direct sequencing; BQT, below quantitation threshold; ND, not determined. Listings sharing superscripted symbols (asterisk [*] and dagger [†]) indicate that these heteroplasmies were present in the same individual.
In a forensic database (Armed Forces DNA Identification Laboratory database) of 2263 HV1 sequences with racial proportions similar to those represented in this study (position 16390 not included in database).
In 16/37 instances of heteroplasmy, the majority species within the individual corresponded to the variant that is predominant in the population (table 2). In all but 2/13 instances of heteroplasmy at position 16093, T was the minority species, despite the fact that ∼94% of individuals worldwide manifest a T at that position (Armed Forces DNA Identification Laboratory database). The relative proportion of the minority species was often low: in 29/36 heteroplasmies, it was <20% of the mtDNA population, and, in 20 heteroplasmies, it was <10% (the minority relative proportion was not determined for the sample with 16189 C/T heteroplasmy).
In addition to determination of the position of heteroplasmy by means of sequence analysis of bands excised from DGGE gels, PCR products displaying heteroplasmy were directly sequenced on both strands by automated sequencing using dye-labeled terminators. In only three instances did we believe that the heteroplasmic position was clearly evident by means of direct sequencing and that it would have been flagged as a likely instance of heteroplasmy (table 2). This is due to the low levels of minor heteroplasmic variants in many cases, but it was also true for some heteroplasmic mixtures of more-equal proportions. As noted above, at a number of nucleotide positions, it was readily apparent that the sequence signal from one variant was intrinsically stronger than that of the other variant, resulting in relative peak heights that did not reflect the actual proportions of the variants.
Discussion
In this study, a DGGE assay was used to analyze the heteroplasmic variation in a large population sample of the mtDNA HV1 region and to characterize the spectrum of sites where heteroplasmy most frequently occurs. Of significance, this assay has provided what we believe to be essentially complete detection of all heteroplasmic variants that occur at a level ⩾5% (and, for most positions, at a level <5%). The results indicate that heteroplasmy within this detection window is quite common, occurring in ∼14% of the population and at a broad spectrum of sites. The incidence found in the present study is considerably higher than that reported in previous studies of HV1 heteroplasmy that used sequencing (Bendall et al. 1996) and SSCP analysis (Gocke et al. 1998), which reported frequencies of 1.48% and 2.53%, respectively.
One area in which heteroplasmy detection may not have been complete in the present study was proximal to the HV1 C-stretch region in individuals who carried a C at position 16189. These 30 individuals displayed pronounced length heteroplasmy in the C-stretch that creates a complex DGGE pattern that would have obscured sequence heteroplasmy in this region. To get around this to the extent possible, for these samples we used an alternate DGGE primer set—F15989GC/R16175—that would detect additional sequence heteroplasmy, although not beyond position 16155. The DGGE primer set used for the second half of HV1 (F16144GC/R16410) also would likely fail to detect heteroplasmy close to position 16189, because of the proximity of the GC-clamped primer to this area. It should be noted that no sequence heteroplasmy was detected in any of the 30 individuals with C-stretch–induced length heteroplasmy. We cannot unequivocally establish whether this was the result of the reduced ability to detect heteroplasmy in this restricted region in individuals with the 16189C polymorphism or whether these individuals were, in essence, homoplasmic. Howell and Bogolin Smejkal (2000) have reported hypermutation of T at position 16192 when there is also a C polymorphism at position 16189. The two instances of 16192 heteroplasmy that we observed did have T as the majority variant, but they were not linked to a 16189C polymorphism. However, the reduced ability of our DGGE assay to detect heteroplasmy at position 16192 in those individuals with 16189C and resultant length heteroplasmy may be why we did not observe the effect reported by Howell and Bogolin Smejkal (2000).
Our sequencing results underscore the predominant reason why heteroplasmy in the CR had been undetected for so long and why we have lacked a good estimate for its frequency in the population. Direct sequencing can provide a poor means for detection of heteroplasmy, unless extensive confirmatory experiments are performed, and most population studies have not been conducted with heteroplasmy detection in mind. This is true not only because low levels of heteroplasmy are difficult to distinguish from background but also because problems can be encountered with terminator-labeled sequencing chemistry, as a result of sequence-dependent signal intensity (Parker et al. 1995, 1996). This phenomenon was quite pronounced at position 16093 (with C having a substantially stronger inherent signal than T) and is probably responsible for the failure to convincingly detect heteroplasmy from direct sequence data at other positions, even when the proportion of the minor variant was shown by DGGE to be 20%–25% (e.g., positions 16309, 16301, and 16234).
In considering the results of this study, one should bear in mind that we are assaying PCR-amplified fragments, as opposed to cellular mtDNA itself. That said, it is difficult to envision experimental artifacts that could give rise to these results without the presence of heteroplasmy in the endogenous mtDNA population. Point mutations generated from misincorporation during the PCR process are expected to be greatly reduced by the use of a high-fidelity polymerase (Pfu) (Kunkel and Loeb 1981), and they can be ruled out by the reproducibility of our results. Contamination of the blood samples themselves could produce reproducible results, but we reject this as a general explanation, since mixtures were seen only at one or two positions (the latter in only two samples). Randomly selected white individuals differ from one another at an average of four sites in the HV1 region (Stoneking 1994), and this value is larger for some of the other racial groups involved. This same reasoning would eliminate low levels of paternally inherited mtDNA as a plausible cause of the results.
In principle, another possible cause of the apparent mixtures we observed with DGGE is the detection of nuclear pseudogenes. However, there are two reasons why we do not believe that nuclear pseudogenes are contributing to our data. The first reason is, again, that the mixtures in any one individual occur at only one or (rarely) two positions. A nuclear pseudogene would segregate in the population independently from maternally transmitted mtDNA lineages. Different mtDNA lineages differ, on average, at four sites, and there would be no reason to expect independently segregating pseudogenes to correlate, in sequence similarity, with the mtDNA types of individuals in which they are detected. The second reason is that there is a known nuclear pseudogene that is widely present in the general population (Zischler et al. 1995; Thomas et al. 1996). This pseudogene is highly distinctive, differing from the Cambridge reference sequence at 27 positions in the 16164–16365 region alone. Although our DGGE assay is capable of resolving this pseudogene from mitochondrial sequences, we detected the pseudogene in only two individuals in our population survey. In both instances, there was an F16144GC primer mismatch caused by a polymorphism in the individual’s authentic mtDNA sequence (not shown), resulting in reduced efficiency of amplification of the mitochondrial component and preferential coamplification of the pseudogene. In the absence of an amplification primer mismatch, this common pseudogene was not detected in any of our samples. This indicates that, under normal circumstances, the abundance of mtDNA copies in a cell overshadows any existing nuclear pseudogenes that may coamplify, causing the nuclear sequences to go undetected by our DGGE assay.
We conclude that the mixtures revealed by DGGE represent variation in the endogenous mtDNA of the individuals (or mtDNA lineages) studied. This is consistent with the fact that the sites at which heteroplasmy was observed have a higher-than-average rate of evolutionary substitution. This would be expected if mtDNA mutation were giving rise to the observed heteroplasmy, since site-by-site mutation rates in HV1 are highly heterogeneous (Hasegawa et al. 1993; Wakeley 1993; Excoffier and Yang 1999; Meyer et al. 1999) (table 2). Given the relative rates reported by Meyer et al. (1999), the average relative rate of evolution for sites where heteroplasmy was detected is 3.1 times greater than the average relative rate over the entire HV1. If position 16093 is omitted, the average relative rates of the heteroplasmic sites remain essentially the same (the reported relative rate for position 16093 is itself 3.0 times the average). Very similar numbers are obtained with the relative rates reported by Excoffier and Yang (1999). We note that position 16104 has the lowest estimated rate of evolution among the heteroplasmic sites (roughly half the overall average rate), yet it nonetheless showed two instances of heteroplasmy. It is interesting that both individuals had T—which occurs rarely in the overall population—as the predominant nucleotide. The significance of this, if any, is unclear.
The most striking observation in our study was the high frequency and directionality of heteroplasmy at position 16093. A total of 13 instances of heteroplasmy were detected at 16093, and all but 2 had T—the majority population type—as the clear minority component (one individual had C as the minority component, and one had essentially equal proportions of C and T). In population sequence databases, ∼6% of the population carries a C, rather than a T, at 16093. In our study set, 5% were heteroplasmic at 16093, and all but two of these individuals would have been typed as “C” in a standard sequencing study. This suggests that a mechanistic bias determines variation at 16093. It seems that C can readily be manifested as the majority base within an individual's mtDNA pool but that complete fixation for C is rare. It is possible that developmental selection acts against mtDNA pools that are fixed for C at 16093. Alternatively, the base C at 16093 may be hypermutable, so that a subpopulation of mtDNAs carrying 16093T are continually generated within individuals originally fixed for 16093C. Similar hypermutability in the reverse direction (T→C) is not supported by our data. Unusual mutational activity at position 16093 is supported by previous observations of intergenerational “substitution” (a large shift in predominant type) (Parsons et al. 1997) and frequent heteroplasmy at 16093 (Sullivan et al. 1996; Armed Forces DNA Identification Laboratory, unpublished data).
In this study, all samples were from blood. Thus, we do not know whether the heteroplasmies we detected were due to somatic mutations within an individual or were inherited through the germline. It is possible that both processes contributed. However, there is other evidence that suggests that heteroplasmy within the CR is not rarely an inherited condition. Parsons et al. (1997) reported three instances of intergenerational substitution in HV1, including 16093, and the results of subsequent DGGE studies indicated that these actually involved heteroplasmic shifts of extreme ratio (not shown). In addition, a comprehensive analysis of HV1 heteroplasmy among multiple postmortem tissues from 22 fetuses and 21 adults indicated a common occurrence of low-level heteroplasmy shared among many tissues, pointing toward the conclusion that the heteroplasmy was inherited or, at least, originated very early in development (Tully 1998). We do not have precise data on the ages of many of the individuals in our current population study, although we believe that none were of the advanced ages represented in recently published reports, which found aging-related accumulation of specific heteroplasmic mutations in the HV2 region of the CR (Michikawa et al. 1999; Calloway et al. 2000). Also, no longitudinal data were collected in our study. It remains possible that the high-frequency heteroplasmies (e.g., 16093) found in the present study could represent aging-associated accumulation of heteroplasmy. However, in our previous study of postmortem fetal and adult tissues, C/T heteroplasmy at position 16093 was observed in individuals aged 29–52 years and in an 18-wk-old fetus (Tully 1998).
It has repeatedly been hypothesized that the transmission of a woman's mtDNA population to her offspring is associated with a genetic bottleneck (Howell et al. 1992; Poulton and Marchington 1996; Marchington et al. 1997; Poulton et al. 1998). Offspring of a heteroplasmic mother can display marked differences in their heteroplasmic ratios, sometimes to the point of apparent homoplasmic base substitutions (Parsons et al. 1997; Sekiguchi et al. 1999). In an investigation of the segregation of point-mutation heteroplasmy in humans, Bendall et al. (1996) traced heteroplasmy in HV1 through multiple mother-to-child transmissions in a number of lineages and were able to derive estimates of the most-probable effective bottleneck sizes from various transmissions (range 1–46).
Bendall et al. (1996) also used the population frequency of observed HV1 heteroplasmy and bottleneck size estimates to calculate the “mutation rate” of the HV1 region (in this instance, “mutation rate” refers to the rate at which homoplasmic substitutions appear in the population, as considered in most evolutionary phylogenetic studies). In that study, the incidence of heteroplasmy was estimated, by inspection of direct sequence data, to be 7 (1.48%) of 473 (that this value is much lower than that which we report here is to be expected, given that a general sequence survey was used to estimate heteroplasmy rates). On the basis of this number and a range of bottleneck size estimates and thresholds for heteroplasmy detection, a mutation rate of 1.2×10-6 to 2.7×10-5 site/generation was obtained. It was concluded that these values were in good accord with previous phylogenetic estimates of the HV1 evolutionary rate.
We can apply the same calculation to our data, by selecting a heteroplasmy-detection threshold of 5% (see table 5 in Bendall et al. 1996) and by counting only those observations that were measured at levels ⩾5%. This results in a calculated mutation rate of 1.8×10-5 to 1.3×10-4 site/generation (this range includes sampling error as well as a range of bottleneck sizes from table 5 in a study by Bendall et al. [1996]). If we omit position 16093, in recognition of the fact that something unusual is occurring there with regard to heteroplasmy and fixation, the range becomes 1.0×10-5 to 8.1×10-5 site/generation. The latter estimate is higher than typical phylogenetic rate estimates and is very close to the range of 2.4×10-5 to 8.0×10-5 site/generation that Parsons et al. (1997) reported as an empirically estimated substitution rate for HV1 and HV2. However, to these considerations, we would add the caveat that the calculation of Bendall et al. (1996) uses a probability of .5 that observed heteroplasmies will become fixed for the mutant variant. In cases of low-level heteroplasmy, we believe that the minority component is more likely to be the new mutation and that the chance that the new mutation will become fixed would therefore be less than half. If we go to the extreme case of adopting the premise that the new mutation is the minority variant in every instance, and if we perform the calculations assuming the probability of fixation is equal to the minority proportion of the mixture, then the range is 2.6×10-6 to 2.1×10-5 site/generation. This returns us to a range that is more similar to frequently reported rates (Parsons et al. 1997; Jazin et al. 1998; Parsons and Holland 1998; Sigurdardottir et al. 2000); however, it is based on a premise that would tend to underestimate the rate, and it does not take into account the contribution of the heteroplasmic sites with a measured proportion <.05. (We note that this cutoff of .05 is made for purposes of direct comparison with the analysis of Bendall et al. [1996] and that the accuracy of our ratio quantitation [fig. 2] is such that the cutoff should be considered to be an approximation). Further resolution of the rate and manner of CR evolution will likely result from a synthesis of additional information on mutational variation within an individual (such as that provided by the present study), additional information on the variation in the germline between immediate generations, and the patterns of change inferred at the larger population level, as shown in Meyer et al. (1999) and Excoffier and Yang (1999).
The widespread and rather frequent occurrence of heteroplasmy throughout HV1 has implications for forensic testing that is based on comparisons of mtDNA sequences. The effect of heteroplasmy is well illustrated by the case of Tsar Nicholas II. The mtDNA from skeletal remains believed to be those of the Tsar was shown to be heteroplasmic in HV1, but no heteroplasmy was evident in known living maternal relatives of the Tsar (Gill et al. 1994). This perceived “mismatch” caused some confusion until heteroplasmy was also confirmed in the skeletal remains of the Tsar’s brother (Ivanov et al. 1996). In that case, co-occurrence of heteroplasmy provided an additional level of variation, thereby greatly strengthening the identification. The results of the present systematic survey of HV1 heteroplasmy are consistent with general experience in the practical arena of forensic testing, where heteroplasmy is now encountered with regularity. The effect of heteroplasmy is of particular importance in cases where an unknown sample and a comparative reference sample are separated by the action of a genetic bottleneck between generations or between single hairs (Sullivan et al. 1996; Bendall et al. 1997; Holland and Parsons 1999; Grzybowski 2000). Apparent single-base differences, or heteroplasmy in only one of two compared samples, can be due to differential segregation of heteroplasmy from the same biological source. Quantitative knowledge of the rate and pattern of HV1 heteroplasmy provided in this study can be used to guide interpretation in such cases. For example, heteroplasmic differences at position 16093 can be considered to be of little significance because of the high frequency of heteroplasmy there, whereas differences at several positions known to have very low rates of heteroplasmy would provide much stronger evidence that the samples are not from the same biological source or lineage (forensic treatment in such cases is reviewed by Holland and Parsons [1999]).
Our results indicate that heteroplasmy in HV1 of the human mtDNA CR is more common in the general human population than was previously believed. Until recently, intraindividual variation in this important segment of DNA was unknown or was considered to be of minimal significance (Stoneking 1996). The picture that is now emerging is one in which the study of mtDNA population genetics and evolution must view the mtDNA pool both within an individual and among individuals in a maternally related lineage as a dynamic source of variation. The challenge now is to understand how the complex “microevolutionary” processes of mtDNA mutation, segregation, and transmission affect the more-traditional population genetics patterns of mtDNA variation that have long been studied. It may be that a better understanding of these underlying processes will provide new insight and will substantially extend our ability to draw conclusions from mtDNA population genetics and phylogenetic analyses.
Acknowledgments
The authors gratefully acknowledge the Armed Forces DNA Identification Laboratory, the Armed Forces Center for Medical and Molecular Genetics, the University of Maryland Division of Human Genetics, and Cellmark Diagnostics, for their contributions to this work. The authors are also grateful to Dr. Barbara C. Levin and Dr. Dennis Reeder from NIST, for their review of this manuscript and for their helpful comments and discussion, and to two anonymous reviewers, for their helpful suggestions. We thank Sonja Meyer and Arndt von Haessler, and Laurent Excoffier, for providing data in tabular form from their respective published studies on mutation rates.
Footnotes
Disclaimer: The opinions and assertions expressed herein are solely those of the authors and are not to be construed as official or the views of the United States Department of Defense, the United States Department of the Army, or the United States Department of Commerce. This paper is a contribution of the United States National Institute of Standards and Technology (NIST) and the United States Department of Defense and is not subject to copyright. Certain commercial equipment, instruments, or companies are identified in this paper to specify the experimental procedure. Such identification does not imply recommendation or endorsement by the government, nor does it imply that the materials or equipment identified are the best available for this purpose.
Electronic-Database Information
The URL for data in this article is as follows:
- United States National Institutes of Health Image Web Page, http://rsb.info.nih.gov/nih-image/ (for Image program used to estimate heteroplasmic proportions)
References
- Abrams ES, Murdaugh SE, Lerman LS (1990) Comprehensive detection of single base changes in human genomic DNA using denaturing gradient gel electrophoresis and a GC clamp. Genomics 7:463–475 [DOI] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MHL, Coulson AR, Drouin J, Eperon IC, et al (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465 [DOI] [PubMed] [Google Scholar]
- Bendall KE, Macaulay VA, Baker JR, Sykes BC (1996) Heteroplasmic point mutations in the human mtDNA control region. Am J Hum Genet 59:1276–1287 [PMC free article] [PubMed] [Google Scholar]
- Bendall KE, Macaulay VA, Sykes BC (1997) Variable levels of a heteroplasmic point mutation in individual hair roots. Am J Hum Genet 61:1303–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bendall KE, Sykes BC (1995) Length heteroplasmy in the first hypervariable segment of the human mtDNA control region. Am J Hum Genet 57:248–256 [PMC free article] [PubMed] [Google Scholar]
- Bodenteich A, Mitchell LG, Merril CR (1991) A lifetime of retinal light exposure does not appear to increase mitochondrial mutations. Gene 108:305–309 [DOI] [PubMed] [Google Scholar]
- Calloway CD, Reynolds RL, Herrin GL Jr, Anderson WW (2000) The frequency of heteroplasmy in the HVII region of mtDNA differs across tissue types and increases with age. Am J Hum Genet 66:1384–1397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clayton DA (1992) Transcription and replication of animal mitochondrial DNA. Int Rev Cytol 141:217–232 [DOI] [PubMed] [Google Scholar]
- Colton T (1974) Inference on proportions. In: Statistics in medicine. Little, Brown; Boston; pp 151–188 [Google Scholar]
- Comas D, Paabo S, Bertranpetit J (1995) Heteroplasmy in the control region of human mitochondrial DNA. Genome Res 5:89–90 [DOI] [PubMed] [Google Scholar]
- Danan D, Sternberg D, Van Steirteghem A, Cazeneuve C, Duquesnoy P, Besmond C, Goosens M, et al (1999) Evaluation of parental mitochondrial inheritance born after intracytoplasmic sperm injection. Am J Hum Genet 65:463–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Excoffier L, Yang Z (1999) Substitution rate variation among sites in the mitochondrial hypervariable region I of humans and chimpanzees. Mol Biol Evol 16:1357–1368 [DOI] [PubMed] [Google Scholar]
- Gill P, Ivanov PL, Klimpton C, Piercy R, Benson N, Tully G, Evett I, et al (1994) Identification of the remains of the Romanov family by DNA analysis. Nat Genet 6:130–135 [DOI] [PubMed] [Google Scholar]
- Gocke CD, Benko FA, Rogan PK (1998) Transmission of mitochondrial DNA heteroplasmy in normal pedigrees. Hum Genet 102:182–186 [DOI] [PubMed] [Google Scholar]
- Greenberg BD, Newbold JE, Sugino A (1983) Intraspecific nucleotide sequence variability surrounding the origin of replication in human mitochondrial DNA. Gene 21:33–49 [DOI] [PubMed] [Google Scholar]
- Grzybowski T (2000) Extremely high levels of human mitochondrial DNA heteroplasmy in single hair roots. Electrophoresis 21:548–553 [DOI] [PubMed] [Google Scholar]
- Handt O, Meyer S, von Haeseler A (1998) Compilation of human mtDNA control region sequences. Nucleic Acids Res 26:126–129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanekamp JS, Thilly WG, Chaudhry MA (1996) Screening for human mitochondrial DNA polymorphisms with denaturing gradient gel electrophoresis. Hum Genet 98:243–245 [DOI] [PubMed] [Google Scholar]
- Hasegawa M, Di Renzo A, Kocher TD, Wilson AC (1993) Toward a more accurate time scale for the human mitochondrial DNA tree. J Mol Evol 37:347–354 [DOI] [PubMed] [Google Scholar]
- Hauswirth WW, Laipis PJ (1982) Mitochondrial DNA polymorphism in a maternal lineage of Holstein cows. Proc Natl Acad Sci USA 79:4686–4690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holland MM, Fisher DL, Mitchell LG, Rodriguez WC, Canik JJ, Merril CR, Weedn VW (1993) Mitochondrial DNA sequence analysis of human skeletal remains: identification of remains from the Vietnam War. J Forensic Sci 38:542–553 [PubMed] [Google Scholar]
- Holland MM, Parsons TJ (1999) Mitochondrial DNA sequence analysis—validation and use for forensic casework. Forensic Science Review 11:21–50 [PubMed] [Google Scholar]
- Horai S, Hayasaka K (1990) Intraspecific nucleotide sequence differences in the major noncoding region of human mitochondrial DNA. Am J Hum Genet 46:828–842 [PMC free article] [PubMed] [Google Scholar]
- Howell N, Bogolin Smejkal C (2000) Persistent heteroplasmy of a mutation in the human mtDNA control region: hypermutation as an apparent consequence of simple-repeat expansion/contraction. Am J Hum Genet 66:1589–1598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howell N, Halvorson S, Kubacka I, McCullough DA, Bindoff LA, Turnbull DM (1992) Mitochondrial gene segregation in mammals: is the bottleneck always narrow? Hum Genet 90:117–120 [DOI] [PubMed] [Google Scholar]
- Howell N, Kubacka I, Mackey DA (1996) How rapidly does the human mitochondrial genome evolve? Am J Hum Genet 59:501–509 [PMC free article] [PubMed] [Google Scholar]
- Ivanov Pl, Wadhams MJ, Roby RK, Holland MM, Weedn VW, Parsons TJ (1996) Mitochondrial DNA sequence heteroplasmy in the Grand Duke of Russia Georgij Romanov establishes the authenticity of the remains of Tsar Nicholas II. Nat Genet 12:417–420 [DOI] [PubMed] [Google Scholar]
- Jazin EE, Cavelier L, Eriksson I, Oreland L, Gyllensten U (1996) Human brain contains high levels of heteroplasmy in the noncoding regions of mitochondrial DNA. Proc Natl Acad Sci USA 93:12382–12387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jazin EE, Soodyall P, Jalonen P, Lindholm E, Stoneking M, Gyllensten U (1998) Mitochondrial DNA mutation rate revisited: hot spots and polymorphism. Nat Genet 18:109–110 [DOI] [PubMed] [Google Scholar]
- Jenuth JP, Peterson AC, Fu K, Shoubridge EA (1996) Random genetic drift in the female germline explains the rapid segregation of mammalian mitochondrial DNA. Nat Genet 14:146–151 [DOI] [PubMed] [Google Scholar]
- Koehler CM, Lindberg GL, Brown DR, Beitz DC, Freeman AE, Mayfield JE, Myers AM (1991) Replacement of bovine mitochondrial DNA by a sequence variant within one generation. Genetics 129:247–255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Loeb LA (1981) Fidelity of mammalian DNA polymerases. Science 213:765–767 [DOI] [PubMed] [Google Scholar]
- Laubscher KH, Glew RH, Lee RE, Okinaka RT (1994) Use of denaturing gradient gel electrophoresis to identify mutant sequences in the ß-glucocerebrosidase gene. Hum Mutat 2:411–415 [DOI] [PubMed] [Google Scholar]
- Levin BC, Cheng H, Holland MM, Reeder DJ (1996) Heteroplasmy responsible for difficulty experienced in sequencing the human mitochondrial DNA HV1 region containing the C-stretch. Paper presented at the Seventh International Symposium on Human Identification, Scottsdale, AZ, September 19–21 [Google Scholar]
- Lombes A, Diaz C, Romero NB, Ziegler F, Fardeau M (1992) Analysis of the tissue distribution and inheritance of heteroplasmic mitochondrial DNA point mutation by denaturant gradient gel electrophoresis in MERRF syndrome. Neuromuscul Disord 2:323–330 [DOI] [PubMed] [Google Scholar]
- Marchington DR, Hartshorne GM, Barlow D, Poulton J (1997) Homopolymeric tract heteroplasmy in mtDNA from tissues and single oocytes: support for a genetic bottleneck. Am J Hum Genet 60:408–416 [PMC free article] [PubMed] [Google Scholar]
- Meyer S, Weiss G, von Haeseler A (1999) Pattern of nucleotide substitution and rate heterogeneity in the hypervariable regions I and II of human mtDNA. Genetics 152:1103–1110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michikawa Y, Hofhaus G, Lerman LS, Attardi G (1997) Comprehensive, rapid and sensitive detection of sequence variants of human mitochondrial tRNA genes. Nucleic Acids Res 25:2455–2463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michikawa Y, Mazzucchelli F, Bresolin N, Scarlato G, Attardi G (1999) Aging-dependent large accumulation of point mutations in the human mtDNA control region for replication. Science 286:774–779 [DOI] [PubMed] [Google Scholar]
- Monnat RJ Jr, Loeb LA (1985) Nucleotide sequence preservation of human mitochondrial DNA. Proc Natl Acad Sci USA 82:2895–2899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monnat RJ Jr, Reay DT (1986) Nucleotide sequence identity of mitochondrial DNA from different human tissues. Gene 43:205–211 [DOI] [PubMed] [Google Scholar]
- Myers RM, Fischer SG, Lerman LS, Maniatis T (1985) Nearly all single base substitutions in DNA fragments joined to a GC-clamp can be detected by denaturing gradient gel electrophoresis. Nucleic Acids Res 13:3131–3145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olivo PD, Van de Walle MJ, Laipis PJ, Hauswirth WW (1983) Nucleotide sequence evidence for rapid genotypic shifts in the bovine mitochondrial D-loop. Nature 306:400–402 [DOI] [PubMed] [Google Scholar]
- Parker LT, Deng Q, Zakeri H, Carlson C, Nickerson DA, Kwok PY (1995) Peak height variations in automated sequencing of PCR products using Taq dye-terminator chemistry. Biotechniques 19:116–121 [PubMed] [Google Scholar]
- Parker LT, Zakeri H, Deng Q, Spurgeon S, Kwok PY, Nickerson DA (1996) AmpliTaq DNA polymerase, FS dye-terminator sequencing: analysis of peak height patterns. Biotechniques 21:694–699 [DOI] [PubMed] [Google Scholar]
- Parsons TJ, Holland MM (1998) Reply to Jazin et al. Nat Genet 18:110 [Google Scholar]
- Parsons TJ, Muniec DS, Sullivan K, Woodyatt N, Alliston-Greiner R, Wilson MR, Berry DL, et al (1997) A high observed substitution rate in the human mitochondrial DNA control region. Nat Genet 15:363–367 [DOI] [PubMed] [Google Scholar]
- Poulton J, Macaulay V, Marchington DR (1998) Mitochondrial genetics '98: is the bottleneck cracked? Am J Hum Genet 62:752–757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poulton J, Marchington DR (1996) Prospects for DNA-based prenatal diagnosis of mitochondrial disorders. Prenat Diagn 16:1247–1256 [DOI] [PubMed] [Google Scholar]
- Richards M, Corte-Real H, Forster P, Macauley V, Wilkinson-Herbots H, Demaine A, Papiha S, et al (1996) Paleolithic and neolithic lineages in the European gene pool. Am J Hum Genet 59:185–203 [PMC free article] [PubMed] [Google Scholar]
- Sekiguchi K, Kasai K, Levin BC (1999) Human mitochondrial DNA heteroplasmic variation among thirteen maternally-related family members. Paper presented at the Tenth International Symposium on Human Identification. Orlando, FL, September 29–October 2 [Google Scholar]
- Sheffield VC, Cox DR, Lerman LS, Myers RM (1989) Attachment of a 40–base-pair G+C rich sequence (GC-clamp) to genomic DNA fragments by the polymerase chain reaction results in improved detection of single-base changes. Proc Natl Acad Sci USA 86:232–236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sigurdardottir S, Helgason A, Gulcher JR, Stefansson K, Donnelly P (2000) The mutation rate in the human mtDNA control region. Am J Hum Genet 66:1599–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steighner RJ, Tully LA, Karjala J, Coble M, Holland MM (1999) Comparative identity and homogeneity testing of the mtDNA HV1 region using denaturing gradient gel electrophoresis. J Forensic Sci 44:1186–1198 [PubMed] [Google Scholar]
- Stoneking M (1994) Mitochondrial DNA and human evolution. J Bioenerg Biomembr 26:251–259 [DOI] [PubMed] [Google Scholar]
- ——— (1996) Mitochondrial DNA heteroplasmy: out of the closet. Biol Chem 377:603–604 [PubMed] [Google Scholar]
- Sullivan KM, Alliston-Greiner R, Archampong FIA, Piercy R, Tully G, Gill P, Lloyd-Davies C (1996) A single difference in mtDNA control region sequence observed between hair shaft and reference samples from a single donor. Paper presented at the Seventh International Symposium on Human Identification, Scottsdale, AZ, September 19–21 [Google Scholar]
- Thomas R, Zischler H, Paabo S, Stoneking M (1996) Novel mitochondrial DNA insertion and its usefulness for human population studies. Hum Biol 68:847–854 [PubMed] [Google Scholar]
- Traystman MD, Higuchi M, Kasper CK, Antonarakis E, Kazazian HH Jr (1990) Use of denaturing gradient gel electrophoresis to detect point mutations in the factor VIII gene. Genomics 6:293–301 [DOI] [PubMed] [Google Scholar]
- Tully LA (1998) Examination of the use of forensic DNA typing from two perspectives: I. Mitochondrial DNA heteroplasmy. II. The role of DNA typing in criminal investigations. PhD thesis, University of Maryland, Baltimore [Google Scholar]
- Vigilant L, Pennington R, Harpending H, Kocher TD, Wilson AC (1989) Mitochondrial DNA sequence in single hairs from a southern African population. Proc Natl Acad Sci USA 86:9350–9354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vigilant L, Stoneking M, Harpending H, Hawkes K, Wilson AC (1991) African populations and the evolution of human mitochondrial DNA. Science 253:1503–1507 [DOI] [PubMed] [Google Scholar]
- Wakeley J (1993) Substitution rate variation among sites in hypervariable region 1 of human mitochondrial DNA. J Mol Evol 37:613–623 [DOI] [PubMed] [Google Scholar]
- Wallace DC (1995) 1994 William Allan Award Address: mitochondrial DNA variation in human evolution, degenerative disease, and aging. Am J Hum Genet 57:201–223 [PMC free article] [PubMed] [Google Scholar]
- Walsh PS, Metzger DA, Higuchi R (1991) Chelex 100 as a medium for simple extraction of DNA for PCR- based typing from forensic material. Biotechniques 10:506–513 [PubMed] [Google Scholar]
- Wilson MR, Polansky D, Replogle J, DiZinno JA, Budowle B (1997) A family exhibiting heteroplasmy in the human mitochondrial DNA control region reveals both somatic mosaicism and pronounced segregation of mitotypes. Hum Genet 100:167–171 [DOI] [PubMed] [Google Scholar]
- Yoon KL, Modica-Napolitano JS, Ernst SG, Aprille JR (1991) Denaturing gradient gel method for mapping single base changes in human mitochondrial DNA. Anal Biochem 196:427–432 [DOI] [PubMed] [Google Scholar]
- Zischler H, Gelsert H, von Haeseler A, Paabo S (1995) A nuclear “fossil” of the mitochondrial D-loop and the origin of modern humans. Nature 378:489–492 [DOI] [PubMed] [Google Scholar]