Abstract
Citrobacter spp. are gram-negative commensal bacteria that infrequently cause serious nosocomial infections in compromised hosts. They are often resistant to cephalosporins due to overexpression of their chromosomal β-lactamase. During a recent study of multidrug-resistant Enterobacteriaceae (MDRE) in solid-organ transplant patients, we found that almost half of patients colonized with MDRE carried one or more cefpodoxime-resistant Citrobacter freundii, Citrobacter braakii, or Citrobacter amalonaticus strains. Pulsed-field gel electrophoresis showed that 36 unique strains of Citrobacter were present among 32 patients. Genetic and phenotypic analysis of the resistance mechanisms of these bacteria showed that the extended-spectrum β-lactamase (ESBL) SHV-5 or SHV-12 was encoded by 8 strains (26%) and expressed by 7 strains (19%). A number of strains were resistant to other drug classes, including aminoglycosides (28%), trimethoprim-sulfamethoxazole (31%), and fluoroquinolones (8%). PCR and DNA analysis of these multiresistant strains revealed the presence of class I integrons, including the first integrons reported for C. braakii and C. amalonaticus. The integrons encoded aminoglycoside resistance, trimethoprim resistance, or both. Despite the prevalence of MDR Citrobacter spp. in our solid-organ transplant patients, only a single infection with a colonizing strain was recorded over 18 months. Low-virulence Citrobacter spp., which can persist in the host for long periods, could influence pathogen evolution by accumulation of genes encoding resistance to multiple antimicrobial classes.
Although Citrobacter species are infrequent nosocomial pathogens, local or systemic breaches of host defenses can allow them to cause a range of infections. These include urinary tract infections, neonatal sepsis, brain abscess (18, 21), meningitis (21, 23), bloodstream infections (39), intra-abdominal sepsis (37), and pneumonia (26, 28, 42). Invasive Citrobacter infections are associated with a high mortality rate, with 33 to 48% of patients succumbing to Citrobacter bacteremia (10, 37). A recent survey of bacteremic bone marrow transplant recipients found that chromosomal β-lactamase (AmpC)-producing organisms, including Citrobacter, were associated with excess mortality compared with other gram-negative species (6).
The high mortality rate associated with Citrobacter infections may be due in part to ineffective empirical antibiotic therapy. Citrobacter species are commonly resistant to extended-spectrum cephalosporins. A recent survey of 34,530 Enterobacteriaceae isolates from the SENTRY database found that 10 to 23% of Citrobacter spp. were resistant to ceftazidime (CAZ) (20). An earlier, U.S. survey of susceptibility to piperacillin-tazobactam found that 30% of Citrobacter freundii isolates were resistant (4). Increasing resistance to quinolones and aminoglycosides, in addition to β-lactams, was found in a series of C. freundii isolates collected in Taiwan between 1987 and 1998 (40). The majority of these isolates were resistant to extended-spectrum cephalosporins and antipseudomonal penicillins.
Chromosomal, inducible AmpC β-lactamases in this bacterial population are well described and are an important mechanism of resistance to β-lactams. SHV- and TEM-derived extended-spectrum β-lactamases (ESBLs) have also been described for Citrobacter species in the context of outbreaks of clonal strains and/or plasmids (8, 9, 12, 17, 30-32). Outside of these reports, there have been no systematic surveys for ESBLs among AmpC-producing organisms. Screening for ESBLs in this population is complicated, as most screening methods rely on synergy with β-lactamase inhibitors, which may be masked in the presence of the inhibitor-resistant chromosomal β-lactamase.
Multidrug resistance in the Enterobacteriaceae has been linked with the carriage of integrons, genetic elements capable of assimilating antibiotic resistance genes as cassettes through site-specific recombination catalyzed by a specific integrase enzyme. In particular, aminoglycoside and antifolate resistances are significantly associated with integron carriage in the Enterobacteriaceae (41). ESBL genes, particularly those derived from TEM and SHV, are not frequently found within integrons. However, ESBLs and integrons can be present on the same large conjugative plasmids, mediating coselection for multiple resistances by single drug classes.
We recently completed a study of the natural history and epidemiology of multidrug-resistant Enterobacteriaceae isolates among solid-organ transplant recipients (15). During that study, the largest group of multiresistant Enterobacteriaceae identified belonged to the genus Citrobacter. In this work, we characterize the molecular mechanisms of antibiotic resistance in these Citrobacter isolates. The presence of both ESBLs and integrons in these bacteria was a marker of multidrug resistance and suggests that Citrobacter spp. may serve as potential sources of resistance determinants for more virulent organisms.
MATERIALS AND METHODS
Bacterial strains.
During a study of ESBL colonization in the nonoutbreak setting (15), newly admitted solid-organ transplant recipients were screened prospectively for Enterobacteriaceae resistant to extended-spectrum cephalosporins with screening surveillance rectal swabs on MacConkey agar containing 2 μg of cefpodoxime (MacPOD)/ml. Resistance was confirmed by VITEK (bioMérieux) using an ESBL card with a breakpoint of 8 μg of cefpodoxime (CPD)/ml and by broth microdilution (15). Swabs were obtained upon admission and weekly until discharge. Two hundred eighty-seven patients were screened over 18 months. Clinical specimens requested during routine care were collected and plated on MacPOD plates. In total, 143 (125 colonizing and 18 clinical) strains of CPD-resistant Enterobacteriaceae were grown from 69 of 287 patients (24%) (15). Using VITEK, the organisms were identified to the species level, and their antimicrobial susceptibility profiles were determined. The largest group, Citrobacter spp. (57 of 143 strains [39.8%]), was further analyzed for this study. All strains were stored frozen at −70°C as glycerol stocks and passaged once on sheep blood agar plates before further characterization.
PFGE.
A pulsed-field gel electrophoresis (PFGE)-typing protocol adapted from the method of Gautom (16) was used. Total DNA preparations embedded in 1.6% low-melt agarose (Bio-Rad) plugs were digested with SfiI restriction enzyme (New England Biolabs) at 50°C overnight as per the manufacturer's recommendations. Chromosomal DNA was then separated in a 1% PFGE-grade agarose (Bio-Rad) gel with 0.5× Tris-borate-EDTA running buffer supplemented with 50 μM thiourea (34) using a CHEF DRII PFGE system (Bio-Rad). The program parameters were the following: initial time, 5 s; final time, 35 s; run time, 20 h; ratio, 1; voltage, 200 V; temperature, 12°C. The gel was poststained with 2 μg of ethidium bromide/ml in double-distilled water for 30 min and then destained for 1 h in double-distilled water. The bands were then visualized with UV light and photographed. Samples that differed by more than three bands upon visual inspection were considered to be separate strains (1).
PCR detection of TEM and SHV genes.
PCR primers used for amplification of TEM and SHV genes were designed by first aligning available sequences downloaded from GenBank (National Center for Biotechnology Information) with ClustalW (National Research Council of Canada [http://www.cbr.nrc.ca/services/clustalw_form.html]). Conserved regions at the 5′ and 3′ ends of the genes, excluding areas in which previous mutations had been found, were chosen as primer-binding sites. Primers for TEM gene amplification were the following: TEMup (5′ TAT TCA ACA TTT CCG TGT C), TEMdown (5′ CTG TCT ATT TCG TTC ATC C), SHVup (5′ TTA TAT TCG CCT GTG TAT TA), and SHVdown (5′ ATG CGC TGT GCT TTGTTA T). Total DNA was extracted from overnight cultures for use as a template using Instagene (Bio-Rad) following the manufacturer's instructions. The PCR volume was 50 μl total, containing 1 μl of template DNA, 2 μl of 10 mM deoxynucleoside triphosphates, 50 pmol (each) of primer, 5 μl of 10× PCR buffer, 10 μl of Q solution, and 0.5 μl of HotStart Taq (Qiagen). The PCR program, using a Perkin-Elmer 2400 thermocycler, consisted of 15 min at 95°C, 30 cycles of 30 s at 95°C, 30 s at 60°C, and 45 s at 72°C, followed by a final extension of 7 min at 72°C. Fifty microliters of each reaction mixture was run on a 1% agarose gel. If a band of the predicted size (≈800 bp for both TEM and SHV products) could not be visualized, PCR was repeated with an annealing temperature of 55°C. Bands of the expected size were excised from the gel and gel purified with a Qiagen gel purification kit (Qiagen).
Cloning and sequencing of TEM- and SHV-specific gene products.
TEM and SHV PCR products were either sequenced directly using the TEM- and SHV-specific primers or cloned into pCR2.1 TOPO (Invitrogen) according to the manufacturer's instructions. The cloning mixture was transformed into chemically competent Escherichia coli TOP10 cells (Invitrogen) and plated on Luria-Bertani (LB) agar plates containing 50 μg of ampicillin/ml and 2% 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (Bioshop, Mississauga, Ontario, Canada). Insertion of the TEM or SHV gene product in the plasmid was verified by a rapid plasmid-extraction technique (11). Recombinants were grown overnight at 37°C, and plasmids were isolated with the Qiagen plasmid miniprep kit. Two separate cloned amplification products were sequenced on both strands using M13 primers. The sequencing was performed at the Molecular Biology Core Facility at York University, North York, Canada, using the dye-deoxy-terminator method on an ABI377 sequencer (Perkin-Elmer).
Isoelectric focusing of β-lactamases.
Isoelectric focusing of cell lysates and nitrocefin detection of β-lactamases were performed as described previously (27, 35) with modifications. Briefly, cells from a 2-ml overnight culture made from a single colony in LB broth were harvested by microcentrifugation (12,000 × g; 10 min) and resuspended in 500 μl of 1% glycine plus 50 mM phosphate buffer (pH 7.5) on ice. The cell suspension was transferred to FastProtein Blue tubes (BIO 101, Inc.) and shaken in a Fast Prep machine (BIO 101, Inc.) at speed 6 for 30 s to release the cell contents. Samples were centrifuged at 4°C in a microcentrifuge for 30 min at 13,000 × g, and the supernatant was transferred to a new tube. All samples were standardized before being loaded on isoelectric focusing (IEF) gels by first testing the β-lactamase activity of the protein suspension. Seventeen microliters of protein extract was added to 50 μl of nitrocefin solution (50 μg/ml in 1% glycine plus 50% glycerol), and the time required for the solution to turn from yellow to dark pink was recorded. For a conversion rate of 30 to 45 s, 5 μl of sample was loaded. Standardized samples were run in precast IEF mini-gels, pH 3 to 10 (Bio-Rad), in a Mini-Protean II unit at 4°C at 100 V for 1 h, 250 V for 1 h, and 500 V for 30 min. β-Lactamase activity and pI were determined by pouring molten 3% agarose containing nitrocefin (1 mg/ml) over the gel and comparing the bands to standards run on the same gel. The β-lactamase standards used were extracts of strains containing TEM-1, pI 5.4; SHV-1, pI 7.6; SHV-3, pI 7.0; SHV-4, pI 7.8; and SHV-5, pI 8.2 (kindly provided by G. Jacoby, Lahey Clinic).
Plasmid analysis.
Plasmids were isolated from a 5-ml overnight culture in LB medium using a modified alkaline lysis protocol (5). Undigested plasmid DNA was separated on a 1% agarose gel stained with 2 μg of ethidium bromide/ml and photographed.
PCR amplification and DNA sequencing of integrons.
PCR was used to detect class I integrons. Primers intI1up (5′ ATG AAA ACC GCC ACT GCG CCG TTA) and intI1down (5′ ATC GAA ATC CAG ATC CTT GAC CC), directed against the intI1 gene, encoding the class I integrase enzyme (33), were used with chromosomal DNA templates from aminoglycoside-resistant or SHV-positive strains. Strains positive for intI1 were reamplified with the intI1 downstream primer in conjunction with a primer directed to the 3′ conserved sequence of the class I integrons (5′AAA GCA GAC TTG ACC TGA) (33). The amplification products were digested separately with EcoRI, Sau3AI, RsaI, and TaqI to identify unique patterns. Amplification products with unique restriction fingerprints were sequenced to identify resistance cassettes.
Nucleotide sequence accession numbers.
Integron sequences described in this study can be obtained from GenBank under accession numbers AF458082 (CF-4), AY069972 (CF-6), AF458081 (CF-12), and AF486817 (CA-1).
RESULTS
Identification of ESBL-producing Citrobacter spp. in solid-organ transplant patients.
Two hundred eighty-seven newly admitted solid-organ transplant patients were screened for CPD-resistant Enterobacteriaceae colonization or infection (15). Sixty-nine patients were colonized (125 strains) or infected (18 strains) with CPD-resistant Enterobacteriaceae. The genera isolated included Escherichia, Klebsiella, Citrobacter, and Enterobacter. The largest group consisted of 57 strains of Citrobacter spp. (1 of Citrobacter amalonaticus, 10 of Citrobacter braakii, and 46 of C. freundii), divided between 55 rectal isolates and 2 clinical isolates. Citrobacter spp. therefore represented 39.9% (57 of 143) of CPD-resistant Enterobacteriaceae recovered, and of 69 patients from whom CPD-resistant Enterobacteriaceae were recovered, 32 (46.4%) were colonized with one or more CPD-resistant Citrobacter spp.
PFGE fingerprinting of Citrobacter isolates.
PFGE showed that 36 unique PFGE types were present among the 57 isolates. Isolates were designated as duplicates if they were recovered from the same patient on more than one occasion and shared the same PFGE profile. Using this definition, there were a total of 38 nonduplicate isolates. Strain CF-15 was found in three patients (Table 1). Patient P14 had CF-15, a second strain of C. freundii (CF-14), and a strain of C. amalonaticus (CA-1), while four additional patients (P15, P19, P26, and P27) were each cocolonized by two unique strains of Citrobacter (Table 1). The majority of patients (29 of 32 or 90.6%) were colonized by one or more unique PFGE types.
TABLE 1.
Characteristics of cefpodoxime-resistant Citrobacter spp. isolated from solid-organ transplant patients
| PFGE typea | Patient no. | Resistance to antibioticc
|
Inte- gron | |||||
|---|---|---|---|---|---|---|---|---|
| CAZ | FOX | GEN | TOB | CIP | SXT | |||
| CF-1 | P1 | R | R | |||||
| CF-2 | P2 | R | ||||||
| CF-3 | P3 | R | ||||||
| CF-4 | P4 | R | R | R | R | IV | ||
| CF-5 | P5 | R | R | R | R | od | ||
| CF-6 | P6 | R | R | R | R | R | V | |
| CF-7 | P7 | R | R | R | R | R | II | |
| CF-8 | P8 | R | R | R | R | I | ||
| CF-9 | P9 | R | ||||||
| CF-10 | P10 | R | ||||||
| CF-11 | P11 | R | ||||||
| CF-12 | P12 | R | R | R | R | I | ||
| CF-13 | P13 | R | Rb | |||||
| CF-14 | P14 | R | R | R | R | R | R | I |
| CF-15 | P14, P15, P16 | R | R | R | R | R | o | |
| CF-16 | P17 | R | R | R | R | R | I | |
| CF-17 | P18 | R | R | R | o | |||
| CF-18 | P19 | |||||||
| CF-19 | P19 | R | Rb | |||||
| CF-20 | P20 | R | ||||||
| CF-21 | P21 | R | R | |||||
| CF-22 | P22 | R | ||||||
| CF-23 | P23 | R | ||||||
| CF-24 | P24 | R | Rb | |||||
| CF-25 | P25 | R | ||||||
| CF-26 | P26 | R | ||||||
| CF-27 | P26 | R | ||||||
| CF-28 | P27 | o | ||||||
| CF-29 | P27 | |||||||
| CB-1 | P28 | R | ||||||
| CB-2 | P29 | R | ||||||
| CB-3 | P30 | R | R | |||||
| CB-4 | P31 | R | ||||||
| CB-5 | P32 | R | ||||||
| CB-6 | P15 | R | R | R | o | |||
| CA-1 | P14 | R | R | R | R | R | III | |
CF, C. freundii; CB, C. braakii; CA, C. amalonaticus.
Tested against cefotetan instead of FOX.
R, resistant. NCCLS breakpoints for Enterobacteriaceae are the following: for CAZ, ≥32 μg/ml; for FOX, ≥32 μg/ml; for GEN, ≥16 μg/ml; for TOB, ≥16 μg/ml; for CIP, ≥4 μg/ml; for SXT, ≥4/76 μg/ml (29a ). GEN, gentamicin; TOB, tobramycin.
The class I integrase gene only, not a complete integron, could be amplified by PCR.
There were only two CPD-resistant Citrobacter spp. isolated from clinical specimens during the study period. One isolate was from a urine specimen (C. braakii); the other was a sputum isolate (C. freundii). The urinary isolate was obtained from a patient previously colonized with the same organism, whereas the patient with the sputum isolate had not previously been colonized with that strain.
Antimicrobial susceptibility patterns.
Susceptibility testing of Citrobacter isolates demonstrated resistance to a broad range of antibiotics (Table 1). While all strains were resistant to CPD, most were resistant to other cephalosporins as well. Eighteen of thirty-six strains (50.0%) were resistant to CAZ, while 32 of 36 strains (88.9%) were resistant to the cephamycin cefoxitin (FOX) or cefotetan (CTT). Twelve of thirty-six strains (33.3%) were resistant to one or more additional classes of antibiotics (aminoglycosides, fluoroquinolones, and/or trimethoprim-sulfamethoxazole [SXT]), with the most common resistances being trimethoprim-sulfamethoxazole (11 of 36 or 30.6%) and aminoglycosides (10 of 36 or 27.8%). Previous or concomitant antibiotic therapy was identified as a significant risk factor for carriage of an antibiotic-resistant strain (15).
Identification of SHV and TEM ESBLs in CPD-resistant Citrobacter spp.
Total DNA from each isolate was used as a template for PCR with SHV and TEM consensus primers. Gene amplification products of the expected size for SHV (∼800 bp) were obtained from 8 of 36 (22.2%) PFGE types, while TEM products (∼800 bp) were obtained from 6 of 36 (16.7%) PFGE types (Table 2). Duplicate isolates of individual PFGE profiles gave identical results with PCR.
TABLE 2.
Detection of SHV and TEM β-lactamases in Citrobacter spp. by PCR and IEF
| PFGE type | Result of analysis
|
||
|---|---|---|---|
| SHV PCR | TEM PCR | IEF (pI) | |
| CF-5 | SHV-12 | 5.4, 8.2 | |
| CF-8 | SHV-12 | TEM-1 | 5.4, 8.2 |
| CF-12 | SHV-12 | TEM-1 | 5.4, 8.2 |
| CF-14 | SHV-12 | 8.2 | |
| CF-15 | SHV-5 | 5.4, 8.2 | |
| CF-16 | SHV-12 | TEM-1 | 5.4, 8.2 |
| CF-28 | SHV-5 | ∼9 | |
| CA-1 | SHV-12 | TEM-1 | 5.4, 6.3, 8.2 |
| CF-6 | TEM-1 | NDa | |
| CF-7 | TEM-1 | ND | |
ND, not determined.
DNA sequence analysis of the amplified genes showed that the SHV products were SHV-5 and SHV-12, while all of the TEM products were non-ESBL TEM-1. To determine whether the ESBLs detected by PCR were expressed, isoelectric focusing (IEF) was performed, followed by nitrocefin detection. IEF analysis showed that seven of eight SHV-containing strains produced an enzyme with a pI of 8.2, the predicted isoelectric point for SHV-5 and SHV-12. The exception was CF-28, which produced a band with a pI of >8.2, in the range expected for the chromosomal AmpC enzyme of C. freundii (pI = 8.6). Unlike the other ESBL-producing strains in this study, CF-28 was sensitive to both CAZ and ceftriaxone, suggesting that the level of expression of SHV-5, if present, is very low. This strain was considered susceptible to FOX using NCCLS breakpoints (Table 1), although it was CPD resistant.
Interestingly, the C. amalonaticus strain produced an uncharacterized β-lactamase of pI 6.3, consistent with that of the previously described ESBLs TEM-3, TEM-16, TEM-18, and TEM-22 (2, 3, 19, 24, 38), in addition to TEM-1 and SHV-12. However, only TEM-1 could be amplified from this strain by PCR upon repeated attempts. It is possible that there is insufficient homology in the regions of primer binding to amplify the associated gene or that the detected enzyme is a non-TEM cephalosporinase. These results emphasize the value of IEF for the detection of other β-lactamases that may be produced by a particular strain and show that PCR with limited primer sets may underestimate the prevalence of ESBL genes.
All but one of the SHV-5- and SHV-12-containing isolates (the exception being CF-28; see above) demonstrated resistance to several antibiotic classes in addition to β-lactams. All were resistant to SXT and aminoglycosides, and three were also resistant to ciprofloxacin (CIP).
Identification of integron sequences in multidrug-resistant Citrobacter spp.
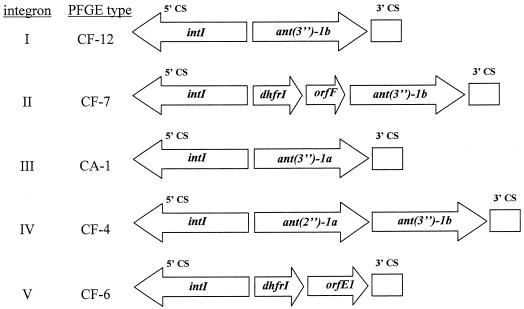
To investigate whether multidrug resistance in the above-discussed Citrobacter strains was associated with integrons, we performed PCR using primers directed to the integrase gene of class I integrons, commonly found in Enterobacteriaceae. All aminoglycoside-resistant strains were positive for intI1, as were all SXT-resistant and CIP-resistant strains. While CIP resistance is typically the result of target mutations, it has previously been noted to be significantly associated with integron carriage in the Enterobacteriaceae (25). In this study, CIP resistance was not significantly associated with integron carriage due to the number of integron-positive, CIP-susceptible strains (10 of 13) (Table 3). Total DNA from integrase-positive strains was then amplified with primers directed to the 5′ and 3′ conserved sequences (CS) of class I integrons (33). Only 8 of 13 integrase-positive strains yielded amplification products with the class I 5′CS and 3′CS primers, suggesting that the remaining 5 strains either lack the 3′CS or have insufficient homology to the 3′CS primer to produce a product. PCR products from this second reaction were fingerprinted by endonuclease digestion, revealing five unique restriction patterns among the eight strains. The five unique products were sequenced, and the results are shown in Fig. 1.
TABLE 3.
Antimicrobial resistance associated with integron carriage
| Antibiotic | Total no. of strains (n = 36) | No. of resistant, integron- positiveb strains | No. of resistant, integron- negative strains | No. of susceptible, integron- positiveb strains | P valuec |
|---|---|---|---|---|---|
| CAZ | 18 | 12 | 6 | 1 | 0.001 |
| Cephamycins (FOX or CTT) | 32 | 11 | 21 | 2 | 0.745 |
| GEN | 8 | 8 | 0 | 5 | 0.002 |
| TOB | 8 | 8 | 0 | 5 | 0.002 |
| Aminoglycosidesa | 10 | 10 | 0 | 3 | <0.001 |
| CIP | 3 | 3 | 0 | 10 | 0.267 |
| SXT | 11 | 11 | 0 | 2 | <0.001 |
Resistant to GEN, TOB, or both.
Strains in which the class I integrase gene was detected by PCR.
Mann-Whitney U one-tailed test.
FIG. 1.
Arrangement of genes within class I integrons identified in Citrobacter spp. Integron numbers and pulsed-field types correspond to those in Table 1. Genes identified by DNA sequence analysis are shown as labeled arrows. The locations of the 5′ and 3′ conserved sequences are shown. intI, integrase gene; ant, aminoglycoside acetyltransferase gene; dhfr, dihydrofolate reductase gene; orf, open reading frame.
Four of five integron types (II to V) were unique to individual Citrobacter strains from different patients, while the remaining integron (I) was common to four separate strains from four individual patients. All strains with the common integron also carried SHV-12.
Three distinct aminoglycoside resistance genes were identified in four integrons, one (from CF-4) of which contained the rare configuration of two separate aminoglycoside-modifying genes in tandem (Fig. 1). The integron from CF-7 contained both a dihydrofolate reductase gene (dhfr1) and ant(3′′)-1b, while the integron from CF-6 contained only dhfr1, even though this strain is resistant to aminoglycosides. These results show that multiple mechanisms of resistance are present among these Citrobacter isolates.
DISCUSSION
Citrobacter spp. represented the largest group (39.8%) of CPD-resistant Enterobacteriaceae colonizing transplant recipients (15). Almost half (46.4%) of transplant patients colonized with a CPD-resistant Enterobacteriaceae strain had at least one CPD-resistant Citrobacter strain. Despite the high prevalence of colonization, there was only one infection with a colonizing drug-resistant Citrobacter strain over the 18-month period, suggesting that Citrobacter spp. are of low virulence even in this heavily immunosuppressed population. The extremely low prevalence of Citrobacter infections in population-based studies (0.8% of gram-negative infections in the SENTRY database) (20) supports the concept that these organisms are less pathogenic than other Enterobacteriaceae.
NCCLS guidelines for ESBL screening (double disk potentiation) in the Enterobacteriaceae apply to Klebsiella and E. coli species only (29). Several studies have found CPD resistance to be the most sensitive screening test for ESBLs in Klebsiella pneumoniae and E. coli (13, 22). For Citrobacter and Enterobacter, coexpression of the chromosomal AmpC β-lactamase can confer CPD, cephamycin, and β-lactamase inhibitor resistance, masking the presence of SHV- and TEM-derived ESBLs.
In this study, 26% of CPD-resistant Citrobacter isolates carried ESBL genes, 88% of which were expressed, as determined by nitrocefin detection following IEF. Two other studies done in a nonoutbreak setting found rates of ESBL carriage among Citrobacter spp. to be 7% (3 of 42) (7) and 4% (3 of 68) (13). In these studies, ESBL screening was done by double disk potentiation, suggesting that the proportion of SHV- and TEM-containing Citrobacter spp. may have been underestimated.
Thirteen of thirty-six (36%) isolates were positive for intI1 by PCR, including the first integrase genes described for C. amalonaticus and C. braakii. However, complete integrons could be amplified from only 8 of 13 strains. There is meager literature on integrons in Citrobacter spp., but their prevalence among gram-negative bacteria in general appears to be rising. A recent study of E. coli, Klebsiella, and Enterobacter isolates from unselected blood cultures reported an increase in integron prevalence from 4.7 to 17.4% over a 7-year period (36).
In this study, integron carriage was linked with the presence of ESBLs (SHV-5 and SHV-12), although the SHV genes were not found within the integron structure. In addition, both integron carriage and the presence of an ESBL were highly correlated with multiple antibiotic resistances (Table 3). In some cases, the resistance patterns of individual isolates could be related to the resistance-related genes found. In others, however, there was broader resistance demonstrated than would be inferred from genotypic information. For example, all aminoglycoside-resistant isolates were intI1 positive, but not all integrons in these strains could be shown to contain aminoglycoside resistance determinants.
Other studies have reported integrons to be associated with broad antimicrobial resistance even when they do not encode multiple resistance determinants (41). In this study, integron carriage was significantly correlated with CAZ resistance (P ⩽ 0.001), even when CAZ hydrolases were not encoded by the integron. Similar findings have been reported for other integron-containing Enterobacteriaceae (25). The most likely explanation for this phenomenon is that the resistance determinants were found on multiresistant plasmids. Indeed, all multiply resistant strains harbored large plasmids that could not be transferred by mating with a lab strain of E. coli (not shown). In this study, the combination of immunosuppressed hosts and exposure to a broad range of antibiotics (15) may have contributed to selection for strains that had accumulated multiple resistance determinants. Also, when multiresistance plasmids are present, selection for resistance to multiple antibiotics can occur through use of a single agent. A recent population-based study of SXT resistance among E. coli strains found evidence of multiresistance plasmids and the coselection of SXT resistance through the use of other antibiotic classes (14). This phenomenon may also explain the simultaneous expression of redundant resistance determinants, such as ESBL and AmpC β-lactamases.
Citrobacter and similar species may play a unique role in bacterial evolution. They are of low virulence and thus can persist in a host population for long periods. Over time, they could accumulate resistance determinants which may then be available for transfer to more virulent organisms. In addition to specific resistance genes, they accumulate the means of gene capture and dissemination, i.e., integrons, transposons, and plasmids. This can set the stage for an outbreak if, for example, resistance and virulence determinants are located on the same plasmid or if an inherently virulent organism acquires a resistance plasmid and spreads among hosts.
In summary, we have found that antibiotic-resistant, low-virulence Citrobacter species are common colonizers of immunosuppressed patients exposed to multiple antimicrobial agents. These bacteria demonstrate broad drug resistance encoded by a diverse array of genetic mechanisms. It is possible that Citrobacter and similar organisms may contribute to the evolution of bacterial pathogens by acting as persistent sources of resistance genes. The presence of low-virulence, resistant bacteria in hospitalized patients may complicate surveillance and infection control efforts.
Acknowledgments
C. Pepperell and J. V. Kus contributed equally to this work.
We thank Amer Johri, Kim Pettem, Minh-Hien Le, Pat Clark, and Helen Didier for excellent technical assistance. We thank Mike Mulvey and Dave Boyd for assistance with IEF experiments, George Jacoby for supplying ESBL strains, and Jim Brunton for helpful discussions.
This work was supported by a grant from the Physician Services Foundation Inc.
REFERENCES
- 1.Arbeit, R. 1995. Laboratory procedures for the epidemiologic analysis of microorganisms, p. 190-208. In P. R. Murray (ed.), Manual of clinical microbiology, 6th ed. ASM Press, Washington, D.C.
- 2.Arlet, G., S. Goussard, P. Courvalin, and A. Philippon. 1999. Sequences of the genes for the TEM-20, TEM-21, TEM-22, and TEM-29 extended-spectrum beta-lactamases. Antimicrob. Agents Chemother. 43:969-971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arlet, G., M. Rouveau, G. Fournier, P. H. Lagrange, and A. Philippon. 1993. Novel, plasmid-encoded, TEM-derived extended-spectrum beta-lactamase in Klebsiella pneumoniae conferring higher resistance to aztreonam than to extended-spectrum cephalosporins. Antimicrob. Agents Chemother. 37:2020-2023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baron, E. J., and R. N. Jones. 1995. National survey of the in vitro spectrum of piperacillin-tazobactam tested against more than 40,000 aerobic clinical isolates from 236 medical centers. Diagn. Microbiol. Infect. Dis. 21:141-151. [DOI] [PubMed] [Google Scholar]
- 5.Birnboim, H. C. 1983. A rapid alkaline extraction method for the isolation of plasmid DNA. Methods Enzymol. 100:243-255. [DOI] [PubMed] [Google Scholar]
- 6.Collin, B. A., H. L. Leather, J. R. Wingard, and R. Ramphal. 2001. Evolution, incidence, and susceptibility of bacterial bloodstream isolates from 519 bone marrow transplant patients. Clin. Infect. Dis. 33:947-953. [DOI] [PubMed] [Google Scholar]
- 7.Coudron, P. E., E. S. Moland, and C. C. Sanders. 1997. Occurrence and detection of extended-spectrum beta-lactamases in members of the family Enterobacteriaceae at a veterans medical center: seek and you may find. J. Clin. Microbiol. 35:2593-2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.D'Agata, E., L. Venkataraman, P. DeGirolami, L. Weigel, M. Samore, and F. Tenover. 1998. The molecular and clinical epidemiology of Enterobacteriaceae-producing extended-spectrum beta-lactamase in a tertiary care hospital. J. Infect. 36:279-285. [DOI] [PubMed] [Google Scholar]
- 9.de Champs, C., D. Sirot, C. Chanal, M. C. Poupart, M. P. Dumas, and J. Sirot. 1991. Concomitant dissemination of three extended-spectrum beta-lactamases among different Enterobacteriaceae isolated in a French hospital. J. Antimicrob. Chemother. 27:441-457. [DOI] [PubMed] [Google Scholar]
- 10.Drelichman, V., and J. D. Band. 1985. Bacteremias due to Citrobacter diversus and Citrobacter freundii. Incidence, risk factors, and clinical outcome. Arch. Intern. Med. 145:1808-1810. [PubMed] [Google Scholar]
- 11.Eckhardt, T. 1978. A rapid method for the identification of plasmid desoxyribonucleic acid in bacteria. Plasmid 1:584-588. [DOI] [PubMed] [Google Scholar]
- 12.El Harrif-Heraud, Z., C. Arpin, S. Benliman, and C. Quentin. 1997. Molecular epidemiology of a nosocomial outbreak due to SHV-4-producing strains of Citrobacter diversus. J. Clin. Microbiol. 35:2561-2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Emery, C. L., and L. A. Weymouth. 1997. Detection and clinical significance of extended-spectrum beta-lactamases in a tertiary-care medical center. J. Clin. Microbiol. 35:2061-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Enne, V. I., D. M. Livermore, P. Stephens, and L. M. Hall. 2001. Persistence of sulphonamide resistance in Escherichia coli in the UK despite national prescribing restriction. Lancet 357:1325-1328. [DOI] [PubMed] [Google Scholar]
- 15.Gardam, M., L. L. Burrows, J. V. Kus, J. Brunton, D. E. Low, J. M. Conly, and A. Humar. Is surveillance for multidrug resistant Enterobacteriaceae an effective infection control strategy in the absence of an outbreak? J. Infect. Dis., in press. [DOI] [PubMed]
- 16.Gautom, R. K. 1997. Rapid pulsed-field gel electrophoresis protocol for typing of Escherichia coli O157:H7 and other gram-negative organisms in 1 day. J. Clin. Microbiol. 35:2977-2980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gniadkowski, M., I. Schneider, A. Palucha, R. Jungwirth, B. Mikiewicz, and A. Bauernfeind. 1998. Cefotaxime-resistant Enterobacteriaceae isolates from a hospital in Warsaw, Poland: identification of a new CTX-M-3 cefotaxime-hydrolyzing beta-lactamase that is closely related to the CTX-M-1/MEN-1 enzyme. Antimicrob. Agents Chemother. 42:827-832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Graham, D. R., and J. D. Band. 1981. Citrobacter diversus brain abscess and meningitis in neonates. JAMA 245:1923-1925. [PubMed] [Google Scholar]
- 19.Gutmann, L., M. D. Kitzis, D. Billot-Klein, F. Goldstein, G. Tran Van Nhieu, T. Lu, J. Carlet, E. Collatz, and R. Williamson. 1988. Plasmid-mediated beta-lactamase (TEM-7) involved in resistance to ceftazidime and aztreonam. Rev. Infect. Dis. 10:860-866. [DOI] [PubMed] [Google Scholar]
- 20.Jones, R. N., S. G. Jenkins, D. J. Hoban, M. A. Pfaller, and R. Ramphal. 2000. In vitro efficacy of six cephalosporins tested against Enterobacteriaceae isolated at 38 North American medical centres participating in the SENTRY Antimicrobial Surveillance Program, 1997-1998. Int. J. Antimicrob. Agents 15:111-118. [DOI] [PubMed] [Google Scholar]
- 21.Kline, M. W. 1988. Citrobacter meningitis and brain abscess in infancy: epidemiology, pathogenesis, and treatment. J. Pediatr. 113:430-434. [DOI] [PubMed] [Google Scholar]
- 22.Livermore, D. M., and D. F. Brown. 2001. Detection of beta-lactamase-mediated resistance. J. Antimicrob. Chemother. 48(Suppl. 1):59-64. [DOI] [PubMed] [Google Scholar]
- 23.Lu, C. H., W. N. Chang, Y. C. Chuang, and H. W. Chang. 1999. Gram-negative bacillary meningitis in adult post-neurosurgical patients. Surg. Neurol. 52:438-444. [DOI] [PubMed] [Google Scholar]
- 24.Mabilat, C., and P. Courvalin. 1990. Development of “oligotyping” for characterization and molecular epidemiology of TEM beta-lactamases in members of the family Enterobacteriaceae. Antimicrob. Agents Chemother. 34:2210-2216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martinez-Freijo, P., A. C. Fluit, F. J. Schmitz, V. S. Grek, J. Verhoef, and M. E. Jones. 1998. Class I integrons in Gram-negative isolates from different European hospitals and association with decreased susceptibility to multiple antibiotic compounds. J. Antimicrob. Chemother. 42:689-696. [DOI] [PubMed] [Google Scholar]
- 26.Mateos Rodriguez, F., E. Perez Moro, M. P. Atienza Morales, and J. L. Beato Perez. 2000. Community-acquired bacteremic pneumonia due to Citrobacter diversus. An. Med. Int. 17:165-166. [PubMed] [Google Scholar]
- 27.Mathew, A., A. M. Harris, M. J. Marshall, and G. W. Ross. 1975. The use of analytical isoelectric focusing for detection and identification of beta-lactamases. J. Gen. Microbiol. 88:169-178. [DOI] [PubMed] [Google Scholar]
- 28.Matsui, S. 1994. A case of severe pneumonia in an elderly man, caused by Citrobacter freundii suspected to have a low susceptibility to imipenem/cilastatin sodium. Nihon Kyobu Shikkan Gakkai Zasshi 32:1078-1082. [PubMed] [Google Scholar]
- 29.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial disk susceptibility tests, 7th ed. Approved standard M2-A7. NCCLS, Wayne, Pa.
- 29a.National Committee for Clinical Laboratory Standards. 2000. Performance standards for antimicrobial susceptibility testing. Approved standard M100-S12. NCCLS, Wayne, Pa.
- 30.Neuwirth, C., E. Siebor, J. Lopez, A. Pechinot, and A. Kazmierczak. 1996. Outbreak of TEM-24-producing Enterobacter aerogenes in an intensive care unit and dissemination of the extended-spectrum beta-lactamase to other members of the family Enterobacteriaceae. J. Clin. Microbiol. 34:76-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Palucha, A., B. Mikiewicz, W. Hryniewicz, and M. Gniadkowski. 1999. Concurrent outbreaks of extended-spectrum beta-lactamase-producing organisms of the family Enterobacteriaceae in a Warsaw hospital. J. Antimicrob. Chemother. 44:489-499. [DOI] [PubMed] [Google Scholar]
- 32.Petit, A., G. Gerbaud, D. Sirot, P. Courvalin, and J. Sirot. 1990. Molecular epidemiology of TEM-3 (CTX-1) beta-lactamase. Antimicrob. Agents Chemother. 34:219-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ploy, M. C., F. Denis, P. Courvalin, and T. Lambert. 2000. Molecular characterization of integrons in Acinetobacter baumannii: description of a hybrid class 2 integron. Antimicrob. Agents Chemother. 44:2684-2688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Romling, U., and B. Tummler. 2000. Achieving 100% typeability of Pseudomonas aeruginosa by pulsed-field gel electrophoresis. J. Clin. Microbiol. 38:464-465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sanders, C. C., W. E. Sanders, Jr., and E. S. Moland. 1986. Characterization of beta-lactamases in situ on polyacrylamide gels. Antimicrob. Agents Chemother. 30:951-952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schmitz, F. J., D. Hafner, R. Geisel, P. Follmann, C. Kirschke, J. Verhoef, K. Kohrer, and A. C. Fluit. 2001. Increased prevalence of class I integrons in Escherichia coli, Klebsiella species, and Enterobacter species isolates over a 7-year period in a German university hospital. J. Clin. Microbiol. 39:3724-3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shih, C. C., Y. C. Chen, S. C. Chang, K. T. Luh, and W. C. Hsieh. 1996. Bacteremia due to Citrobacter species: significance of primary intraabdominal infection. Clin. Infect. Dis. 23:543-549. [DOI] [PubMed] [Google Scholar]
- 38.Sougakoff, W., S. Goussard, G. Gerbaud, and P. Courvalin. 1988. Plasmid-mediated resistance to third-generation cephalosporins caused by point mutations in TEM-type penicillinase genes. Rev. Infect. Dis. 10:879-884. [DOI] [PubMed] [Google Scholar]
- 39.Tellez, I., G. S. Chrysant, I. Omer, and W. E. Dismukes. 2000. Citrobacter diversus endocarditis. Am. J. Med. Sci. 320:408-410. [DOI] [PubMed] [Google Scholar]
- 40.Wang, J. T., S. C. Chang, Y. C. Chen, and K. T. Luh. 2000. Comparison of antimicrobial susceptibility of Citrobacter freundii isolates in two different time periods. J. Microbiol. Immunol. Infect. 33:258-262. [PubMed] [Google Scholar]
- 41.White, P. A., C. J. McIver, and W. D. Rawlinson. 2001. Integrons and gene cassettes in the Enterobacteriaceae. Antimicrob. Agents Chemother. 45:2658--2661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zuniga, J., P. Gonzalez, A. Henriquez, and A. Fernandez. 1991. Bacteremic pneumonia caused by Citrobacter diversus: report of a case. Rev. Med. Chile 119:303-307. [PubMed] [Google Scholar]